Abstract
We used a microarray study in order to compare the time course expression profiles of two Chlamydomonas reinhardtii strains, namely the high H2 producing mutant stm6glc4 and its parental WT strain during H2 production induced by sulfur starvation. Major cellular reorganizations in photosynthetic apparatus, sulfur and carbon metabolism upon H2 production were confirmed as common to both strains. More importantly, our results pointed out factors which lead to the higher H2 production in the mutant including a higher starch accumulation in the aerobic phase and a lower competition between the H2ase pathway and alternative electron sinks within the H2 production phase. Key candidate genes of interest with differential expression pattern include LHCSR3, essential for efficient energy quenching (qE). The reduced LHCSR3 protein expression in mutant stm6glc4 could be closely related to the high-light sensitive phenotype. H2 measurements carried out with the LHCSR3 knock-out mutant npq4 however clearly demonstrated that a complete loss of this protein has almost no impact on H2 yields under moderate light conditions. The nuclear gene disrupted in the high H2 producing mutant stm6glc4 encodes for the mitochondrial transcription termination factor (mTERF) MOC1, whose expression strongly increases during –S-induced H2 production in WT strains. Studies under phototrophic high-light conditions demonstrated that the presence of functional MOC1 is a prerequisite for proper LHCSR3 expression. Furthermore knock-down of MOC1 in a WT strain was shown to improve the total H2 yield significantly suggesting that this strategy could be applied to further enhance H2 production in other strains already displaying a high H2 production capacity. By combining our array data with previously published metabolomics data we can now explain some of the phenotypic characteristics which lead to an elevated H2 production in stm6glc4.
Introduction
The sulfur starvation method [1] for continuous hydrogen production in the green alga Chlamydomonas reinhardtii has received a lot of attention in the last decade as it improved the obtainable hydrogen yield significantly [2]. Under anaerobic conditions, C. reinhardtii and a number of other photosynthetic microorganisms can produce H2 via hydrogenase enzymes [3]. The production of H2 re-oxidizes reduced ferredoxin thereby maintaining essential ATP production [4]. Under illuminated conditions, H2 production is normally short-lived due to the inhibitory effects of O2 produced by photosynthesis on hydrogenase expression and activity [5]. By depriving the algae of sulfur, the photosynthesis to respiration ratio is decreased to less than one, effectively removing the dissolved O2 in the sealed culture yielding conditions supportive of anaerobic H2 production [1]. During S-deprived H2 production, major reorganizations of cellular structures and metabolic pathways occur within C. reinhardtii to aid survival [6]–[11]. First, the cell is reported to switch into the enhanced S acquisition/assimilation mode and as a result the transcript abundance of responsible enzymes greatly increases [6], [9]. In parallel, photosynthesis is down-regulated in response to the lower assimilation capacity. The decrease in photosynthesis was observed widely in light harvesting proteins, reaction centers and components of the electron transport chain as well as in components of the Calvin cycle when transcript [6] or protein levels [7], [8] of respective genes were analyzed. Enhanced protein degradation was also evident while certain proteins with lower S content are proposed to replace the function of their counterparts [6], [9]. Induction of anaerobiosis through sulfur depletion also triggers starch and lipid accumulation as shown in metabolomic studies on S-deprived H2 production [10], [11]. Upon the establishment of anaerobiosis due to the continuous net O2 consumption, additional sets of changes occur. Aerobic metabolic processes including citric acid cycle and oxidative phosphorylation are suppressed and replaced by fermentative pathways including H2 production [6], [10]–[12]. Due to the complexity of S-starvation induced H2 production, many factors have influences on the final H2 productivity. Reduced carbon sources such as starch or acetate are required for H2ase expression as their consumption is needed to drive C. reinhardtii cultures into anaerobiosis before H2 production can occur [12]–[15]. The starting pH was shown to have strong influences on H2 production with an optimum pH of 7.3 [16]. Availability of alternative electron sinks like carbon fixation as well as the activity of reductive enzymatic reactions coupled to ferredoxin also determine H2 productivity since both can reduce electron flow through the H2ase pathway [17]–[19]. The duration of the aerobic phase also determines the onset of H2 production and also affects how much energy is stored, consumed or made available to HYDA. Previous observations suggest that a short aerobic phase is desirable for S-deprived H2 production since it reduces the consumption of energy storage compounds such as starch and lipids. However, eliminating the aerobic phase altogether led to lower productivities [17], presumably due to the lack of the essential energy accumulation phase associated with fully functional oxygenic photosynthesis. As has been shown recently, enhanced oxygen consumption by introduction of leghemoglobin and ferrochelatase into Chlamydomonas is a means to improve hydrogen production [20].
S-starvation induced H2 production has been studied using different high-throughput technologies including transcriptomics [6], proteomics [7] and metabolomics [10]–[11]. Together with other studies on individual aspects of H2 production, these studies have contributed to the increasingly complete picture of the H2 production mechanism in C. reinhardtii with the identification of important target genes and pathways which can be used for future improvements. In this study we took one step further, using microarray to analyze and compare the time-course global expression profiles of two C. reinhardtii strains under S-deprived H2 production. A time-resolved transcriptome study takes into account that the H2 production process is composed of two main phases. In phase 1 anaerobiosis is established thereby activating the H2ase pathway and in phase 2 H2 production is sustained by the consumption of intracellular energy stores generating electrons which feed into the H2ase pathway [11]. In both phases regulative mechanisms occur, which represent potential targets for genetic engineering approaches designed to improve photobiological H2 production. An improved knowledge about the temporal aspects of transcriptome changes required to trigger and sustain H2 production in C. reinhardtii should enable more targeted genetic engineering strategies (e.g. by using inducible overexpression or knock-down systems). In addition they provide further insights into the transcriptomic differences between the high H2 producing strain stm6glc4 [21], [22] and a wild type strain in order to understand why this mutant produces higher amounts of H2 within a given time.
Results and Discussion
The acclimatory response to sulfur deprivation differs largely between stm6glc4 and wild type
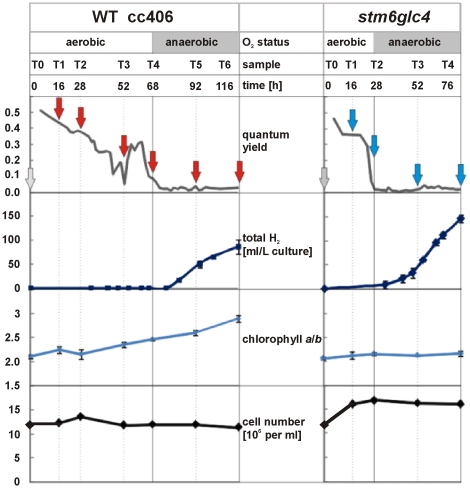
In this study, we took samples at various time points during the course of S-deprived H2 production. Samples were simultaneously taken for parallel metabolome analyses reported in a previous study [11]. Figure 1 depicts the sampling points and how quantum yield (ΦPSII), total H2 yield, chlorophyll a/b ratio and cell number changed in the two strains during the course of the experiment. Samples taken at the indicated time points were compared with the corresponding reference sample (T0), which was taken immediately prior to S-starvation and transcript abundance in both samples was determined by microarray analysis.
Figure 1. Changes in quantum yield, total H2, chlorophyll a/b ratio and cell number of the two strains stm6glc4 and WT cc406 during the course of S-deprived H2 production.
Arrows below the different time points indicate where samples were taken for microarray analysis.
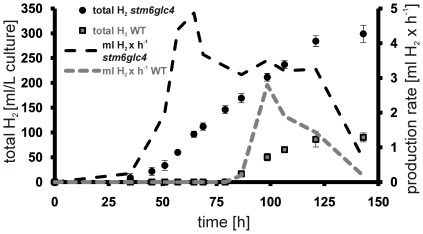
As can be seen in Figure 1, patterns of quantum yield change differ significantly between wild type (WT) and high H2 production mutant stm6glc4. In stm6glc4 ΦPSII declined sharply to below 0.1 within 28 h after the start of S-depletion while it decreased more slowly in the WT. Since the ΦPSII drop correlates strongly with the drop of oxygen in the culture [12], the pattern of ΦPSII changes indicates that the anaerobic phase started much earlier in stm6glc4 than in WT (after 28 h compared to 68 h) and as a result, H2 production started much earlier in the mutant. This offers a significant advantage for the development of high efficiency H2 production processes. However, it has to be emphasized that efficient H2 production requires a residual activity of PSII, since the PSII-dependent H2ase pathway represents a vital part of the entire process [13]. Furthermore at the end of the experiment, the total amount of H2 produced by stm6glc4 was over 3–5 times higher than the amount produced by the WT (Figure 2). In addition to the faster transition to anaerobiosis, peak H2 production rate was also higher in stm6glc4 compared to WT (Figure 2, right y-axis; 4.9 vs. 2.8 ml•h−1). Within the same first 48 h of the anaerobic phase, stm6glc4 had already produced 50% more H2.
Figure 2. Total volume of H2 produced by stm6glc4 (black circles) and WT (grey squares) during the course of the experiment.
The total volume of H2 produced during the experiment expressed as ml/L culture is shown on the left y-axis with the duration of the experiment indicated on the x-axis. Production rates given as ml per hour are indicated on the right y-axis and presented as dotted lines.
In both strains, cell number increased slightly after sulfur depletion was induced, and then slowly decreased toward the end of the experiment. Surprisingly, while chlorophyll a to b ratio (Fig. 1, chl a/b) increased steadily in WT from about 2 to around 2.8, that value remained rather constant at around 2 in stm6glc4, indicating different adaption response to S-deprivation.
Microarray analysis reveals numerous genes which are differentially regulated in the mutant
The detailed analysis of all microarray data including four time points in stm6glc4 and six time points in WT led to the identification of 410 nuclear encoded genes displaying a more than 2 fold differential transcript change for at least one of the time points (T1–T4 for stm6Glc4 and T1–T6 for WT) after sulfur deprivation in comparison to the sulfur-deplete T0 condition (151 genes in stm6glc4 and 342 genes in WT). Among them, 189 genes could be assigned into certain functional groups while the majority of the remaining genes are not functionally annotated.
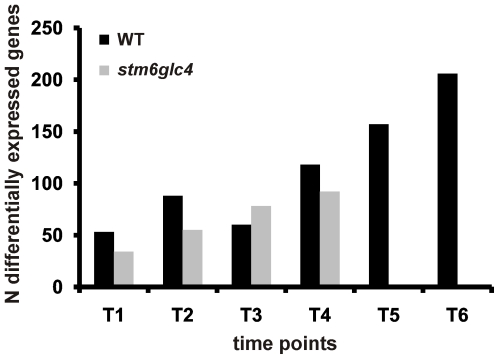
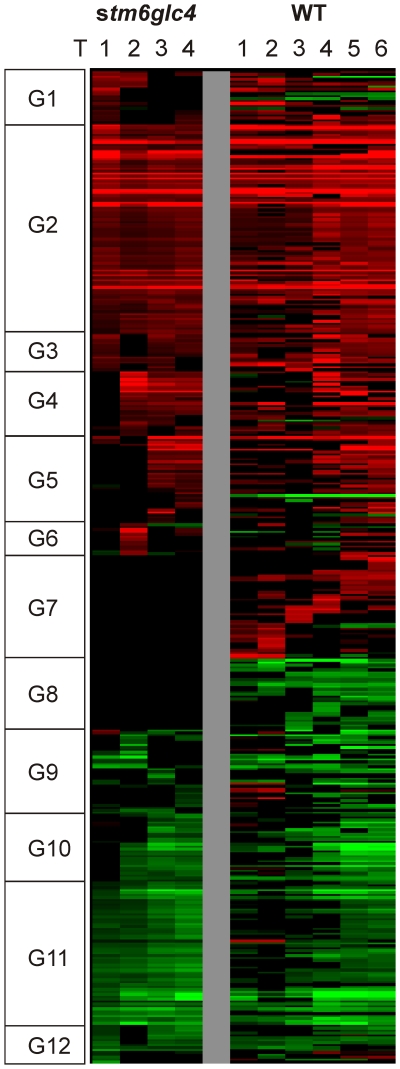
In both strains, the number of differentially expressed genes generally increased during the course of the experiment (Figure 3), reflecting the increasing “physiological distance” from the reference physiological state at T0. Due to two additional time points T5 and T6, the number of differentially expressed genes was significantly higher in WT, which might have been caused by a longer duration of sulfur deprivation in this case. Interestingly the number of genes differentially expressed in WT at time points T1/T2 is significantly higher than the corresponding number for stm6glc4 at these sampling points. This points at a dampened gene regulatory response in stm6glc4, which could partly explain the higher susceptibility of stm6glc4 to –S-induced photo-damage, as can be seen by the precipitous drop in photosynthetic quantum yield. To determine the time-dependent gene expression log2 ratios were first grouped by hierarchical clustering and clustered data then visualized by heat mapping (Figure 4). Based primarily on the transcript variations from stm6glc4 data sets, 12 main expression pattern groups were identified (G1–G12). The heat map provides an overview of different expression patterns observed among the differentially expressed genes (DEGs) as well as the contrast between stm6glc4 and WT. The list of all DEGs with their transcript abundance change at each time point is presented in Table S1.
Figure 3. Number of differentially expressed genes identified at each time point for both strains (stm6glc4 and WT) by microarray analyses.
Figure 4. Heat map of all differentially expressed genes which were hierarchically clustered based on stm6glc4 data set.
Changes in transcript abundance compared to T0 are indicated by different colors: Red (increase), Green (decrease) and Black (unchanged). Genes were categorized into 12 different groups (G1–12) based on their expression patterns.
Sulfur deprivation induced H2 production is a biphasic process. Within the first phase sulfur depletion impairs the PSII repair cycle and causes a declined oxygen evolving activity. Consequently oxygen evolution decreases constantly until oxygen consumption by mitochondrial respiration exceeds the rates of production [1].
The initial phase is therefore characterized by the establishment of anaerobic conditions. Once anaerobiosis has been established the H2ase pathway is activated. In addition to comparing the overall transcriptomic differences between WT and stm6glc4 we were also able to distinguish between the two different stages in order to take the biphasic character of H2 production into account. Genes specifically up- or down-regulated in only the mutant or the WT were chosen from our array data to correlate distinctive transcriptomic changes with the phenotypical differences between WT and the mutant. Genes displaying a WT- or mutant-specific expression pattern during the establishment of anaerobiosis (Fig. 1 T1/T2) are listed in Table 1. Processes up-regulated in both strains during the first 28 h after sulfur deprivation include sulfur acquisition/recycling as well as carotenoid biosynthesis (Table 1). The up-regulation of genes encoding proteins implicated in the acquisition of sulfur is a direct consequence of sulfur depletion and has already been described [9]. An up-regulation of carotenoid biosynthesis in response to sulfur depletion has previously been demonstrated for other green algae like Dunaliella bardawil [23]. Although transcript abundance of all PSII associated major light-harvesting genes (LHCBM) except for LHCBM9 is significantly reduced [6], the expression of stress-related LHC genes (Table 1; LHCSR1/3) is induced after withdrawal of sulfur [9], [24]. Interestingly the encoded proteins might have a higher carotenoid content compared to LHCBMs in vivo as suggested by refolding studies in the presence of various pigments [25].
Table 1. Wild Type-specific expression changes in T1 and T2.
| Upregulated | ||||
| Process | Gene/Locus | Description | ID | Group |
| photosynthesis | LHCSR3/Cre08.g367400 | stress-related LHC protein | 8770.D | 1 |
| LHCSR1/Cre08.g365900 | stress-related LHC protein | 251.A | 7 | |
| Cre07.g320450/Cre07.g320400 | CBR-like ELIP | 9621.E | 7 | |
| carbon metabolism | GWD2/Cre07.g332300 | R1 protein, α-glucan water dikinase | 1457.C | 9 |
| GND1 a/b/Cre12.g526800 | 6-phosphogluconate dehydrogenase | 337.A | 4 | |
| nitrogen metabolism | Cre11.g477200 | NmrA-like protein | 5077.C | 5 |
| AMT1;1/Cre03.g159254 | ammonium transporter | 199.A | 9 | |
| CO2 concentrating mechanism (CCM) | LCR1/Cre02.g136800 | low-CO2 inducible Myb transcription factor | 266.A | 7 |
| HLA3/Cre02.g097800 | Ci uptake | 21.A | 9 | |
| Amino acid metabolism: | OAT1/Cre11.g474800 | Ornithine transaminase | 6901.C | 5 |
| Lipid metabolism: | Cre01.g035350 | Trans-2-enoyl-CoA reductase | 1311.C | 7 |
| Nucleotide metabolism | Cre03.g184400 | NUDIX_Hydrolase_19 | 5676.C | 7 |
| Transport: | CCP2/Cre04.g222750 | putative mitochondrial carrier | 128.A | 7 |
| Metabolism of cofactors: | Cre08.g359700 | Lipoate synthase | 2624.C | 7 |
| Vesicular transport | Cre06.g289700 | TRAPP component | 8048.D | 7 |
| Transcription/Translation | Cre01.g031050 | SPT5 transcription elongation factor | 5127.C | 7 |
| Cre02.g102400 | DNA-directed RNA polymerase SU | 6049.C | 7 | |
| Cre12.g528900 | PUA like RNA binding protein | 3412.C | 9 | |
| Proteolyis | Cre16.g663350 | clp protease ATP-binding subunit | 4348.C | 7 |
| Cell division | Cre12.g519700 | YihA/EngB-like GTPase | 7786.D | 7 |
| Downregulated | ||||
| Transcription/Translation: | Cre02.g130150 | SAP-domain containing | 5599.C | 8 |
| Ca2+ homeostasis/signaling | ACA2/Cre12.g505350 | calcium-transporting ATPase | 4472.C | 8 |
List of genes showing either a specific down- or upregulation in one of both examined strains. Genes contained in the list displayed a 2fold down- or upregulation for at least one of the time points T1/T2. Differentially expressed genes are sorted according to the cellular processes involved as deduced from their functional annotation. Gene names are given along with the corresponding locus names (Phytozome 7.0; http://www.phytozome.net/) and a description of their function. Indicated gene IDs correspond to those given in the gal file (http://www.chlamy.org/galfile.xls/) for Chlamydomonas olinucleotide array v2.0 [50]. Heat map group assignments (Figure 4) for each gene are given as well.
Among the genes showing a differential regulation within T1/T2 were those encoding for stress-related LHC (light-harvesting proteins) proteins namely LHCSR1, LHCSR3 and a putative Cbr-like ELIP protein. LHCSR3 showed a strongly increased transcript level in the WT (12 fold in T1 vs. T0; Table S1) whereas the increase in stm6glc4 was very moderate (2 fold in T1 vs. T0; Table S1). The physiological relevance and especially the impact of the dramatic expression induction of LHCSR3 on H2 production are still unknown. LHCSR1 shows an identity of 87% to LHCSR3 but in contrast to LHCSR3, which is essential for energy-dependent quenching (qE) as demonstrated by the characterization of the knock-out mutant npq4 [26], little is known about the physiological function of LHCSR1. The transcript of LHCSR1 is exclusively up-regulated in the WT and no expression changes upon sulfur depletion were observed for the mutant (Table S1) yielding in an about 50 fold higher transcript level in WT cells compared to stm6glc4 cells in the phase after S-depletion and before H2 production (Figure S1; LHCSR1; preH2). Calcium and a plant-specific Calcium Sensor (CAS) calcium binding protein seem to be involved in the expression regulation of LHCSR3 [27] and interestingly one of the genes differentially regulated between WT and mutant encodes a protein potentially functioning within Ca2+ homeostasis and signaling (ACA2; Table 1). In contrast to the mutant, which did not show any differential regulation of the gene ACA2, a significant down-regulation occurred in the WT. BLAST analyses performed with ACA2 indicated a high homology to PIIb-type ATPases from A. thaliana (UniProtKB Q9M2L4/Q9LU41) located in the plasma membrane, vacuole, plastid envelope, or endoplasmatic reticulum and some evidence exists for the requirement of P-ATPases for proper stress-responsiveness [28].
The Cbr (carotene biosynthesis-related)-like ELIP (early light-induced protein) is a homolog (35% identity) of an ELIP-like protein identified in Dunaliella bardawil [29] named Cbr (UniProtKB P27516), for which zeaxanthin binding and a photoprotective role was proposed [30]. Similar to LHCSR1 mRNA the transcript of the Cbr-like ELIP gene showed a considerably lower steady-state level (only 2.35±0.25% of WT level) in the mutant within T1/T2 (Figure S1; Cbr-like ELIP; preH2). Another transcript exclusively up-regulated in the WT encodes for a putative α-glucan water dikinase (Table 1 and Figure S1 GWD2; EC 2.7.9.4). GWDs phosphorylate starch at the C6 position of amylopectin-related glucosyl residues [31] and rates of starch phosphorylation were shown to be increased during net starch breakdown in C. reinhardtii [32]. The exclusive up-regulation of GWD2 in the WT in response to sulfur depletion (4 fold induction T2 vs. T0; Table S1) and a 7 fold higher steady state mRNA level compared to the mutant (Figure S1) could provide an explanation for the different extents of –S-induced starch accumulation observed between WT and stm6glc4 within T1/T2 [11]. A lower total amount of starch and a reduced net starch synthesis in the WT could be due to a higher activity of a starch phosphorylating enzyme leading to higher breakdown rates thus decreasing net synthesis. Import of glucose supplied in the media in the case of stm6glc4 has to be considered as a contributing factor for the higher starch accumulation in the mutant. However, increased starch accumulation could also be noted for the parental strain stm6, which is not equipped with a hexose uptake system, if it was grown in acetate-containing TAP media [33].
We also compared the transcriptome of H2 producing (Fig. 1: T3/4 stm6glc4 and T5/6 WT) cells from WT and mutant strain like it was conducted for the T1/T2 phase (Table 2). In contrast to LHCSR3 the other two stress-related light-harvesting proteins LHCSR1 and Cbr-like ELIP which were also exclusively up-regulated in the WT during T1/2 still showed a significant up-regulation compared to T0 during H2 production (Table 2 and Figure S1; LHCSR1 (H2) and CBR-like ELIP (H2)). The PIIb-type ATPase ACA2 which was down-regulated in the WT during T1/T2 was also expressed at a reduced level while cells were producing H2 (Table 2; ACA2). Expression of ACA1 was up-regulated in the WT in T5/6 whereas no differential gene expression could be detected in the mutant within this phase (Table 2; ACA1). ACA1 like ACA2 displays similarity to A. thaliana PIIb-type ATPases (UniProtKB: Q9SZR1; Q9M2L4). Among the genes up-regulated (2.9 fold T6 vs T0; Table S1) in the WT but not differentially expressed in the mutant was a gene similar (20.1% identity) to CGDL15 (Gene ID: 5718701; UniProtKB A8IWH9), which encodes for a protein harbouring a lipase 3 domain (Table 2; Cre03.g155250). The mRNA steady-state level of this gene in the WT was about 3 fold higher than in the mutant under H2 producing conditions (Figure S1). Class 3 lipases are triacylglycerol lipases (EC 3.1.1.3) and interestingly the exclusive up-regulation of the putative lipase correlates well with a reduction of the lipid content during H2 production in WT cells and with an unchanged level in the stm6glc4 as determined by Nile red staining [11].
Table 2. Wild Type-specific expression changes in T5 and T6.
| Upregulated | ||||
| Process | Gene/Locus | Description | ID | Group |
| Photosynthesis | LHCSR1/Cre08.g365900 | stress-related LHC protein | 251.A | 7 |
| Cre07.g320450 | CBR-like ELIP | 9621.E | 7 | |
| Carbon metabolism | Cre16.g692800 | Aldo-keto reductase | 698.C | 7 |
| Cre17.g726700 | putative acetate-CoA ligase | 3329.C | 7 | |
| Amino acid metabolism: | HPD2/Cre02.g136100 | 4-hydroxyphenylpyruvate dioxygenase | 9.A | 7 |
| Lipid metabolism: | Cre01.g035350 | Trans-2-enoyl-CoA reductase | 1311.C | 7 |
| Cre03.g155250 | similar to CGLD15 (TAG lipase-like) | 3844.C | 12 | |
| Transport: | ZIP6/Cre06.g299600 | zinc-iron transporter | 2440.C | 7 |
| Signalling: | Cre12.g520000 | Ankyrin-repeat containing protein | 1171.C | 1 |
| Calcium homeostasis/signalling: | ACA1/Cre09.g388850 | calmodulin binding calcium transporting ATPase | 9169.E | 1 |
| Transcription/Translation | Cre01.g031050 | SPT5 transcription elongation factor | 5127.C | 7 |
| Cre04.g226400 | Histone-like transcription factor | 9318.E | 7 | |
| Cre06.g273900 | Histone 2A | 3023.C | 7 | |
| Protein folding | Cre13.g603950 | peptidyl-prolyl cis-trans isomerase | 2173.C | 7 |
| Proteolyis | RSE2/Cre01.g056650 | intramembrane metalloprotease | 2786.C | 7 |
| Cell division | Cre12.g519700 | YihA/EngB-like GTPase | 7786.D | 7 |
| HSP22F/Cre14.g617400 | heat-shock response | 9317.E | 6 | |
| Sulfur acquisition/recycling | SIR1/Cre08.g365700 | ferredoxin-sulfite reductase | 8577.D 3620.C | 4 |
| Downregulated | ||||
| CO2 concentrating mechanism (CCM): | CAH2/Cre04.g223050 | carbonic anhydrase, α type, periplasmic | 38.A | 8 |
| LCR1/Cre02.g136800 | low-CO2 inducible Myb transcription factor | 266.A | 7 | |
| CAH4/Cre05.g248400 | mitochondrial carbonic anhydrase | 91.A | 9 | |
| Transcription/Translation: | Cre02.g130150 | SAP-domain containing | 5599.C | 8 |
| Cre12.g504700 | Histone H2B | 360.A | 1 | |
| Cre06.g273900 | Histone H2A | 9199.E | 8 | |
| RPL15/Cre02.g091100 | Ribosomal protein L15 | 9459.E | 8 | |
| Ca2+ homeostasis/signaling | ACA2/Cre12.g505350 | calcium-transporting ATPase | 4472.C | 8 |
| Chaperones | LCI15/Cre16.g685050 | putative metallochaperon | 285.A | 9 |
| Vesicular transport | Cre06.g289700 | TRAPP component | 8048.D | 7 |
| Cre06.g290100 | SNARE protein | 476.A | 1 | |
| Signalling | CYG54/Cre12.g489900 | Adenylate/guanylate cyclase | 4565.C | 8 |
| AGG2/Cre17.g738000 | phototactic protein | 9625.E | 8 | |
| Photosynthesis | LHCA5/Cre10.g425900 | PS I light harvesting protein | 213.A | 8 |
| CTH1B/Cre12.g510050 | Copper target 1 protein MgPME cyclase | 19.A | 1 | |
| Cell wall | PHC12/Cre11.g472250 | cell wall protein pherophorin C12 | 2719.C | 8 |
| GP2/Cre06.g258800 | Hydroxyproline-rich glycoprotein | 14.A | 8 | |
| Carbon metabolism | LCI9/Cre02.g130700 | putative α-amylase | 6737.C | 8 |
| Lipid metabolism | Cre03.g164350 | putative Lysophospholipase II | 7018.C | 9 |
| Cre12.g512300 | Lipoxygenase/oxylipin synthesis | 7494.C | 9 | |
| Others | Cre18.g745350 | CAP10-like | 5866.C | 11 |
List of genes showing either a specific down- or upregulation in one of both examined strains. Genes contained in the list displayed a 2fold down- or upregulation for at least one of the time points T3/T4 in stm6glc4 or T5/T6 in the WT. Differentially expressed genes are sorted according to the cellular processes involved as deduced from their functional annotation. Gene names are given along with the corresponding locus names (Phytozome 7.0; http://www.phytozome.net/) and a description of their function. Indicated gene IDs correspond to those given in the gal file (http://www.chlamy.org/galfile.xls/) for Chlamydomonas olinucleotide array v2.0 [50]. Heat map group assignments (Figure 4) for each gene are given as well.
Several reductive pathways in the plastid rely on reduced ferredoxin as an electron donor [34]. These pathways can therefore withdraw electrons from the H2ase pathway thus reducing the rates of H2 production. An interesting finding in this regard was the stronger up-regulation of a ferredoxin sulfite reductase in the WT (Table S1; Figure S1) during H2 production, which was not observed for the mutant (Table 2; SIR1). This enzyme uses reduced ferredoxin as an electron donor [35] thereby potentially competing for electrons otherwise used by the H2ase pathway which could partially explain the higher rates of H2 production in stm6glc4. A function of this enzyme in the absence of sulphur in the media during H2 producing conditions could be the reduction of sulfite generated within sulfur redistribution pathways, which can be seen as an essential survival strategy for cells deprived of sulphur [9].
Transcriptomic differences between WT and stm6glc4 at T0
Gene expression in stm6glc4 and WT was also analyzed at T0 (T0 stm6glc4 vs. T0 WT), and four genes encoding for proteins belonging to carbon metabolism showed increased transcript levels in the mutant (Table S2). The strongest difference in the T0 expression level was found for the gene ICL1 that codes for the enzyme isocitrate lyase (EC 4.1.3.1) whose steady-state transcript level was over 30 fold (microarray data) or 2.6 fold (RT-Q-PCR, Figure S1) more abundant in the mutant. This enzyme catalyzes the formation of succinate and glyoxylate from isocitrate in the initial step of the glyoxylate cycle [36], which enables Chlamydomonas to grow on acetate as a sole carbon source. A stronger expression of this enzyme indicates that the mutant consumes more acetate via the glyoxylate cycle, which is in good agreement with our metabolomics data demonstrating a higher acetate consumption of the mutant during transition to anaerobiosis when oxidative phosphorylation is still operational [11]. stm6glc4 also showed stronger expression of ascorbate peroxidase (APX1) under normal conditions compared to WT suggesting that the mutant might suffer more from oxidative stress, which is consistent with an earlier finding that the parental strain stm6 displays elevated levels of lipid hydroperoxides already in standard growth light [21].
The reduced LHCSR3 expression in mutant stm6glc4 is closely related to the high-light sensitive phenotype
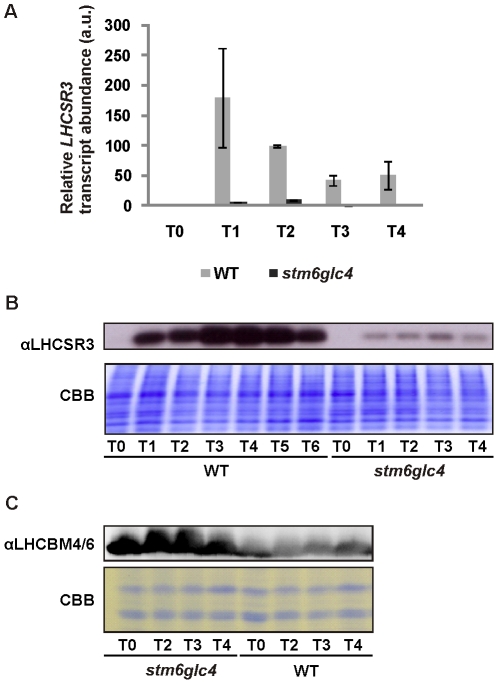
One intriguing result was the difference in the level of expression of LHCSR3 between WT and stm6glc4. Consequently we decided to analyze this differential regulation in more detail. LHCSR3 has been reported to play an important role in non photochemical quenching (NPQ) [26]. The marked difference regarding the mRNA level of LHCSR3 between stm6glc4 and WT as observed in our microarray experiments was further confirmed by real-time PCR (Fig. 5 A) and by immuno-blotting studies (Fig. 5 B; αLHCSR3). In both strains, LHCSR3 protein level increased after induction of anaerobiosis by S-starvation but the level was significantly higher in WT throughout the course of the experiment compared to that in stm6glc4. As suggested by the Chl a/b data (Fig. 1) recorded during H2 production for WT and stm6glc4 the mutant showed a smaller LHCII antenna phenotype indicated by lower levels of the LHCBM isoforms 4 and 6 (Fig. 5 C; αLHCBM4/6) during the entire course of the experiment, suggestive of considerable differences between the stress-acclimation responses displayed by WT and mutant.
Figure 5. Expression of LHCSR3 in WT and stm6glc4 under H2 production conditions.
A: Real-time RT-Q-PCR analysis of LHCSR3 mRNA expression in the WT (grey bars) and mutant stm6glc4 (black bars) before (T0) and after sulphur deprivation (T1–T4). The indicated time points correspond to those used for microarray sample collection (see Fig. 1). Standard deviations are derived from measurements with three technical replicates. B: Immunoblot analysis of LHCSR3 accumulation in WT and stm6glc4 during H2 production. Protein extracts were derived from the samples taken during microarray sample collection and the indicated time points correspond to those shown in Figure 1. Along with the immunodetection of LHCSR3 (αLHCSR3) a Coomassie-stained gel is shown to assess protein loading. C: Immunodetection of the major light-harvesting protein isoforms 4 and 6 (αLHCBM4/6) in protein extracts from WT and stm6glc4. Samples for protein extraction were taken along with the sample collection for RNA extraction prior to microarray analysis. The indicated time points are identical to the time points used for the microarray studies.
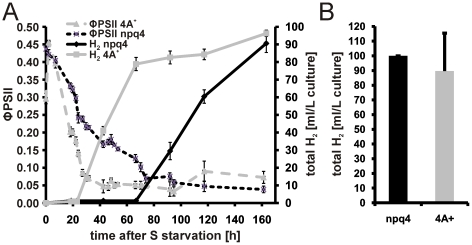
It was previously demonstrated that knock-out of LHCSR3 results in a reduced fitness of Chlamydomonas cells grown in a changing light environment indicating a prominent role of energy-dependent quenching (qE) for cell survival under outdoor conditions [26]. Against the background that expression of LHCSR3 is rapidly induced after withdrawal of sulfur and reaches very high steady state levels it is possible that LHCSR3 accumulation under –S conditions serves to protect PSII against photodamage, especially if it is considered that an impairment of D1 de novo synthesis after sulfur depletion makes PSII more susceptible against light-induced damage [37]. Therefore one potential explanation for the earlier onset of H2 production in stm6glc4 compared to the WT was that the lack of LHCSR3 accumulation makes PSII more prone to damage, which results in a faster transition to anaerobiosis as a precondition for an activation of the H2ase pathway. Consequently we investigated whether a complete loss of LHCSR3, as in the knock-out mutant npq4 [26], [38], has any impact on the decline of PSII activity after S deprivation or on the H2 production capacity. The absence of LHCSR3 had no significant effect on the kinetics of ΦPSII decline (Fig. 6A) or the total H2 production rates (Fig. 6 B) as deduced from H2 experiments performed with npq4 and WT strain (4A+ [26]). In conclusion the amount of LHCSR3 protein is not a decisive factor for the H2 production capacity under –S conditions combined with the moderate light intensity (300 µmol photons m−2 s−1) used within the present work.
Figure 6. Comparison of np4 and the WT strain 4A+ regarding their H2 production capacity and quantum yield kinetics after sulphur depletion.
A: Representative H2 measurement with np4 and 4A+. Changes in the photosynthetic quantum yield (ΦPSII) were recorded and plotted on the left y-axis. The total amount of H2 produced (total H2 [ml/L culture]) during the experiment is indicated on the right y-axis and the duration of the measurement (time after S starvation) is given on the x-axis. Black lines represent the mutant np4 and grey lines the WT 4A+ (continuous line H2 yield; dotted line ΦPSII). Standard deviations derived from three technical replicates are indicated as error bars. B: Total H2 yields from np4 (black bar) and 4A+ (grey bar) are given as mean values calculated from three independent biological replicates using three technical replicates per measurement. Error bars indicate standard deviations.
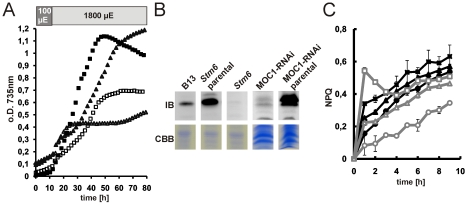
However, LHCSR3 has been previously shown to accumulate particularly in high light [25]–[27] and a loss of this protein causes a reduced fitness if cells are challenged by varying light intensities [26]. We consequently investigated the relationship between the genotype of stm6 (loss of the nucleus-encoded mitochondrial protein MOC1 [21]), which is the parental strain of stm6glc4 and the expression of LHCSR3 under high light conditions. LHCSR3 expression data obtained with the MOC1 deletion mutant stm6glc4 and a WT cell line after sulfur starvation (Fig. 5B) demonstrated that a loss of MOC1 protein has a profound effect on the extent of LHCSR3 accumulation, nicely emphasizing the relevance of functional inter-organelle signaling between mitochondria and chloroplasts under certain stress conditions [21]. MOC1 has been suggested to play an important role in the regulation of oxidative phosphorylation in the light and in preparing the mitochondria as a redox valve for the chloroplast thereby reducing the risk of ROS damage in particular under increasing high light conditions [21]. The knock-out of MOC1 leads to a reduced growth rate of stm6 in high light and minimal media compared to the complemented strain B13 (Fig. 7A). A similar growth defect could be observed for a MOC1-RNAi strain under identical conditions (Fig. 7A). Under these conditions the levels of LHCSR3 protein were almost undetectable in stm6 and the MOC1-RNAi strain, whereas in the parental strains (cc1618 for stm6 and cc124 for MOC1-RNAi) and in the MOC1-complemented strain B13 LHCSR3 accumulated under highlight (Fig. 7B). As was shown in a recent study, active photosynthetic electron transfer is required for proper LHCSR3 accumulation [27], so that the low amount of LHCSR3 in high-light grown stm6 cells could indicate an impairment of photosynthetic electron transfer. In line with the finding that stm6 fails to accumulate LHCSR3 under high light the MOC1-free mutant displayed a low NPQ capacity, if compared to B13 and its parental strain (Fig. 7C).
Figure 7. LHCSR3 expression and non-photochemical quenching capacity in stm6, B13, MOC1-RNAi and parental WT strains.
A: Phototrophic high light growth (1800 µE) of Stm6 (open triangles), B13 (filled triangles), MOC1-RNAi (open squares) and its parental strain cc124 (filled squares) in minimal media (HSM). The light regime (15 h 100 µE; 65 h 1800 µE) is indicated with light or dark grey shaded boxes at the top of the panel. Shown is one representative growth out of three independent growth experiments. B: Immunoblot analysis (αLHCSR3) of LHCSR3 protein expression in phototrophically grown stm6, B13, stm6 parental (cc1618), MOC1-RNAi and MOC1-RNAi parental (cc124) cells. Cells were first cultivated in 100 µE for 15 h before the light intensity was increased to 1800 µE. Samples for protein extraction were taken after eight hours of high-light treatment. Protein loading is shown with a Coomassie stain (CBB). C: Determination of the non-photochemical quenching (NPQ) capacity in stm6 (circles), B13 (triangles) and stm6 parental (squares). Cultures were either incubated in low light (filled symbols; black lines) or high light (open symbols; grey lines) prior to the measurement of NPQ over 10 min.
These results clearly demonstrate that impaired mitochondrial function affects non-photochemical quenching in the plastid and is therefore another example for the intense inter-organelle crosstalk between chloroplasts and mitochondria [39]. A correlation between impaired non-photochemical quenching and perturbed mitochondrial function has recently been reported for the CMSII mutant from tobacco, which lacks functional complex I [40]. Apart from the accumulation of key quenching proteins such as LHCSR3 cyclic electron flow (CEF) around PSI was shown to be required for normal NPQ activity [41] and interestingly one of the phenotypical characteristics of stm6 and its derivative strains is a reduced cyclic electron flow (CEF) around PSI [33].
MOC1 is required for functional LHCSR3 expression and MOC1 levels are inversely correlated with H2 production capacity
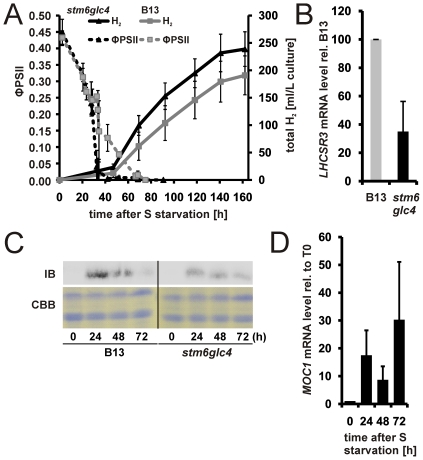
To correlate differences between H2 production in WT and stm6glc4 with the presence or absence of functional MOC1 we also investigated the effect of stm6 complementation on H2 production. The rescued cell line B13, which expresses MOC1 at WT-levels (not shown), was compared to stm6glc4 concerning its H2 production capacity (Fig. 8A) and the photosynthetic quantum yield (ΦPSII) was traced during the H2 production experiment (Fig. 8A). Re-introduction of a functional MOC1 gene had an adverse effect on the H2 production rate (Fig. 8A) clearly showing that MOC1 indeed has a strong impact on –S-induced H2 production. The –S- induced drop in PSII activity in B13 was almost as fast as in stm6glc4 and both ΦPSII curves had an overall similar shape (Fig. 8A). Although the kinetics of induced PSII damage was similar between the two strains significant differences were seen in the expression levels of LHCSR3. Real-time data (Fig. 8 B) and immunoblot studies (Fig. 8C) showed that re-introduction of functional MOC1 into stm6 led to a recovery of the stress-induced expression of LHCRS3. However, recovery of LHCSR3 expression was not accompanied by an increased protection of PSII against photodamage under H2 production conditions as seen by the still rapid drop of ΦPSII in B13 and an early onset of H2 production comparable to that observed for stm6glc4. This shows again that the level of LHCSR3 accumulation has little effect on the resistance of PSII against photodamage during –S induced H2 production at least in moderate light. In good agreement with the hypothesis that MOC1 is deeply implicated in the acclimation processes triggered by sulfur depletion and consecutive anaerobiosis, MOC1 mRNA levels display a steady and strong increase during H2 production (Fig. 8 D). In line with the finding that B13 shows a ΦPSII decline kinetics comparable to that of stm6glc4 (Fig. 8 A), the highest mRNA level is only observed long after production of H2 has been initiated and not during transition to anaerobiosis.
Figure 8. Complementation of stm6glc4 with a functional MOC1 gene reduces hydrogen production and restores LHCSR3 accumulation.
A: Representative H2 measurement with B13 and stm6glc4. The change in the photosynthetic quantum yield (ΦPSII; left y-axis; dotted lines) was recorded along with the total amount of H2 produced by both cultures (total H2; right y-axis; continuous lines). On the x-axis the time after sulfur depletion is indicated. B13 (squares) is shown in grey and stm6glc4 (triangles) in black. B: Abundance of LHCSR3 mRNA after 40 hours of sulphur starvation in B13 (grey bar) and stm6glc4 (black bar) determined by RT-Q-PCR. The mRNA level of B13 was set to 100% and standard deviations indicated by error bars represent three different experiments. C: Immunodetection of LHCSR3 in samples of B13 and stm6glc4 after sulphur depletion with indicated duration times. The upper panel shows an anti-LHCSR3 immunoblot (IB) and the lower one a Coomassie-stained gel serving as a loading control (CBB). D: RT-Q-PCR analysis of the MOC1 mRNA expression in WT samples taken at different time points after (24, 48 72 h) and before (0 h) sulphur depletion. Expression is given relative to T0, which was set to 1.
As a conclusion our data demonstrate that the absence of MOC1 perturbs regulative mechanisms underlying the strong induction of LHCSR3 expression in response to sulfur depletion found in wild type cells. Given that the recovery of LHCSR3 expression in B13 has little effect on the protection of PSII against –S induced photo-damage we deduce that there is no clear correlation between the available amount of LHCSR3 and the susceptibility to PSII damage under the H2 production conditions used in the present study.
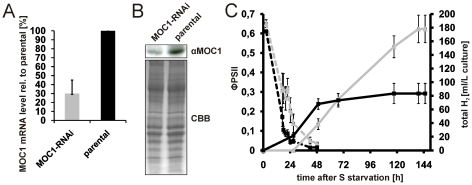
As suggested by differences seen regarding the H2 production capacity of B13 and mutant stm6glc4 and underlined by the considerable increase of MOC1 mRNA content during H2 production it can be concluded that the available amount of MOC1 has a strong impact on H2 production. We therefore decided to prove this hypothesis by application of amiRNA-mediated knock-down of MOC1. The pChlamyRNA3int construct [42] was used to knock-down the expression of MOC1 in the WT strain cc124. The transformant showing the largest reduction of MOC1 expression on the mRNA (≈70±15%; Fig. 9A) and protein level (≈52±13% (n = 3); Fig. 9B) was chosen for further analysis. H2 production in the RNAi strain was significantly increased by ≈125% (Fig. 9C; total H2; continuous grey line) compared to the parental strain (continuous black line) without displaying any differences regarding the rate of PSII activity decline in response to sulfur depletion (Fig. 9C, ΦPSII MOC1-RNAi (dotted grey line) vs. parental (dotted black line)). We therefore conclude that a reduced amount of MOC1 improves H2 production by exerting its effects at later stages of H2 production.
Figure 9. Effects on H2 production caused by amiRNA-mediated knock-down of MOC1.
A: MOC1 mRNA expression in the MOC1 knock-down strain (MOC1-RNAi; grey bar) and the WT parental strain (parental; black bar) as determined by RT-Q-PCR. MOC1 expression in the knock-down strain is given relative to the expression in the parental strain (set to 100%). Error bars indicate the standard deviation from three biological experiments. B: Representative immunoblot experiment (αMOC1) using an antiserum raised against MOC1 and the MOC1 knock-down strain (MOC1-RNAi) as well as its parental strain (parental). A Coomassie brilliant blue stain serves a loading control (CBB). C: Representative H2 measurement with the MOC1 knock-down (grey curves) and parental strain (black curves). Total H2 production (right y-axis; total H2; continuous lines) and the change of the photosynthetic quantum yield (left y-axis; ΦPSII; dotted lines) were recorded. The duration of the sulphur deplete condition is given on the x-axis. Indicated standard deviations (error bars) are derived from three technical replicates per strain. D: Result from four independent H2 experiments with the MOC1 knock-down strain (MOC1-RNAi; grey bar) and parental strain (parental; black bar). The H2 capacity (total volume of H2 produced) is calculated relative to the parental strain which was set to 100%. Error bars represent the standard deviation from four biological experiments with three technical replicates per experiment.
Summary
In the present study, the transcriptome of a mutant displaying a high H2 production capacity was compared to a WT strain in order to determine which differences in expression patterns may contribute to an earlier onset, and higher rate of H2 production in the mutant stm6glc4 (Figure 10). Informative differences in the expression of certain genes between mutant and WT could already be detected at T0 before the cells were resuspended in sulfur deplete media. The expression of ICL1, required for efficient acetate catabolism was strongly increased in the mutant providing a potential explanation for the higher acetate consumption of the mutant while anaerobiosis is being established [11]. An increased acetate consumption could be accompanied by higher rates of respiration caused by a better provision of reducing equivalents, and indeed the dissolved oxygen concentration in mixotrophic stm6 cultures is lower than in wildtype strains [33], which can be explained by a combination of higher cyanide-insensitive respiration [21] and a lower number of active PSII complexes [33] in the mutant. Higher rates of respiration caused shorten the aerobic phase [12], [14] and indeed stm6glc4 starts producing H2 much earlier than the WT strain.
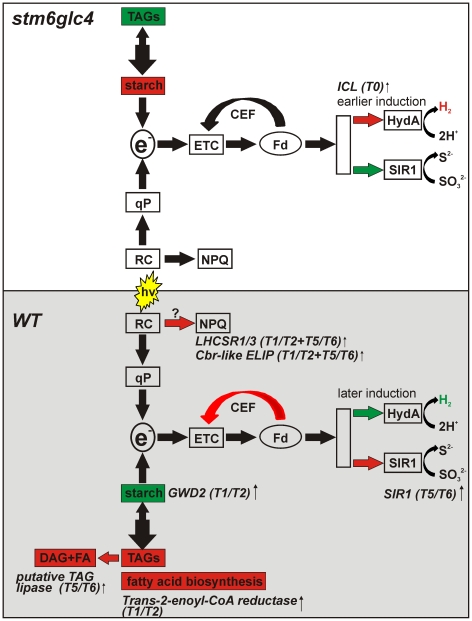
Figure 10. Summary of the transcriptomic differences between WT (grey shaded box) and mutant stm6glc4 (white box) which contribute to an increased H2 production capacity of the mutant.
A relative increase in the size of metabolite pools is indicated by a red and a decrease by a green colour. The metabolomic data used in the model were taken from Doebbe et al. [11]. The upregulation of physiological processes or pathways is also indicated by red and a downregulation by a green colour. The names of differentially expressed genes (see Table 1 and 2) are given along with the time points for which a WT- or mutant-specific regulation was identified. Abbreviations/symbols: TAGs (triacylglycerides); e− (electrons fed into the H2ase pathway); ETC (electron transport chain); CEF (cyclic electron flow around PSI); Fd (ferredoxin); HydA (H2ase isoform A); SIR1 (ferredoxin-dependent sulfite reductase); qP (photochemical quenching); RC (reaction center of PSII); NPQ (non-photochemical quenching); DAG (diacylglycerol); FA (fatty acid).
Chlamydomonas cells accumulate energy storage compounds during the aerobic –S phase preceding the onset of H2 production. The most prominent compounds produced are starch and triacylglycerides (TAGs). TAGs and starch are synthesized by competing biosynthetic pathways [43] and their relevance for photobiological H2 production differs greatly. In contrast to starch, TAGs are not likely to be a substrate for H2 production, due to the impaired β-oxidation of fatty acids in an anaerobic environment, such as the conditions during H2 production. Starch degradation could be used to generate reducing equivalents for the non-photochemical reduction of the plastoquinon pool, via the plastidic NAD(P)H–PQ oxidoreductase Nda-2 [44], although the actual significance of this PSII-independent pathway for H2 production is still a matter of debate [13].
WT and mutant strains showed strong differences in their preference for one of the major energy storage compounds. The WT accumulated more TAGs than the mutant, whereas the mutant synthesized more starch and less TAGs [11]. In line with the increased TAG content in the WT the amount fatty acids produced during the aerobic phase was much higher in the WT than in the mutant [11]. An increased content of C18 fatty acids in the WT [11] during the aerobic and anaerobic phase is in good agreement with the up-regulation of a trans-2-enoyl-CoA-reductase in T1/T2 (Figure S1, T2ECR, preH2) and T5/T6, since this enzyme is needed for the elongation of fatty acids [45]. During the H2 production phase the WT uses more TAGs than the mutant and a putative TAG lipase showing stronger expression in the WT might be involved in this process. The lower accumulation of starch in WT cells compared to those of the mutant could be caused by a higher expression of a glucan water-dikinase (GWD2) an enzyme implicated in the breakdown of starch [31]–[32].
The stress-related LHC protein LHCSR3 was much stronger induced in the WT than in stm6glc4. This protein has already been shown to be required for energy-dependent quenching so that protection of PSII under high-light conditions represents a potential function of this protein. However, the increased susceptibility of PSII to –S induced photodamage, and the resulting shortened aerobic phase in the mutant, which accumulates a significantly lower amount of LHCSR3, cannot be explained by the inability to accumulate LHCSR3 amounts found in the WT. Under the moderate light conditions used in this study a complete loss of the protein, which is the case for mutant strain npq4, does not result in a more rapid transition from aerobic to anaerobic conditions. Furthermore no increase in the rate of H2 production in the anaerobic phase could be observed for the knock out mutant indicating either that energy dissipation by non-photochemical quenching (qE) does not reduce the electron flow through the H2ase pathway, or that qE is not triggered under the experimental conditions used in this study. High energy state quenching (qE) requires a low luminal pH [46] and biochemical analysis of LHCSR3 indicated that protonation of several residues in LHCSR3 is a precondition for active quenching [25]. The moderate light intensity used in our H2 experiments could have been insufficient to form a ΔpH enabling high energy state quenching. Apart from LHCSR3 the major light-harvesting protein LHCBM1 was demonstrated to represent an essential component of the non-photochemical quenching mechanism in C. reinhartii [47]. The expression of LHCBM1 is constantly and strongly down-regulated during the entire course of the –S induced H2 production process [6], so that a critical factor is almost absent although LHCSR3 is expressed at high amounts. Nevertheless it cannot be ruled out that the level of LHCSR3 present in the cell has an impact on H2 production at higher light intensities. Under phototrophic high light conditions in sulfur replete media a clear correlation between the presence of MOC1 and the ability to induce LHCSR3 expression could be noted (Fig. 9B). Strains expressing a low amount or no MOC1 at all showed impaired growth under high light (Fig. 9A), which was accompanied by a lack of LHCSR3 induction (Fig. 9B) and a reduced NPQ capacity (Fig. 9C).
Several reductive reactions in the plastid rely on the use of ferredoxin thus representing pathways potentially competing with proton reduction catalyzed by H2ase. A stronger mRNA expression of the enzyme SIR1 could provide part of the explanation for a lower rate of H2 production in the WT. In a previous study it was reported that stm6 the parental strain of stm6glc4 used in the present work is blocked in state 1 and not capable of performing cyclic electron transport around PSI [33] causing an increased supply of substrate to the H2ase.
Conclusion
Our study reveals distinctive expression patterns in WT and stm6glc4 strains during sulfur deprivation, strongly confirming previous results showing major cellular reorganizations. We also show that beside the mostly similar responses, stm6glc4 revealed distinctive expression patterns. A preference of starch over lipids as an energy storage compound and higher rates of acetate assimilation in the aerobic phase can be deduced from our metabolomics data [11]. Our microarray data indicated a higher expression of the gene GWD2 exclusively found in the WT. The encoded enzyme might be implicated in starch breakdown, thus explaining why stm6glc4 accumulates more starch in the aerobic phase. In regard to acetate assimilation and respiration during the aerobic phase preceding H2 we identified a higher mRNA amount for the gene ICL1 in the mutant at T0, just before cultures have been depleted of sulfur. Reduced cyclic electron flow [33] and less competition by ferredoxin-dependent sulfite reduction as implied by the lower expression of SIR1 contribute to the increased H2 production capacity of mutant stm6glc4. Our study revealed potential targets for the genetic engineering of Chlamydomonas strains aiming at higher H2 production capacities. Among these targets are enzymes potentially involved in starch breakdown within the aerobic phase (GWD2; glucan water-dikinase), competing ferredoxin-dependent enzymes (SIR1) and enzymes needed for efficient acetate assimilation (ICL1).
Materials and Methods
Strains, growth and H2 production conditions
The following Chlamydomonas reinhardtii strains were used: WT (CC-406 cw15 mt−), stm6 and stm6glc4. Stm6 was created from CC-1618 (arg7 cw15 mt−) by transformation with pARG7.8 [21]. B13 is the MOC1 complemented strain of stm6 and was created by transforming stm6 with a MOC1 carrying cosmid [21]. stm6glc4 is a derivative strain of mutant stm6 and was generated by transformation with a vector encoding a hexose uptake symporter HUP1 gene from Chlorella kessleri [22].
For knockdown of MOC1 and generation of the MOC1-RNAi strain artificial microRNA sequences were designed using Web Micro RNA designer 3 (http://wmd3.weigelworld.org):
forward:5′-ctagtCCGCTCGACATTCACACAATAtctcgctgatcggcaccatgggggtggtggtgatcagcgctaTATTCTGTGAATGTCGAGCGGg-3′
reverse:5′ctagcCCGCTCGACATTCACAGAATAtagcgctgatcaccaccacccccatggtgccgatcagcgagaTATTGTGTGAATGTCGAGCGGa-3′.
Oligonucleotide sequences were cloned into vector pChlamiRNAi3int according to Molnar et al. [42] to generate vector pRNAi6.18A. 2 µg of plasmid pRNAi6.18A was absorbed onto 550 nm gold particles according to the manufacturer's instructions (Seashell technologies) and a gene gun (BIO-RAD-Model PDS-1000/He Biolistic® Particle Delivery System; Bio-Rad Laboratories) was used to transform C. reinhardtii strain CC-124 (mt− 137c) by biolistic bombardment. Transformants were selected on TAP paromomycin 10 µg ml−1 plates and screened for stable knock-down of MOC1 by immunblot analyses and candidates were further examined by RT-Q-PCR using MOC1-specific primers. Non-photochemical quenching mutant npq4 was created by transformation of CC-425 (arg7-8 cw15 mt+ sr-u-2-60) with pJD67 [38]. Mutant npq4 has been determined as having a knockout in LHCSR3 [26] while 4A+ is a WT strain [48] used as a control in previous studies on npq4 [26].
Culturing and H2 production were carried out as described previously [11]. Strains lacking a hexose uptake transporter were cultured in TAP [49] and stm6glc4 in TAP +1 mM glucose for optimal H2 production [22]. For the high-light growth experiment in HSM media [49] and 2% CO2 a photobioreactor FMT150 from PSI (Brno, Czech Republic) was used.
Sample collection
For strain stm6glc4, samples from four time points were collected at 16 h, 28 h, 52 h and 76 h after sulfur starvation (T1, T2, T3 and T4 respectively). T1 represents the oxygen consuming phase while T2 marks the beginning of the anaerobic phase. At T3 and T4, the production of H2 was observed. For strain WT, samples were collected at slightly different time points due to slower net oxygen consumption. The first four time points T1, T2, T3 and T4 were collected at 16 h, 28 h, 52 h and 68 h, respectively. T4 marks the start of anaerobic phase as indicated by the drop of quantum yield below 0.1. In addition, two more samples were collected at 92 h (T5) and 116 h (T6) to cover the H2 production phase which started later in WT (see Figure 1). The samples were compared with the T0 reference samples harvested from late stationary phase cultures of the corresponding strains before S-starvation.
RNA preparation
Samples taken from the bioreactors were immediately centrifuged (3000× g, 2 minutes at room temperature). Fresh cell pellets were lysed immediately with RNA Lysis Buffer (SV Total RNA Isolation System, Promega) and RNA was isolated according to the supplied manual.
Microarray preparation and obtaining of data
Chlamydomonas microarray slides version 2 [50] were obtained from Dr. Author Grossman (Stanford University, USA). A recent study confirmed the specificity of 8760 features (out of 10000) according to the recent annotation [51]. Our analysis of the feature specificity according to the new annotation (Augustus 10.2) confirmed the usability of 7120 features, and just these features were included in the analysis. Microarray analysis was carried out essentially as described previously [6]. Three biological replicates for each time point were analyzed and global lowess normalization of the raw microarray data was carried out, by using the median spot intensities [6]. For each time point, a paired two-way t-test was conducted to determine significant changes in gene expression (threshold: p>0.05, within at least 4 out of 6 replica). Raw and normalized data were deposited in the GEO database (GSE30252) and followed MIAME requirements. Hierarchical clustering for both strains was performed by centroid linkage using the log2 ratios of the differentially expressed genes. Visualization was performed on Java TreeView [52].
Quantitative real-time RT-PCR
Quantitative real time RT-PCR analysis for all genes presented in Figure 10 was carried out as previously described [53]. Briefly, Real-time RT-Q-PCR was carried out using the SensiMix One-Step kit (Quantace) in conjunction with the DNA Engine Opticon system (Bio-Rad). For each sample the cycle threshold (Ct) values for the reference/housekeeping gene (18S rRNA) and target genes were used to calculate the relative amount of target mRNA according to the equation rA = E −[Ct(target gene)−Ct(reference gene)] with E representing the PCR efficiency according to Rasmussen [54]. Real-time data represent the median values and standard deviations of three technical replicates for each gene and time point. The primer sequences in Table 3 were used.
Table 3. Sequences of the primers used for RT-Q-PCR analyses of selected genes.
| Gene/Locus (Accession No)/Description | Sequence 5′→3′ |
| ICL1/Cre06.g282800/Isocitrate lyase | ICL1 for: tacaactgctcgccctcttt/rev: tgaacatgccgtagttgagc |
| T2ECR/Cre01.g035350/Trans-2-enoyl-CoA-reductase | T2ECR for: agaggcagtcatccagatcg/rev: gtcctccttgagcttgtgct |
| GWD2/Cre07.g332300/R1 protein, α-glucan water dikinase | GWD2 for: atcgagcccttcaagcacta/rev: aggatgtcgtacatggtccag |
| LHCSR1/Cre08.g365900/Stress-related LHC protein | LHCSR1 for: gccatctaccacttccagca/rev: ggctcgtagtcgtccttcag |
| Cre07.g320450/Cre07.g320400/Cbr-like ELIP | CBR for: aagtacgttgacggcgaaat/rev: gggagtccacgttcagagag |
| Cre03.g155250/similar to CGLD15 (TAG lipase-like) | TAGlip for: aacaagcggctgtatgctg/rev: cacatgagctgcagaagca |
| SIR1/Cre08.g365700/ferredoxin-sulfite reductase | SIR1 for: tgcagctcatgaagttccac/rev. aggtcgtccatcaccaggta |
| 18S rRNA/AY665726, 18S ribosomal RNA | 18S rRNA for: cctgcggcttaatttgactc/rev: accggaatcaacctgacaag |
Pigment measurement
Pigments were extracted from Chlamydomonas reinhardtii cells in 80% (v/v) acetone. The insoluble fraction was precipitated by centrifugation (2 min, 20,000× g) before measuring chlorophyll concentration and chl a/b ratio according to Arnon [55].
Immunoblot analysis of LHCSR3 and LHCBM4/6 accumulation
For LHCSR3 immunodetection experiments samples were either taken from H2 producing stm6glc4 or WT cultures at the indicated time points after S depletion (Fig. 5) or from phototrophic growth experiments conducted in PSI bioreactors using HSM minimal media (Fig. 7). For the high-light experiment in HSM cells were first cultivated in 100 µmol photons•m−2•s−1 for 15 h before the light intensity was increased to 1800 µmol photons•m−2•s−1. Samples for protein extraction were taken after eight hours of high-light treatment. LHCBM4/6 protein amounts were determined in samples taken from H2 producing cultures. The antisera raised against LHCSR3 and LHCBM4/6 were a kind gift from M. Hippler (IBBP, Münster University).
Chlorophyll fluorescence measurements
Photosynthetic quantum yield ΦPSII was measured directly on bioreactors' surface with a MINI-PAM (Waltz, Germany) saturating pulse of 3,000 µmol photons•m−2•s−1.
Measurement of non-photochemical quenching
Non photochemical quenching was measured as described in [46].
SDS PAGE
Protein analyses were carried out using standard procedures as described in [56].
Supporting Information
Real-time RT-Q-PCR analysis of the mRNA levels of selected genes in stm6glc4 and WT. RNA samples of the second microarray experiment were analyzed by RT-Q-PCR in order to determine the relative amount of selected transcripts in stm6glc4 compared to WT (set to 100%). Three different time points according to Figure 1 were analyzed: T0 (before sulfur deprivation), preH2 (after sulfur depletion and before H2 production; T2 WT; T1 stm6glc4) and H2 (H2 production phase; T5 WT; T3 stm6glc4). Standard deviations are given as error bars and were calculated from three technical replicates. Genes: ICL1 (isocitrate lyase; Cre06.g282800; Table S2), T2ECR (Trans-2-enoyl-CoA-reductase; Cre01.g035350; Table 1 and 2), GWD2 (R1 protein, α-glucan water dikinase; Cre07.g332300; Table 1), LHCSR1 (stress-related LHC protein, Cre08.g365900, Table 1 and 2), Cbr-like ELIP (Cre07.g320450/Cre07.g320400; Table 1 and 2), TAG-lipase (similar to CGLD15/putative TAG lipase; Table 2), SIR1 (ferredoxin-sulfite reductase; Cre08.g365700; Table 2).
(TIF)
A complete list of all microarray data. Expression relative to T0 is given for both strains and all examined time points. GeneID numbers correspond to those contained in the gal file (http://www.chlamy.org/galfile.xls, [50]) and heat map group assignments (Figure 4) for each gene are given as well. An increase in the mRNA abundance (Tn/T0>1) is indicated by red highlighting and a decrease (Tn/T0<1) by green highlighting. Available functional assignments (column “Annotation”) are given and were used for sorting (column “Group”).
(XLS)
Genes showing either a specific down- or upregulation in one of both examined strains at T0. Differentially expressed genes are sorted according to the cellular processes involved as deduced from their functional annotation. Gene names are given along with the corresponding locus names (Phytozome 7.0; http://www.phytozome.net/) and a description of their function.
(DOCX)
Acknowledgments
We are grateful to M. Hippler for the provision of antisera and K. Niyogi for providing us with strains npq4 and 4A+.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The authors would like to thank the EU/Energy FP7 project SOLAR-H2 (contract 212508, OK), the German Federal Ministry of Science BMBF-ForSys-Partner project (contract 0315265A, OK), the Australian Research Council (DP0877147, BH) as well as the Engineering and Physical Sciences Research Council (EP/F00270X/1, PN. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Melis A, Zhang L, Forestier M, Ghirardi ML, Seibert M. Sustained photobiological hydrogen gas production upon reversible inactivation of oxygen evolution in the green alga Chlamydomonas reinhardtii. Plant Physiol. 2000;122:127–136. doi: 10.1104/pp.122.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kruse O, Hankamer B. Microalgal hydrogen production. Curr Opin Biotechnol. 2010;21:238–243. doi: 10.1016/j.copbio.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Ghirardi ML, Posewitz MC, Maness P-C, Dubini A, Yu J, et al. Hydrogenases and hydrogen photoproduction in oxygenic photosynthetic organisms. Annu Rev Plant Biol. 2007;58:71–91. doi: 10.1146/annurev.arplant.58.032806.103848. [DOI] [PubMed] [Google Scholar]
- 4.Rupprecht J, Hankamer B, Mussgnug JH, Ananyev G, Dismukes C, et al. Perspectives and advances of biological H2 production in microorganisms. Appl Microbiol Biotechnol. 2006;72:442–449. doi: 10.1007/s00253-006-0528-x. [DOI] [PubMed] [Google Scholar]
- 5.Happe T, Kaminski A. Differential regulation of the Fe-hydrogenase during anaerobic adaptation in the green alga Chlamydomonas reinhardtii. Eur J Biochem. 2002;269:1022–1032. doi: 10.1046/j.0014-2956.2001.02743.x. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen AV, Thomas-Hall SR, Malnoe A, Timmins M, Mussgnug JH, et al. The transcriptome of photo-biological hydrogen production induced by sulphur deprivation in the green alga Chlamydomonas reinhardtii. Eukaryot Cell. 2008;7:1965–1979. doi: 10.1128/EC.00418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen M, Zhao L, Sun Y-L, Cui S-X, Zhang L-F, et al. Proteomic analysis of hydrogen photoproduction in sulfur-deprived Chlamydomonas cells. J Proteome Res. 2010;9:3854–3866. doi: 10.1021/pr100076c. [DOI] [PubMed] [Google Scholar]
- 8.Zhang L, Happe T, Melis A. Biochemical and morphological characterization of sulfur-deprived and H2 producing Chlamydomonas reinhardtii (green alga). Planta. 2002;214:552–561. doi: 10.1007/s004250100660. [DOI] [PubMed] [Google Scholar]
- 9.González-Ballester D, Casero D, Cokus S, Pellegrini M, Merchant SS, et al. RNA-Seq analysis of sulfur-deprived Chlamydomonas cells reveals aspects of acclimation critical for cell survival. Plant Cell. 2010;22:2058–2084. doi: 10.1105/tpc.109.071167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Timmins M, Zhou W, Rupprecht J, Lim L, Thomas-Hall SR, et al. The metabolome of Chlamydomonas reinhardtii following induction of anaerobic H2 production by sulfur depletion. J Biol Chem. 2009;284:23415–23425. doi: 10.1074/jbc.M109.003541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doebbe A, Keck M, La Russa M, Mussgnug JH, Hankamer B, et al. The interplay of proton, electron and metabolite supply for photosynthetic H2 production in C. reinhardtii. J Biol Chem. 2010;285:30247–30260. doi: 10.1074/jbc.M110.122812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kosourov S, Patrusheva E, Ghirardi ML, Seibert M, Tsygankov A. A comparison of hydrogen photoproduction by sulfur-deprived Chlamydomonas reinhardtii under different growth conditions. J Biotechnol. 2007;128:776–787. doi: 10.1016/j.jbiotec.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 13.Chochois V, Dauvillee D, Beyly A, Tolleter D, Cuine S, et al. Hydrogen production in Chlamydomonas: Photosystem II-dependent and -independent pathways differ in their requirement for starch metabolism. Plant Physiol. 2009;151:631–640. doi: 10.1104/pp.109.144576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fouchard S, Hemschemeier A, Caruana A, Pruvost K, Legrand J, et al. Autotrophic and mixotrophic hydrogen photoproduction in sulfur-deprived Chlamydomonas cells. Appl Environ Microbiol. 2005;71:6199–6205. doi: 10.1128/AEM.71.10.6199-6205.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Posewitz MC, Smolinski SL, Kanakagiri S, Melis A, Seibert M, et al. Hydrogen photoproduction is attenuated by disruption of an isoamylase gene in Chlamydomonas reinhardtii. Plant Cell. 2004;16:2151–2163. doi: 10.1105/tpc.104.021972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kosourov S, Seibert M, Ghirardi ML. Effects of extracellular pH on the metabolic pathways in sulfur-deprived, H2-producing Chlamydomonas reinhardtii cultures. Plant Cell Physiol. 2003;44:146–155. doi: 10.1093/pcp/pcg020. [DOI] [PubMed] [Google Scholar]
- 17.Hemschemeier A, Fouchard S, Cournac L, Peltier G, Happe T. Hydrogen production by Chlamydomonas reinhardtii: an elaborate interplay of electron sources and sinks. Planta. 2008;227:397–407. doi: 10.1007/s00425-007-0626-8. [DOI] [PubMed] [Google Scholar]
- 18.Hemschemeier A, Happe T. Alternative photosynthetic electron transport pathways during anaerobiosis in the green alga Chlamydomonas reinhardtii. Biochim Biophys Acta. 2011;1807:919–926. doi: 10.1016/j.bbabio.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 19.White AL, Melis A. Biochemistry of hydrogen metabolism in Chlamydomonas reinhardtii wild type and a Rubisco-less mutant. Int J Hydrogen Energy. 2006;31:455–464. [Google Scholar]
- 20.Wu S, Xu L, Huang R, Wang Q. Improved biohydrogen production with an expression of codon-optimized hemH and lba genes in the chloroplast of Chlamydomonas reinhardtii. Bioresour Technol. 2011;102:2610–2616. doi: 10.1016/j.biortech.2010.09.123. [DOI] [PubMed] [Google Scholar]
- 21.Schoenfeld C, Wobbe L, Borgstaedt R, Kienast A, Nixon PJ, et al. The Nucleus-encoded protein MOC1 is essential for mitochondrial light acclimation in Chlamydomonas reinhardtii. J Biol Chem. 2004;279:50366–50374. doi: 10.1074/jbc.M408477200. [DOI] [PubMed] [Google Scholar]
- 22.Doebbe A, Rupprecht J, Beckmann J, Mussgnug JH, Hallmann A, et al. Functional integration of the HUP1 hexose symporter gene into the genome of C. reinhardtii: Impacts on biological H2 production. J Biotechnol. 2007;131:27–33. doi: 10.1016/j.jbiotec.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 23.Ben-Amotz A, Avron M. On the factors which determine massive β-carotene accumulation in the halotolerant alga Dunaliella bardawil. Plant Physiol. 1983;72:593–597. doi: 10.1104/pp.72.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z, Shrager J, Jain M, Chang C-W, Vallon O, et al. Insights into the survival of Chlamydomonas reinhardtii during sulfur starvation based on microarray analysis of gene expression. Eukaryot Cell. 2004;3:1331–1348. doi: 10.1128/EC.3.5.1331-1348.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonente G, Ballottari M, Truong TB, Morosinotto T, Ahn TK, et al. Analysis of LhcSR3, a protein essential for feedback de-excitation in the green alga Chlamydomonas reinhardtii. PLoS Biol. 2011;18:e1000577. doi: 10.1371/journal.pbio.1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peers G, Truong TB, Ostendorf E, Busch A, Elrad D, et al. An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature. 2009;462:518–521. doi: 10.1038/nature08587. [DOI] [PubMed] [Google Scholar]
- 27.Petroutsos D, Busch A, Janßen I, Trompelt K, Bergner SV, et al. The chloroplast calcium sensor CAS is required for photoacclimation in Chlamydomonas reinhardtii. Plant Cell. 2011;23:2950–2963. doi: 10.1105/tpc.111.087973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kudla J, Batistič O, Hashimoto K. Calcium signals: The lead currency of plant information processing. Plant Cell. 2010;22:541–563. doi: 10.1105/tpc.109.072686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lers A, Levy H, Zamir A. Co-regulation of a gene homologous to early light-induced genes in higher plants and beta-carotene biosynthesis in the alga Dunaliella bardawil. J Biol Chem. 1991;266:13698–13705. [PubMed] [Google Scholar]
- 30.Banet G, Pick U, Zamir A. Light-harvesting complex II pigments and proteins in association with Cbr, a homolog of higher-plant early light-inducible proteins in the unicellular green alga Dunaliella. Planta. 2000;210:947–955. doi: 10.1007/s004250050702. [DOI] [PubMed] [Google Scholar]
- 31.Ritte G, Heydenreich M, Mahlow S, Haebel S, Kotting O, et al. Phosphorylation of C6- and C3-positions of glucosyl residues in starch is catalysed by distinct dikinases. Febs Letters. 2006;580:4872–4876. doi: 10.1016/j.febslet.2006.07.085. [DOI] [PubMed] [Google Scholar]
- 32.Ritte G, Scharf A, Eckermann N, Haebel S, Steup M. Phosphorylation of transitory starch is increased during degradation. Plant Physiol. 2004;135:2068–2077. doi: 10.1104/pp.104.041301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kruse O, Rupprecht J, Bader K-P, Thomas-Hall S, Schenk PM, et al. Improved photobiological H2 production in engineered green algal cells. J Biol Chem. 2005;280:34170–34177. doi: 10.1074/jbc.M503840200. [DOI] [PubMed] [Google Scholar]
- 34.Knaff DB, Hirasawa M. Ferredoxin-dependent chloroplast enzymes. Biochim Biophys Acta. 1991;1056:93–125. doi: 10.1016/s0005-2728(05)80277-4. [DOI] [PubMed] [Google Scholar]
- 35.Nakayama M, Akashi T, Hase T. Plant sulfite reductase: molecular structure, catalytic function and interaction with ferredoxin. J Inorg Biochem. 2000;82:27–32. doi: 10.1016/s0162-0134(00)00138-0. [DOI] [PubMed] [Google Scholar]
- 36.Kornberg HL, Krebs HA. Synthesis of cell constituents from C2-Units by a modified tricarboxylic acid cycle. Nature. 1957;179:988–991. doi: 10.1038/179988a0. [DOI] [PubMed] [Google Scholar]
- 37.Wykoff DD, Davies JP, Melis A, Grossman AR. The regulation of photosynthetic electron transport during nutrient deprivation in Chlamydomonas reinhardtii. Plant Physiol. 1998;117:129–139. doi: 10.1104/pp.117.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niyogi KK, Bjorkman O, Grossman AR. Chlamydomonas xanthophyll cycle mutants identified by video imaging of chlorophyll fluorescence quenching. Plant Cell. 1997;9:1369–1380. doi: 10.1105/tpc.9.8.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raghavendra AS, Padmasree K. Beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Trends Plant Sci. 2003;8:546–553. doi: 10.1016/j.tplants.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 40.Cardol P, De Paepe R, Franck F, Forti G, Finazzi G. The onset of NPQ and ΔμH+ upon illumination of tobacco plants studied through the influence of mitochondrial electron transport. Biochim Biophys Acta. 2010;1797:177–188. doi: 10.1016/j.bbabio.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 41.Tolleter D, Ghysels B, Alric J, Petroutsos D, Tolstygina I, et al. Control of hydrogen photoproduction by the proton gradient generated by cyclic electron flow in Chlamydomonas reinhardtii. Plant Cell. 2011;23:2619–2630. doi: 10.1105/tpc.111.086876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molnar A, Bassett A, Thuenemann E, Schwach F, Karkare S, et al. Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J. 2009;58:165–174. doi: 10.1111/j.1365-313X.2008.03767.x. [DOI] [PubMed] [Google Scholar]
- 43.Wang ZT, Ullrich N, Joo S, Waffenschmidt S, Goodenough U. Algal Lipid bodies: stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryotic Cell. 2009;8:1856–1868. doi: 10.1128/EC.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jans F, Mignolet E, Houyoux P-A, Cardol P, Ghysels B, et al. A type II NAD(P)H dehydrogenase mediates light-independent plastoquinone reduction in the chloroplast of Chlamydomonas. Proc Natl Acad Sci. 2008;105:20546–20551. doi: 10.1073/pnas.0806896105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishimaki T, Yamanaka H, Mizugaki M. Studies on the metabolism of unsaturated fatty acids. XIV. Purification and properties of NADPH-dependent trans-2-enoyl-CoA reductase of Escherichia coli K-12. J Biochem. 1984;95:1315–1321. doi: 10.1093/oxfordjournals.jbchem.a134737. [DOI] [PubMed] [Google Scholar]
- 46.Finazzi G, Johnson GN, Dall'Osto L, Zito F, Bonente G, et al. Nonphotochemical quenching of chlorophyll fluorescence in Chlamydomonas reinhardtii. Biochemistry. 2006;45:1490–1498. doi: 10.1021/bi0521588. [DOI] [PubMed] [Google Scholar]
- 47.Elrad D, Niyogi KK, Grossman AR. A major light-harvesting polypeptide of photosystem II functions in thermal dissipation. Plant Cell. 2002;14:1801–1816. doi: 10.1105/tpc.002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dent RM, Haglund CM, Chin BL, Kobayashi MC, Niyogi KK. Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of Chlamydomonas reinhardtii. Plant Physiol. 2005;137:545–556. doi: 10.1104/pp.104.055244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harris E. The Chlamydomonas Sourcebook. Academic Press, San Diego. San Diego: 1989. [Google Scholar]
- 50.Eberhard S, Jain M, Im C, Pollock S, Shrager J, et al. Generation of an oligonucleotide array for analysis of gene expression in Chlamydomonas reinhardtii. Curr Genet. 2006;49:106–124. doi: 10.1007/s00294-005-0041-2. [DOI] [PubMed] [Google Scholar]
- 51.Voß B, Meinecke L, Kurz T, Al-Babili S, Beck CF, et al. Hemin and magnesium-protoporphyrin IX induce global changes in gene expression in Chlamydomonas reinhardtii. Plant Physiol. 2011;155:892–905. doi: 10.1104/pp.110.158683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saldanha AJ. Java Treeview-extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 53.Wobbe L, Blifernez O, Schwarz C, Mussgnug JH, Nickelsen J, et al. Cysteine modification of a specific repressor protein controls the translational status of nucleus-encoded LHCII mRNAs in Chlamydomonas. Proc Natl Acad Sci. 2009;106:13290–13295. doi: 10.1073/pnas.0900670106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rasmussen R. Quantification on the LightCycler instrument. In: Meuer S, Wittwer C, Nakagawara K, editors. Rapid Cycle Real-Time PCR: Methods and Applications. Heidelberg: Springer-Verlag Press; 2001. pp. 21–34. [Google Scholar]
- 55.Arnon DI. Copper enzymes in isolated chloroplasts. polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press); 1989. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Real-time RT-Q-PCR analysis of the mRNA levels of selected genes in stm6glc4 and WT. RNA samples of the second microarray experiment were analyzed by RT-Q-PCR in order to determine the relative amount of selected transcripts in stm6glc4 compared to WT (set to 100%). Three different time points according to Figure 1 were analyzed: T0 (before sulfur deprivation), preH2 (after sulfur depletion and before H2 production; T2 WT; T1 stm6glc4) and H2 (H2 production phase; T5 WT; T3 stm6glc4). Standard deviations are given as error bars and were calculated from three technical replicates. Genes: ICL1 (isocitrate lyase; Cre06.g282800; Table S2), T2ECR (Trans-2-enoyl-CoA-reductase; Cre01.g035350; Table 1 and 2), GWD2 (R1 protein, α-glucan water dikinase; Cre07.g332300; Table 1), LHCSR1 (stress-related LHC protein, Cre08.g365900, Table 1 and 2), Cbr-like ELIP (Cre07.g320450/Cre07.g320400; Table 1 and 2), TAG-lipase (similar to CGLD15/putative TAG lipase; Table 2), SIR1 (ferredoxin-sulfite reductase; Cre08.g365700; Table 2).
(TIF)
A complete list of all microarray data. Expression relative to T0 is given for both strains and all examined time points. GeneID numbers correspond to those contained in the gal file (http://www.chlamy.org/galfile.xls, [50]) and heat map group assignments (Figure 4) for each gene are given as well. An increase in the mRNA abundance (Tn/T0>1) is indicated by red highlighting and a decrease (Tn/T0<1) by green highlighting. Available functional assignments (column “Annotation”) are given and were used for sorting (column “Group”).
(XLS)
Genes showing either a specific down- or upregulation in one of both examined strains at T0. Differentially expressed genes are sorted according to the cellular processes involved as deduced from their functional annotation. Gene names are given along with the corresponding locus names (Phytozome 7.0; http://www.phytozome.net/) and a description of their function.
(DOCX)