Abstract
Background
We previously demonstrated that the Arabidopsis thaliana AtMYB60 protein is an R2R3MYB transcription factor required for stomatal opening. AtMYB60 is specifically expressed in guard cells and down-regulated at the transcriptional levels by the phytohormone ABA.
Results
To investigate the molecular mechanisms governing AtMYB60 expression, its promoter was dissected through deletion and mutagenesis analyses. By studying different versions of AtMYB60 promoter::GUS reporter fusions in transgenic plants we were able to demonstrate a modular organization for the AtMYB60 promoter. Particularly we defined: a minimal promoter sufficient to confer guard cell-specific activity to the reporter gene; the distinct roles of different DOF-binding sites organised in a cluster in the minimal promoter in determining guard cell-specific expression; the promoter regions responsible for the enhancement of activity in guard cells; a promoter region responsible for the negative transcriptional regulation by ABA. Moreover from the analysis of single and multiple mutants we could rule out the involvement of a group of DOF proteins, known as CDFs, already characterised for their involvement in flowering time, in the regulation of AtMYB60 expression.
Conclusions
These findings shed light on the regulation of gene expression in guard cells and provide new promoter modules as useful tools for manipulating gene expression in guard cells, both for physiological studies and future biotechnological applications.
Background
Land plants uptake carbon dioxide for photosynthesis and lose water vapour by transpiration through stomatal pores, present on the surface of leaves and stems. The opening and closure of the pore is mediated by turgor-driven volume changes of two surrounding guard cells, whose pressure is dynamically adjusted according to environmental and hormonal cues. In response to abiotic stresses, such as drought or high salinity, one of the most rapid responses of plants is the closure of stomata, mediated by the hormone abscisic acid (ABA), to prevent excessive water loss by transpiration (reviewed in [1]).
The genetic manipulation of stomatal activity is emerging as a promising approach to reduce the water requirement of crops, and to enhance productivity under stress conditions [2]. Proper engineering of stomatal responses requires the use of guard cell-specific promoters, or the identification of guard cell-specific mutants, to avoid undesirable side effects on plant growth and productivity.
Several promoters that confer guard cell-specific gene expression or enhanced gene expression in guard cells have been isolated through different methods: functional characterization of single genes [3-9]; large scale gene- or enhancer-trap screens [10-12]. Moreover transcriptomic and proteomic studies have identified additional candidates [13-16]. Nevertheless the majority of these promoters are not guard cell-specific, as they drive the expression of reporter genes in other cell types, including the vascular tissues [6,10,17,18], flower organs [8,9] or starch containing cells [5], significantly reducing the number of true guard cell-specific full size promoters [3,10,14,19,20]. Most importantly, a detailed experimental analysis of guard cell-specific promoters has been performed only in very few cases [11,12,14].
A true guard cell-specific promoter is driving expression of the Arabidopsis AtMYB60 (At1g08810) gene [10,19,21,22]. We have previously shown that AtMYB60 is expressed in guard cells [10], and the complete 5' and 3' intergenic genomic regions of this gene, cloned respectively upstream and downstream to reporter genes, were able to drive specific expression in guard cells [10,19]. Guard cell specificity of the AtMYB60 promoter has been also demonstrated by Nagy et al. (2009) and by Meyer et al (2010), who used this promoter to complement the mrp5-1 mutant phenotype exclusively in guard cells, and to specifically express the AtLMT12 protein at high levels in guard cells, respectively.
Very little information is available concerning promoter cis-elements regulating guard cell-specific expression [8,10-12,14,16]. DOF-binding sites have been suggested to have a role in such a regulation [8,10-12]. DOF (DNA binding with One Finger) proteins are plant specific transcription factors involved in light, phytohormones and pathogen signalling and responses as well as seed development (reviewed by [23]). A role for [T/A]AAAG DOF-binding sites in mediating gene expression in guard cells has been experimentally defined only for the potato KST1 gene [8]. However, in Arabidopsis the role of DOF-motifs in controlling guard cell expression is still controversial [10-12]. The study performed on the potato KST1 promoter [8] and the bioinformatic analysis performed on several guard-cell specific Arabidopsis promoters [10] suggest that the presence of clusters of DOF cis-elements, rather than their absolute number, is important to confer guard cell-specificity to a promoter region [10]. Yet, the role of DOF-binding sites in driving guard cell expression in Arabidopsis and the hypothesis of cluster organization remains to be experimentally investigated.
The guard-cell specific AtMYB60 promoter presents several DOF clusters, making it an ideal model to test the hypothesis that DOF clusters are important for guard cell-specific expression. Moreover the AtMYB60 expression is modulated by different environmental cues such as light, dark and drought stress [19], suggesting the presence of different cis-elements controlling these transcriptional responses. In this report we aimed to isolate the cis-elements responsible for the AtMYB60 guard cell specific expression. We generated Arabidopsis transgenic lines carrying truncated or mutagenised AtMYB60 promoter versions fused to the GUS reporter gene. Using a combination of histochemical and expression analysis we were able to identify a minimal promoter necessary and sufficient to drive guard cell specific expression. Using the same tools, we were also able to map a region required for ABA-mediated repression.
Results
In-silico analysis of the AtMYB60 promoter
In a previous study, we demonstrated that the complete 5' and 3' AtMYB60 intergenic genomic regions - cloned upstream and downstream of the β-glucoronidase (GUS) reporter gene, respectively - could specifically drive strong GUS activity in stomata of Arabidopsis seedlings and adult plants [19]. No GUS signals were detected in any other cell type or in tissues devoid of stomata [19].
To investigate the possible cis-acting elements that regulate AtMYB60 expression, we surveyed the genomic region upstream of the AtMYB60 translational start codon for the presence of known transcription factor binding sites using the PLACE software [24]. Our analysis produced a significant enrichment in the [A/T]AAAG motifs in the AtMYB60 promoter compared to the average distribution of [A/T]AAAG oligos in intergenic regions throughout the Arabidopsis genome (P < 0.01) (Figure 1). Interestingly, these [A/T]AAAG motifs, have been shown to be involved in the regulation of guard cell expression of the potato potassium channel KST1 gene [8]. Also, clusters of [A/T]AAAG motifs, required for the binding of DOF-type transcription factors [25], were over represented in different guard cells-specific promoters [6,10,12]. In particular, Galbiati and colleagues suggested, as guard cell-specific cis-element, a cluster of at least three [A/T]AAAG motifs located on the same strand within a region of 100 bp [10]. Using the criteria previously described by Galbiati and collaborators (2008), we found three of these guard cell-specific clusters in the 5' intergenic region of the AtMYB60 gene (Figure 1), suggesting a conserved mechanism for guard cell specific expression.
Figure 1.
Nucleotide sequence of the 5'-region of the AtMYB60 gene. Nucleotides are numbered on the left with the translational start site designated as +1. The ATG is in bold. The 5' UTR is in lower case letters. The DOF-binding sites are grey boxed, the W-box, considered in the text, is white boxed. Clusters of DOF-binding sites, as defined by Galbiati and colleagues (2008), are underlined. The CAAGTTG motif described as a putative cis-element for ABA repression ([16]) is dotted underlined.
Identification of the AtMYB60 minimal promoter
To gain more insights into the cis-elements that regulate the AtMYB60 expression in guard cells, we produced a set of Arabidopsis transgenic lines carrying the complete 1,307 bp 5' intergenic region upstream of the translational start codon fused to the reporter GUS (construct -1,307::GUS, Figure 2A). GUS staining analysis of 15 independent T2 lines revealed that this region contains all the cis-acting elements required for expression of the reporter in stomata (Figure 2B), while no GUS signals were detected in any other cell type or in tissues devoid of stomata (Additional file 1).
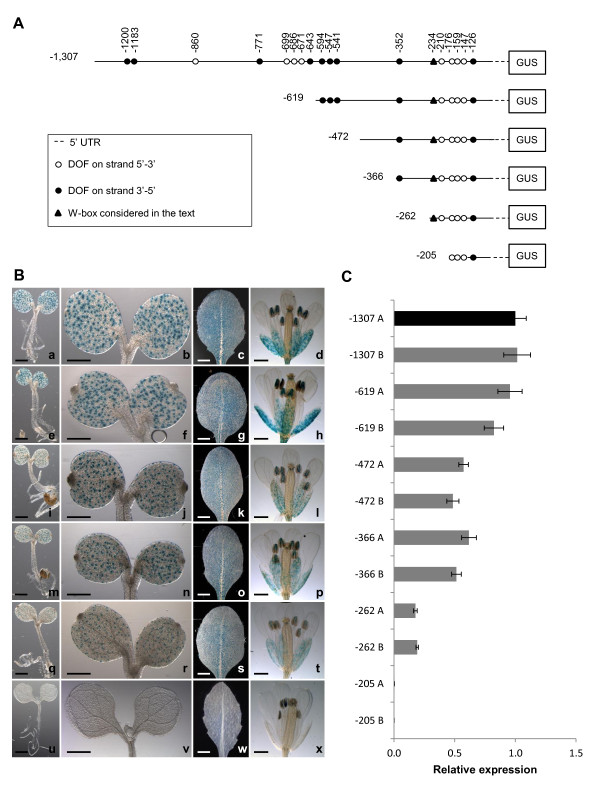
Figure 2.
Deletion analysis of the AtMYB60 upstream region. A, Schematic diagrams of different deletions of AtMYB60 upstream region fused to the GUS reporter gene. The positions of the different DOF-binding sites and of the W-box, described in the text, are shown. B, Histochemical assay for GUS activity in seedlings, rosette leaves and flowers of plants transformed with -1,307::GUS (a-d), -619::GUS (e-h), -472::GUS (i-l), -366::GUS (m-p), -262::GUS (q-t) and -205::GUS (u-x) constructs. The analysis of independent lines harbouring the same construct showed identical patterns of GUS staining. Samples were incubated in the staining solution for 16 hours for all the lines, with the exception of line -205::GUS, for which the staining was prolonged to 48 hours. Scale bars represent 1 mm. C, Relative expression level of the GUS reporter gene in the different transgenic lines harbouring the -1,307::GUS (-1,307 A and B), -619::GUS (-619 A and B), -472::GUS, -366::GUS (-366 A and B), -262::GUS (-262 A and B) or -205::GUS (-205 A and B) constructs. Two lines for each construct were analysed by Real Time RT-PCR. The transcript amount in the line -1,307 A was arbitrarily set to 1 (black column) and used to normalize the relative expression levels in each line. The ACTIN2 gene (At3g18780) was used as a control.
Next, we made a series of 5' deletions of the -1,307 bp genomic region to define the minimum sequence length required for the expression in guard cells (Figure 2A). These truncated promoters (fused to the GUS gene) were stably transferred to Arabidopsis and 10 to 15 independent T2 transgenic lines were analysed in detail. Deletions of the distal part of the 1,307 bp region to position -619 (construct -619::GUS), -472 (-472::GUS), or -366 (-366::GUS) from the ATG codon, did not alter expression of the reporter in guard cells located on both vegetative and floral organs (Figure 2B). Further deletions (to position -262) indicated that the 262 bp proximal region was sufficient to drive expression of the reporter in stomata (Figure 2B). However, the removal of the region between -262 bp and -205 bp (construct -205::GUS) completely abolished GUS activity in guard cell (Figure 2B). Transgenic lines carrying the -205::GUS fusion did not show GUS staining in any other cell type, even after prolonged staining (up to 48 h, Figure 2B). This finding suggests that the 57 bp region located between positions -262 and -205 contains cis-elements essential for expression in stomatal guard cells. Based on these results, we defined the -262 bp region upstream of the ATG codon as the minimal promoter of the AtMYB60 gene.
To thoroughly investigate quantitative differences in GUS expression among lines carrying different deletion:reporter constructs, we determined the relative amount of GUS transcript by quantitative RT-PCR (qRT-PCR). mRNA samples derived from two representative independent lines (A and B) were analysed for each construct (Figure 2C). Lines harbouring the 1,307 bp 5' intergenic region or the -619 deletion fused to the reporter, did not show any significant differences in their GUS transcript accumulation. Conversely, deletions to position -472 and -366 resulted in a two-fold decrease in GUS expression compared to the -1,307::GUS line, while deletion to position -262 resulted in a five-fold decrease (Figure 2C, p < 0.01). These results indicate that one or more sequences with function of enhancer are present in the genomic region between -619 bp and -472 bp and between -472 and -262 from the ATG of AtMYB60. In accordance with the results obtained from the histochemical analysis, qRT-PCR experiments did not detect significant GUS transcripts accumulation in lines carrying the -205::GUS fusion.
Site-directed mutagenesis of the AtMYB60 minimal promoter
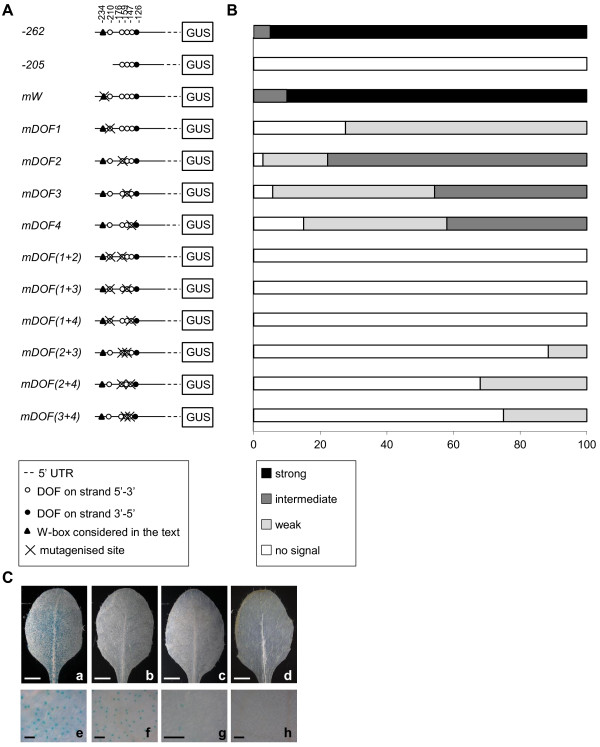
Promoter deletion experiments indicate that the AtMYB60 minimal promoter region (construct -262::GUS) contains all the cis-acting elements required to sustain expression of a reporter gene in guard cells. This region encompasses the [A/T]AAAG cluster proximal to the ATG codon, which consists of four AAAAG DOF-binding sites (Figures 1 and 3A). In addition, the PLACE software identified in this region a single W-box, corresponding to the binding site of WRKY transcription factors [26], located upstream of the [A/T]AAAG cluster (Figure 3A). To address the functional significance of the individual cis-elements present in the AtMYB60 minimal promoter, we evaluated the effects of targeted nucleotide substitutions on GUS expression (Figure 3A). Mutated versions of the minimal promoter were generated by PCR and fused to GUS and at least 30 T2 independent transgenic lines for each mutated promoter::GUS combination were visually scored and classified to reflect their relative guard-cell specific GUS staining. A representative example of each category is provided in Figure 3C.
Figure 3.
Role of DOF-binding sites in the minimal promoter in driving GUS activity in guard cells. A, Schematic diagrams of constructs -262::GUS, -205::GUS and of constructs containing mutagenised version of the minimal promoter in different DOF-binding sites and in the W-box at position -234. B, Percentage of lines for each construct showing strong (column segment in black), intermediate (in dark grey), weak (light grey) or no signal (white). C, A leaf from a line harbouring the -262::GUS construct (a and a particular in e), shown as an example of strong GUS activity. In the following pictures examples of different lines harbouring the mDOF3::GUS construct showing respectively an intermediate (b and f), a weak (c and g) and no GUS activity (d and h). Scale bars represent 1 mm (a-d) or 0.1 mm (e-h).
We initially tested the role of the single W-box cis-element, by replacing the consensus sequence TTGAC, with the non-functional TTGAA motif [27]. Lines carrying the mutated W-box (mW::GUS) showed similar levels of GUS expression to the wild-type promoter, indicating that W-box does not contribute to mediate gene expression in guard cells (Figure 3B). Next, we produced mutant promoters in which single DOF motifs within the [A/T]AAAG cluster were converted to the unrelated CGCGA sequence. Inactivation of the most distal AAAAG site relative to the ATG (hereinafter referred to as DOF1) resulted in a dramatic decrease of GUS expression (mDOF1::GUS construct, Figure 4B). 30% of the lines carrying the mDOF1::GUS construct did not show GUS expression, whereas the remaining 70% only showed weak staining, thus indicating a crucial role for DOF1 in regulating AtMYB60 expression in guard cells (Figure 3B). Mutations of the second, third or fourth most proximal AAAAG site (hereinafter referred to as DOF2, DOF3 and DOF4, respectively), resulted in a reduced GUS expression, although to a lesser extent than the one in the DOF1 (Figure 4B, mDOF2::GUS, mDOF3::GUS and mDOF4::GUS plants). In particular, none of the 30 mDOF2::GUS transgenic lines displayed strong expression of the reporter, nearly 70% showed intermediate expression, 25% showed weak expression and the remaining 5% did not show any GUS staining (Figure 3B). A comparable distribution among strong, intermediate and weak lines was obtained from the analysis of the mDOF3::GUS and mDOF4::GUS plants (Figure 3B).
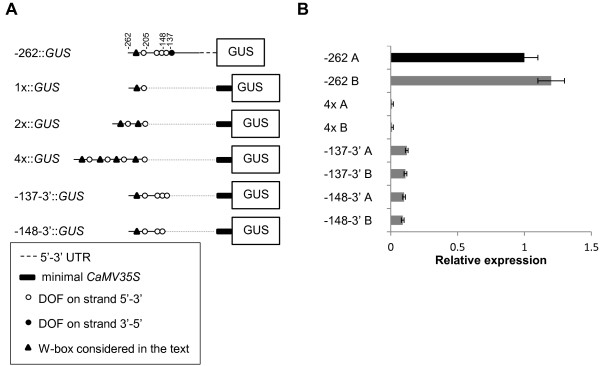
Figure 4.
Oligomerisation of the 57 bp sequence and 3' deletions of the AtMYB60 minimal promoter. A, Schematic diagrams of the constructs. In the 4x::GUS construct the fragment of 57 bp between -262 and -205 in tandem array of four copies was fused to the minimal promoter CaMV 35S (min 35S in the scheme, portion between -46 and +1) upstream of the GUS reporter gene. In the constructs -137-3'::GUS and -148-3'::GUS, 3' deleted versions of the AtMYB60 minimal promoter were fused to the same portion of the CaMV 35S. (B) Relative expression level of the GUS reporter gene in the different transgenic lines harbouring the constructs. Two lines for each construct were analysed by Real Time RT-PCR. The transcript amount in the line -1307 A was arbitrarily set to 1 (black column) and used to normalize the relative expression levels in each line. The ACTIN2 gene was used as a control. The symbols are the same described in Figure 2. The dotted lines indicate the regions deleted in AtMYB60 minimal promoter sequences.
To establish whether DOF-binding sites could exert additive roles in mediating gene expression in stomata we produced a second series of promoters, in which two AAAAG motifs were mutated simultaneously. Mutations of DOF1 and DOF2 (mDOF(1+2)::GUS), DOF1 and DOF3 (mDOF(1+3)::GUS) or DOF1 and DOF4 (mDOF(1+4)::GUS) completely inactivated the minimal promoter, as GUS expression was abolished in all the mDOF(1+2)::GUS, mDOF(1+3)::GUS and mDOF(1+4)::GUS lines analysed (Figure 3B). Interestingly, the concurrent mutation of DOF2 and DOF3 (mDOF(2+3)::GUS) resulted in a strong, but yet not complete, inactivation of the promoter activity in guard cells, as 15% of the mDOF(2+3)::GUS lines displayed weak expression of the reporter in stomata. Likewise, concomitant inactivation of either DOF2 and DOF4, or DOF3 and DOF4 did not completely eliminate GUS expression in guard cell (Figure 3B). Taken together, these results indicate that the putative [A/T]AAAG DOF-binding sites located in the AtMYB60 promoter are necessary to mediate its expression in guard cells.
A single DOF cluster is sufficient to drive low expression in guard cell
Our deletion analysis of the AtMYB60 promoter indicates that the 57 bp region between positions -262 and -205 is essential for gene expression in stomatal guard cells (Figure 2). This region contains the DOF1 cis-element required for guard cell expression as shown by mutagenesis analysis results (Figure 3). To establish whether this 57 bp region was sufficient to activate expression in guard cells, we fused one (1x::GUS construct), two (2x::GUS) and four tandem copies (4x::GUS) of the 57 bp fragment to the minimal CaMV35S promoter [28] upstream of the GUS reporter gene (Figure 4A), effectively reconstructing an artificial DOF cluster containing one, two or four copies of the DOF1 element. However, we did not observe GUS activity in any of the 30 independent stable transformants produced for each construct, even after prolonged staining (data not shown). These data were confirmed by qRT-PCR analysis of independent lines carrying the 4x::GUS fusion (Figure 4B), indicating that the multimerisation of the DOF1 site per se is not sufficient to drive gene expression in guard cell. This might derive from an inappropriate organization and/or spatial distribution of the different DOF elements in the context of the minimal promoter. To test this hypothesis we made two 3' deletions of the AtMYB60 minimal promoter: the -148-3'::GUS and -137-3'::GUS constructs containing the first three and four DOF-binding sites respectively of the most proximal cluster fused upstream of the minimal CaMV35S promoter (Figure 4B). Our initial histochemical analysis did not reveal any GUS positive lines (data not shown). To substantiate this result we also performed a qRT-PCR analysis on fifteen independent lines for each construct. Interestingly, eight lines out of fifteen showed a low but significant GUS transcript accumulation compared to the full length minimal promoter (Figure 4B). These results suggest that the presence of the cluster containing three or four DOF-binding sites is sufficient to drive GUS activity in guard cells, even though at a very low level. This finding implies that other cis-elements present downstream of position -137 are required for the full functionality of the minimal promoter.
The guard cell-related CDF1, CDF2, CDF3 and CDF5 DOF-type transcription factors do not regulate AtMYB60 expression in stomata
Target mutagenesis experiments of the AtMYB60 promoter demonstrated that [A/T]AAAG DNA consensus motifs are essential cis-acting elements in the regulation of AtMYB60 expression in guard cells. Consequently, their cognate DOF proteins represent the most likely candidates as trans-acting factors. As the Arabidopsis genome contains 36 DOF-coding genes [23], candidate DOF transcription factors involved in the regulation of AtMYB60 expression should fulfil two criteria: they should be expressed in guard cells and the loss of their gene function should abolish or significantly down-regulate the expression of AtMYB60 in this cell type.
The CYCLING DOF FACTOR 1 (CDF1, At5g62430) gene, involved in the regulation of photoperiodic flowering, has been shown to be highly expressed in the vascular tissue and guard cells [29]. We thus investigated the expression of the AtMYB60 gene in the loss-of-function cdf1-R allele. As shown in Additional file 2 we did not detect significant differences in the accumulation of AtMYB60 transcripts in homozygous cdf1-R plants compared with the wild type.
It is important to note that in photoperiodic flowering, CDF1 acts redundantly with three other DOF proteins, namely CDF2 (At5g39660), CDF3 (At3g47500) and CDF5 (At1g69570) [30], belonging to the same phylogenetic group II [31]. Similarly to CDF1, promoter::GUS analyses revealed that CDF2, CDF3 and CDF5 are strongly expressed in guard cells.
We thus analysed the expression of AtMYB60 in single, double, triple and quadruple cdf mutants to determine the possible role of these additional candidate CDF proteins. As for cdf1-R mutant, the level of expression of AtMYB60 was not significantly reduced in the cdf2-1, cdf3-1 and cdf5-1 single mutants (Additional file 2). Likewise, AtMYB60 expression was not altered in any of the double, triple or quadruple mutant combinations, indicating that, despite their expression in guard cells, these four CDF proteins are not trans-regulators of AtMYB60 expression in stomata (Additional file 2).
Identification of a promoter region that negatively responds to ABA
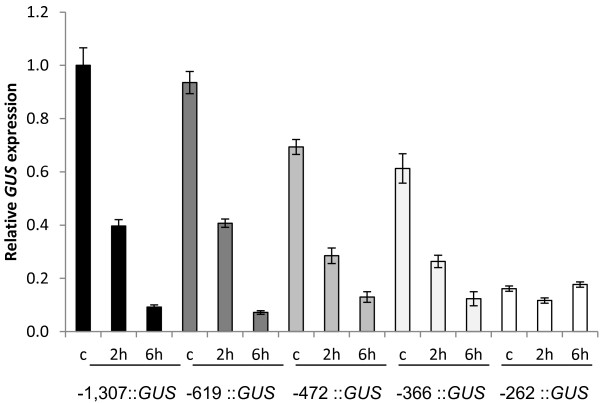
We previously reported that transcript accumulation of the AtMYB60 gene is rapidly down-regulated by exogenous applications of the hormone ABA, which plays a fundamental role in regulating gene expression in response to drought stress [19]. To identify the promoter region responsible for the ABA-mediated AtMYB60 down-regulation, we applied ABA to the previously described transgenic lines harbouring serial deletions of the AtMYB60 promoter (Figure 2). Quantitative RT-PCR analysis revealed a similar decrease in GUS transcript levels in transgenic lines carrying the full length as well as the -619, -472 and -366::GUS fusions (Figure 5). The kinetic of down-regulation of the GUS transcript was comparable to the one observed for the endogenous gene AtMYB60 [19], indicating that -619, -472 and -366 promoters maintain the sequences responsible for transcriptional down-regulation by ABA. Also, these results suggest that the CAAGTTG motif, present in the AtMYB60 promoter between -619 and -613 (dotted underlined in Figure 1), and recently described as overrepresented in ABA-repressed genes [16], does not play a significant role in the ABA-dependent repression of AtMYB60 expression. Rather, qRT-PCR experiments performed on different independent lines carrying the -246::GUS construct showed that the minimal promoter sequence lacks the region responsible for negative regulation by ABA, as these lines did not show changes in GUS expression in response to the hormone as shown in Figure 5.
Figure 5.
Expression of the GUS gene in different transgenic lines in response to ABA treatment. Two lines for each construct shown in Figure 2 were analysed by Real Time RT-PCR. c represent the control samples. The transcript amount in the sample -1307 A control was arbitrarily set to 1 and used to normalize the relative expression levels in each line. The ACTIN2 gene was used as a control.
Taken together these data indicate that, although the minimal promoter maintains the cis-elements necessary for guard cell expression, it lacks the motifs that mediate the negative regulation by ABA, becoming ABA-insensitive. We can thus conclude that the region between -366 and -262 contains elements necessary for ABA down-regulation.
Discussion
Very few guard cell-specific promoters have been described to date [3,10,14,19,20]. Independent studies demonstrated that the AtMYB60 promoter can be considered guard cell-specific, being sufficient to drive expression of reporter genes specifically in guard cells [19,21]. Moreover this promoter has also been used to complement a mutant phenotype specifically in guard cells [21], and to investigate subcellular localization exclusively in guard cells [22]. In this study we identified the AtMYB60 minimal promoter that is necessary and sufficient to drive guard cell-specific expression.
DOF-binding sites are required for AtMYB60 guard-cells expression
Our in silico analysis identified three DOF site clusters (Figure 1). Initial deletion studies revealed a prominent role for the most proximal DOF cluster (relative to the ATG start codon). Site-directed mutagenesis showed that the distal most DOF-binding site (DOF1 at position -210, Figure 3) plays a major role in driving guard cell expression compared to other DOF motifs of the same cluster (DOF2 at position -176, DOF3 at -159 and DOF4 at -147, Figure 3). These other DOF elements play partially additive roles, as clearly demonstrated by the combined mutagenesis of these sites and DOF1 site which resulted in a drastically reduced GUS activity (Figure 3). DOF-binding sites are thus key determinants in mediating guard cell expression, in accordance with the DOF cluster hypothesis we previously formulated [10]. A suggestion for a similar involvement of DOF cis-elements in Arabidopsis derives from the work of Gardner and colleagues (2009) that identified DOF motifs in a region controlling guard cell expression. Other authors identified a region enriched in DOF-binding sites in the guard cell-specific pGC1 promoter, although the mutation of a single DOF site did not impair promoter activity [14]. Interestingly, a DOF cluster organization is present in the promoter of the grape VvMYB60 gene, a putative ortholog of AtMYB60, indicating a conservation of the cluster structure during the evolution among AtMYB60 orthologs [32]. The results reported by Plesch and colleagues (2001) on the DOF motif organisation in the potato KST1 promoter highlight a more general evolutionary conservation of this module in the control of guard cell-specific activity of promoters.
Although we cannot rule out the possibility that other unknown transcription factors might interact with those same cis-elements, DOF factors represent likely candidates as AtMYB60 regulators. The most parsimonious hypothesis resulting from combining our results indicates that DOF proteins act as positive regulators of AtMYB60. The potato StDOF1 protein has been shown to bind in vitro to the guard cell specific promoter of KST1 [8], while no data are available for any Arabidopsis DOF proteins. Among the Arabidopsis DOF genes, CDF1, CDF2, CDF3, and CDF5 (CDFs) are expressed in guard cells [29]. However, singles and multiple cdf mutants show a wild-type pattern of AtMYB60 expression, ruling out their involvement in AtMYB60 regulation (Additional file 2). The majority of Arabidopsis DOF genes are expressed in guard cells [33,34] and may thus act redundantly, as already demonstrated among members of this family [30]. All these aspects do not facilitate the identification of obvious candidates as AtMYB60 regulators. We are trying to identify the DOF genes involved in the regulation of AtMYB60 by analysis of its expression in mutants of genes preferentially expressed in the guard cells (http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi[33]).
Multiple cis-elements participate to enhance AtMYB60 guard-cells expression
Transcriptional GUS fusions, harbouring different deletions of the 5' intergenic region to position -262 from the ATG, conferred GUS activity exclusively in guard cells (Figures 2 and Additional file 1). The activity of these promoter regions is in apparent discrepancy with the detection of AtMYB60 gene expression in seeds, as revealed by available microarray analysis data [33,34] and in roots, as recently reported [35]. One hypothesis to explain this incongruity could be the presence of other regulatory regions present outside the complete 5' and 3' intergenic regions flanking the AtMYB60 coding sequence. Intron sequences, for example, may be involved in such a regulation, as previously demonstrated for different plant genes ([36] and references herein).
While guard-cell specific expression was invariably maintained by functional AtMYB60 promoter variants, the levels of expression varied considerably. In addition to DOF-binding sites, other cis-elements are required to boost the AtMYB60 expression. Indeed, an artificial DOF1 binding site repeated in single or multiple copies could not drive guard cell expression (Figure 4A). The incorporation of the entire proximal DOF cluster (e.g. -137-3'::GUS) resulted in a small but significant guard cell transcriptional activity. Thus, other cis-elements downstream of position -137 are required for full activity of the minimal promoter. It is known that cis-elements other than DOF-binding sites are involved in the regulation of guard cell expression. In the case of the guard cell-specific AtPDR3 gene no [A/T]AAAG clusters were identified in a 1000-bp region upstream of the ATG codon, suggesting the presence of other regulatory units [10].
Modular organization of the AtMYB60 promoter
In this study we also investigated the regulation of the AtMYB60 promoter activity in response to ABA. ABA treatments induce global changes in gene expression in Arabidopsis [16,37-40]. Transcriptomic analyses revealed extensive regulation of gene expression by ABA also in guard cells [13,14,16]. While cis-elements that positively regulate the response to ABA have been functionally characterised (for a review, see [41]), those that negatively regulate the response to ABA are largely unknown. A CAA[G/C]TTG motif has been shown to be over-represented in ABA-repressed gene promoters and thus proposed for such a role [16,39]. The AtMYB60 promoter contains one CAAGTTG motif between -619 and -613 from the ATG, yet our results do not support its proposed role as negative regulator of ABA response. Conversely, a region between positions -366 to -262 contained the entire requirement for the ABA-mediated repression Figures 5 and 6. It has been proposed that evolution may have favoured the differentiation of mechanisms for ABA down-regulation rather than up-regulation, rendering more difficult for any ABA-repression motif to achieve statistical significance [16]. Our data may provide a valuable model system to clarify the mechanism mediating ABA repression.
Figure 6.
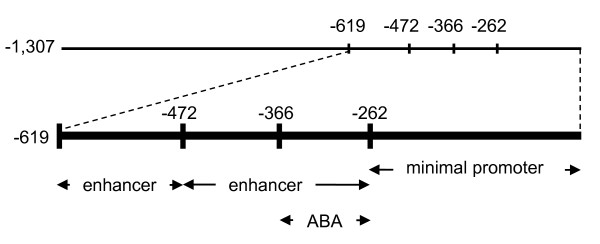
Modular organization of the AtMYB60 promoter. Different portions of the AtMYB60 promoter defined through deletion analysis are shown. ABA indicates the region responsible for the negative regulation by ABA treatment.
Our data suggests a modular organization for the AtMYB60 promoter as summarised in Figure 6. Through a serial deletion analysis, we defined the AtMYB60 minimal promoter, sufficient to induce guard cell-specific activity (construct -262::GUS, Figure 2). A 57 bp region, located between position -262 and position -205, is necessary to confer GUS activity in guard cells (Figure 2A). We also identified two regions that enhance the expression of the GUS gene between -619 bp and -472 bp and between -472 and -262 (Figure 2B and 2C). Besides providing pieces of evidence for such modular organization, our work indicates that the different portions of the AtMYB60 promoter may prove useful for manipulating gene expression in guard cells, with the possibility to obtain different level of expression. Moreover, the minimal promoter (whose activity is not influenced by ABA) can be used for ABA-independent expression of target genes in guard cells
Interestingly, both the full length and the minimal promoters maintain their guard cell-specific activity in heterologous systems, such as the crop species tomato and tobacco (Francia, personal communication), thus indicating the conservation of this cell-specific regulatory mechanism among different plant species. Moreover, preliminary results suggest that the AtMYB60 minimal promoter can be combined with other cis-regulatory modules to produce functional guard cell-specific chimeric promoters (Francia, personal communication). As a whole our data demonstrate that both the full length and the minimal AtMYB60 promoters provide a valuable tool to manipulate gene expression specifically in guard cells, both for physiological studies and downstream biotechnological applications.
Conclusions
Our work provides strong evidence for the involvement of [A/T]AAAG elements in the regulation of the AtMYB60 expression, illustrating their functional cluster organization. Future work will concentrate on the analysis of candidate DOF transcription factors that control this mechanism. Finally we identify a region of the AtMYB60 promoter required for the negative regulation by ABA, offering the possibility to discover novel cis-elements for this kind of regulation.
Methods
Plant Material
All plant material described was in the Col-0 accession. The cdf1-R line (35S::CDF1-RNAi #23) was kindly provided by Takato Imaizumi [29]. The cdf2-1, cdf3-1 and cdf5-1 null alleles are T-DNA insertion line. Single, double, triple and quadruple cdf mutants have been previously described [30].
Construction of AtMYB60 promoter::GUS fusions
5'-deletions of the 5' intergenic genomic region upstream of the AtMYB60 gene were generated by PCR amplification from plasmid p1.3-2.2::GUS, previously described [19], using different forward primers and a single reverse primer. Forward and reverse primers incorporated a HindIII and a BamHI, respectively. The PCR fragments were cloned into the pCR4-TOPO vector (Invitrogen Corporation, Carlsbad, CA), cut with HindIII and BamHI and ligated upstream of the uidA coding sequence in the pBI101.3 binary vector (Clontech, Palo Alto, CA, USA). The resulting plasmids were renamed -1307::GUS, -619::GUS, -472::GUS, -366::GUS, -262::GUS and -205::GUS (Figure 2).
Chimeric promoters containing different 3'-deleted fragments of the AtMYB60 minimal promoter and 46-bp CaMV 35S promoter were produced by amplifying the sequence of the CaMV 35S promoter from -46 to +1 [28] from plasmid pBI121 (Clontech, Palo Alto, CA, USA), using the forward primer 35SXba containing a XbaI site and the reverse primer 35SBam with a BamHI site. The PCR product was cloned into the pCR4-TOPO vector and the XbaI-BamHI fragment was cloned into the pBI101.3 vector (renamed 35Smin-pBI101.3). The regions from -262 to -137 and from -262 to -148 of the AtMYB60 minimal promoter were amplified by PCR from plasmid p1.3-2.2::GUS, using the reverse primers p60R6 and p60R7 incorporating a XbaI site and a single forward primer p60F3 with the HindIII site. The corresponding PCR products were cloned into the pCR4-TOPO vector and the HindIII-XbaI fragments were cloned into the 35Smin-pBI101.3 vector to give the -137-3'::GUS and -148-3'::GUS vectors, respectively (Figure 3).
Chimeric promoters containing different copies of the region between -262 and -205 of the AtMYB60 promoter were obtained by synthesising one copy of this sequence, using the forward primer p60F3 with a HindIII site and the reverse primer p60R3 with an XbaI site. The resulting PCR product was cloned into the pCR4-TOPO vector and the HindIII-XbaI fragment was ligated into the 35Smin-pBI101.3 vector (construct 1x::GUS). A second copy of this region was generated using the primers p60F3 and p60R5b, both incorporating a HindIII site; the fragment HindIII-HindIII was cloned into the construct 1x::GUS, generating the construct 2x::GUS. This plasmid was used as a template to generate two other copies of the sequence from -262 to -205 using the primers p60F11 and p60R3 incorporating an XbaI site. The fragment XbaI-XbaI was cloned into the plasmid 2x::GUS, to generate the construct 4x::GUS. All the oligonucleotide sequences are reported in Table 1. PCR products were sequenced and the correct orientation of the fragment into the final vector was verified by restriction.
Table 1.
Sequence of oligonucleotides used in this study
| Name | Sequence |
|---|---|
| p60F1 | AAGCTTCACAAGGACACAAGGACA |
| p60 F2b | AAGCTTCAAGTTGCAGTGAATGA |
| p60F8b | AAGCTTTAACGAGCTCCTTTTATGG |
| p60F9 | AAGCTTCCATTTATGAGTTGATTATCA |
| p60F3 | AAGCTTCGTGTGGAGATCAACAT |
| p60F5 | AAGCTTGCAGAGTGACTCGTGA |
| p60R5 | TCTCGGATCCTCTAGATCTCTCTG |
| p60R6 | TCTAGAGAAGAACTTTTAAATCTGC |
| p60R7 | TCTAGAAAATCTGCTTTTTCTTGAC |
| p60R5b | AAGCTTCTTTTCCATTAACTATTTTG |
| p60F11 | TCTAGACGTGTGGAGATCAACAT |
| p60R3 | TCTAGACTTTTCCATTAACTATTTTG |
| 35SXba | TCTAGACAAGACCCTTCCTC |
| 35SBam | GGATCCTCCTCTCCAAATGA |
| mp60DOF1F1 | AGTTAATGGcgcgaGCAGAGTGACTCGTGA |
| mp60DOF2F1 | TGGCAGATCCcgcgaAGGTTGTCAAGAAAA |
| mp60DOF3F2 | TGTCAAGAcgcgaCAGATTTAAAAGTTCTT |
| mp60DOF4F2 | CAAGAAAAAGCAGATTTcgcgaTTCTTC |
| mp60WRKYF1 | AAGCTTCGTGTGGAGATCAACATATCTTCGTTAATTGAaTACGCAAAATA |
| GUSRTF1 | TACGGCAAAGTGTGGGTCAATAATCA |
| GUSRTR1 | CAGGTGTTCGGCGTGGTGTAGAG |
| ATACT2F | TGCTTCTCCATTTGTTTGTTTC |
| ATACT2R | GGCATCAATTCGATCACTCA |
| qRT-MYB60-F | CATGAAGATGGTGATCATGAGG |
| qRT-MYB60-R | TTCCATTTGACCCCCAGTAG |
| PP2a-F | CAGCAACGAATTGTGTTTGG |
| PP2a-R | AAATACGCCCAACGAACAAA |
Italic and lower case letters indicate restriction and mutagenised sites, respectively
Site-directed mutagenesis analysis
Base mutations of the different DOF sites were generated using the megaprimer method [42]. For the mutagenised versions of the AtMYB60 minimal promoter different megaprimers were PCR amplified from plasmid p1.3-2.2::GUS, using as forward primers mp60DOF1F1, mp60DOF2F1, mp60DOF3F2 and mp60DOF4F2 and the single reverse primer p60R5. The megaprimers were gel purified and used in a second PCR reaction on plasmid p1.3-2.2::GUS with the primer p60F3. The PCR products were cloned into pCR4-TOPO and sequenced before cloning into pBI101.3 vector using the restriction sites HindIII and BamHI to generate the following constructs: mDOF1::GUS, mDOF2::GUS, mDOF3::GUS, mDOF4::GUS. To generate multiple mutagenised sites the templates for the second PCR amplification were plasmids already carrying a first mutagenised DOF site. In the case of the preparation of the construct mW::GUS the megaprimer method was not necessary, as the site to mutagenise is in a position 5' terminal into the minimal promoter and a single PCR reaction was performed with primers mp60WRKYF1 and p60R5, the PCR product was then cloned with the procedure already described.
All the oligonucleotide sequences are reported in Table 1.
Arabidopsis transformation and growth conditions
Wild-type Columbia (Col-0) plants were transformed using the Agrobacterium tumefaciens strain GV3101 carrying the constructs described above with the floral dip method [43]. Transformed lines were selected on kanamycin and single-insertion lines were selected for further analyses. Analyses of transgenic lines were performed on T2 or on homozygous T3 plants grown under long-day conditions (16 hr light; 8 hr darkat 100 μmol m-2 sec-1) at 22°C in a growth chamber. Seeds were germinated in Petri dishes containing Murashige and Skoog medium, 1% w/v sucrose and 0.8% w/v agar for seedling analysis or directly on soil for adult plant organ analysis. The ABA treatment was performed as previously described [19].
GUS activity assays and histochemical staining
For detection of GUS activity, tissues were fixed for 2 h in 90% (v/v) acetone at -20°C, incubated for 16-48 hours, at 37°C, in 0.05% (w/v) X-glucoronic acid, 0.1% (v/v) Triton X-100, and 0.5mM ferrocyanidine in 50 mM phosphate buffer (pH 7) and subsequently cleared in 70% (v/v) ethanol. Seedlings and flowers were cleared with a chloral hydrate:glycerol:water solution (8:1:2, v/v). Samples were examined using a Leica M205 FA stereomicroscope (Leica Microsystems GmbH Wetzlar, Germany) and a Zeiss Axiophot D1 microscope (Carl Zeiss MicroImaging, LLC Thornwood, New York, USA). Stereomicroscope images were recorded using the Leica LAS software version 2.8.1. Microscope images were recorded with an AxioCam MRc5 camera (Zeiss) using the AxioVision program (version 5.0).
Quantification of mRNA expression
RNA isolation, reverse transcription, qRT-PCR reactions and data analysis were performed as previously described [30]. GUS expression was analysed using primers GUSRT-F1 and GUSRT-R1, ACTIN2 gene (primers ATACT2F, ATACT2R) was used as a reference for normalization. AtMYB60 expression in different cdf mutants was analysed using primers qRT-MYB60-F and qRT-MYB60-R. PP2A gene, corresponding to At1g13320 (primers PP2a-F and PP2a-R) was used as a reference for normalization [44]. All primer sequences are reported in Table 1.
Authors' contributions
EC carried out the construction of promoter-reporter plasmids, plant transformation and drafted the manuscript. EC, AA and LC did transgenic Arabidopsis analysis. FF carried out cdf mutant analysis. CT, MG, and GC conceived the study, participated in its design and coordination. MG, LC and FF helped to draft the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Analysis of GUS activity in seeds at different developmental stages in 1,307::GUS line. A: open silique showing signal only in stomata and not in developing seeds. B: mature-green-stage seed (13 DAP). C: a 24 h imbibed seed. D: embryo isolated from a 24 h imbibed seed. The same results were obtained in all transgenic lines described in Figure 2. Scale bars represent 0.1 mm.
Relative expression of the AtMYB60 gene in the different cdf mutants. cdf1-R is an RNAi line ([29]). The other single and multiple mutants have been previously described ([30]). The PP2a (At1g13320) gene was used as a control [44].
Contributor Information
Eleonora Cominelli, Email: cominelli@ibba.cnr.it.
Massimo Galbiati, Email: massimo.galbiati@unimi.it.
Alessandra Albertini, Email: alessandra.albertini@unimi.it.
Fabio Fornara, Email: fabio.fornara@unimi.it.
Lucio Conti, Email: lucio.conti@unimi.it.
George Coupland, Email: coupland@mpipz.mpg.de.
Chiara Tonelli, Email: chiara.tonelli@unimi.it.
Acknowledgements
This work was supported by the Italian "Progetto AGER, bando Viticoltura da Vino" (SERRES 2010-2015), by the BIOGESTECA 15083/RCC project, funded by Regione Lombardia and by the AGRISOST project, funded by Fondazione Umberto Veronesi per il Progresso delle Scienze, Milano, Italy.
References
- Kim TH, Böhmer M, Hu H, Nishimura N, Schroeder JI. Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol. 2010;61:13.11–13.31. doi: 10.1146/annurev-arplant-042809-112226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder JI, Kwak JM, Allen GJ. Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature. 2001;410:327–330. doi: 10.1038/35066500. [DOI] [PubMed] [Google Scholar]
- Gray JE, Holroyd GH, van der Lee FM, Bahrami AR, Sijmons PC, Woodward FI, Schuch W, Hetherington AM. The HIC signalling pathway links CO2 perception to stomatal development. Nature. 2000;408:713–716. doi: 10.1038/35047071. [DOI] [PubMed] [Google Scholar]
- Kopka J, Provart NJ, Muller-Rober B. Potato guard cells respond to drying soil by a complex change in the expression of genes related to carbon metabolism and turgor regulation. Plant J. 1997;11:871–882. doi: 10.1046/j.1365-313X.1997.11040871.x. [DOI] [PubMed] [Google Scholar]
- Muller-Rober B, La Cognata U, Sonnewald U, Willmitzer L. A truncated version of an ADP-glucose pyrophosphorylase promoter from potato specifies guard cell-selective expression in transgenic plants. Plant Cell. 1994;6:601–612. [PMC free article] [PubMed] [Google Scholar]
- Nakamura RL, McKendree WL, Hirsch RE, Sedbrook JC, Gaber RF, Sussman MR. Expression of an Arabidopsis potassium channel gene in guard cells. Plant Physiol. 1995;109:371–374. doi: 10.1104/pp.109.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander M, Svensson J, Palva ET, Welin BV. Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Mol Biol. 2001;45:263–279. doi: 10.1023/A:1006469128280. [DOI] [PubMed] [Google Scholar]
- Plesch G, Ehrhardt T, Mueller-Roeber B. Involvement of TAAAG elements suggests a role for Dof transcription factors in guard cell-specific gene expression. Plant J. 2001;28:455–464. doi: 10.1046/j.1365-313x.2001.01166.x. [DOI] [PubMed] [Google Scholar]
- Terryn N, Arias MB, Engler G, Tire C, Villarroel R, Van Montagu M, Inze D. rha1, a gene encoding a small GTP binding protein from Arabidopsis, is expressed primarily in developing guard cells. Plant Cell. 1993;5:1761–1769. doi: 10.1105/tpc.5.12.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbiati M, Simoni L, Pavesi G, Cominelli E, Francia P, Vavasseur A, Nelson T, Bevan M, Tonelli C. Gene trap lines identify Arabidopsis genes expressed in stomatal guard cells. Plant J. 2008;53:750–762. doi: 10.1111/j.1365-313X.2007.03371.x. [DOI] [PubMed] [Google Scholar]
- Gardner MJ, Baker AJ, Assie JM, Poethig RS, Haseloff JP, Webb AA. GAL4 GFP enhancer trap lines for analysis of stomatal guard cell development and gene expression. J Exp Bot. 2009;60:213–226. doi: 10.1093/jxb/ern292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesch G, Kamann E, Mueller-Roeber B. Cloning of regulatory sequences mediating guard-cell-specific gene expression. Gene. 2000;249:83–89. doi: 10.1016/S0378-1119(00)00150-5. [DOI] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI. Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell. 2004;16:596–615. doi: 10.1105/tpc.019000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Costa A, Leonhardt N, Siegel RS, Schroeder JI. Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods. 2008;4:6. doi: 10.1186/1746-4811-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Zhang W, Stanley BA, Assmann SM. Functional proteomics of Arabidopsis thaliana guard cells uncovers new stomatal signaling pathways. Plant Cell. 2008;20:3210–3226. doi: 10.1105/tpc.108.063263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RS, Pandey S, Li S, Gookin TE, Zhao Z, Albert R, Assmann SM. Common and unique elements of the ABA-regulated transcriptome of Arabidopsis guard cells. BMC Genomics. 2011;12:216. doi: 10.1186/1471-2164-12-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husebye H, Chadchawan S, Winge P, Thangstad OP, Bones AM. Guard cell- and phloem idioblast-specific expression of thioglucoside glucohydrolase 1 (myrosinase) in Arabidopsis. Plant Physiol. 2002;128:1180–1188. doi: 10.1104/pp.010925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang YK, Dubos C, Dodd IC, Holroyd GH, Hetherington AM, Campbell MM. AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr Biol. 2005;15:1201–1206. doi: 10.1016/j.cub.2005.06.041. [DOI] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol. 2005;15:1196–1200. doi: 10.1016/j.cub.2005.05.048. [DOI] [PubMed] [Google Scholar]
- Francia P, Simoni L, Cominelli E, Tonelli C, Galbiati M. Gene trap-based identification of a guard cell promoter in Arabidopsis. Plant Signal Behav. 2008;3:684–686. doi: 10.4161/psb.3.9.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy R, Grob H, Weder B, Green P, Klein M, Frelet-Barrand A, Schjoerring JK, Brearley C, Martinoia E. The Arabidopsis ATP-binding cassette protein AtMRP5/AtABCC5 is a high affinity inositol hexakisphosphate transporter involved in guard cell signaling and phytate storage. J Biol Chem. 2009;284:33614–33622. doi: 10.1074/jbc.M109.030247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Mumm P, Imes D, Endler A, Weder B, Al-Rasheid KA, Geiger D, Marten I, Martinoia E, Hedrich R. AtALMT12 represents an R-type anion channel required for stomatal movement in Arabidopsis guard cells. Plant J. 2010;63:1054–1062. doi: 10.1111/j.1365-313X.2010.04302.x. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol. 2004;45:386–391. doi: 10.1093/pcp/pch055. [DOI] [PubMed] [Google Scholar]
- PLACE. A Database of Plant Cis-acting Regulatory DNA Elements. http://www.dna.affrc.go.jp/PLACE/ [DOI] [PMC free article] [PubMed]
- Yanagisawa S, Schmidt RJ. Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 1999;17:209–214. doi: 10.1046/j.1365-313X.1999.00363.x. [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- Yu D, Chen C, Chen Z. Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell. 2001;13:1527–1540. doi: 10.1105/TPC.010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velten J, Morey KJ, Cazzonelli CI. Plant viral intergenic DNA sequence repeats with transcription enhancing activity. Virol J. 2005;2:16. doi: 10.1186/1743-422X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science. 2005;309:293–297. doi: 10.1126/science.1110586. [DOI] [PubMed] [Google Scholar]
- Fornara F, Panigrahi KC, Gissot L, Sauerbrunn N, Ruhl M, Jarillo JA, Coupland G. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell. 2009;17:75–86. doi: 10.1016/j.devcel.2009.06.015. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. The Dof family of plant transcription factors. Trends Plant Sci. 2002;7:555–560. doi: 10.1016/S1360-1385(02)02362-2. [DOI] [PubMed] [Google Scholar]
- Galbiati M, Matus JT, Francia P, Rusconi F, Canon P, Medina C, Conti L, Cominelli E, Tonelli C, Arce-Johnson P. The grapevine guard cell-related VvMYB60 transcription factor is involved in the regulation of stomatal activity and is differentially expressed in response to ABA and osmotic stress. BMC Plant Biol. 2011;11:142. doi: 10.1186/1471-2229-11-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An "Electronic Fluorescent Pictograph" browser for exploring and analyzing large-scale biological data sets. PLoS One. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis eFP Browser. http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi
- Oh JE, Kwon Y, Kim JH, Noh H, Hong SW, Lee H. A dual role for MYB60 in stomatal regulation and root growth of Arabidopsis thaliana under drought stress. Plant Mol Biol. 2011;77:91–103. doi: 10.1007/s11103-011-9796-7. [DOI] [PubMed] [Google Scholar]
- Schauer SE, Schluter PM, Baskar R, Gheyselinck J, Bolanos A, Curtis MD, Grossniklaus U. Intronic regulatory elements determine the divergent expression patterns of AGAMOUS-LIKE6 subfamily members in Arabidopsis. Plant J. 2009;59:987–1000. doi: 10.1111/j.1365-313X.2009.03928.x. [DOI] [PubMed] [Google Scholar]
- Seki M, Ishida J, Narusaka M, Fujita M, Nanjo T, Umezawa T, Kamiya A, Nakajima M, Enju A, Sakurai T. et al. Monitoring the expression pattern of around 7,000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct Integr Genomics. 2002;2:282–291. doi: 10.1007/s10142-002-0070-6. [DOI] [PubMed] [Google Scholar]
- Zeller G, Henz SR, Widmer CK, Sachsenberg T, Ratsch G, Weigel D, Laubinger S. Stress-induced changes in the Arabidopsis thaliana transcriptome analyzed using whole-genome tiling arrays. Plant J. 2009;58:1068–1082. doi: 10.1111/j.1365-313X.2009.03835.x. [DOI] [PubMed] [Google Scholar]
- Choudhury A, Lahiri A. Comparative analysis of abscisic acid-regulated transcriptomes in Arabidopsis. Plant Biol. 2011;13:28–35. doi: 10.1111/j.1438-8677.2010.00340.x. [DOI] [PubMed] [Google Scholar]
- Matsui A, Ishida J, Morosawa T, Mochizuki Y, Kaminuma E, Endo TA, Okamoto M, Nambara E, Nakajima M, Kawashima M. et al. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 2008;49:1135–1149. doi: 10.1093/pcp/pcn101. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005;10:88–94. doi: 10.1016/j.tplants.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Kammann M, Laufs J, Schell J, Gronenborn B. Rapid insertional mutagenesis of DNA by polymerase chain reaction (PCR) Nucleic Acids Res. 1989;17:5404. doi: 10.1093/nar/17.13.5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Hong SM, Bahn SC, Lyu A, Jung HS, Ahn JH. Identification and testing of superior reference genes for a starting pool of transcript normalization in Arabidopsis. Plant Cell Physiol. 2010;51:1694–1706. doi: 10.1093/pcp/pcq128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of GUS activity in seeds at different developmental stages in 1,307::GUS line. A: open silique showing signal only in stomata and not in developing seeds. B: mature-green-stage seed (13 DAP). C: a 24 h imbibed seed. D: embryo isolated from a 24 h imbibed seed. The same results were obtained in all transgenic lines described in Figure 2. Scale bars represent 0.1 mm.
Relative expression of the AtMYB60 gene in the different cdf mutants. cdf1-R is an RNAi line ([29]). The other single and multiple mutants have been previously described ([30]). The PP2a (At1g13320) gene was used as a control [44].