Abstract
Polynucleotide kinase/phosphatase (PNKP) is a critical mammalian DNA repair enzyme that generates 5′-phosphate and 3′-hydroxyl groups at damaged DNA termini that are required for subsequent processing by DNA ligases and polymerases. The PNKP phosphatase domain recognizes 3′-phosphate termini within DNA nicks, gaps, or at double- or single-strand breaks. Here we present a mechanistic rationale for the recognition of damaged DNA termini by the PNKP phosphatase domain. The crystal structures of PNKP bound to single-stranded DNA substrates reveals a narrow active site cleft that accommodates a single-stranded substrate in a sequence-independent manner. Biochemical studies suggest that the terminal base pairs of double-stranded substrates near the 3′-phosphate are destabilized by PNKP to allow substrate access to the active site. A positively charged surface distinct from the active site specifically facilitates interactions with double-stranded substrates, providing a complex DNA binding surface that enables the recognition of diverse substrates.
Keywords: DNA substrate recognition, HAD family, , protein-DNA complexes, X-ray crystallography
DNA damage induced by ionizing radiation, as well as DNA repair intermediates generated during base excision repair, often result in the formation of DNA ends containing 3′-phosphate/5′-hydroxyl termini. Because DNA polymerases and ligases require 3′-hydroxyl/5′-phosphate termini to complete DNA repair, cells have evolved enzymes to recognize and process these damaged DNA ends. Mammalian polynucleotide kinase/phosphatase (PNKP) contains 3′-phosphatase and 5′-kinase activities present in two distinct active sites (1). The critical role of PNKP in various DNA repair processes is underlined by the fact that RNAi knockdown of PNKP leads to marked sensitization of human cells to spontaneous mutation as well as to various genotoxic agents (2). A particularly important role is attributed to the phosphatase activity as small molecules that selectively inhibit the phosphatase but not the kinase activity of PNKP sensitize human cells to DNA damage in a PNKP-dependent manner (3, 4). Mutations in the PNKP phosphatase domain are associated with the developmental neurological disorder MCSZ, revealing a critical role for PNKP in human neurological development (5).
The PNKP phosphatase domain plays a central role in several critical DNA repair processes. For example, PNKP participates in base excision repair (BER) in partnership with the NEIL (endonuclease VIII-like) DNA glycosylases. NEILs exhibit β,δ-AP (aminopurine) lyase activity, which produces single nucleotide gap DNAs containing 3′-phosphate termini that are subsequently dephosphorylated by PNKP prior to gap filling and ligation (6, 7). PNKP is also associated with the repair of double-strand breaks through the nonhomologous end joining (NHEJ) pathway (8, 9), in a manner that relies on the interaction of PNKP with the NHEJ scaffolding protein, XRCC4 (10). Consistent with its role in these DNA repair pathways, PNKP recognizes 3′-phosphate termini in the context of DNA gaps and nicks, as well as double- and single-strand breaks (11). While recent structural and biochemical approaches have begun to shed light on how the kinase active site recognizes its substrates (12), the way in which substrates are recognized by the distinct phosphatase active site are not well understood. Here we present the results of crystallographic and biochemical studies of PNKP phosphatase and its interactions with DNA substrates. Our results reveal a deep phosphatase active site cleft that accommodates single-stranded substrates in a sequence-independent manner. We present biochemical data that support a model for the recognition of double-stranded substrates in which base pairing near the 3′-phosphate is destabilized by the enzyme. We suggest that PNKP distorts target DNA structures to access damaged substrate DNA ends, thus providing a molecular mechanism for the involvement of PNKP in the repair of both single- and double-strand breaks.
Results and Discussion
Structure Determination of PNKP Catalytic Domain Bound to 3′-Phosphate DNA Substrates.
In order to provide a structural view of PNKP phosphatase—substrate interactions, we set out crystallize the murine PNKP catalytic domain, encompassing both the phosphatase and kinase domains, in complex with DNA substrates containing 3′-phosphate termini. Substrate binding was stabilized by mutation of Asp170 to alanine (PNKPD170A). Asp170 is predicted to attack the 3′-phosphate, generating a covalent phospho-aspartate intermediate in the first step of the reaction (13). We were successful in crystallizing this domain in the apo form, and bound to five nucleotide single-stranded DNAs bearing either C, T, or A at the 3′ position, and these structures were determined to resolutions between 1.7 and 2.1 Å (Table S1, Materials and Methods, and Fig. S1).
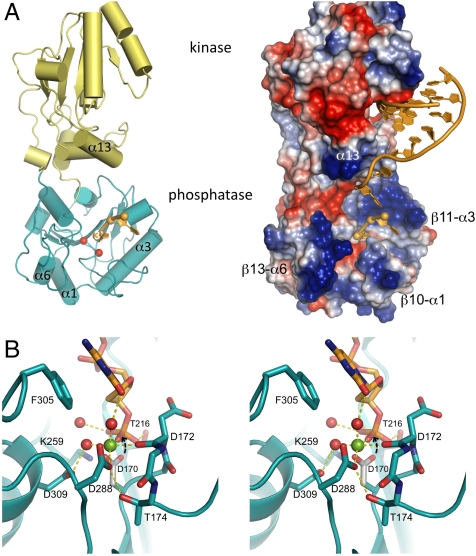
The structures reveal a phosphatase active site that binds the single-stranded substrates on the same side of the enzyme as the kinase active site (Fig. 1A). The narrow active site groove is bounded by walls composed of the β10-α1, β11-α3, β12-α4, and the β13-α6 regions (Fig. 1A and Fig. S2). The kinase domain overhangs the phosphatase active site, in particular α13, which is believed to play a role in recognition of kinase substrates (12). The two nucleotides of substrate DNA visible in these structures traverse ∼13 Å from the surface of the phosphatase domain to the catalytic center. The 3′-most base is accommodated in an open channel formed between the β10-α1 and β13-α6 loops.
Fig. 1.
Structure of PNKP bound to DNA substrates. (A) Overview of PNKP—DNA interactions in ribbon (left) and electrostatic surface representations (right). The PNKP phosphatase domain is teal, the kinase domain is yellow, and DNA substrates orange. In addition to the phosphatase DNA substrate, a model for the docking of a minimal DNA substrate to the kinase domain (12) is shown in the right box. Selected protein elements that bound the phosphatase active site are labeled, and the positions of the catalytic Asp170 and Asp172 residues in the phosphatase are indicated by red spheres. (B) Stereo view of the PNKPD170A phosphatase active site bound to a substrate DNA containing a 3′ cytosine. Residues interacting with the 3′ nucleotide as well as the active site Mg++ are labeled. Asp170 is modeled (gray) based on its orientation in the wild type structure (11) and its predicted in-line attack on the substrate 3′-phosphate is indicated with an arrow.
Mechanism of Catalysis.
The PNKP phosphatase domain adopts a haloacid dehydrogenase (HAD) fold (14) commonly found in many phosphatases, and contains conserved active site residues that suggest the enzyme utilizes a two step mechanism that is characteristic of this family (13). Key to catalysis is an active site Mg++ ion, which binds and stabilizes the growing negative charge on the substrate phosphate during catalysis, as well as binding and positioning one of the critical active site aspartate residues. This Mg++ was not visible in the previous structure of full-length PNKP, however Mg++ is visible in the high resolution apoPNKPD170A and PNKPD170A—substrate complexes. The Mg++ is bound by Asp288 carboxylate, the backbone carbonyl of Asp172, and likely also the side chain carboxylate of Asp170, which is not present in these structures (Fig. 1B). Filling out the octahedral coordination shell are two well ordered water molecules and the 3′-phosphate. The 3′-phosphate is also bound by Lys259, Thr216, and the main chain NH of Leu171. In this way, the substrate phosphate is held in place for nucleophilic attack of the Asp170 carboxylate in the first step of the reaction to generate the covalent phospho-aspartate intermediate. The phospho-aspartate is hydrolyzed in the second step, likely catalyzed by Asp172, which deprotonates the attacking water molecule.
Recognition of Single-Stranded DNA Substrates by PNKP.
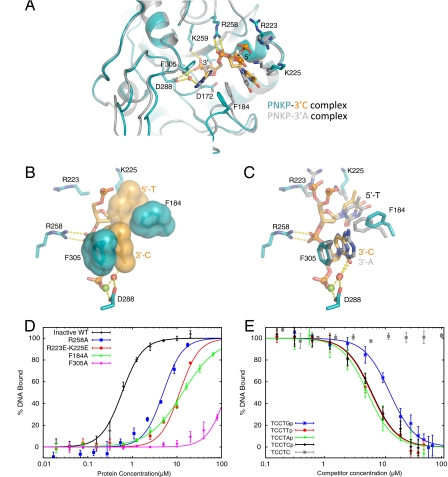
The structures of PNKP bound to each of the three ssDNA substrates reveal a number of protein-DNA contacts involving the terminal two nucleotides (Fig. 2A). The base of the 3′-most nucleotide contacts Phe305 through a stacking interaction that is similar in the three substrate structures determined (Fig. 2 B and C). The importance of this contact was assessed using a fluorescence polarization (FP) assay in which the substrate pentanucleotide 3′-phosphate DNA was 5′-labeled with fluorescein (FAM-DNA, Materials and Methods). Titration of this DNA with PNKPD170A yielded a dissociation constant (KD) of 600 ± 30 nM, however mutation of Phe305 to alanine (F305A) almost completely abolished binding (Fig. 2D). In addition to the recognition of the 3′-phosphate, the 5′-phosphate of the 3′ terminal nucleotide is also bound through a bidentate interaction with Arg258. Mutation of this residue to alanine (R258A) reduces substrate binding affinity ∼10-fold, indicating a significant role in substrate binding. The next nucleotide adopts different conformations in the three different structures, in spite of the fact that it is a thymine nucleotide in each of the structures. This variation is likely due to differences in the packing environments in the different crystal forms, which result in small conformational shifts in the β10-α1 and β13-α6 regions (Fig. 2A). The 5′-phosphate of this residue makes long-range electrostatic interactions with Arg223 and Lys225, as demonstrated by the fact that mutation of these residues to glutamate (R223E-K225E) markedly reduces substrate binding affinity (Fig. 2D). The thymine base of this nucleotide makes hydrophobic contacts to Phe184 (Fig. 2 B and C). Mutation of Phe184 to alanine (F184A) dramatically reduces substrate binding affinity, however this effect may also arise from a loss of contact of Phe184 with the 3′ terminal base (Fig. 2D). The recognition of the penultimate nucleotide explains the finding that pCp is dephosphorylated much less efficiently than pCpCp (11).
Fig. 2.
Recognition of single-stranded substrate DNA by the PNKP phosphatase domain. (A) Superposition of the complexes of PNKPD170A with a 3′-cytosine substrate (teal and orange) with the complex containing a 3′-adenosine (gray). Substrate contacting residues are shown as sticks and hydrogen bonding interactions are shown with dotted lines. (B) Close-up view of interactions of PNKPD170A with the 3′-cytosine substrate. Surfaces are displayed for Phe184, Phe305 and the substrate bases to illustrate stacking interactions that stabilize this complex. The red sphere indicates a water that forms part of the Mg++ coordination shell that also hydrogen bonds to the O2 of the 3′ base. (C) Similar to (B), with the 3′-adenosine complex superimposed in gray. (D) Determination of the binding of PNKPD170A and mutants to 5′-FAM-TTCTCp-3′ by fluorescence polarization (FP) spectroscopy. PNKPD170A (inactive WT) or the indicated variant was titrated against the FAM-labeled DNA and the data was fit to a sigmoidal binding curve (SI Text). (E) Influence of the identity of the 3′ base on PNKPD170A-substrate interactions. FP competition experiments were carried out in which PNKPD170A-FAM-DNA complexes were challenged with the indicated unlabeled DNAs.
PNKP does not directly interact with the Watson-Crick edge of the 3′ terminal base, instead, water molecules hydrogen bond to this surface. The one conserved water-mediated interaction with the base involves a water that constitutes part of the octahedral coordination shell of the catalytic Mg++. This water forms a hydrogen bond with the O2 atoms of cytosine or thymine bases at the 3′ position, or an equivalent hydrogen bond with the N3 of adenine (Fig. 2C), suggesting that PNKP binds 3′-phosphate substrates in a manner that is independent of the identity of the 3′ base. We used DNAs containing A, C, T, or G bases at the 3′ position as competitors of the PNKPD170A-FAM-DNA complex in FP experiments. Each of the DNAs was able to compete for binding to PNKPD170A, dependent on the presence of a 3′-phosphate in the competitor DNA (Fig. 2E). The results indicate that 3′-phosphorylated DNAs containing A, C, or T at the 3′ position bind PNKP with essentially identical affinities, consistent with the lack of sequence-specific interactions between PNKP and these residues in the crystal structures. The DNA containing G at the 3′ position was an approximately threefold weaker competitor, suggesting that PNKP may bind substrates containing a 3′-guanine base less well than other substrates.
Recognition of Double-Stranded Substrates Involves Destabilization of Base Pairing.
PNKP plays well characterized roles in NEIL-dependent BER and NHEJ, and, consistent with these cellular functions, the PNKP phosphatase domain dephosphorylates 3′-phosphate termini present at DNA gaps, nicks, and double-strand breaks with similar efficiencies (11). Our structural data, however, reveal a narrow and deep active site cleft that can accommodate single but not double-stranded substrates. We propose that PNKP interacts with double-stranded substrates by destabilizing base pairing near the 3′-phosphate terminus, thereby releasing the substrate strand to allow access to the phosphatase active site. We propose that Phe184, which protrudes from a protein surface feature we term “the Phe wedge,” plays a critical role in the initiation of base-pair destabilization through hydrophobic interactions with the substrate bases, destabilizing intrahelical base stacking in the substrate (Fig. 3 A and B). Phe305, which is buried deeper in the active site, could play a role in stabilizing the liberated single-strand in the catalytic conformation through stacking interactions with the 3′-terminal base. Distinct from the active site cleft is a second positively charged region composed of residues from the Phe wedge (Lys182) as well as the β13-α6 loop (residues 300–303). We propose that these residues interact with the partner strand in a double-stranded substrate, providing additional binding energy in the initial recognition of the duplex DNA before base-pair destabilization, as well as interactions with the released nondamaged strand after base-pair destabilization.
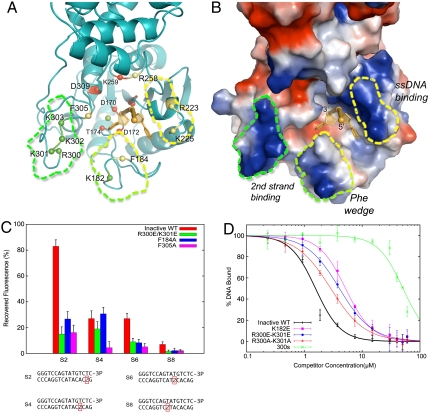
Fig. 3.
Interactions of PNKP with dsDNA substrates involves base-pair destabilization. (A) Ribbons view of a PNKPD170A—ssDNA complex in which residues directly involved in catalysis, Mg++ binding, or 3′-phosphate binding, are red, those involved in other interactions with the ssDNA substrate are yellow, and residues proposed to interact with the second, undamaged strand in a two-stranded complex are colored green. (B) Electrostatic surface representation of the same view as in (B), with the ssDNA binding surface, Phe wedge and 2nd strand binding surfaces indicated. (C) Demonstration of PNKP-dependent destabilization of base-pairing in double-stranded substrates by 2AP spectroscopy. 2AP base-pair destabilization was assessed through the recovery of 2AP fluorescence upon binding PNKPD170A compared to a single-stranded control. The four 2AP substrates used are indicated at the bottom, and results for PNKPD170A (inactive WT), as well as the indicated PNKPD170A mutants are shown. (D) Interactions between PNKPD170A or the mutants indicated and a dsDNA substrate were assessed in a FP competition assay in which a single-stranded FAM-DNA was challenged with an unlabeled dsDNA. 300s refers to a mutant in which residues 300–303 are all mutated to glutamate.
To detect base-pair destabilization in double-stranded PNKP substrates, we tested the ability of PNKP to modulate the fluorescence of double-stranded DNA substrates harboring 2-aminopurine (2AP). 2AP is a fluorescent base analog that is quenched by base stacking interactions (15) and has been used to study base-pair destabilization induced by protein-DNA interactions (16). We created a set of blunt-ended double-stranded substrates containing a 3′-phosphate terminus on one strand and a single 2AP at positions 2, 4, 6, or 8 with respect to the blunt end in the complementary strand. We measured 2AP fluorescence in each of these samples bound to PNKPD170A, corrected for background fluorescence from the PNKPD170A-dsDNA complex. The results were expressed as a % recovery of fluorescence in each sample normalized to the fluorescence of the 2AP-labeled single-strand (Fig. 3C and Materials and Methods). The results clearly indicate that PNKP binding induces a dramatic (83%) reduction in fluorescence quenching in the substrate containing 2AP at position 2 (S2, Fig. 3C). Substrates containing 2AP further from the end of the substrate (at positions 4 and 6) showed a less pronounced, 27% reduction in 2AP quenching, while the substrate harboring 2AP at position 8 exhibited very little reduction in quenching. These results are consistent with destabilization of base pairing in the terminal 2 base pairs of the blunt-ended substrate, while the following base pairs up to position 6 may be partially destabilized. Base pairing further from the DNA end (position 8) is not perturbed by PNKP interactions. We next tested the ability of PNKPD170A harboring additional mutations to destabilize substrate base pairing. PNKPD170A/F305A elicited only a small (16%) reduction in fluorescence quenching in the 2AP S2 substrate, and little detectable reduction in quenching in any of the other substrates, suggesting a key role for Phe305 and its stacking interactions with the base of the 3′-phosphorylated residue in base-pair destabilization (Fig. 3C). PNKPD170A/F184A also induced only a small reduction in quenching S2, however quenching in the S4 substrate was similar to PNKPD170A. Fluorescence recovery for the S6 and S8 substrates was minimal. Together, these results suggest that both phenylalanine residues in the active site cleft are important for substrate base-pair destabilization.
To investigate the role of the putative second strand binding surfaces (Fig. 3 A and B) in substrate recognition, we generated several mutants in these residues and tested their substrate binding and base-pair destabilization activities. All of the mutants generated in this region (residues 182, 300–303) displayed levels of single-stranded substrate binding similar to PNKPD170A (Fig. S3). To test the impact of these mutations on double-strand substrate binding, we assessed the ability of a double-stranded DNA containing a blunt, 3′-phosphorylated end, to compete with a single-stranded FAM-DNA in an FP assay (Fig. 3D). In a control experiment, the double-stranded DNA was found to compete approximately sixfold more efficiently for PNKP binding than the unlabeled version of the single-strand, suggesting that double-stranded substrates are bound more tightly by PNKP than single-stranded substrates (Fig. S4). Mutation of both Arg300 and Lys301 to alanine resulted in a small but reproducible reduction in the competition of the dsDNA compared to the wild type PNKPD170A control. Mutation of the same residues to glutamate resulted in a further reduction in competition, suggesting electrostatic interactions with dsDNA. Mutation of positively charged residues 300–303 to glutamate (300s, Fig. 3D), resulted in a dramatic loss of dsDNA competition, even though this variant bound to the single-stranded FAM-DNA with nearly wild type affinity (Fig. S3). Lys182 from the Phe wedge may also play a role in dsDNA binding because mutation of this residue to glutamate also reduced dsDNA competition (Fig. 3D). Finally, we used our 2AP fluorescence assay to test the ability of the PNKPD170A/R300E/R301E mutant to destabilize substrate base pairing. The results indeed indicate a dramatic reduction in 2AP fluorescence recovery in the S2 DNA compared to the wild type (Fig. 3C). This result indicates that these residues are directly involved in destabilization of the duplex DNA, because this mutant binds single-stranded DNA with nearly wild type affinity.
A Model for PNKP-Induced Substrate Distortion.
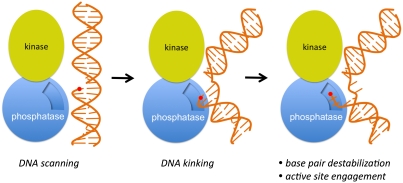
We suggest the following model for DNA damage detection and catalysis (Fig. 4). We propose that the large, nearly contiguous substrate binding surface across the PNKP kinase and phosphatase domains (Fig. 1A) interacts nonspecifically with DNA and could possibly scan for regions of flexibility indicative of DNA strand breaks. Distortion of the DNA at the strand break could allow further interrogation of the site of damage, facilitating access of the 3′-terminus to the groove at the interface of the kinase and phosphatase domains that yields access to the phosphatase active site. PNKP-driven destabilization of 2–3 base pairs at the site would allow access to the phosphatase catalytic center and dephosphorylation of the 3′-end. To compensate for the unfavorable free energy associated with base-pair disruption, the complementary, undamaged strand may interact with the second strand binding region (Fig. 3 A and B). The importance of this region is underlined by its conservation throughout metazoan PNKP enzymes (Fig. S2). Interestingly, this region is absent in prokaryotic T4 PNKP. While structural and mutagenesis data in the T4 system (17) suggest a similar mechanism of recognition of single-stranded nucleic acids, the recognition and processing of double-stranded substrates may involve a mechanism distinct from that employed by the mammalian enzymes. The mechanism presented here predicts that nicked or gapped substrates may interact with or sterically occlude interactions of a second substrate at the kinase active site. In agreement with this model, it has been shown that the binding of such structures to PNKP bearing mutations at phosphatase active site strongly inhibits productive substrate binding by the kinase domain (18).
Fig. 4.
A model for the detection and recognition of 3′-phosphate termini in dsDNA substrates by PNKP. PNKP may initially scan DNA via nonspecific interactions that involve the positively charged surfaces of both the kinase and phosphatase domains located on the same side of the protein (left). Encounter of a flexible lesion such as a single nucleotide gap containing a 3′-phosphate terminus (red circle) could allow distortion of the substrate, enhancing interactions with PNKP (middle). Engagement of the phosphatase active site requires destabilization of limited base pairs proximal to the 3′-phosphate end, prior to catalysis.
An evolving theme in selective recognition of damaged DNA by repair enzymes is the binding-assisted distortion of the damaged DNA. For example, the recently determined structures of the 5′-flap endonuclease FEN-1 (19), and the related 5′-exonuclease hExo1 (20) bound to DNAs suggest a binding mechanism in which the substrate is kinked with limited base-pair disruption near the site of cleavage. Such distortion is required for access to the backbone of the DNA and in-line attack of the phosphodiester bonds that are central to the mechanisms of both PNKP and the 5′-endonucleases. Such binding mechanisms result in significant affinity of the repair enzyme for the product, which has been suggested to be critical for the ordered processing of DNA lesions by multiple enzymes in a manner that protects potentially mutagenic DNA repair intermediates (21). Consistent with this model, PNKP has been shown to have significant affinity for the products of both its kinase and phosphatase domains (22). Interactions with the single-strand repair scaffold protein, XRCC1 or the NHEJ scaffold protein, XRCC4, facilitate PNKP product release, consistent with the idea that these scaffold proteins organize multistep, coordinated repair of DNA damage (23–25).
Materials and Methods
Fluorescence Polarization Spectroscopy.
The binding affinity of the PNKPD170A to DNA was evaluated by fluorescence polarization spectroscopy using a 3′-phosphorylated fluorescent ssDNA (5′-FAM-TCCTCp-3′, designated FAM-DNA) synthesized by Integrated DNA Technologies (IDT). To test the binding affinity of other 3′-phosphorylated DNA substrates, a competition assay was used. In this assay, we measured the ability of 3′-phosphorylated DNAs (see SI Text for sequences) to compete off the fluorescein-labeled DNA.
Fluorescence Spectroscopic Studies of 2AP-Modified DNA Complexes.
Steady-state fluorescence of DNAs containing 2AP was measured at room temperature on a PTI spectrofluorometer. The 2AP was excited at 315 nm and the fluorescence was monitored at 370 nm with 5-nm spectral resolution for excitation and emission. A complete description of this experiment is provided in SI Text.
Crystallization.
The PNKPD170A catalytic fragment, either in the apo state or bound to ssDNA substrates, was crystallized at room temperature by hanging drop vapor diffusion. Crystals of apo PNKPD170A were obtained by mixing an equal volume of protein at 4 mg/mL (1 μL) and well solution composed of 0.1 M Bis-Tris pH 7, 20% PEG 3350. For PNKPD170A complexed with DNA, protein was incubated with 3′-phosphorylated ssDNAs (5′-TTCTXp-3′; X = A,T,C) in a 1∶3 ratio on ice for 2 h. Crystals grew in similar conditions to that of the apo PNKPD170A (25% PEG3350 and pH range 6.5–7) with 10% t-butanol as additive. Details for protein expression, purification, crystallographic data collection and processing, and for structure determination and refinement are found in SI Text.
Supplementary Material
Acknowledgments.
We thank P. Grochulski (Canadian Light Source, Saskatoon, SK), S. Classen (Advandced Light Source, Berkeley, CA) and M. Soltis (Stanford Synchrotron Radiation Laboratory, Stanford, CA) for help with diffraction data collection and M. Weinfeld for helpful discussions and critical reading of this manuscript. This work was supported by grants from the Canadian Cancer Society, Alberta Cancer Foundation, and the National Institutes of Health. J.N.M.G. acknowledges the support of the HHMI International Scholarship program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.A.T. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3U7E, 3U7F, 3U7G, and 3U7H).
See Commentary on page 20855.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112036108/-/DCSupplemental.
References
- 1.Weinfeld M, Mani RS, Abdou I, Aceytuno RD, Glover JN. Tidying up loose ends: the role of polynucleotide kinase/phosphatase in DNA strand break repair. Trends Biochem Sci. 2011;36:262–271. doi: 10.1016/j.tibs.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rasouli-Nia A, Karimi-Busheri F, Weinfeld M. Stable down-regulation of human polynucleotide kinase enhances spontaneous mutation frequency and sensitizes cells to genotoxic agents. Proc Natl Acad Sci USA. 2004;101:6905–6910. doi: 10.1073/pnas.0400099101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freschauf GK, et al. Identification of a small molecule inhibitor of the human DNA repair enzyme polynucleotide kinase/phosphatase. Cancer Res. 2009;69:7739–7746. doi: 10.1158/0008-5472.CAN-09-1805. [DOI] [PubMed] [Google Scholar]
- 4.Freschauf GK, et al. Mechanism of action of an imidopiperidine inhibitor of human polynucleotide kinase/phosphatase. J Biol Chem. 2010;285:2351–2360. doi: 10.1074/jbc.M109.055764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen J, et al. Mutations in PNKP cause microcephaly, seizures and defects in DNA repair. Nat Genet. 2010;42:245–249. doi: 10.1038/ng.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiederhold L, et al. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Das A, et al. NEIL2-initiated, APE-independent repair of oxidized bases in DNA: evidence for a repair complex in human cells. DNA Repair. 2006;5:1439–1448. doi: 10.1016/j.dnarep.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karimi-Busheri F, Rasouli-Nia A, Allalunis-Turner J, Weinfeld M. Human polynucleotide kinase participates in repair of DNA double-strand breaks by nonhomologous end joining but not homologous recombination. Cancer Res. 2007;67:6619–6625. doi: 10.1158/0008-5472.CAN-07-0480. [DOI] [PubMed] [Google Scholar]
- 9.Chappell C, Hanakahi LA, Karimi-Busheri F, Weinfeld M, West SC. Involvement of human polynucleotide kinase in double-strand break repair by non-homologous end joining. Embo J. 2002;21:2827–2832. doi: 10.1093/emboj/21.11.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koch CA, et al. Xrcc4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. Embo J. 2004;23:3874–3885. doi: 10.1038/sj.emboj.7600375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernstein NK, et al. The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase. Mol Cell. 2005;17:657–670. doi: 10.1016/j.molcel.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 12.Bernstein NK, et al. Mechanism of DNA substrate recognition by the mammalian DNA repair enzyme, polynucleotide kinase. Nucleic Acids Res. 2009;37:6161–6173. doi: 10.1093/nar/gkp597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernstein NK, et al. Polynucleotide kinase as a potential target for enhancing cytotoxicity by ionizing radiation and topoisomerase I inhibitors. Anti-Cancer Agents in Medicinal Chemistry. 2008;8:358–367. doi: 10.2174/187152008784220311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koonin EV, Tatusov RL. Computer analysis of bacterial haloacid dehalogenases defines a large superfamily of hydrolases with diverse specificity. Application of an iterative approach to database search. J Mol Biol. 1994;244:125–132. doi: 10.1006/jmbi.1994.1711. [DOI] [PubMed] [Google Scholar]
- 15.Guest CR, Hochstrasser RA, Sowers LC, Millar DP. Dynamics of mismatched base pairs in DNA. Biochemistry. 1991;30:3271–3279. doi: 10.1021/bi00227a015. [DOI] [PubMed] [Google Scholar]
- 16.Reha-Krantz LJ. The use of 2-aminopurine fluorescence to study DNA polymerase function. Methods in molecular biology. 2009;521:381–396. doi: 10.1007/978-1-60327-815-7_21. [DOI] [PubMed] [Google Scholar]
- 17.Zhu H, Smith P, Wang LK, Shuman S. Structure-function analysis of the 3′ phosphatase component of T4 polynucleotide kinase/phosphatase. Virology. 2007;366:126–136. doi: 10.1016/j.virol.2007.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobson CJ, Allinson SL. The phosphatase activity of mammalian polynucleotide kinase takes precedence over its kinase activity in repair of single strand breaks. Nucleic Acids Res. 2006;34:2230–2237. doi: 10.1093/nar/gkl275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsutakawa SE, et al. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 Superfamily. Cell. 2011;145:198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orans J, et al. Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell. 2011;145:212–223. doi: 10.1016/j.cell.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson SH, Kunkel TA. Passing the baton in base excision repair. Nat Struct Biol. 2000;7:176–178. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- 22.Mani RS, Karimi-Busheri F, Fanta M, Cass CE, Weinfeld M. Spectroscopic studies of DNA and ATP binding to human polynucleotide kinase: evidence for a ternary complex. Biochemistry. 2003;42:12077–12084. doi: 10.1021/bi030127b. [DOI] [PubMed] [Google Scholar]
- 23.Lu M, et al. Independent mechanisms of stimulation of polynucleotide kinase/phosphatase by phosphorylated and non-phosphorylated XRCC1. Nucleic Acids Res. 2010;38:510–521. doi: 10.1093/nar/gkp1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mani RS, et al. XRCC1 stimulates polynucleotide kinase by enhancing its damage discrimination and displacement from DNA repair intermediates. J Biol Chem. 2007;282:28004–28013. doi: 10.1074/jbc.M704867200. [DOI] [PubMed] [Google Scholar]
- 25.Mani RS, et al. Dual modes of interaction between XRCC4 and polynucleotide kinase/phosphatase: implications for nonhomologous end joining. J Biol Chem. 2010;285:37619–37629. doi: 10.1074/jbc.M109.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.