Abstract
High Gleason score is currently the best prognostic indicator for poor prognosis in prostate cancer. However, a significant number of patients with low Gleason scores develop aggressive disease as well. In an effort to understand molecular signatures associated with poor outcome in prostate cancer, we analyzed a microarray dataset characterizing 281 prostate cancers from a Swedish watchful-waiting cohort. Patients were classified on the basis of their mRNA microarray signature profiles indicating embryonic stem cell expression patterns (stemness), inactivation of the tumor suppressors p53 and PTEN, activation of several oncogenic pathways, and the TMPRSS2–ERG fusion. Unsupervised clustering identified a subset of tumors manifesting stem-like signatures together with p53 and PTEN inactivation, which had very poor survival outcome, a second group with intermediate survival outcome, characterized by the TMPRSS2–ERG fusion, and three groups with benign outcome. The stratification was validated on a second independent dataset of 150 tumor and metastatic samples from a clinical cohort at Memorial Sloan–Kettering Cancer Center. This classification is independent of Gleason score and therefore provides useful unique molecular profiles for prostate cancer prognosis, helping to predict poor outcome in patients with low or average Gleason scores.
Keywords: transcriptional profile, stem-ness, prognostic value
Microarrays, measuring the levels of many mRNA species in a cancerous tissue, have been useful in defining subclasses of breast cancers (1, 2). These subsets are further defined by their mutational profiles and the different prognostic outcomes of each subclass. A molecular definition is commonly accomplished by a supervised or unsupervised cluster analysis of the levels of mRNAs from the different genes under study. Ben-Porath et al. (3) introduced a different approach for the analysis of gene expression microarrays by studying the concordant expression of sets of genes that were found to be commonly over- or underexpressed in embryonic stem cells (ESCs) and thereby defined a gene signature characteristic for that cell or tissue type. They demonstrated that this ESC signature predominantly corresponded to the subclass of breast tumors termed the basal or triple-negative tumors and to the Her-2 positive tumors. Mizuno et al. (4) confirmed this observation and further demonstrated that tumors with the ESC signature almost always had p53 mutations or an inactive p53 function. These are typically tumors with poor clinical outcome, commonly associated with relapse and high mortality. In addition, Mizuno et al. (4) defined expression signatures for induced pluripotent stem cells (iPSCs) and the polycomb repressive complex-2 (PRC2). They showed that there is an overlap between the ESC and iPSC signatures and p53 mutant tumors, whereas expression of the PRC2 signature is enhanced in breast tumors with a wild-type p53 gene. Thus, these expression signatures defined subsets of breast or lung cancers that reflected both the mutation profiles and clinical outcomes (4). A similar result was independently obtained in liver cancers with p53 mutations and a poor outcome (5). The association of p53 mutations and an ESC profile also appears to be related to the efficient production of iPS cells from fibroblasts after the introduction of four transcription factors, Myc, Klf4, Oct4, and Sox2 (6). In the absence of p53 activity, Oct4 and Sox2 can increase the efficiency of producing iPS cells from 0.1% up to 80% and accomplish this in 5–6 d instead of 2 wk (7–11). Thus, p53 appears to block or reduce the production of an ESC or iPSC phenotype in either normal or cancerous cells that start with a differentiated phenotype.
If the ESC signature observed in several types of cancers commonly indicates a poor prognostic outcome, it should be possible to apply this same approach to other cancer types, particularly to the analysis of prostate cancers. Although a large number of men develop prostate cancers at young and older ages, ∼15% of these men appear to have a poor prognosis, whereas many can be left untreated or minimally treated with a good outcome (12). At present, the best prognostic indicator for outcome of a prostate cancer is the Gleason score determined by the pathology of the tumor section. A high Gleason score of 8, 9, or 10 commonly indicates poor survival; however, a significant number of patients with a lower Gleason score of 6 or 7 do develop aggressive disease and also have poor survival. Molecular lesions associated with poor survival have been identified. The loss of one or both copies of the tumor suppressor PTEN is found in a large percentage of prostate cancers, leading to an increased activity in the PI3kinase pathway (13, 14). The TMPRSS2–ERG chromosome fusion has been observed in 50–60% of prostate tumors (13, 15–17). This fusion of an androgen–responsive promoter with the ERG transcription factor (an ETS family member) may lead to increased cell migration (18), promotion of an epithelial–mesenchyme transition (EMT), and proliferation (18–21), but the details remain unclear. P53 mutations have been reported in 3–20% of prostate cancers at diagnosis (22–24) and are often correlated with tumor recurrence, castration resistance, and grade of the tumor (22, 23). Overexpression of MYC due to amplifications has also been observed in a fraction of prostate tumors, leading to increased proliferative signals (24–26). Despite all this information, there is no reliable method for determining the rate of relapse of a tumor and the outcome of survival at the time of diagnosis.
For this reason we used a newly published microarray dataset characterizing 281 prostate cancers from a watchful-waiting cohort recruited in Sweden between 1977 and 1999 (27), together with the associated clinical information including age, Gleason scores, cancer-specific mortality, and survival times. The tumors were tested for a set of different transcriptional signature profiles curated from the literature to represent stemness features and other known lesions in prostate cancer, followed by an unsupervised clustering analysis. We repeated this profiling procedure with 150 prostate tumor samples from an independent dataset (24) collected from patients at Memorial Sloan–Kettering Cancer Center between 2000 and 2006. This cohort contained significantly more early stage, low grade tumors than the Swedish cohort and patients of much younger age, reflecting modern clinical patient statistics. On the basis of the analysis of these datasets, one of which provides untreated long-term survival outcome and one of which represents a patient cohort as seen in the clinic today, we establish a subdivision of prostate cancers into molecular subtypes and demonstrate their prognostic impact.
Results
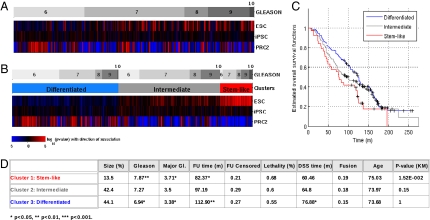
On the basis of our previous work (4) where an ESC signature was closely correlated with both p53 mutations and a very poor prognostic outcome, we examined whether any of the 281 prostate tumors from Sboner et al. (27) expressed an ESC, iPSC, or PRC2 signature. We stratified the tumors by Gleason score (Fig. 1A, Upper) and analyzed the gene signature patterns across the Gleason score-defined groups (Fig. 1A, ESC, iPSC, and PRC2 bars). Signature scores for each signature on each tumor sample were calculated by gene set enrichment analysis (28, 29). Concordant overexpression of all genes in a signature (compared with the mean expression of all genes) leads to a high positive score and indicates the presence of the signature in the tumor, whereas concordant underexpression of all signature genes leads to a negative score and is indicative of the absence of the signature in the tumor. Throughout this article, a red color indicates that the signature was found in the tumor, and a blue color indicates its absence. The ESC signature was identified in 38 (13%) of the tumors and a significant part of them had high Gleason scores of 8, 9, or 10 (Fig. 1B). The iPSC signature was present in ∼30% of the tumors and was distributed throughout the range of Gleason scores. This pattern is somewhat different from that observed in breast tumors, where the ESC and iPSC signatures are more strongly correlated (4). The PRC2 signature was observed in 44% of the tumors and tended to be located in the tumors with a low Gleason score. Unsupervised clustering into three groups on the basis of the ESC, iPSC, and PRC2 signatures subdivided the prostate tumors into an ESC cluster, a PRC2 cluster, and an intermediate cluster with no significant enrichment for either of these signatures (Fig. 1B). Whereas all three groups contain Gleason 6–10 tumors, the average Gleason score was significantly lower in the PRC2 tumors (6.94) than in the ESC tumors (7.87, P = 0.002, Fig. 1D). Furthermore, ESC tumors had poor survival compared with the PRC2 tumors, with survival in the intermediate class falling between them (Fig. 1C).
Fig. 1.
Signature analysis for stemness signatures (ESC, iPSC, PRC2) reveals stem-like tumors that show a distribution of Gleason scores with significantly increased mean. A total of 281 samples from the Sboner et al. (27) dataset (GSE16560) were analyzed. (A) Heatmap displaying signature scores of the stemness signatures characterizing embryonic stem cells (ESC), induced pluripotent stem cells (iPSC), and PRC2 activity as differentiation signature, after clustering the samples by Gleason score. Colors represent significance of score assignment, with red representing positive enrichment scores and blue negative ones. (B) An unsupervised clustering into three groups on the basis of the signature scores reveals clusters representing stem-like tumors, differentiated tumors, and a third, intermediate group with moderate, weakly significant expression of stem-like features. (C) Kaplan–Meier estimate of overall survival times for the three clusters. (D) Average values of clinical variables for the three clusters. Follow-up (FU) time is listed together with censoring (patients alive at closing of the study). Lethality refers to disease-specific mortality and DSS time denotes disease-specific survival time. Fusion refers to the actual TMPRSS2–ERG fusion status (FISH), and the last column shows P values for the split of curves in the Kaplan–Meier plots associated with the test group versus all other groups having higher mean survival times. Significance was calculated using Fisher's exact test and Student's t test and is indicated by asterisks.
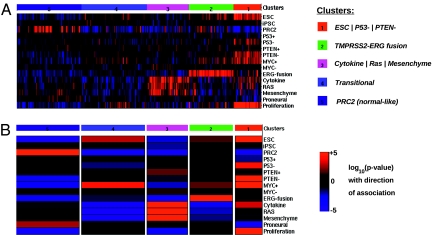
To explore the relevance of these observations in the context of common molecular alterations in prostate tumors, and to obtain additional understanding of pathway mechanisms within the given groups, we further stratified the set of 281 prostate tumors using a library of diverse gene expression signatures (Fig. 2A, Right). These signatures were derived from a number of microarray datasets where defined mutations or alterations were known to occur (Tables S1A and S1B). In particular, we formulated signatures indicating functionality of p53 and PTEN, presence of the TMPRSS2–ERG gene fusion, proliferation and MYC target activation, RAS pathway activity, and inflammatory signals. An unsupervised cluster analysis was then carried out to determine how many different clusters were produced from the 281 prostate tumors and which signatures were present in a cluster. Five clusters of prostate cancer were obtained and their profiles are shown in Fig. 2A. The enrichment (red) or depletion (blue) of a signature within a cluster is further emphasized by the heat map in Fig. 2B, where the color intensity is indicative of a high statistical association (P ≤ 1.0e-05). The five clusters are named for the signatures that are significantly enriched within them: (i) ESC | P53− | PTEN− (also referred to as stem-like), (ii) TMPRSS2–ERG fusion, (iii) Cytokine | RAS | Mesenchyme (also referred to as inflammatory-like), (iv) transitional, and (v) PRC2 (also referred to as differentiated-like) (Fig. 2B).
Fig. 2.
Clustering by signature profiles reveals distinct molecular subtypes. Samples from Sboner et al. (27) (GSE16560) were clustered using a customized prostate-specific library of expression signatures. Five clusters or subtypes were found by unsupervised clustering. (A) Heatmap of signature scores for single signatures, displaying the distinct molecular features of the subtypes. Shown is the average score for each signature group. Colors indicate significance of the score assignment (red, positive; blue, negative). (B) Schematic representation of the signature profiles of the clusters. Columns represent clusters, colors represent significance of the overall association of a signature with the cluster, red represents positive association with the signature, and blue represents negative association. Width of columns is relative to size of the cluster.
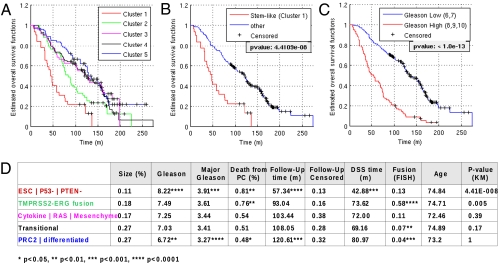
To understand the clinical relevance of this molecular classification, we next studied their association with Gleason score, overall survival, and lethality (Fig. 3), where lethality is defined as a confirmed death from prostate cancer (27). The stem-like group ESC | P53− | PTEN− was associated with the poorest overall survival (Fig. 3A, P = 4.41e-08), with a mean overall survival time of 57 mo, 81% lethal cases and mean disease-specific survival time of 43 mo. This group was followed by the TMPRSS2–ERG fusion group, with a significantly lower survival outcome compared with the remaining three groups (Fig. 3A, P = 0.005), a mean overall survival of 93 mo, and significantly increased lethality rate of 76%. The remaining groups have statistically similar survival outcomes, with a mean overall survival >103 mo and <55% lethal cases. The difference in mortality risk within the ESC | P53− | PTEN− group and the TMPRSS2–ERG fusion group versus the three remaining benign groups was further illustrated in the hazard ratios according to the Cox proportional hazard model. Patients within the stem-like group carried a 3.2-fold higher risk [95% confidence interval (CI) [2.07, 5.05], P < 1.0e-06], whereas patients within the fusion group still carried a 1.8-fold higher risk (95% CI [1.26, 2.66], P < 0.01). Remarkably, among the 200 patients with Gleason scores of 6 and 7 only, the patients with a stem-like profile carried a 2.7-fold higher risk of dying of the disease (95% CI [1.22, 5.89], P = 0.01), whereas the increase was 1.9-fold in patients with the fusion profile (95% CI [1.18, 3.11], P < 0.01). Notably, age at diagnosis was not a distinguishing factor among the groups. In summary the ESC | P53− | PTEN− group makes up ∼11% of the cohort with highly aggressive prostate cancer and the TMPRSS2–ERG group consists of another 18% with clearly malignant disease, whereas the remaining 61% of patients have comparatively benign forms of prostate cancer with a clearly improved survival prognosis. Together, the ESC | P53− | PTEN− group and the TMPRSS2–ERG group account for 40% of all prostate cancer deaths in this study (16% and 24%, respectively), comparable to 43% accounted for by the group with high Gleason scores. All three risk groups combined contain 62% of all lethal cases. Indeed the ESC | P53− | PTEN− group and the subgroup with a high Gleason score both had very similar predictive power for survival (compare Fig. 3C with 3D).
Fig. 3.
Clinical analysis of clusters shows graded outcomes with the stem-like group having the poorest outcome. Clinical outcome data available for the Swedish watchful-waiting cohort (Sboner et al.) (27) (GSE16560) were analyzed on the distinct subgroups found by signature profiling. (A–C) Kaplan–Meier estimates for survival functions for the different subgroups, including side-by-side comparison of survival analysis based on signature profiling (A and B) and Gleason score (C). Note that the stem-like group in A and B contains 11% of samples, whereas the group with high Gleason scores in C contains 29% of samples. (D) Clinical variables for the subgroups show a highly significant prognostic value for the stem-like subtype. Follow-up (FU) time is listed together with censoring for overall survival, lethality indicates cases with disease-specific death as determined in the original study, and DSS time refers to disease-specific survival time. Fusion refers to actual TMPRR2–ERG fusion status (FISH). Age distribution was insignificant for all groups. The last column shows the P values for differences in the Kaplan–Meier plots associated with the tested group versus all other groups having higher mean survival times. Significance of assignments is indicated by asterisks.
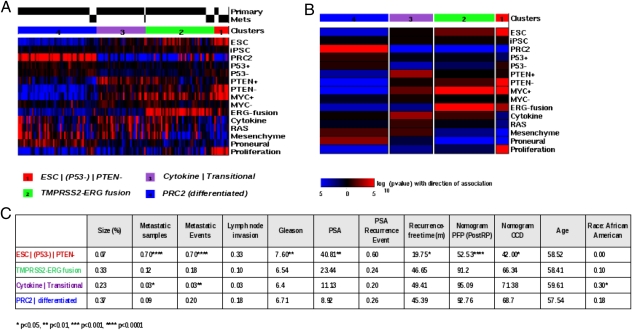
Due to the time frame of the study, the Swedish dataset contained samples of rather untypical nature from a modern clinical perspective. Samples had been taken by transurethral resection of the prostate (TURP) and had been paraffin embedded; RNA had been extracted using cDNA-mediated annealing, selection, extension, and ligation (DASL) and was measured on an Illumina custom chip containing only 6,144 genes. To test the validity of the above results for clinical use, we repeated the signature profile analysis with a second independent dataset published by Taylor et al. (24) (GSE21032), which was collected between 2000 and 2006 at Memorial Sloan–Kettering Cancer Center. Here the samples were taken at radical prostatectomy, fresh frozen, and analyzed on an Affymetrix Human Exon microarray. Statistical parameters for this dataset were quite different from those of the Swedish study, with average Gleason score at biopsy of 6.6 (compared with 7.2), 12% high-grade tumors (compared with 30%), and average age at diagnosis of 58 y (compared with 72 y). Patients were treated with various regimens and outcome was followed for 5 y. Recorded were invasion parameters such as lymph node invasion and metastatic events, prostate-specific antigen (PSA) levels and PSA recurrence-free time, Gleason score and stage, as well as patient population parameters such as age and race. Survival time was recorded but heavily censored with only one death from prostate cancer within the 5-y period and was thus not considered for this study. The set contained in total 131 primary tumors, 19 metastases, 29 adjacent normal samples, and six cell lines. Signature scores were calculated by gene set enrichment analysis and samples were clustered in an unsupervised manner as before. Samples stratified into four stable groups with very similar association patterns as in the Swedish set. Fig. 4A shows signature association patterns of the 150 primary and metastatic tumor samples and Fig. 4B shows overall association with signatures on the four clusters (analogous to Fig. 2 A and B). We detected a group ESC | (P53−) | PTEN− of stem-like tumors analogous to the stem-like group in the first set; although a weak signal for loss of p53 function was present here, it was not significant. This result may have technical reasons (small signature size) or may be related to the earlier age of diagnosis in this cohort (as P53 mutations are known to be late events in prostate cancer) (22, 23). A TMPRSS2–ERG fusion group, a differentiated PRC2 group, and one Cytokine | Transitional group—which can be compared with an admixture of the Cytokine | RAS | Mesenchyme and the transitional subtypes within the first dataset (the algorithm may collapse clusters in smaller datasets due to Bayesian optimization)—reemerged with highly comparable molecular characteristics. Notably, the fusion group contained some samples with MYC activation but no proliferation or stemness. The Cytokine | Transitional group contained 30% samples of African-American ethnicity (P = 0.03), which may also have influenced results. Remarkably, all six cell lines clustered with the ESC | (P53−) | PTEN− group, whereas about half of all adjacent normal samples clustered with the Cytokine | Transitional and the PRC2 group, respectively (Fig. S2). The table in Fig. 4C shows the clinical parameters associated with the four groups. Again the stem-like group was the most aggressive with 70% metastatic samples (P < 1.0E-05), significantly reduced time to PSA recurrence (19.75 mo, P < 0.05), and high Gleason scores (7.6, P < 0.01). The fusion group was intermediate and the Cytokine | Transitional group was benign, whereas the differentiated group (PRC2) included 5 metastatic samples (9%) and had a total of 20% metastatic events, comparable to the fusion group. Group sizes were similar, with the stem-like group being slightly smaller (7% compared with 11%) and the fusion group larger (33% compared with 18%). Size variation might be due to different biases in sample collection (age in cohort, PSA detection). Overall, 17% of all cases with recurrent or metastatic disease were found in the ESC | (P53−) | PTEN− group and 33% in the fusion group, adding up to 50% of all such cases recorded in the 5-y follow-up period (in comparison, the high Gleason score group contains 33% and all combined contain 65% of recurrent or metastatic cases). Considering the extensive differences between the two cohorts, we observed a remarkable correspondence between the molecular profiles and their associated outcomes in both groups.
Fig. 4.
Clustering by signature profiles validates distinct subtypes on an independent clinical cohort. mRNA expression data from 185 samples from Taylor et al. (24) (GSE21034) were used as the validation set. Shown are 150 prostate tumor samples contained in this set, 131 primary tumor samples and 19 metastatic tissue samples (29 adjacent tissue samples and 6 cell line samples not shown). Samples were clustered along signature scores as before. Four clusters were found by unsupervised clustering. Clusters emerged with highly similar association patterns as in the Sboner et al. (27) dataset; compare Fig. 2. (A) Heatmap of signature scores for single signatures, displaying the distinct molecular features of the subtypes. Colors indicate significance of the score assignment (red, positive; blue, negative). (B) Schematic representation of the signature profiles of the clusters. Columns represent clusters, colors represent significance of the overall association of a signature with the cluster, red represents positive association with the signature, and blue represents negative association. Width of columns is relative to size of the cluster. (C) Clinical data were available for these samples. Gleason score and PSA were determined at biopsy (diagnosis). Nomogram prediction values are shown for progression-free probability (PFP) and organ-confined disease (OCD). Average values on clusters were calculated and significance was determined using Fisher's exact test for discrete and Student's t test for continuous variables and is indicated by asterisks.
Discussion
Examination of the survival curves of patients with prostate cancers in a watchful-waiting group reveals prostate cancers that are aggressive and result in death rapidly and others that are indolent and have a more modest impact upon survival. Discriminating which type of prostate cancer a patient has at the time of diagnosis would be very helpful in formulating a plan for aggressive or modest therapy. To date the Gleason score has been the best indicator of overall survival. However, there are a number of patients with Gleason scores of 6 or 7 who will develop aggressive tumors shortly after diagnosis and have poor survival. A method to detect these patients at diagnosis would be helpful.
We have used microarray data from prostate tumors to explore methods to add to the Gleason score and help predict patients with low Gleason scores who will have poor survival. We have combined gene expression signature profiling with unsupervised cluster analysis to identify combinations of signatures in subsets of prostate cancers. We have taken a path from studies in breast cancers where ESC signatures and p53 mutations were highly correlated and predicted the poorest outcomes for overall survival (4, 3). Prostate tumors with the ESC signature were associated with poor survival and were significantly enriched for high Gleason scores. In contrast, differentiated prostate tumors were associated with lower Gleason scores and better survival outcomes. These results corroborate previous associations between ESC-like cancers and survival in breast (3) and liver tumors (5).
Upon addition of several signatures representing common alterations in prostate tumors, the 281 tumors from the watchful-waiting cohort were robustly stratified into five groups or prostate cancer molecular subtypes (Fig. 2). The class with the worst survival outcome and the highest Gleason score contained 11% of the tumors and is characterized by an ESC signature together with p53 and PTEN inactivation signatures and strong proliferation and MYC activation signals (ESC | P53− | PTEN−). This signature combination is similar to the Gleason score in predicting a very poor survival (Fig. 3). The second subclass is composed of tumors manifesting the TMPRSS2–ERG transcriptional signature, representing gene expression patterns associated with this fusion. The enrichment for this signature is corroborated by actual fusion status as measured by FISH (58% fusions, P < 1.0e-07). Recent studies have shown that amplifications of the fusion regions occur in prostate cancer and are associated with higher Gleason scores, whereas presence of the fusion alone does not correlate with increased Gleason score or poorer outcome (30, 31). This result implies that only a subset of the fusion-positive tumors will have a strong transcriptional signal, whereas some fusion-negative tumors will have the signal due to gene amplifications. This outcome fits with our observation in this subclass. The subclass is composed of 18% of the tumors in the dataset and also has a poorer survival outcome than the remaining group. The three remaining groups, containing 61% of tumors, have statistically similar Gleason scores and overall survival outcomes. Among these there is a group of prostate tumors enriched for PRC2 signature, indicative of differentiation, which manifests a trend of better outcome compared with the other two groups. Finally, we note that these results are independent of patient age in the cohort and, although the ESC | P53− | PTEN− group is enriched for high Gleason scores (55%), this molecular signature and Gleason score-based classifications are clearly not identical and not dependent as variables.
These results were validated on a second cohort that closely resembles modern clinical patient populations. Even though sampling method and data extraction were different, stratification into a stem-like group with ESC | (P53−) | PTEN− signature profile and high MYC activation, a fusion group with TMPRSS2–ERG fusion downstream signals, an inflammatory/transitional group, and a differentiated group reemerged in an unsupervised manner. Clinical analysis confirmed the stem-like group as having the worst prognosis. Among the other groups, the fusion group tended toward a poorer prognosis, whereas the inflammatory/transitional group had benign characteristics. Interestingly, the latter was correlated with ethnicity (30% African-American background), corroborating reports of higher rates of inflammatory disease in prostate cancer in African-Americans (32, 33). It overall resembled an admixture of the two corresponding groups in the Swedish set. The differentiated group contained some metastatic tissue samples among mostly benign primary tumor samples and clinical parameters thus tended toward poorer prognosis. Unfortunately, the total number of metastatic samples (5) is too small to allow significant conclusions. It should be emphasized that this second cohort was fundamentally different from the watchful-waiting cohort in almost all parameters. Patients were younger and of mixed ethnicity, had been selected through PSA detection at earlier stages of tumor development, and were treated under different regimens. However, patients with stem-like tumors had a significantly higher risk of recurrent, metastatic, or invasive disease. In contrast to the Swedish study, loss of p53 function was not significantly observed in the stem-like group in this cohort. This finding might well relate to the younger age and overall earlier stage. Recent reports on mouse studies have proposed a progressive collaboration between early MYC amplifications, loss of PTEN, and finally loss of p53, leading to invasive prostate cancer (34, 35). The presence of MYC activation and PTEN loss in the younger group in the Memorial Sloan–Kettering Cancer Center cohort, combined with additional loss of p53 in the older group in the Swedish cohort, supports this model in human prostate cancer. In both cases, the molecular profiles are stem-like. Interestingly, the younger cohort contains ERG fusion-like tumors that have strong MYC activation signals but no proliferation and little or no stemness features. This result suggests that the stem-like subtype might depend on additional activation of the PI3K pathway and/or loss of p53 function, further supporting the hypothesis that p53 counteracts stemness initiation or progression, as well as MYC hyperactivity.
This work is a unique classification of prostate tumors into subgroups with distinct survival outcomes based upon microarray data. The analysis was able to detect two structurally different groups of patients carrying increased risk, with the stem-like tumors being the most aggressive subtype, in two fundamentally different datasets and across different sampling techniques. Under watchful-waiting conditions, patients with stem-like tumors (ESC | P53− | PTEN−) carried a 3.2-fold increased mortality risk (95% CI [2.07, 5.05], P < 1.0E-06) compared with patients in either of the benign subgroups. In particular, patients with Gleason scores of 6 or 7 within this group were 2.7 times more likely to die of the disease than patients with Gleason scores of 6 or 7 in the benign groups (95% CI [1.22, 5.89], P = 0.01). This result suggests that the classification has independent prognostic value and can help to predict adverse outcome in patients with low Gleason scores.
Materials and Methods
Gene Expression Data.
Sample data were downloaded from Gene Expression Omnibus (GEO), series GSE16560 for the Sboner et al. dataset (27), and whole-transcript data were downloaded from GEO series GSE21034 for the Taylor et al. dataset (24).
Gene Expression Signatures.
Expression signatures were collected from the literature. Sources and gene sets are listed in the Tables S1A and S1B. Gene set enrichment analysis (GSEA) (28, 29) was performed for each signature and each tumor sample. Overlap of expression signatures was analyzed and was found insignificant (Table S4 A and B).
Unsupervised Clustering.
Unsupervised clustering was performed applying a Bayesian clustering methodology to the gene expression signature profiles, using as input the gene signature profiles and a library of clustering methods (Fig. S1 and Table S2). The Bayesian clustering method finds the optimal number of groups, the assignment of each sample to a group, and a score quantifying the quality of the clustering (Table S3); for more details see SI Materials and Methods.
Signature Enrichment on Prostate Tumor Groups.
The enrichment or depletion of a signature on a prostate tumor group was determined by applying GSEA (28, 29) with all signatures to the average gene expression of the group.
Significance Tests for Clinical Variables.
Significance of clinical variable values on the clusters was calculated using Fisher's exact test for discrete variables and Student's t test for continuous variables, with the hypothesis that mean values on a cluster differed from the overall mean. Prediction of the TMPRSS2–ERG fusion through signatures had an accuracy of 85% (false positive rate, 35%; false negative rate, 11%). Kaplan–Meier analysis was applied using the Matlab routine kmplot. Risk calculation for mortality in the groups was performed according to the Cox proportional hazard model using the Matlab routine coxphfit.
Correlation Between Gleason Score-Based and Molecular Classifications.
The Sboner et al. (27) sample set was divided into low (<8) versus high Gleason scores and into ESC | P53− | PTEN− versus all others, respectively. Total correlation was computed and compared with the distribution of all possible classifications into two subsets of the same size. The result was insignificant with P = 0.5937.
Datasets used for this study are publicly available under the GEO accession numbers GSE16560 (Sboner et al.) (27) and GSE21034 (Taylor et al.) (24).
Supplementary Material
Footnotes
The authors declare no conflict of interest.
See Commentary on page 20861.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117029108/-/DCSupplemental.
References
- 1.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Weigelt B, et al. Molecular portraits and 70-gene prognosis signature are preserved throughout the metastatic process of breast cancer. Cancer Res. 2005;65:9155–9158. doi: 10.1158/0008-5472.CAN-05-2553. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Porath I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizuno H, Spike BT, Wahl GM, Levine AJ. Inactivation of p53 in breast cancers correlates with stem cell transcriptional signatures. Proc Natl Acad Sci USA. 2010;107:22745–22750. doi: 10.1073/pnas.1017001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woo HG, et al. Association of TP53 mutations with stem cell-like gene expression and survival of patients with hepatocellular carcinoma. Gastroenterology. 2010;140:1063–1070. doi: 10.1053/j.gastro.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 7.Kawamura T, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marión RM, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanna J, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong H, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.American Cancer Society . Cancer Facts & Figures 2009. Atlanta: American Cancer Society; 2009. [Google Scholar]
- 13.Wang SY, et al. Pten deletion leads to the expansion of a prostatic stem/progenitor cell subpopulation and tumor initiation. Proc Natl Acad Sci USA. 2006;103:1480–1485. doi: 10.1073/pnas.0510652103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sircar K, et al. PTEN genomic deletion is associated with p-Akt and AR signalling in poorer outcome, hormone refractory prostate cancer. J Pathol. 2009;218:505–513. doi: 10.1002/path.2559. [DOI] [PubMed] [Google Scholar]
- 15.Clark J, et al. Complex patterns of ETS gene alteration arise during cancer development in the human prostate. Oncogene. 2008;27:1993–2003. doi: 10.1038/sj.onc.1210843. [DOI] [PubMed] [Google Scholar]
- 16.Mosquera JM, et al. Characterization of TMPRSS2-ERG fusion high-grade prostatic intraepithelial neoplasia and potential clinical implications. Clin Cancer Res. 2008;14:3380–3385. doi: 10.1158/1078-0432.CCR-07-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albadine R, et al. TMPRSS2-ERG gene fusion status in minute (minimal) prostatic adenocarcinoma. Mod Pathol. 2009;22:1415–1422. doi: 10.1038/modpathol.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carver BS, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Cai Y, Ren C, Ittmann M. Expression of variant TMPRSS2/ERG fusion messenger RNAs is associated with aggressive prostate cancer. Cancer Res. 2006;66:8347–8351. doi: 10.1158/0008-5472.CAN-06-1966. [DOI] [PubMed] [Google Scholar]
- 20.Klezovitch O, et al. A causal role for ERG in neoplastic transformation of prostate epithelium. Proc Natl Acad Sci USA. 2008;105:2105–2110. doi: 10.1073/pnas.0711711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomlins SA, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10:177–188. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agell L, et al. KLF6 and TP53 mutations are a rare event in prostate cancer: Distinguishing between Taq polymerase artifacts and true mutations. Mod Pathol. 2008;21:1470–1478. doi: 10.1038/modpathol.2008.145. [DOI] [PubMed] [Google Scholar]
- 23.Schlomm T, et al. Clinical significance of p53 alterations in surgically treated prostate cancers. Mod Pathol. 2008;21:1371–1378. doi: 10.1038/modpathol.2008.104. [DOI] [PubMed] [Google Scholar]
- 24.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenkins RB, Qian J, Lieber MM, Bostwick DG. Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization. Cancer Res. 1997;57:524–531. [PubMed] [Google Scholar]
- 26.Gurel B, et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod Pathol. 2008;21:1156–1167. doi: 10.1038/modpathol.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sboner A, et al. Molecular sampling of prostate cancer: A dilemma for predicting disease progression. BMC Med Genomics. 2010;3:8. doi: 10.1186/1755-8794-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 29.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Attard G, et al. Transatlantic Prostate Group Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2008;27:253–263. doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fine SW, et al. TMPRSS2-ERG gene fusion is associated with low Gleason scores and not with high-grade morphological features. Mod Pathol. 2010;23:1325–1333. doi: 10.1038/modpathol.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mason TE, et al. Association of CD14 variant with prostate cancer in African American men. Prostate. 2010;70:262–269. doi: 10.1002/pros.21060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.American Cancer Society . Cancer Facts & Figures for African Americans 2005-2006. Atlanta: American Cancer Society; 2005. pp. 1–28. [Google Scholar]
- 34.Kim J, et al. A mouse model of heterozygous, c-MYC-initiated prostate cancer with loss of Pten and p53. Oncogene. 2011 doi: 10.1038/onc.2011.236. 10.1038/onc.2011.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clegg NJ, et al. MYC cooperates with AKT in prostate tumorigenesis and alters sensitivity to mTOR inhibitors. PLoS ONE. 2011;6(3):1–14. doi: 10.1371/journal.pone.0017449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.