Abstract
Implementation of zinc interventions for subjects suspected of being zinc-deficient is a global need, but is limited due to the absence of reliable biomarkers. To discover molecular signatures of human zinc deficiency, a combination of transcriptome, cytokine, and microRNA analyses was applied to a dietary zinc depletion/repletion protocol with young male human subjects. Concomitant with a decrease in serum zinc concentration, changes in buccal and blood gene transcripts related to zinc homeostasis occurred with zinc depletion. Microarray analyses of whole blood RNA revealed zinc-responsive genes, particularly, those associated with cell cycle regulation and immunity. Responses of potential signature genes of dietary zinc depletion were further assessed by quantitative real-time PCR. The diagnostic properties of specific serum microRNAs for dietary zinc deficiency were identified by acute responses to zinc depletion, which were reversible by subsequent zinc repletion. Depression of immune-stimulated TNFα secretion by blood cells was observed after low zinc consumption and may serve as a functional biomarker. Our findings introduce numerous novel candidate biomarkers for dietary zinc status assessment using a variety of contemporary technologies and which identify changes that occur prior to or with greater sensitivity than the serum zinc concentration which represents the current zinc status assessment marker. In addition, the results of gene network analysis reveal potential clinical outcomes attributable to suboptimal zinc intake including immune function defects and predisposition to cancer. These demonstrate through a controlled depletion/repletion dietary protocol that the illusive zinc biomarker(s) can be identified and applied to assessment and intervention strategies.
Zinc, an essential micronutrient ubiquitously distributed in plants and animals, is required for numerous catalytic, structural, and regulatory functions (1). In humans, documented outcomes of inadequate zinc ingestion include growth retardation, diarrhea, dermatitis, hypogonadism, hematological abnormalities, and impaired immunity (1, 2). Dietary zinc deficiency, with a predicted worldwide prevalence of 31% (3), has been estimated to be responsible for approximately 450,000 global deaths in children under 5 y of age (4). Therapeutic properties of supplemental zinc against infectious diseases substantiate the value of this nutrient for the maintenance of human health (3). Although the biological properties of zinc have been intensively explored since the first characterization of its essentiality (5), a reliable diagnostic tool to assess the dietary zinc status of individual humans or populations is lacking (6). Consequently a biomarker defining zinc status is needed for the identification of individuals or communities that may have low zinc status and those which may benefit from zinc interventions.
Serum or plasma zinc concentrations are widely used for the assessment of zinc status in experimental animal and human models. Nongovernmental organizations have promoted its use in population surveys (7). However, due to its lack of specificity—e.g., responsiveness to stress, starvation, or immunological conditions—its capability to serve as a diagnostic tool for zinc deficiency is limited (6). Recently, both expression of metallothionein (MT) and zinc transporter genes in leukocyte subsets have been shown to be directly proportional to the availability of zinc in vitro (8, 9) and in humans under dietary zinc supplementation (10, 11). These findings reflect the regulatory role of zinc in gene expression and zinc homeostasis and produce the hypothesis that specific genomic changes and specific functional outcomes can be used to identify zinc deficiency, but have yet to be studied.
Our major aim in this study was to identify differential responses of specific blood gene transcripts, circulating microRNAs (miRNA), and cytokines that occur when the dietary intake of zinc is acutely reduced below the dietary requirement and then is replenished. Specimens were collected in a noninvasive manner and via blood sampling, and techniques allowing sample stabilization, high-throughput analyses and bioinformatics were implemented to maximize the value of these methods for zinc status assessment.
Results
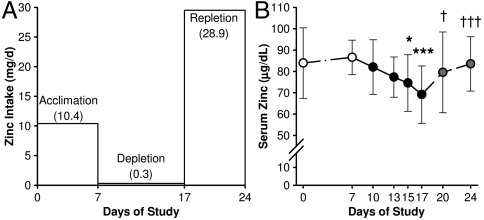
Atomic emission analysis confirmed the zinc content of the acclimation diets as 10.4 mg zinc per day (Fig. 1A) which is about the current Recommended Dietary Allowance of zinc for adult males (1). The male subjects in this study had a mean age and body weight of 24 yr and 77.1 kg, respectively (Table S1). Intersubject variability of serum zinc levels decreased by the end of the acclimation phase (Day 7), and all participants had a serum zinc concentration above the suggested lower cutoff value of 74 μg/dL for low zinc status (12). The low zinc (0.3 mg/d by analysis) and high phytate content (1.4 g/d) of the depletion diet yielded a phytate-to-zinc molar ratio of 462. Commensurate with consumption of the zinc-depletion diet (Day 7–17), subjects had a significant decrease in serum zinc concentrations. This was reversed by subsequent zinc supplementation (Day 18–24) (Fig. 1B). Body weights were constant during the three phases of the study (Table S1). Hematological parameters remained within normal ranges at the end of the zinc-depletion period (Day 17), but with significant changes in erythrocyte and platelet indices (Table S2).
Fig. 1.
Experimental design and serum zinc response induced by short-term acute dietary zinc depletion in humans. (A) Dietary protocol for acute dietary zinc depletion. Controlled zinc intake of 10.4 mg/d was provided during acclimation (Day 0–7) and 0.3 mg/d was provided for the depletion (Day 7–17). A self-selected diet plus a supplement of 15 mg/day was provided for repletion (Day 18–24). (B) Serum zinc concentrations during the effectiveness of the experimental diet for dietary zinc depletion. Values are mean ± standard deviation (SD) (n = 9). Those significantly different compared to baseline (Day 7) and postdepletion levels (Day 17) are noted by asterisks and crosses, respectively (n = 9). For * and †, P < 0.05 and *** and †††, P < 0.001.
Effects of Zinc-Depletion on Zinc-Related Transcript Levels.
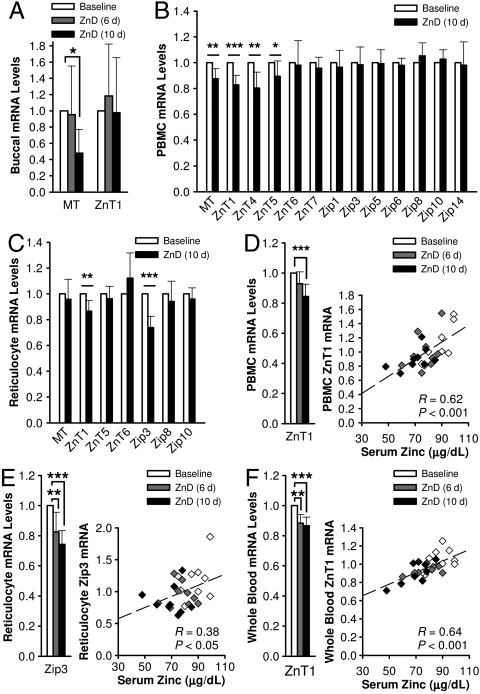
Among the 14 zinc-related transcripts in buccal RNA tested, only those for MT and ZnT1 were detected by qPCR (Fig. 2A). Criteria for determining the reliability of these assays include: amplification curve comparable to reference human RNA and CT values below 35. A significant decrease in MT mRNA abundance was observed in samples collected after 10 d of dietary zinc depletion (Day 17), while no change was found in the ZnT1 transcript levels.
Fig. 2.
Acute dietary zinc depletion influences zinc-related gene transcripts in buccal epithelial and blood cells. (A) Relative MT and ZnT1 mRNA abundance in buccal cell RNA during dietary zinc depletion. Values were normalized to 18S rRNA levels and baseline levels for each individual were set at 1. Analysis of MT and zinc transporter transcripts in (B) PBMC and (C) reticulocyte RNA during dietary zinc depletion. Values were normalized to GAPDH mRNA levels and baseline levels for each individual were set at 1. Correlation of (D) PBMC ZnT1, (E) reticulocyte Zip3, (F) whole blood ZnT1 transcript abundance with serum zinc concentrations during zinc depletion were determined by correlation coefficients (R) from linear regression analyses. White, baseline levels; gray, measures after 6 d of zinc depletion (Day 13); black, measures after 10 d of zinc depletion (Day 17). Values are mean ± SD. Values significantly different to respective baseline levels are indicated by asterisks. (*P < 0.05; **P < 0.01; ***P < 0.001 (n = 9).
Cell type-specific responsiveness of MT and some zinc transporter transcript levels to dietary zinc depletion were observed by qPCR with PBMC and reticulocyte RNA. MT, ZnT1, ZnT4, and ZnT5 mRNA levels significantly decreased in PBMC (Fig. 2B); while reduction in ZnT1 and Zip3 transcripts of purified reticulocytes (Fig. 2C) occurred by the end of acute dietary zinc depletion (Day 17). Of the differentially expressed (DE) zinc transporter transcripts, ZnT1 and Zip3 were the most significantly affected in PBMC and reticulocytes, respectively. Linear regression analysis revealed a significant correlation between serum zinc concentrations and PBMC ZnT1 mRNA and reticulocyte Zip3 mRNA abundance during the depletion period (Fig. 2D, E). The reduction of ZnT1 mRNA levels, observed for both PBMC and reticulocyte RNA, was also measurable in whole blood RNA along with a significant association with serum zinc concentrations as well (Fig. 2F). Of note are the significant reductions in the Zip3 and ZnT1 transcripts in reticulocytes and whole blood, respectively, after 6 d of the zinc depletion (Day 13), which is before the serum zinc had decreased significantly.
Microarray Analysis of Whole Blood RNA.
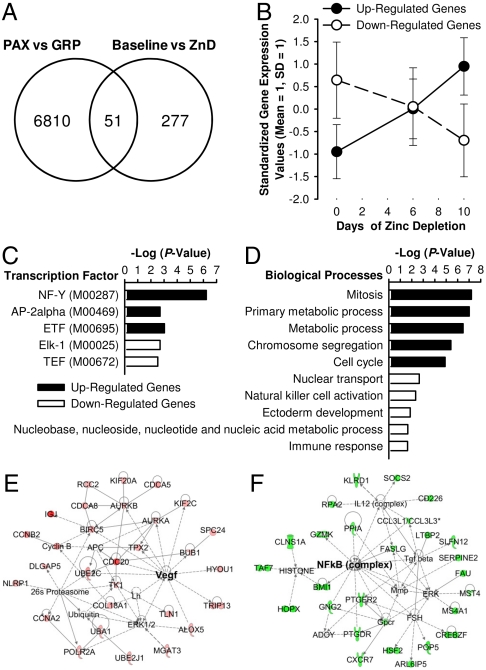
We next transitioned to analyses not targeted at zinc homeostasis. Whole blood RNA isolated using the PAXgene system (PAX) was highly intact as indicated by RNA integrity numbers above 8, and A260/A230 and A260/A280 ratios above 2.00. RNA recovery with high purity after globin transcript depletion was approximately 75%. Globin mRNA removal was verified by electropherograms (see Fig. S1). Using the Illumina BeadChip with 47,231 probes for human genes, differential expression of 328 genes by zinc depletion was identified by a pairwise comparison at P < 0.005 (Table S3). The influence of globin RNA on the detection of DE genes by microarray analysis was identified using pooled samples of Day 7, 13, and 17. Using an upper cutoff threshold of detection (P = 0.05), a total of 6,861 genes detectable in RNA after globin RNA reduction (GRP) were considered absent when the RNA samples were not processed for the removal of globin RNA (Fig. 3A). Among those masked genes, 51 were among the 328 genes determined to be DE by dietary zinc intake. Of particular note is that temporal increases and decreases throughout the 10-d depletion period were observed by the 192 up-regulated and 136 down-regulated genes, respectively, using these pooled samples (Fig. 3B).
Fig. 3.
Characterization of differentially expressed (DE) gene transcripts in whole blood RNA produced by short-term dietary zinc depletion. (A) Effects of globin RNA reduction on the detection of DE genes. RNA from whole blood collected with the PAXgene system (PAX) on Day 7, 13, and 17 were pooled as 3 groups and processed for globin RNA reduction (GRP). Transcripts were considered present when the detection P-value was lower than 0.05 in at least one of each group. (B) Temporal trends of the up-regulated (solid line) and down-regulated (dashed line) gene transcripts during the dietary zinc-depletion phase as identified by microarray experiments with pooled samples. (C) Regulatory elements enriched in the putative promoter regions (-1,000 ∼ +200) of genes responsive to dietary zinc depletion. (D) Top five biological processes represented by the up-regulated (solid bars) and down-regulated (open bars) genes affected by dietary zinc depletion. (E) Top functional network of up-regulated genes were associated with cellular assembly and organization, DNA replication, recombination and repair, and cell cycle. (F) Top functional network of down-regulated genes were associated with cell death, cell-mediated immune response, and cellular development. Red, up-regulated by zinc restriction; green, down-regulated by zinc restriction.
Integration of the transcriptome profiles into molecular and network databases using bioinformatic software enabled the prediction of physiological effects attributed to dietary zinc depletion. Promoter integration in microarray analysis (PRIMA) predictions based on the cis-regulatory elements of the responsive genes identified NF-Y, AP-2α, and ETF as the putative transcription factors mediating dietary zinc effects on the up-regulated genes and similarly the roles of Elk-1 and TEF in the repression of the down-regulated genes (Fig. 3C and Table S4). Gene ontology enrichment analysis of gene clusters, categorized based on their expression pattern using the PANTHER algorithm, identified overrepresentation of functional categories associated with cell cycle regulation for the up-regulated genes and nucleic acid and immune responses among the down-regulated genes (Fig. 3D).
Further characterization of the 328 DE genes using ingenuity pathway analysis (IPA) confirmed the enrichment in functional categories related to cell proliferation by the genes up-regulated during the dietary restriction regimen (Table S5). It is of note that cancer was identified as the disease most highly associated with up-regulated genes (Table S5) while genes associated with biological events related to cell death, cell-mediated immune responses, and cellular development and function were down-regulated by acute dietary zinc depletion (Table S6). Overrepresented functional networks by the up-regulated and down-regulated genes, respectively, imply enhancement in cell proliferative events and repression in cell death, cell-mediated immunity, and cellular development as physiological outcomes of inadequate zinc intake. Network nodes were at Vegf and NFkB (complex), respectively (Fig. 3 E and F).
Gene Transcripts with Potential of being Biomarkers of Zinc Deficiency.
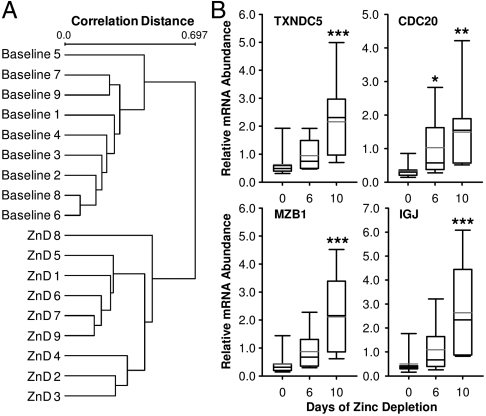
Pairwise comparisons of before- and after-treatment values identified indices responsive to the treatment. However, in order to identify molecular responses with potential for being a biomarker, interindividual variability was taken into account. As shown in Fig. 4A, a clear discrimination between baseline (Day 7) and end of zinc depletion (Day 17) was produced by hierarchical clustering with 108 DE genes determined by unpaired t-test (P < 0.005). After filtering by fold-changes above 2, a total of eight well-characterized genes were determined likely targets for status assessment of dietary zinc intake (Table S6). Separate qPCR assays of IGJ, CDC20, MZB1, and TXNDC5 mRNAs confirmed the responsiveness of these gene transcripts to acute dietary zinc depletion identified by microarray analysis (Fig. 4B).
Fig. 4.
Identification of whole blood gene transcripts holding the potential of being a biomarker of dietary zinc depletion. (A) Average-linkage hierarchical clustering of the arrays based on the gene expression profile of 108 DE genes identified by unpaired t-test at P < 0.005. Correlation distance was determined by the algorithm of (1-Pearson Correlation)/2 using EXPANDER. (B) Relative transcript levels of CDC20, TXNDC5, MZB1, and IGJ during zinc depletion. Values were normalized to GAPDH mRNA levels. Black and gray lines across bars indicate median and mean values, respectively. Whiskers indicate the interquartile range. Values significantly different to respective baseline levels are indicated by asterisks *P < 0.05; **P < 0.01; ***P < 0.001 (n = 9).
Serum MicroRNA Responsive to Dietary Zinc Levels.
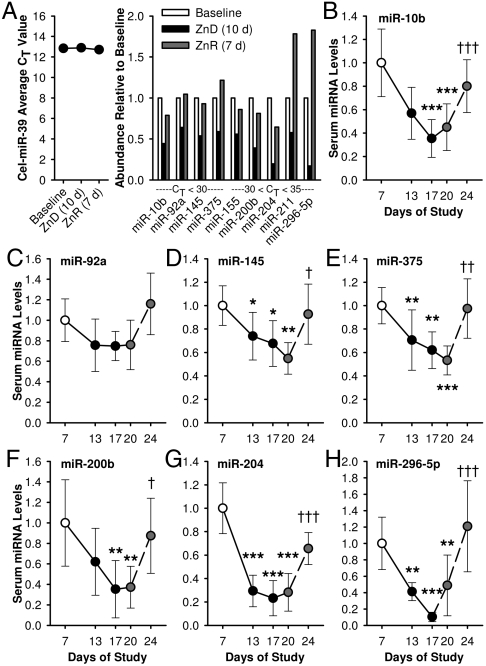
A qPCR-based array for miRNA detection was utilized for the identification of zinc-responsive signature miRNAs circulating in pooled sera. CT values acquired by the exogenous synthetic cel-miR-39 controls validated consistent yields of serum RNA recovery. Among the 85 serum miRNAs measured with the array, a total of 20 were shown to respond to the dietary zinc-depletion regimen with fold-changes above 1.5 and with CT values below 35 (Fig. S2). We identified nine miRNAs (miR-10b, -155, -200b, -296-5p, -375, -92a, -145, -204, and -211) responding to dietary zinc deprivation and repletion in opposite modes (Fig. 5A). Individual qPCR assays confirmed the zinc-responsiveness of these miRNAs (Fig. 5 B–H).
Fig. 5.
Dietary zinc intake influences miRNAs circulating in serum. (A) Identification of zinc-responsive miRNAs in sera using qPCR arrays. Data of sera collected from subjects at Day 7, 17, and 24 are shown (pooled sera of n = 8). Yields of exogenous cel-miR-39 were determined to validate the uniformity of the miRNA isolation method and for normalization. Relative transcript levels of (B) miR-10b, (C) -29a, (D) -145, (E) -375, (F) -200b, (G) -204, and (H) -296-5p in individual serum samples were measured by ligation-mediated qPCR (n = 6). Values were normalized to cel-miR-39 levels. Solid and dashed lines show the temporal patterns of responses to zinc depletion and repletion, respectively. Values are mean ± SD. Values significantly different to baseline and postdepletion values are noted by asterisks, and crosses, respectively. For * and †, P < 0.05; ** and ††, P < 0.01; and *** and †††, P < 0.001 (n = 6)
Inflammatory Cytokine Production by Whole Blood.
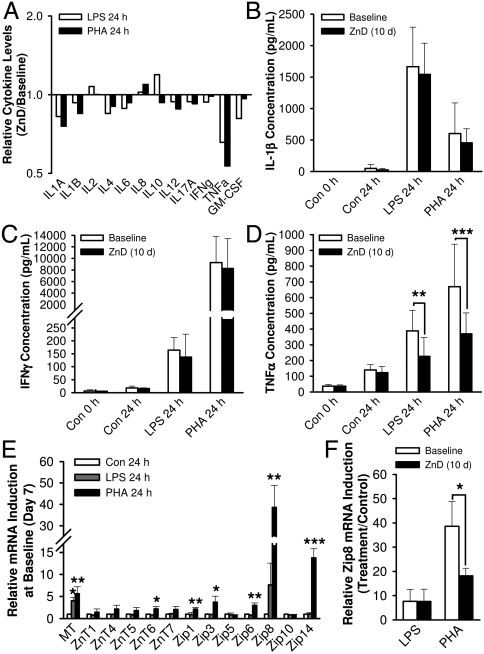
Mitogen-stimulated whole blood cytokine assays were used to test the capability of immune cell activation as a functional approach for identifying dietary zinc deficiency. Cell-free supernatants produced from blood cells collected on Day 7 and Day 17 were pooled and analyzed using an ELISA array composed of 12 human inflammatory cytokines. Among the cytokines tested, only TNFα showed a noticeable effect of dietary zinc depletion when induced by challenge with LPS or PHA (Fig. 6A). In order to confirm this effect of dietary zinc deprivation, supernatants from cells stimulated with LPS or PHA were collected from individual whole blood-samples and were assayed by ELISAs specific for IL-1β, IFNγ, and TNFα. IL-1β secretion after LPS-activation of monocytes (Fig. 6B) and IFNγ secretion stimulated by PHA-activation of lymphocytes (Fig. 6C) revealed no differences after dietary zinc depletion (Day 17). In contrast, cells from whole blood samples collected at Day 17 secreted significantly lower TNFα in response to immunostimulation by PHA and LPS than those collected at the baseline (Fig. 6D), indicating an effect of zinc on a pathway specific to production of this cytokine.
Fig. 6.
Acute dietary zinc depletion influences immune responses induced by LPS or PHA in vitro. (A) Attenuation of LPS- and PHA-induced TNFα release by dietary zinc depletion measured by a cytokine-focused multianalyte ELISA array. Values of zinc-depleted status are expressed as ratios relative to the baseline levels (pooled samples of n = 8). LPS-induced monocyte activation and PHA-induced lymphocyte activation measured by individual ELISA assays of (B) IL-1β and (C) IFNγ, respectively (n = 8). (D) Repression in immunostimulated TNFα production by dietary zinc depletion (n = 8). (E) Whole blood MT and zinc transporter transcripts responsive to LPS- or PHA-induced activation on Day 7 (n = 3). Values were normalized to 18S rRNA levels. (F) Repression in PHA-induced Zip8 transcript levels by acute dietary zinc depletion (n = 3). Values are mean ± SD. Values significantly different to respective baseline and those significantly different compared to baseline and postdepletion values are noted by asterisks *P < 0.05; **P < 0.01; ***P < 0.001, respectively.
Critical roles of zinc in cells required for the immune response suggest zinc transporter expression during immune cell activation may reflect the host’s dietary zinc status. To address this relationship in a simple assay, whole blood samples from the subjects were treated with PHA and LPS with subsequent analysis of zinc homeostatic gene transcripts, MT mRNA levels showed a statistically significant response to both PHA and LPS challenge, while MT, ZnT6, Zip1, Zip3, Zip6, Zip8, and Zip14 transcripts were up-regulated in response to activation by PHA (Fig. 6E). Of those PHA-induced responses, Zip8 expression, likely produced by lymphocytes (10, 13), was dysregulated by acute dietary zinc depletion with an approximate reduction of 50% (Fig. 6F).
Discussion
Zinc transporter and MT expression were the initial targets of interest due to their direct involvement in zinc trafficking and homeostatic regulation and their likelihood to respond to lower zinc availability by dietary restriction (14). A marked decrease in buccal MT mRNA, the prototypical zinc-regulated gene, by low zinc ingestion indicates its potential as a zinc biomarker that features noninvasive sampling. This finding corresponds with previous results where MT expression in blood cells is altered by dietary zinc restriction (15, 16). Differences between the DE transporter genes in PBMC and reticulocyte RNA that we identified indicate the cell type-specific regulation of these genes. Only ZnT1 mRNA, was uniformly affected by the acute zinc-depletion regimen. These data correspond to previous observations where ZnT1 is increased in response to supplemental zinc (10). This demonstrates the ability of ZnT1 to reflect dietary zinc bioavailability during zinc-deprived and -excess conditions. Serum zinc concentrations were measured as an evaluation tool of compliance and of effectiveness of our dietary zinc-depletion protocol. Its significant decrease by zinc depletion for 8 d and its reversal by repletion, respectively, show this index is indicative of dietary zinc ingestion levels. Depression in serum zinc has been observed by others during similar dietary depletion studies (17, 18). However, responsiveness to other conditions such as acute infection or starvation (7) raise questions regarding serum zinc as a biomarker except for healthy individuals.
Here using microarrays, we identified novel genes in cells from whole blood having modulated expression levels after acute dietary zinc depletion. The combination of RNA stabilization and globin transcript removal allowed preservation of in vivo transcriptome profiles and an increase in detection sensitivity of gene transcripts at low abundance (19, 20). Further bioinformatic analysis of the DE genes led to the identification of biological events associated with dietary zinc depletion. It is of note that the overrepresentation of genes involved in cell cycle and cell-mediated immune response identified by the up- and down-regulated genes in whole blood RNA, respectively, remarkably aligns with the findings from in vitro studies with zinc-deprived and -supplemented immune cell models (8, 9). In addition, these correspond to the well-characterized clinical outcomes of prolonged zinc deficiency, i.e., impaired immunity (21, 22) and predisposition to cancer development (23, 24).
Of considerable interest is the identification of vascular endothelial growth factor (Vegf) in the functional networks enriched by the overexpressed genes. Vegf has been considered to be protumorigenic (25, 26). Recently, intracellular zinc deprivation has been shown to result in increased Vegf production by prostate cancer cells (27). No change of Vegf gene expression in the blood cell populations was detected in our microarray experiments, indicating that expression of this growth factor, holding both autocrine and paracrine functions, may occur in other cell types/organs during zinc depletion. Nuclear factor kappa B (NFκB) was identified in the functional network composed of down-regulated genes, under acute dietary zinc depletion. The immunosuppressive effect of suboptimal zinc conditions has been associated with impaired DNA binding activity of this transcription factor (28). Relevant mechanisms include the zinc dependence for NFκB transactivation indicated by lower affinity of its binding motif under zinc deprivation (29–31). These findings imply that low zinc ingestion leads to a modulation in host defense by affecting gene expression associated with immune responses via a direct effect on NFκB activity. By applying a stringent filtering criterion, we identified eight well-characterized gene transcripts as candidate indices of dietary zinc deprivation (Fig. 4 and Table S6). All of these were up-regulated by the dietary zinc restriction protocol. Simultaneous measurement of these highly responsive genes may serve as a transcriptome-level signature for a genomic approach to assess dietary zinc deficiency.
Alteration in serum miRNA profiles have been shown to reflect the developmental and metastatic stage of various cancer types (32). However, the normal biological role of circulating miRNA remains unclear. Its energy-dependent export by tissues and stable form in peripheral blood suggest its potential to function as an intercellular or -organ messenger (33, 34). Among the 88 miRNAs screened with the assay used, miR-204 and miR-296-5p showed the highest responsiveness of down-regulation to dietary zinc depletion. It is of note that both of these miRNAs have shown to suppress oncogene expression (35, 36). These zinc-responsive miRNAs influence tumor progression, and hence may point to zinc deficiency as a predisposing factor for cancer. For example, miR-375 have been shown to circulate in lower levels in sera of patients with esophageal squamous cell carcinoma (37). The increased risk for esophageal cancer by dietary zinc deficiency has been extensively documented (24). Here we show the acute reversal of depletion-induced decrease in serum miR-375 levels by a zinc repletion regimen. The diagnostic potential of modulation of serum miRNA levels by zinc needs to be further explored.
Transcriptional machinery of miRNA gene expression, shares its regulatory components with that of protein-coding transcripts (38). Computational analysis of the regulatory elements of miRNA genes identified putative transcription factors involved in the regulation of miRNA gene expression (39). The zinc-sensing transcription factor, MTF-1, was predicted as one of the five master-regulators of human pre-miRNA expression. It is of note that two other of these predicted master-regulators, NF-Y and AP-2α, were identified as putative transcription factors eliciting the effects of dietary zinc depletion on gene expression by the microarray dataset of the current study (Fig. 3D).
Impaired immunity is one of the most extensively characterized outcomes of zinc deficiency (21, 22). This includes higher risks of morbidity and mortality from infectious diseases including pneumonia and malaria in populations with high prevalence of zinc deficiency (3, 4). Dependence of immune response on zinc transporter activity (13, 40) also underlines the importance of constant zinc levels in the cells conferring immunity. Whole blood cytokine assays, conducted here as an approach to evaluate the effects of low zinc ingestion on the host’s immune response indicate the potential of TNFα production levels induced by blood cell activation as indicative of zinc status. The increase in TNFα expression by monocytes in response to a high zinc intake (10) agrees with its repression by zinc depletion observed in the present study. This activation-dependent zinc-responsiveness of cytokine expression emphasizes the significance of sufficient dietary zinc intake for populations, particularly, at higher risks of infection or immunological dysfunction (3, 4, 41). A previous microarray study with immune cell lines identified tumor necrosis factors as a central regulator of the biological events associated with the genes influenced by zinc availability (9). Additionally, NFκB, a major regulator of TNFα gene expression, was present in the functional network composed of the down-regulated genes identified by our transcriptome analysis. These bioinformatics data suggest that impaired TNFα production may be a consequence of impairment in NFκB activity under low zinc conditions. Mechanistic studies with various immune and cancer cell models (29–31) support our hypothesis of the involvement of NFκB in conferring the zinc effects on immunoactivator-induced TNFα gene expression.
In conclusion, the present study has identified molecular indices that respond to acute dietary zinc depletion and repletion in humans using broad cell and blood sampling methods. Transcripts of blood cell genes associated with cell cycle regulation, host defense, and the regulation of zinc homeostasis hold the potential to indicate the dietary zinc status. Modulation of specific serum miRNAs by dietary zinc restriction and repletion are among the zinc-responsive biomarkers identified in these studies. TNFα release from whole blood under immunostimulation was clearly depressed by suboptimal zinc intake and represents a biomarker based on function. These findings support the established biological roles of zinc in immunity and cell proliferation, and implicate the therapeutic and prophylactic properties of dietary zinc against immune deficiency and cancer-related disorders.
Materials and Methods
Human Subjects.
Male subjects 21–35 yr of age, weighing at least 50 kg, were recruited to participate. Details of the subject characteristics and exclusion criteria are provided in Table S1. The study was approved by both the Institutional Review Board and the UF Clinical Research Center (UFCRC) of the University of Florida. All subjects provided written informed consent prior to enrollment. The study was registered at clinicaltrials.gov as NCT01221129.
Dietary Zinc Restriction and Sample Collection.
A 24-d observational study comprised of acclimation (7 d; 10.4 mg Zn/d), zinc depletion (10 d; 0.3 mg Zn/d), and zinc repletion (7 d; 28.9 mg Zn/d) phases was conducted with healthy male subjects (n = 9) (Tables S7 and S8). Serum zinc concentrations were monitored throughout the 24-d period. On day 0, 6, and 10 of zinc depletion, buccal swabs and whole blood were collected for molecular assays (Fig. S3. RNA profiles of buccal samples and whole blood were stabilized by using FTA paper cards and PAXgene reagents, respectively, during collection.
RNA Isolation and qPCR.
PBMC and erythroid cells were fractionated from EDTA-treated blood by centrifugation with Histopaque-1077 (Sigma). RNA from buccal samples and fractionated blood cells was isolated using TRI reagent (Ambion). qPCR assays for zinc transporter, MT, and normalization transcripts were designed and conducted as previously described (8, 10).
PAXgene RNA Processing and Microarray.
Whole blood RNA was prepared with the PAXgene system (PreAnalytiX). GLOBINclear (Ambion) was used to reduce the globin RNA abundance. RNA was biotinylated and amplified by the Illumina TotalPrep RNA Amplification kit (Ambion). cRNA products were hybridized to Illumina Human HT-12 v4 microarray chips.
Microarray Data Analysis.
Probes with detection P-values lower than 0.05 in all samples were excluded from the dataset prior to analyses. Raw data were quantile normalized, and differentially expressed genes were determined by BRB-ArrayTools (P < 0.005). Functional enrichment and network analyses were conducted with the EXPANDER 5.2 (42), PANTHER (43) and ingenuity pathway analysis softwares.
Whole Blood Cytokine Assay.
Heparinized whole blood was diluted with 4 vol of RPMI-1640 and was incubated with or without LPS (1 mg/L) or PHA (10 mg/L). After 24 h, cell-free medium was collected and treated with protease inhibitor cocktail (Pierce). RNA was extracted from the blood pellets using TRI reagent BD (Molecular Research Center). Cytokine levels were quantified with ELISArrays (SABiosciences), and zinc-related transcripts were assessed by qPCR.
Serum miRNA Assay.
Serum RNA was prepared using QIAzol (Qiagen) and the miRNeasy Mini kit (Qiagen). Synthetic cel-miR-39 was added prior to the RNA isolation process and served as a means of normalization. Zinc-responsive miRNAs were identified with the human serum miRNA PCR array from SABiosciences. Changes in selected miRNA levels were confirmed with RNA from individual serum samples and miScript qPCR assays (Qiagen).
For details of the study design and experimental procedures, see SI Materials and Methods.
Supplementary Material
ACKNOWLEDGMENTS.
We acknowledge the contributions of Dr. Wendy Dahl for the dietary protocol, as well as the valuable advice of Drs. Nancy D. Denslow, Louis A. Lichten, Tolunay B. Aydemir, and Liang Guo during the planning and analytic phases of this study, and Dr. Michael K. May (National Institute of Diabetes and Digestive and Kidney Diseases) for administrative support. We also thank Dr. Desmond Schatz and Teresa d’Angelo and the staff of the UF Clinical Research Center (UFCRC) and the Gene Expression core at the UF Interdisciplinary Center for Biotechnology Research. Research reported here was supported by National Institutes of Health Grants DK31127 and DK94244 (R.J.C.), Boston Family Endowment Funds of the University of Florida Foundation (R.J.C.), a CALS Graduate Student Alumni Award (M-S.R.), and in part by the NIH/NCRR Clinical and Translational Science Award UL1 RR029890 (UFCRC).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117207108/-/DCSupplemental.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE33174).
References
- 1.King J, Cousins RJ. Zinc. In: Shils ME, Shike M, Ross AC, Caballero B, Cousins RJ, editors. Modern Nutrition in Health and Disease. 10th Ed. Baltimore: Lippincott Williams and Wilkins; 2005. pp. 271–285. [Google Scholar]
- 2.Prasad AS. Clinical manifestations of zinc deficiency. Annu Rev Nutr. 1985;5:341–363. doi: 10.1146/annurev.nu.05.070185.002013. [DOI] [PubMed] [Google Scholar]
- 3.Caulfield LE, Black RE. Zinc deficiency. In: Ezzati M, Lopez AD, Rodgers A, Murray CJ, editors. Comparative Quantification of Health Risks. Vol 1. Geneva, Switzerland: World Health Organization; 2004. pp. 257–280. [Google Scholar]
- 4.Black RE, et al. Maternal and child undernutrition: Global and regional exposures and health consequences. Lancet. 2008;371:243–260. doi: 10.1016/S0140-6736(07)61690-0. [DOI] [PubMed] [Google Scholar]
- 5.Prasad AS. Recognition of zinc-deficiency syndrome. Nutrition. 2001;17:67–69. doi: 10.1016/s0899-9007(00)00469-x. [DOI] [PubMed] [Google Scholar]
- 6.King JC. Zinc: An essential but elusive nutrient. Am J Clin Nutr. 2011;94:679S–684S. doi: 10.3945/ajcn.110.005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibson RS, Hess SY, Hotz C, Brown KH. Indicators of zinc status at the population level: A review of the evidence. Brit J Nutr. 2008;99(Suppl 3):S14–23. doi: 10.1017/S0007114508006818. [DOI] [PubMed] [Google Scholar]
- 8.Cousins RJ, et al. A global view of the selectivity of zinc deprivation and excess on genes expressed in human THP-1 mononuclear cells. Proc Natl Acad Sci USA. 2003;100:6952–6957. doi: 10.1073/pnas.0732111100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haase H, et al. Differential gene expression after zinc supplementation and deprivation in human leukocyte subsets. Mol Med. 2007;13:362–370. doi: 10.2119/2007-00049.Haase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aydemir TB, Blanchard RK, Cousins RJ. Zinc supplementation of young men alters metallothionein, zinc transporter, and cytokine gene expression in leukocyte populations. Proc Natl Acad Sci USA. 2006;103:1699–1704. doi: 10.1073/pnas.0510407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao J, Cousins RJ. Metallothionein mRNA in monocytes and peripheral blood mononuclear cells and in cells from dried blood spots increases after zinc supplementation of men. J Nutr. 2000;130:2180–2187. doi: 10.1093/jn/130.9.2180. [DOI] [PubMed] [Google Scholar]
- 12.Hotz C, Peerson JM, Brown KH. Suggested lower cutoffs of serum zinc concentrations for assessing zinc status: Reanalysis of the second National Health and Nutrition Examination Survey data (1976–1980) Am J Clin Nutr. 2003;78:756–764. doi: 10.1093/ajcn/78.4.756. [DOI] [PubMed] [Google Scholar]
- 13.Aydemir TB, Liuzzi JP, McClellan S, Cousins RJ. Zinc transporter ZIP8 (SLC39A8) and zinc influence IFN-gamma expression in activated human T cells. J Leukocyte Biol. 2009;86:337–348. doi: 10.1189/jlb.1208759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lichten LA, Cousins RJ. Mammalian zinc transporters: Nutritional and physiologic regulation. Annu Rev Nutr. 2009;29:153–176. doi: 10.1146/annurev-nutr-033009-083312. [DOI] [PubMed] [Google Scholar]
- 15.Grider A, Bailey LB, Cousins RJ. Erythrocyte metallothionein as an index of zinc status in humans. Proc Natl Acad Sci USA. 1990;87:1259–1262. doi: 10.1073/pnas.87.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allan AK, et al. Lymphocyte metallothionein mRNA responds to marginal zinc intake in human volunteers. Brit J Nutr. 2000;84:747–756. [PubMed] [Google Scholar]
- 17.Gordon PR, Woodruff CW, Anderson HL, O’Dell BL. Effect of acute zinc deprivation on plasma zinc and platelet aggregation in adult males. Am J Clin Nutr. 1982;35:113–119. doi: 10.1093/ajcn/35.1.113. [DOI] [PubMed] [Google Scholar]
- 18.Chung CS, et al. Current dietary zinc intake has a greater effect on fractional zinc absorption than does longer term zinc consumption in healthy adult men. Am J Clin Nutr. 2008;87:1224–1229. doi: 10.1093/ajcn/87.5.1224. [DOI] [PubMed] [Google Scholar]
- 19.Debey S, et al. A highly standardized, robust, and cost-effective method for genome-wide transcriptome analysis of peripheral blood applicable to large-scale clinical trials. Genomics. 2006;87:653–664. doi: 10.1016/j.ygeno.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 20.Tian Z, et al. A practical platform for blood biomarker study by using global gene expression profiling of peripheral whole blood. PLoS One. 2009;4:e5157. doi: 10.1371/journal.pone.0005157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rink L, Kirchner H. Zinc-altered immune function and cytokine production. J Nutr. 2000;130(5S Suppl):1407S–1411S. doi: 10.1093/jn/130.5.1407S. [DOI] [PubMed] [Google Scholar]
- 22.Fraker PJ, King LE. Reprogramming of the immune system during zinc deficiency. Annu Rev Nutr. 2004;24:277–298. doi: 10.1146/annurev.nutr.24.012003.132454. [DOI] [PubMed] [Google Scholar]
- 23.Ho E. Zinc deficiency, DNA damage and cancer risk. J Nutr Biochem. 2004;15:572–578. doi: 10.1016/j.jnutbio.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 24.Fong LY. Zinc in cancer development and prevention. In: Milner JA, Romagnolo DF, editors. Bioactive Compounds and Cancer. 1st Ed. New York: Humana Press; 2010. pp. 497–531. (Nutrition and Health). [Google Scholar]
- 25.Wu W, Shu X, Hovsepyan H, Mosteller RD, Broek D. VEGF receptor expression and signaling in human bladder tumors. Oncogene. 2003;22:3361–3370. doi: 10.1038/sj.onc.1206285. [DOI] [PubMed] [Google Scholar]
- 26.Hasan MR, Ho SH, Owen DA, Tai IT. Inhibition of VEGF induces cellular senescence in colorectal cancer cells. Int J Cancer. 2011;129:2115–2123. doi: 10.1002/ijc.26179. [DOI] [PubMed] [Google Scholar]
- 27.Golovine K, et al. Depletion of intracellular zinc increases expression of tumorigenic cytokines VEGF, IL-6 and IL-8 in prostate cancer cells via NF-kappaB-dependent pathway. Prostate. 2008;68:1443–1449. doi: 10.1002/pros.20810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prasad AS. Effects of zinc deficiency on Th1 and Th2 cytokine shifts. J Infect Dis. 2000;182(Suppl 1):S62–68. doi: 10.1086/315916. [DOI] [PubMed] [Google Scholar]
- 29.Prasad AS, Bao B, Beck FW, Sarkar FH. Zinc activates NF-kappaB in HUT-78 cells. J Lab Clin Med. 2001;138:250–256. doi: 10.1067/mlc.2001.118108. [DOI] [PubMed] [Google Scholar]
- 30.Oteiza PI, Clegg MS, Keen CL. Short-term zinc deficiency affects nuclear factor-kappab nuclear binding activity in rat testes. J Nutr. 2001;131:21–26. doi: 10.1093/jn/131.1.21. [DOI] [PubMed] [Google Scholar]
- 31.Mackenzie GG, Zago MP, Keen CL, Oteiza PI. Low intracellular zinc impairs the translocation of activated NF-kappa B to the nuclei in human neuroblastoma IMR-32 cells. J Biol Chem. 2002;277:34610–34617. doi: 10.1074/jbc.M203616200. [DOI] [PubMed] [Google Scholar]
- 32.Wittmann J, Jack HM. Serum microRNAs as powerful cancer biomarkers. Biochim Biophys Acta. 2010;1806:200–207. doi: 10.1016/j.bbcan.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 33.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 34.Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee Y, et al. Network modeling identifies molecular functions targeted by miR-204 to suppress head and neck tumor metastasis. PLoS Comput Biol. 2010;6:e1000730. doi: 10.1371/journal.pcbi.1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei JJ, et al. Regulation of HMGA1 expression by microRNA-296 affects prostate cancer growth and invasion. Clin Cancer Res. 2011;17:1297–1305. doi: 10.1158/1078-0432.CCR-10-0993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Komatsu S, et al. Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. Brit J Cancer. 2011;105:104–111. doi: 10.1038/bjc.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 39.Lee J, Li Z, Brower-Sinning R, John B. Regulatory circuit of human microRNA biogenesis. PLoS Comput Biol. 2007;3:e67. doi: 10.1371/journal.pcbi.0030067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kitamura H, et al. Toll-like receptor-mediated regulation of zinc homeostasis influences dendritic cell function. Nat Immunol. 2006;7:971–977. doi: 10.1038/ni1373. [DOI] [PubMed] [Google Scholar]
- 41.Prentice AM, et al. New challenges in studying nutrition-disease interactions in the developing world. J Clin Invest. 2008;118:1322–1329. doi: 10.1172/JCI34034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ulitsky I, et al. Expander: From expression microarrays to networks and functions. Nat Protoc. 2010;5(2):303–322. doi: 10.1038/nprot.2009.230. [DOI] [PubMed] [Google Scholar]
- 43.Thomas PD, et al. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003;13(9):2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.