Abstract
Glycogenin initiates the synthesis of a maltosaccharide chain covalently attached to itself on Tyr195 via a stepwise glucosylation reaction, priming glycogen synthesis. We have captured crystallographic snapshots of human glycogenin during its reaction cycle, revealing a dynamic conformational switch between ground and active states mediated by the sugar donor UDP-glucose. This switch includes the ordering of a polypeptide stretch containing Tyr195, and major movement of an approximately 30-residue “lid” segment covering the active site. The rearranged lid guides the nascent maltosaccharide chain into the active site in either an intra- or intersubunit mode dependent upon chain length and steric factors and positions the donor and acceptor sugar groups for catalysis. The Thr83Met mutation, which causes glycogen storage disease XV, is conformationally locked in the ground state and catalytically inactive. Our data highlight the conformational plasticity of glycogenin and coexistence of two modes of glucosylation as integral to its catalytic mechanism.
Keywords: glycogen storage disorder, glycosyltransferase
Glycogenin initiates glycogen biogenesis in eukaryotes by synthesizing a chain of approximately 6–10 α-1,4-linked glucose units attached to itself (1, 2), which acts as a substrate for bulk glycogen synthesis carried out by glycogen synthase and branching enzyme (3). The initiation process has an inherent importance in the deposition of glucose for storage. Defective glycogenin has been genetically linked with glycogen storage disease (GSD) type XV (OMIM 613507), where a Thr83Met mutation, in conjunction with a nonsense mutation, caused glycogen depletion in muscle and cardiac arrhythmia (4).
Glycogenin belongs to the superfamily of glycosyltransferases (GTs, EC 2.4.x.y) that transfer the monosaccharide moiety of an activated sugar donor to diverse acceptor substrates, with either retention or inversion of the donor anomeric carbon configuration (5). Mammals possess two glycogenin isoforms: glycogenin-1, the predominant form in muscle, and glycogenin-2, found mainly in liver. Both forms are annotated as GT-8 subfamily enzymes due to sequence homology with other members, such as Neisseria meningitides α-1,4-galactosyltransferase LgtC (6). However, glycogenin is unusual among GTs in its ability to catalyze a multistep self-glucosylation reaction, using in each step Mn2+ ion as cofactor and UDP-glucose (UDPG) as glucose donor. In the first step, glycogenin transfers glucose from UDPG onto its tyrosine (Tyr195 in human glycogenin-1), forming a C′1-O-tyrosyl linkage (7–9). This glucose is the start of a growing maltosaccharide chain onto which further glucose units are transferred from UDPG in a stepwise nonprocessive manner, with the chain remaining attached to the tyrosine. In each reaction cycle, the enzyme is believed to operate in an ordered bi-bi kinetic mechanism as proposed for many GTs (10), for the sequential binding of the sugar donor followed by the acceptor (Fig. S1).
The structures of rabbit glycogenin-1 (rGYG1), determined in the apo and UDPG-bound forms (11, 12), have unveiled fundamental properties of the enzyme, revealing a dimeric configuration and the classic GT-A fold (5) with the Mn2+-binding DXD motif. Nevertheless, a number of molecular details relating to glycogenin catalysis remain evasive. First, substrate-induced conformational changes to shield the active site, often advocated as integral to the GT catalytic mechanism (5, 10), have not been observed for glycogenin to date. In addition, the possibilities of intrasubunit (13, 14) and intersubunit (15) glucosylation exist for a glycogenin dimer, as the growing maltosaccharide chain attached to one subunit may theoretically proceed into the active site of its own or adjacent subunit for further glucosylation. This remains a subject of debate and is complicated by a reported monomeric form of glycogenin at low micromolar concentration (13). Furthermore, the mechanism by which glycogenin catalyzes the glucosylation reaction with retention of the anomeric carbon configuration is unclear, as are the binding modes for the sugar donor and acceptor, the latter also confounded by the nonconventional UDPG configuration in the rGYG1 structure as compared to other GT-A enzymes (6). Altogether, these anomalies have thus far hindered further understanding of glycogenin function and mechanism. In this work we present crystal structures of human glycogenin-1 (hGYG1) which provide complete snapshots of conformational dynamics along the catalytic cycle and clear visualization of how maltosaccharide chains bind to the enzyme, allowing us to propose a mechanism for the glucosyl transfer. Our data in particular highlight the open-to-closed transition of a lid segment over the active site as an essential feature in glycogenin catalysis, which is deficient in the disease mutant.
Results
Recombinant hGYG1 Production and Structure Determination.
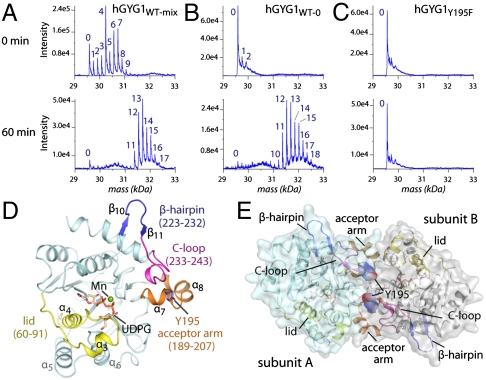
When the catalytic domain of hGYG1 (aa 1–262) was expressed in Escherichia coli BL21(DE3), a mixed population of unglucosylated and endogenously glucosylated species—containing up to nine hexose units—was obtained (hGYG1WT-mix) (Fig. 1A). Expressing hGYG1 in an E. coli strain defective in UDPG synthesis yielded almost sugar-free recombinant protein (hGYG1WT-0) (Fig. 1B). Upon incubation with UDPG and Mn2+, both hGYG1WT-mix and hGYG1WT-0 were catalytically active in vitro and could be self-glucosylated, containing up to 17 attached glucose units (Fig. 1 A and B). In addition, we constructed a Y195F mutant to abolish the site of glucose attachment and, as expected, the purified protein (hGYG1Y195F) was completely sugar-free and could not be glucosylated in vitro (Fig. 1C). Preincubation of hGYG1WT-mix, hGYG1WT-0, and hGYG1Y195F with several ligands, including substrate, cofactor, and products, rendered the enzyme crystallizable in diverse chemical conditions and space groups, allowing structure determination in the 1.5–2.3-Å resolution range (Fig. S2 and Table S1). hGYG1 adopts the GT-A fold (Fig. 1D) previously seen with the rabbit enzyme (11, 12) and in all crystal forms exists as a dimer with the two Tyr195 acceptor residues from both subunits being placed approximately 15.5 Å Cα apart at the twofold interface (Fig. 1E).
Fig. 1.
In vitro glucosylation and overall structure of human GYG1. Mass spectrometry analyses of (A) hGYG1WT-mix, (B) hGYG1WT-0 and (C) hGYG1Y195F proteins after in vitro glucosylation assays at 0 and 60 min, indicating the number of hexose sugars attached. (D) Structure of the hGYG1 protomer displays the typical GT-A fold. (E) Arrangement of a hGYG1 homodimer showing the juxtaposition of the two acceptor arms at the dimer interface.
Conformational Plasticity of Glycogenin During the Catalytic Cycle.
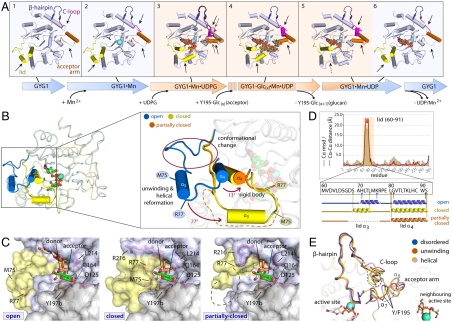
We mapped a number of hGYG1 structures onto the glucosylation reaction path (Fig. 2A) to provide snapshots along the entire reaction coordinates. Close inspection of the hGYG1 structures reveals two distinct states of the enzyme—namely, the ground state (Fig. 2A, blue shade), and active state (Fig. 2A, orange shade). The two states are interchangeable during catalysis and involve conformational rearrangements in three regions that influence active site accessibility (Figs. 1D and 2A): (i) a “lid” segment connecting β3 and β4 (aa 60–91), which comprises a coiled region, short helix (α3), and longer helix (α4); (ii) a helix-turn-helix “acceptor arm” (aa 189–207) harboring the Tyr195 acceptor residue; and (iii) the C-terminal loop (“C loop”; aa 233–243) located close to the acceptor arm, linking a β-hairpin turn and the last helix.
Fig. 2.
Conformational plasticity of hGYG1 during catalysis. (A) Mapping of hGYG1 structures on the glucosyl-transfer reaction, highlighting the ground and active states of the enzyme (blue- and orange-shaded boxes, respectively) and conformational changes on three mobile regions. The structures shown are: hGYG1Y195F·apo (box 1), hGYG1WT-mix·Mn2+ (box 2), hGYG1WT-0/Y195F·Mn2+·UDPG (box 3), hGYG1WT-mix·Mn2+·UDP·Glc4 (box 4), hGYG1WT-0·Mn2+·UDP (monoclinic) (box 5), hGYG1Y195F·Mn2+·UDP (box 6). (B) Displacement of the lid between the superimposed ground (blue) and active (yellow) states, which consists of three components (inset). (C) Surface representation in proximity to the active site in the open, closed and partially closed conformations of the lid (yellow), showing different degree of accessibility to the active site. (D) Pairwise distance calculations (Cα-Cα difference) and secondary structure composition (Cα-rmsd) of the three lid conformations, highlighting the disorder of helix α3 in the partially closed lid. (E) The acceptor arm, disordered in the ground state (blue), adopts either an unwinding coil (orange) or helical conformation (yellow and pink) in the active state. These changes correlate with the conformation of the neighboring C loop and result in different positions of the Tyr195 acceptor residue.
Superimposition of the two states reveals a maximum motion in the lid segment among the three flexible regions (Fig. 2B). In the ground state, the lid adopts an open conformation (blue) positioned away from the active site, leaving it accessible to the exterior (Fig. 2C, open). This ground state is seen in apo-hGYG1 (Fig. 2A, box 1), Mn2+-bound (Fig. 2A, box 2), and Mn2+·UDP-bound (Fig. 2A, box 6) complexes, as well as all reported rGYG1 structures, and represents the enzyme poised for the incoming of substrates before, and the release of products after, a glucosylation cycle. However, for a Michaelis complex (Mn2+·UDPG-bound; Fig. 2A, box 3), or a ternary product complex (Mn2+·UDP and maltosaccharide; Fig. 2A, box 4), the enzyme switches to the active state. Here, the lid adopts a closed conformation shielding the active site pocket and reducing its accessibility (Fig. 2B, yellow). Together with residues Leu214, Asp125, and Tyr197b (b denotes neighboring subunit of a dimer), the closed lid creates a narrow access channel opening up to the dimer interface (Fig. 2C, closed). Additionally, the closed lid, positioned directly adjacent to the acceptor arm of the opposite subunit (Fig. 1E), may confer stability to this structurally flexible region (see later section).
Analysis of the open-to-closed lid rearrangement reveals three mobile components (Fig. 2B): (i) a 13° rigid body rotation of lid helix α4, (ii) a “straightening” of the coiled region to be nearly parallel to the closed-state α4, and (iii) an approximately 19-Å shift of helix α3 combined with approximately 77° rotation. Helix α3 additionally undergoes a change in residue composition from the open (residues 71–76) to closed conformation (residues 69–74) (Fig. 2D). The change in sequence register may require significant disordering of helix α3. The intrinsic flexibility of this segment is evident in a Mn2+·UDP-bound complex (Fig. 2A, box 5) where the lid adopts a “partially closed” conformation with helix α3 completely disordered (Fig. 2B, orange; Fig. 2D, partially closed), thus opening a gap in the active site access channel while the substrate-binding site remains occluded (Fig. 2C, partially closed). This mobility of helix α3 is catalytically significant because two lid residues within it, Arg77 and Met75, which are buried and positionally constrained in the ground state, are released to have more flexibility and traverse approximately 17–23 Å in the closed state (Fig. 2B), such that Arg77 interacts with the UDPG pyrophosphate and Met75 is located in proximity to the UDPG glucose moiety. Together, this conformational rearrangement of the lid segment sequesters the active site and contributes substrate-binding residues for catalysis.
In addition to the lid movement, other less dramatic structural alterations between the two states are observed for the acceptor arm and C loop, which lie in different polypeptide stretches but are topologically juxtaposed (Fig. 2E). In the ground state, the first helix (α7) of the acceptor arm and most of the C loop are disordered, whereas the active state shows both regions ordered. Interestingly, we observed conformational plurality of the acceptor arm into either an unwinding coil or helical (α7) conformation, unlike in rGYG1 where only a helical structure was seen (12). This flexibility likely depends on the corresponding rearrangement of the C loop, especially Pro238, which packs onto residues 193–196 of helix α7 and hence mediates the multiple conformations of the acceptor arm (Fig. S3). Furthermore, the Tyr195 side chain is positioned at various distances (12–14 Å) from the active site and, more importantly, multiple orientations, which provide different trajectories for Tyr195 to attack the UDPG glucose moiety in either of the active sites of the dimer (Fig. S3).
Intra- and Intersubunit Threading of the Nascent Maltosaccharide Chain.
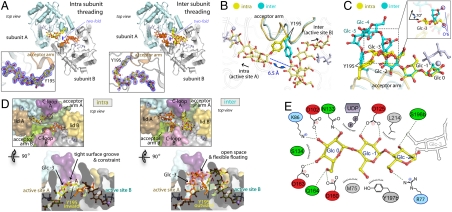
An hGYG1 dimer can potentially catalyze the glucosylation of two nascent maltosaccharide chains, and both intra- (13, 14) and intersubunit (15) mechanisms have been proposed. To investigate this structurally, we cocrystallized hGYG1WT-mix with Mn2+·UDP and determined structures adopting the active state. The electron density allowed us to fully trace two Tyr195 (O)-linked maltotetraose (Glc4) chains in one structure, and two maltohexaose (Glc6) chains in another structure, resembling the presumed product complex of the Glc3 → Glc4 and Glc5 → Glc6 glucosylation cycles, respectively (Fig. 3A). Unexpectedly, the two structures reveal distinct modes of maltosaccharide threading into the dimer active sites as mediated by different conformations of the Tyr195 side chains (Fig. 3B) and sugar chains (Fig. 3C). The Glc4 chains traverse a short path from their respective Tyr195 attachments and extend into the active site of the same subunit (“intramode”). This is accompanied by the Tyr195 side chains pointing inward (Fig. 3D). In contrast, intersubunit threading was observed for the Glc6-linked structure, where the two longer Glc6 chains are accommodated by their Tyr195 attachments now pointing outward, to protrude into the active sites of the opposite subunits in a dimer (“intermode”) (Fig. 3D).
Fig. 3.
Intra- vs. intersubunit threading of hGYG1-bound maltosaccharides. (A) Structures of the maltotetraose-bound (hGYG1WT-mix·Mn2+·UDP·Glc4; left) and maltohexaose-bound (hGYG1WT-mix·Mn2+·UDP·Glc6; right) hGYG1 dimer reveal two distinct intra- and interthreading modes for the substrate sugar chains as supported by 2Fo-Fc electron density map (inset). (B) The Tyr195 side chain exhibits two orientations 6.5 Å apart, which switch between the two threading modes. (C) Superimposition of the Glc4- (yellow) and Glc6-(blue) bound chains demonstrating overlap of Glc 0, Glc -1 and Glc -2 and a 70° kink at the Glc -3 unit (inset) where the two threading modes are diverted. (D) Surface representation of the dimeric interface at the twofold axis, showing how the Glc4 and Glc6 chains are accommodated outside the active site. (E) Schematic illustration of interactions between the Glc 0, Glc -1, Glc -2 units and residues in the active site pocket for both threading modes.
Comparison of the Glc4 and Glc6 chains reveals that the terminal three glucose units (Glc 0, Glc -1, Glc -2; numbered from the nonreducing end of the oligosaccharide chain) occupy near-identical positions in both intra- and intermodes (Fig. 3C) and engage in an intimate association with the active site residues via many hydrogen-bonding interactions (Fig. 3E). The terminal glucose unit (Glc 0) is buried deepest within the active site and forms the most interactions, with its hydroxyl groups hydrogen-bonded to Asn133, Asp102, Lys86, Asp163, Gln164, and Asp160. The Glc -1 unit is anchored by Leu214 and Tyr197b, which optimally position the glucopyranose 2′- and 3′-hydroxyl to form a fork-like hydrogen bond with the Asp125 carboxylate. Toward the entrance of the narrow access channel, the Glc -2 unit engages in fewer interactions, with its hydroxyl groups positioned to hydrogen bond with the lid residue Arg77 and the main-chain carbonyl of Ser196b.
Beyond the terminal three glucose units, the paths for the intra- and interthreading chains are much digressed due to an approximately 70° angular tilt of the Glc -3 glucopyranose ring (Fig. 3C). In the Glc4 chains (intramode), Glc -3 is covalently linked to Tyr195 and packs tightly along the surface groove of the protein (Fig. 3D and Fig. S4) within hydrogen-bonding distance of main-chain Leu214. The longer Glc6 chains (intermode), however, are kinked at Glc -3, allowing the chains to ascend into the open space at the dimer interface and form fewer protein contacts before descending back to link Glc -5 with Tyr195, forming a semicircular track (Fig. 3D and Fig. S4).
Donor and Acceptor Subsites Provide Insights into Glucosyl Transfer.
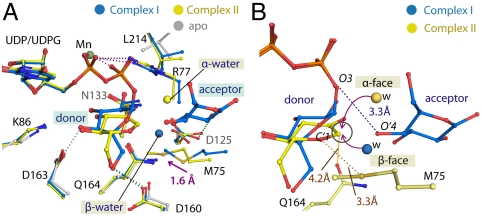
The Glc 0 and Glc -1 units in the enzyme-product complexes map the donor and acceptor subsites in hGYG1, respectively, as they overlap well with those of LgtC (6). We further cocrystallized hGYG1 with Mn2+ and UDPG to observe how the donor and acceptor molecules are positioned during catalysis and, unexpectedly, obtained two complexes (complexes I and II) with different ligand constituents at these subsites (details in Fig. S5). At the donor subsite, in complex I we observed the glucose moiety of UDPG adopting the classic “folded-back” geometry tucked below the UDP pyrophosphates with the UDPO3-glucoseC′1 scissile bond intact, while in complex II we observed a glucosyl species displaced downward (relative to the UDPG glucose moiety) with its C′1 atom 2.6 Å from the UDP β-phosphate O3, indicative of the scissile bond being cleaved (Fig. 4). We propose the glucosyl species to be 2-hydroxyglucal, the product from an enzyme-catalyzed elimination of the unstable oxocarbenium intermediate (details in Fig. S5). At the acceptor subsite two types of ligands were observed: a water molecule located on the opposite side (β-) or same side (α-) of the O3-C′1 bond (β-water and α-water); and a free glucose molecule, feasibly positioned as an acceptor, with its O′4 hydroxyl group equi-planar to the glucosyl species at the donor site (Fig. 4). These ligands suggest various possible attacking directions relative to the anomeric C′1 atom (Fig. 4B).
Fig. 4.
Mapping of donor and acceptor subsites during glucosyl transfer. (A) Various ligands trapped in the active site of hGYG1WT-0·Mn2+·UDPG (complex I, blue) and hGYG1Y195F·Mn2+·UDPG (complex II, yellow). At the donor subsite, the glucose moiety of intact UDPG in complex I, and a glucosyl species in complex II illustrate the capability of this subsite to accommodate the glucopyranose ring at various positions during catalysis. At the acceptor subsite, three ligand identities were observed: a glucose molecule and β-water in complex I, and an α-water in complex II. A 1.6-Å movement of the Met75 side chain is observed between complexes I and II (arrow), providing a mechanism to exclude the β-water molecule and stabilize the glucosyl species. (B) Possible trajectories of nucleophilic attack on the C′1 atom (circled) of the glucosyl species in complex II by the α- and β-water, either above or below the plane of the donor glucopyranose ring, respectively. The O′4 atom of the glucose molecule at the acceptor subsite of complex I is 3.3 Å from the UDP β-phosphate, representing a good attacking position for the equi-planar glucosyl species that is stabilized by Met75 and Gln164 at the donor subsite.
Superimposing all hGYG1 structures shows that residues lining the donor and acceptor subsites are static, with the exception of Leu214 that slides deep into the access channel to configure UDPG in the catalytically competent, folded-back geometry, as well as lid residues Met75 and Arg77, which are brought into the active site upon lid closure. Furthermore, comparing complexes I and II, we observed an additional “tightening” of the Met75 side chain by approximately 1.6 Å closer toward the donor subsite, with its sulfur atom 3.3 Å away from the O′5 of the glucosyl species (Fig. 4B, and see Fig. S6).
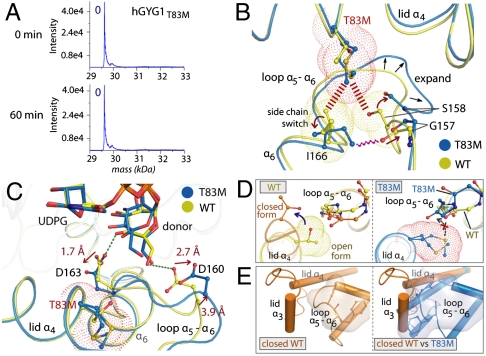
Thr83Met Mutant Is Structurally Ablated in Forming the Active State.
We set out to determine the molecular basis for the Thr83Met mutation linked with GSD type XV (4). Unlike wild-type protein, hGYG1T83M is not endogenously glucosylated when expressed in E. coli BL21(DE3), and remains unglucosylated when supplemented with UDPG and Mn2+ in vitro (Fig. 5A), indicating an impairment of glucosylation. We cocrystallized hGYG1T83M with various ligands and found that, although crystals were obtained under diverse growth conditions, they belonged only to space groups that yielded the ground state of the enzyme (Fig. S2), suggesting the active state could not be formed. In wild-type hGYG1, Thr83 resides at the lid helix α4, packing close to the neighboring loop α5–6 (residues 156–163) and the start of helix α6 (Fig. 5B). Compared to wild-type, hGYG1T83M displays an approximately 5.0-Å outward displacement of loop α5–6 as well as positional changes for Il166, Ser158, and Gly157 (Fig. 5B) to avoid steric clashes due to the bulky Met83 side chain. The rearrangement of loop α5–6 in turn displaces residues Asp160 and Asp163 in the donor subsite (Fig. 5C), residues that have direct roles in fixating the glucose donor for catalysis, culminating in the glucose moiety of UDPG being less ordered than in wild type. Thr83Met also interferes with the rigid body rotation of helix α4 during lid closure, potentially due to the extended Met side chain protruding into a region underneath the neighboring loop α5–6 (Fig. 5D). Furthermore, the displaced loop α5–6 may also prevent the lid helix α3 to achieve its closed conformation and position due to steric clashes (Fig. 5E). Altogether, these changes culminate in the Thr83Met mutant being structurally ablated in transitioning to the active state of the enzyme.
Fig. 5.
Structural basis for the inactivating Thr83Met mutation. (A) In vitro glucosylation assay at 0 and 60 min for hGYG1T83M. (B) The Thr83Met mutation (blue) creates steric constraints (red dashed lines) relative to the wild-type enzyme (yellow). Hence, Ile166 changes rotamer configuration and loop α5–6 extends outward (black arrows) to avoid Ile166 clashing with Gly157 and Ser158. (C) Expansion of loop α5–α6 displaces residues Asp160 and Asp163 (Cα shifts of approximately 3.9 and approximately 0.7 Å; side-chain shifts of approximately 2.7 and approximately 1.7 Å, respectively) away from their wild-type positions. (D and E) The open-to-closed transition of the lid is disrupted in the hGYG1T83M mutant. Wild-type protein allows rigid body rotation of lid α4 (D, left), a movement sterically hindered by T83M (D, right). The expanded T83M α5–α6 loop also blocks helix α3 from reaching its expected position (compare E, left and right).
Discussion
Lid Closure Is Integral to Glycogenin Catalysis.
Protein dynamics play a key role in function and catalysis, transforming an enzyme into a catalytically competent state via conformational changes. In GTs, these changes purportedly involve a sugar-induced interdomain or loop movement (5, 10). In this work, we demonstrate that the conformational plasticity of hGYG1, likely controlled by the presence of the sugar donor UDPG, is integral to its catalysis (Fig. 6A). Essential to this conformational mechanism is an open-to-closed transition of the lid segment, as demonstrated by the GSD XV mutant in which the lid is locked in the open conformation and glucosyl transfer is defective. We anticipate that lid transition may exist in other GT enzymes. For example, the ternary complex of LgtC, belonging to the same GT-8 subfamily as hGYG1, superimposes well with the hGYG1 active state conformation and reveals an equivalent lid-like region that sequesters the donor-bound active site. We therefore believe that a ground state conformation of LgtC would exist for the apo-enzyme, although it has not been demonstrated to date.
Fig. 6.
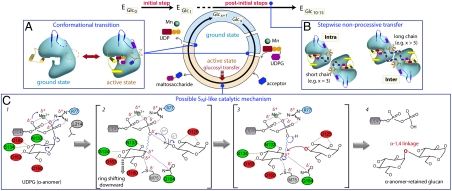
Proposed model of glucosyl transfer catalyzed by glycogenin. Schematic illustration of three salient catalytic features of hGYG1 uncovered in this study. (A) Each glucosyl-transfer cycle requires a transition between the ground and active states, which involves conformational changes in the lid (yellow), acceptor arm (orange), and C loop (purple). (B) In the postinitial steps, the growing maltosaccharide chains (blue hexagons) in the hGYG1 dimer are guided to the active site in either an intra- or intersubunit threading mode dependent on chain length, for further glucosylation in a stepwise manner. (C) The SNi-like retaining glucosyl-transfer mechanism involves lengthening of the scissile UDPO3-glucoseC′1 bond (squiggly line) and formation of an oxocarbenium-like intermediate, followed by nucleophilic attack by the deprotonated 4′-hydroxyl from the glucose acceptor toward the anomeric C′1 atom on the glucose donor.
The catalytic significance of lid closure is manifold. It sequesters the active site and creates an access channel to guide the nascent maltosaccharide chain into the active site, thereby fixating the sugar donor and acceptor groups at their cognate subsites, and mediating the right geometry for glucosyl transfer. It moves into place otherwise distant residues, such as Met75 and Arg77, which have direct interactions with the sugar donor, to complete the essential catalytic machinery. Furthermore, the closed lid in one monomer can stabilize the nearby acceptor arm of the opposite subunit in a dimer. Interestingly, we also observed a partially closed lid conformation, which provides a mechanism at the dimer interface to open up space for the entry as well as exit of donor products and acceptors and may therefore depict a general intermediate step in the open-to-closed transition (and vice versa) of the lid. Our reaction snapshots of hGYG1 place it among very few GT-A enzymes where more than one enzymatic state has been structurally captured and a comparison of the conformational movements in these enzymes reveals their varied nature (Fig. S7).
Role of the Acceptor Arm in Switching Between Intra- and Intersubunit Glucosyl Transfer.
During the glucosyl transfer cycles, the growing maltosaccharide chain is tasked to guide the terminal two glucose units (Glc -1 and Glc -2) via the access channel into the active site. Our maltosaccharide-bound enzyme-product complexes support the existence of both intra- and intersubunit glucosylation modes in dimeric glycogenin, allowing the maltosaccharide chain to approach either active site in the dimer, in a manner that best suits its chain length and conformation (Fig. 6B). Glycogenin attached with shorter maltosaccharide chains (e.g., Glc3) can undergo an intrasubunit glucosyl transfer, made possible by accommodating a single additional glucose after Glc -2 along the surface groove of the protein, toward the active site of the same subunit. This is sterically incompatible with a longer sugar chain (e.g., Glc5 or longer), which is unable to traverse the same surface path due to spatial constraints at the dimer interface. To overcome these constraints, the enzyme can switch to an intersubunit mode: a change dependent on Tyr195 in the acceptor arm shifting its conformation and the longer maltosaccharide chain assuming a semicircular path in the more open solvent exterior before protruding into the active site of the neighboring subunit to position the terminal two glucose units. Despite the expected higher entropic energy incurred in this mode, it is more effective in accommodating two longer glucose chains within the dimer and has the additional advantage that the Tyr195-attached Glc -5 units from both chains, being only approximately 4 Å apart, may stabilize each other. We anticipate that subsequent glucosyl transfer steps (after Glc6) would remain in the intermode, and the two growing maltosaccharide chains may eventually adopt curved structures in the open space, much akin to the helical model proposed for glycogen (16).
As for the initial glucosyl transfer steps (e.g., Glc0 → Glc1, Glc1 → Glc2), the Tyr195 side chain in the unglucosylated hGYG1 structures we captured is too distant (12–14 Å) from either subunits’ active site to predict whether it would adopt an intra- or intersubunit transfer mode. Therefore, further movement of the acceptor arm toward the active site, likely to require even more unwinding of the acceptor arm than captured in our structures, would be necessary for catalysis. We anticipate that a structure of hGYG1 complexed with a “dead-end” sugar donor, trapping the enzyme after one round of glucosyl transfer, may provide insight into the initial step.
A possible SNi-like mechanism for glucosyl-transfer.
The subsites and geometries for the sugar donors and acceptors, defined in the hGYG1 active state structures, render the donor anomeric C′1 spatially accessible to the acceptor O′4 hydroxyl group, a finding consistent with the existing GT literature (5). This contrasts with the scenario depicted by the rGYG1 holo structure, prepared by soaking UDPG into apo-rGYG1 crystals, in which the lid did not close and the donor glucose moiety did not adopt the folded-back configuration and was therefore not readily accessible from the direction of the nascent glucose chain (11, 12). This discrepancy reinforces a requirement of the closed lid in the active state to mediate the donor sugar geometry for catalysis.
The retaining mechanism of glucosyl transfer in glycogenin, and other GTs, remains a subject of debate. Considering a double displacement model widely proposed for GTs (17), two possible nucleophilic residues in hGYG1 that could attack on the β-face of the donor glucopyranose ring to form a covalent glycosyl-enzyme intermediate are Asp160 and Gln164, based on previously suggested candidates for rGYG1 (Asp159) (11) and LgtC (Gln189) (6), respectively. However, they are too distant from the C′1 anomer (4.5 and 6.0 Å for Gln164 and Asp160, respectively) in both the ground and active states, and Asp160 more likely assumes the role of positioning the donor 6′-hydroxyl group. Mutations of these residues in rGYG1 and LgtC also did not completely abolish glycosyltransferase activity, as would be expected for a catalytically essential nucleophile (12, 18).
In contrast, the active site environment of hGYG1 lends support to an alternative SNi-like mechanism, which requires direct nucleophilic substitution on the same α-face as the UDP leaving group (5, 19, 20) (Fig. 6C). First, the lid residue Arg77, which interacts with the UDP β-phosphate, can help withdraw electrons away from the scissile UDPO3-glucoseC′1 bond and stabilize partial charges (step 1), consistent with the significantly reduced glucosylation efficiency of a hGYG1 R77A mutant we observed. Second, an oxocarbenium-like intermediate can form in the transition state after a downward positional shift of the distorted glucopyranose ring (21) to the position of our observed glucal species. The cationic characters at glucose C′1 and O′5 in an oxocarbenium ion can be stabilized by Gln164 from the β-face and the displaced Met75 sulfur, respectively (step 2). Third, the acceptor 4′-hydroxyl group can be deprotonated by the departing UDP phosphate that is within hydrogen-bonding distance, thus generating a nucleophile that is above or equi-planar to the donor C′1 atom, to form an attack with retention of anomeric configuration (steps 3 and 4).
To conclude, structural snapshots of hGYG1 along the reaction cycle have demonstrated the conformational plasticity of both the enzyme and maltosaccharide substrate during glucosylation. The elucidation of this mechanism, which forms the underlying basis for the GSD type XV mutation, allows us to appreciate the essential role glycogenin plays in glycogen synthesis and serves as a starting point for further characterization and therapeutic development toward this new disorder. Beyond glycogenin, our data provide significant insights to the current mechanistic knowledge of the diverse, yet poorly understood, glycosyltransferase superfamily.
Materials and Methods
Human GYG1 (hGYG1) aa 1–262 (IMAGE clone: 3504538) was subcloned into the pNIC28-Bsa4 vector (GenBank accession no. EF198106) incorporating an N-terminal TEV-cleavable His6-tag. All mutants were generated from this plasmid using the QuickChange kit (Stratagene). The corresponding plasmids were transformed either into E. coli BL21(DE3)-R3 cells, or into the M226 strain (CGSC #4997) lacking UDP-glucose pyrophophorylase activity after λDE3 lysogenization (Novagen) and expressed and purified by nickel affinity and gel filtration chromatography. In vitro glucosylation and detection by mass spectrometry were performed as described (22). Purified hGYG1 (10 mg/mL) was cocrystallized with various ligands by sitting drop vapor diffusion at 20 °C. X-ray diffraction data were collected in-house or at the Diamond Light Source. Structures were determined by molecular replacement. Detailed method information is provided in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank staff at the Diamond Light Source for help in synchrotron data collection. The Structural Genomics Consortium is a registered charity (number 1097737; funding details in www.thesgc.org).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3U2T, 3U2U, 3U2V, 3T7O, 3T7M, 3T7N, 3RMW, 3RMV, 3Q4S, 3U2W, 3QVB).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113921108/-/DCSupplemental.
References
- 1.Lomako J, Lomako WM, Whelan WJ. A self-glucosylating protein is the primer for rabbit muscle glycogen biosynthesis. FASEB J. 1988;2:3097–3103. doi: 10.1096/fasebj.2.15.2973423. [DOI] [PubMed] [Google Scholar]
- 2.Smythe C, Cohen P. The discovery of glycogenin and the priming mechanism for glycogen biogenesis. Eur J Biochem. 1991;200:625–631. doi: 10.1111/j.1432-1033.1991.tb16225.x. [DOI] [PubMed] [Google Scholar]
- 3.Preiss J, Walsh DA. The comparative biochemistry of glycogen and starch. In: Ginsburg V, editor. Biology of Carbohydrates. Vol. 1. New York: John Wiley and Sons; 1981. pp. 199–314. [Google Scholar]
- 4.Moslemi AR, et al. Glycogenin-1 deficiency and inactivated priming of glycogen synthesis. N Engl J Med. 2010;362:1203–1210. doi: 10.1056/NEJMoa0900661. [DOI] [PubMed] [Google Scholar]
- 5.Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: Structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 6.Persson K, et al. Crystal structure of the retaining galactosyltransferase LgtC from Neisseria meningitidis in complex with donor and acceptor sugar analogs. Nat Struct Biol. 2001;8:166–175. doi: 10.1038/84168. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez IR, Whelan WJ. A novel glycosyl-amino acid linkage: Rabbit-muscle glycogen is covalently linked to a protein via tyrosine. Biochem Biophys Res Commun. 1985;132:829–836. doi: 10.1016/0006-291x(85)91206-9. [DOI] [PubMed] [Google Scholar]
- 8.Cao Y, Mahrenholz AM, DePaoli-Roach AA, Roach PJ. Characterization of rabbit skeletal muscle glycogenin. Tyrosine 194 is essential for function. J Biol Chem. 1993;268:14687–14693. [PubMed] [Google Scholar]
- 9.Smythe C, Caudwell FB, Ferguson M, Cohen P. Isolation and structural analysis of a peptide containing the novel tyrosyl-glucose linkage in glycogenin. EMBO J. 1988;7:2681–2686. doi: 10.1002/j.1460-2075.1988.tb03121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qasba PK, Ramakrishnan B, Boeggeman E. Substrate-induced conformational changes in glycosyltransferases. Trends Biochem Sci. 2005;30:53–62. doi: 10.1016/j.tibs.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Gibbons BJ, Roach PJ, Hurley TD. Crystal structure of the autocatalytic initiator of glycogen biosynthesis, glycogenin. J Mol Biol. 2002;319:463–477. doi: 10.1016/S0022-2836(02)00305-4. [DOI] [PubMed] [Google Scholar]
- 12.Hurley TD, Stout S, Miner E, Zhou J, Roach PJ. Requirements for catalysis in mammalian glycogenin. J Biol Chem. 2005;280:23892–23899. doi: 10.1074/jbc.M502344200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bazan S, Issoglio FM, Carrizo ME, Curtino JA. The intramolecular autoglucosylation of monomeric glycogenin. Biochem Biophys Res Commun. 2008;371:328–332. doi: 10.1016/j.bbrc.2008.04.076. [DOI] [PubMed] [Google Scholar]
- 14.Romero JM, Issoglio FM, Carrizo ME, Curtino JA. Evidence for glycogenin autoglucosylation cessation by inaccessibility of the acquired maltosaccharide. Biochem Biophys Res Commun. 2008;374:704–708. doi: 10.1016/j.bbrc.2008.07.114. [DOI] [PubMed] [Google Scholar]
- 15.Cao Y, Steinrauf LK, Roach PJ. Mechanism of glycogenin self-glucosylation. Arch Biochem Biophys. 1995;319:293–298. doi: 10.1006/abbi.1995.1295. [DOI] [PubMed] [Google Scholar]
- 16.Goldsmith E, Sprang S, Fletterick R. Structure of maltoheptaose by difference Fourier methods and a model for glycogen. J Mol Biol. 1982;156:411–427. doi: 10.1016/0022-2836(82)90336-9. [DOI] [PubMed] [Google Scholar]
- 17.Soya N, Fang Y, Palcic MM, Klassen JS. Trapping and characterization of covalent intermediates of mutant retaining glycosyltransferases. Glycobiology. 2011;21:547–552. doi: 10.1093/glycob/cwq190. [DOI] [PubMed] [Google Scholar]
- 18.Lairson LL, et al. Intermediate trapping on a mutant retaining alpha-galactosyltransferase identifies an unexpected aspartate residue. J Biol Chem. 2004;279:28339–28344. doi: 10.1074/jbc.M400451200. [DOI] [PubMed] [Google Scholar]
- 19.Goedl C, Nidetzky B. Sucrose phosphorylase harbouring a redesigned, glycosyltransferase-like active site exhibits retaining glucosyl transfer in the absence of a covalent intermediate. Chembiochem. 2009;10:2333–2337. doi: 10.1002/cbic.200900429. [DOI] [PubMed] [Google Scholar]
- 20.Lee SS, et al. Mechanistic evidence for a front-side, SNi-type reaction in a retaining glycosyltransferase (Translated from eng) Nat Chem Biol. 2011;7:631–638. doi: 10.1038/nchembio.628. (in eng) [DOI] [PubMed] [Google Scholar]
- 21.Tanaka Y, Tao W, Blanchard JS, Hehre EJ. Transition state structures for the hydrolysis of alpha-D-glucopyranosyl fluoride by retaining and inverting reactions of glycosylases. J Biol Chem. 1994;269:32306–32312. [PubMed] [Google Scholar]
- 22.Hurley TD, Walls C, Bennett JR, Roach PJ, Wang M. Direct detection of glycogenin reaction products during glycogen initiation. Biochem Biophys Res Commun. 2006;348:374–378. doi: 10.1016/j.bbrc.2006.07.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.