Abstract
Because mutations are inevitable, the genome of each cell in a multicellular organism becomes unique and therefore encodes a record of its ancestry. Here we couple arbitrary single primer PCR with “next generation” DNA sequencing in order to catalog mutations and deconvolve the phylogeny of cultured mouse cells. This study helps pave the way toward construction of retrospective cell fate maps based on mutations accumulating in genomes of somatic cells.
Cells accumulate mutations. Daughter cells inherit these mutations and acquire their own, such that genomes record mitotic history. If genomes from single cells could be sequenced, then it should be possible to infer cellular ancestry1. We and others have shown that mutational hotspots, consisting of repetitive DNA, can be employed to trace cellular lineage1–7. Related approaches track DNA methylation8 and mitochondrial DNA mutations9,10. To obtain sufficient DNA quantities, it has been necessary to use many cells1,2, clonally expand single cells ex vivo6,7, or perform whole genome amplification (WGA)2,3. However, scrutinizing our own2 and other datasets3 we found WGA to be unreliable. WGA was used recently to deep-sequence individual tumor cells and catalog copy number variants informative for lineage tracing11, but it remains uncertain if this will work for normal tissues lacking large-scale, cancer-specific genomic alterations.
The C. elegans cell fate map has yielded remarkable insight into developmental biology12. Fate maps for organisms containing massively greater cell numbers could similarly prove useful. Here we employ arbitrary single primer PCR and next generation sequencing to analyze DNA from limited starting material and identify base substitution mutations at randomly sampled positions of the genome, in order to decode lineages of cultured mouse cells.
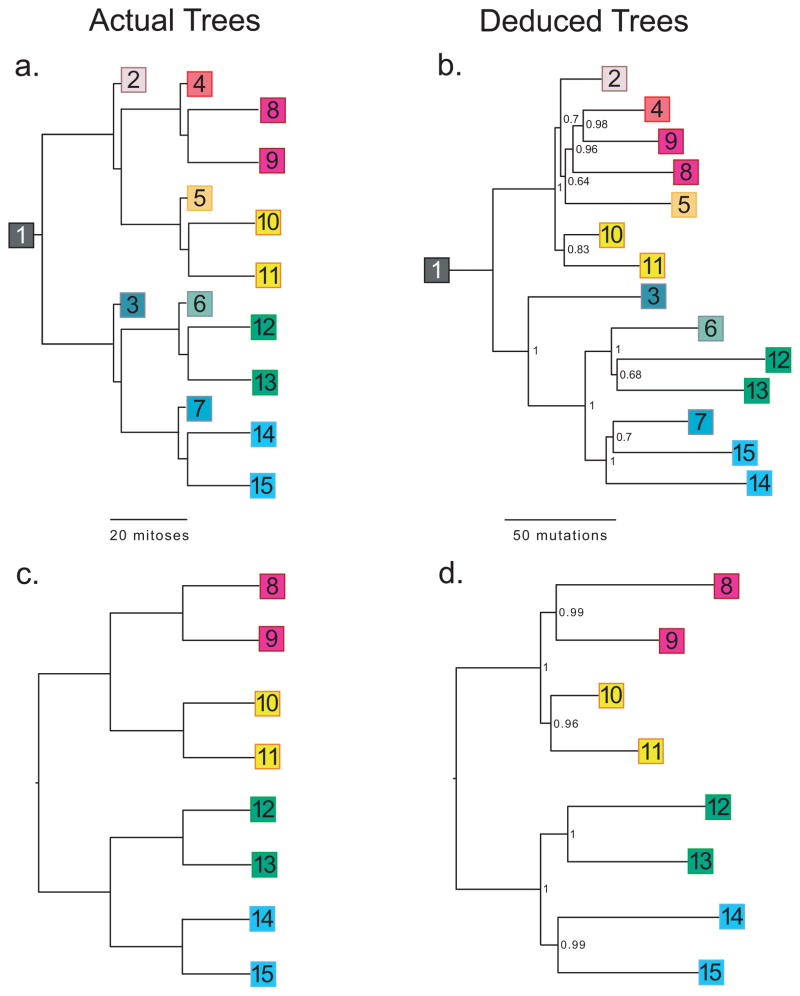
We devised test systems where we passaged cells through defined orders resembling trees (Fig. 1a). Noting an estimated 40 cell doublings between fertilization and birth in mice1, we cultured a single fibroblast for ~20 doublings on a Petri dish, and then isolated single cells from which we seeded a second tier of dishes, and so on, repeating this process 4 times in total. Starting from one cell, over 89 days, we generated 15 dishes. To increase mutations, the starting cell came from a cancer-prone “mutator” mouse deficient for Mlh1 mismatch DNA repair and DNA polymerase delta proofreading activities13. Although sterile, these mice develop normally.
Figure 1.
Cell lineages. (a) A single mouse fibroblast was seeded onto a Petri dish. After approximately 20 doublings, a single cell was used to seed each of the next tier of dishes, and so on. (b) Lineage reconstructed from 592 mutations identified from sequencing of single primer arbitrary PCR products from DNA extracted from all 15 dishes. (c) Simplified lineage tree, similar to Fig. 1a, but showing only the terminal nodes. (d) Lineage reconstructed from the 667 mutations present in only the terminal nodes. Numbers in deduced trees are Bayesian posterior probabilities.
Mutations occurring within genes could alter cell growth and prove problematic for inferring lineage. We therefore chose “arbitrarily-primed PCR”14 to sample random segments of the genome. If the genome’s composition were random, then PCR employing a single oligonucleotide of arbitrary-yet-defined sequence should predictably amplify a portion of the genome solely as functions of primer length and cycle extension time. However, the genome is not random, so we modeled and experimentally evaluated a range of primer lengths and sequences, in order to optimize next generation sequencing capacity. Our aim was to extract maximal sequence from all 15 sampled nodes of the tree in a single run of the ABI SOLiD platform. In silico, we evaluated arbitrary sequence oligonucleotides to predict those for which PCR would yield ~10 Mb of total sequence (with a maximum amplicon of 2 Kb, reasoning length was controllable by cycle extension time). From several sequences identified through modeling and validated by pilot deep sequencing, we selected a 10-mer oligonucleotide (5′-GGGGGGGGAG-3′). Gel electrophoresis reveals a similarly appearing smear for PCR samples from all 15 nodes of the tree (Supplementary Fig. 1). PCR initially employed 50 ng of DNA (≈8,730 cells); however, serial dilution demonstrates equivalent results with template corresponding to ~100 cells (Supplementary Fig. 2).
We sequenced PCR products from each sample using 15/16th of the capacity (dual 8-segment flow cells) of an ABI SOLiD sequencer. 23 Gb of sequence was generated, of which 72% mapped to the reference genome.
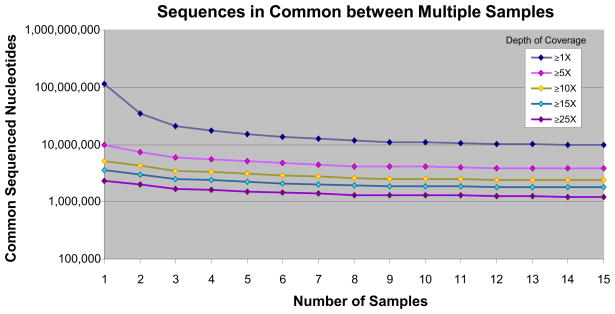
At depth ≥1×, a mean 1.6 Gb of mapped sequence was generated per sample, of which a mean 116 Mb corresponded to unique basepairs. At depth ≥1×, 9.7 Mb (0.37% of the genome) were sequenced commonly across all 15 samples, at mean depth of coverage of 89×.
To determine sample-to-sample consistency, we plotted unique nucleotides sequenced common to all samples as numbers of samples increased, from 1–15 (Fig. 2). Curves vary only over a small range and level off as samples are added. We uploaded all sequence data to the UCSC Genome Browser. Read-depth histograms reveal consistent amplification from one sample to the next (representative sample, Fig. 3), as confirmed by Pearson correlation coefficients for pairwise comparisons (Supplementary Table 1). Arbitrary PCR across whole chromosomes also shows sample-to-sample consistency (Supplementary Fig. 3). We observe that arbitrary PCR reproducibly amplifies sequences common to all samples, but each sample has variable representation at low depths of coverage of unique sequences arising from non-specific priming. Consequently, the majority (78%) of mapped reads is present in all 15 samples, given high depths of coverage at commonly sequenced positions.
Figure 2.
Sample-to-sample reproducibility. Total quantity of unique genomic sequence shared among all samples, at various minimum depths of coverage, as number of samples increases from 1 to 15 (for example, at ≥1× depth of coverage, there are ~10,000,000 unique genomic positions that are sequenced in common to all 15 samples).
Figure 3.
Genome browser snapshot. Shown are histogram plots of an ~3 Kb amplicon on chromosome 1 corresponding to mapped reads from arbitrary PCR for the first 6 samples (bottom six graphs). Other tracks include (from top to bottom): known genes (coverage overlaps with exons and introns of Il19), position of identified mutations (vertical red line at right end of plots) that are found in at least one sample, minimum fold-coverage common to all 15 samples, and mean fold-coverage for all 15 samples. Note that PCR is highly consistent from one sample to the next. Also note low depth-of-coverage reads unique to each sample (flanking the amplicon).
When sequence variation is detected between samples, it may correspond to PCR or sequencing errors or to mutations not transmitted to progeny because they occurred in a subpopulation of cells during clonal expansion, rather than being present when the cell was used to seed the dish. In consideration of these complications we devised a Bayesian method for mutation detection, based on generalizing a standard approach to the case of multiple DNA sequence samples from a common population15, in which mutation analysis is performed collectively on all related samples.
We identified 592 mutations, of which 315 (53%) demonstrate segregation consistent with the known tree (Supplementary Table 2). Variants whose segregation was inconsistent with the known tree could represent PCR and/or sequencing artifacts or could be mutations for which sequencing depth was insufficient to permit detection.
We then attempted to reconstruct the experimental phylogeny from all 592 mutations (including the inconsistent ones). With all 15 samples, using either Bayesian (Fig. 1b) or neighbor-joining approaches (Supplementary Fig. 4a), inferred phylogenies are 79% and 75.7% identical, respectively, to the known tree (quantified per method of Nye16). The Bayesian reconstruction contains 3 errors. Samples 4, 5, and 7 should each be one branch closer to the root. The number of mutations supporting each bifurcation is stochastically distributed (Supplementary Fig. 5); no informative mutations appear to distinguish dishes 5 and 2, which probably contributes to the error.
In adapting this approach toward constructing fate maps from cells extracted from an individual organism, only terminal nodes will actually be available (Fig. 1c). Intermediate nodes, corresponding to progenitor cells for which only daughter cells persist in mature tissues, no longer exist. As a test, we determined if information from just the 8 terminal cells was sufficient to reconstruct phylogeny. We identified 667 mutations, of which 520 (78%) demonstrate segregation consistent with the known tree (Supplementary Table 2). From these mutations, we perfectly reconstructed the tree’s known lineage using either Bayesian (Fig. 1d) or neighbor-joining methods (Supplementary Fig. 4b), p = 7.4×10−6, noting 135,135 possible 8-cell histories. Even though this represents a more challenging problem, we achieve greater accuracy using only terminal nodes because there is more common sequence and greater mitotic distance, with a greater likelihood of mutation, separating sampled nodes.
By counting mutations whose segregation is consistent with the known lineage, we calculated a rate of 10−6 mutations/nucleotide/division, compared with a mutation rate of 1.4×10−6 measured through Luria–Delbrück fluctuation analysis (Supplementary Table 3). In contrast, the reported rate for the DNA polymerase used for PCR is lower, at 4×10−7 errors/nucleotide (http://www.genomics.agilent.com/files/Manual/600385.pdf). Even still, particular mutations introduced by the polymerase (or during sequencing) are expected to occur randomly and not be present in more than one sample, and thus should not influence reconstruction of the lineage.
We performed modeling studies where we sought the minimal number of segregating mutations capable of accurately reconstructing lineage. Simulations imply that trees can be recovered from as few as 40 mutations. To evaluate for error tolerance, we introduced 300 mutations at random positions. Injection of errors did not alter tree topology, because of the unlikely occurrence of random mutations at the same position in multiple samples. We evaluated how mutation rate and number of nucleotides targeted for sequencing influence ability to resolve cells separated by given numbers of cell divisions (Supplementary Table 4). We found the only critical parameter is total number of mutations. This number, N, is given by (r-1)μdt, where r = nodes of the tree, μ = mutation rate, d = number of doublings, and t = number of nucleotides sequenced. For the actual mutation rate (10−6) and target size (~10 Mb at ≥1× coverage common to all samples), we infer capacity to resolve cells separated by as few as 4 divisions. Ability to resolve cell lineage scales linearly with both mutation rate and target size. Therefore, to dissect lineage in organisms with lower mutation rates, one can simply increase DNA sequence target size (by adding additional primers, shortening primer length, or increasing cycle extension time).
As DNA sequencing technology evolves, it will become economically feasible to sequence progressively larger portions of the genome from greater numbers of single cells. Until then, we show here that from just a few cells (or single cells briefly clonally expanded ex vivo—which can be facilitated with a conditional immortalizing oncogene7), mutations found at arbitrarily sampled genomic positions yield information sufficient for inferring cell lineage. Complementary approaches are based on in vivo image and retrospective clonal analysis17. We propose that such studies could provide a better understanding of how cells divide during development and differentiation and in the formation of cancer.
METHODS
Methods and any associated references are available in the online version of the paper.
METHODS
Cell Culture
A spontaneously immortalized embryonic fibroblast cell line was derived from a female Mlh1Δ/ Δ/Pold1e/+ mouse. The mouse was obtained from a cross of heterozygotes for Mlh1Δ (gift of M. Liskay18)and Polde (maintained in the C57BL/6J strain). Cells were grown in DMEM plus 10% FBS with penicillin G (100 U/ml) plus streptomycin (100 μg/ml) at 37°C with 5% CO2 in a humidified incubator. Single cells were isolated by limiting dilution and clonally expanded on a 6 cm dish until confluent (an average of 19 divisions (range 18.1–20.1), calculated as log2 of total cells, as counted with a hemocytometer). ¾ of the cells from each plate were harvested for DNA extraction using the 5prime ArchivePure DNA Cell/Tissue kit. Of the remaining ¼, a small aliquot was again plated at limiting dilution to obtain single cells for clonally isolated subculture. The remainder was preserved in liquid nitrogen. This process was repeated until 4 passages were obtained.
Fluctuation Analysis
Rates of spontaneous mutation to ouabain resistance were measured by fluctuation experiments as described previously19. Mutation rates (mutants per cell division) were calculated from the number of ouabain-resistant colonies in each replica by the maximum likelihood method20 using the newtonLDPlating function in Salvador 2.3 software to estimate m21. 95% Confidence intervals (indicated in parentheses, Supplementary Table 3) were calculated in Salvador 2.3 using the CILDPlating function. Two independent fluctuation experiments were conducted and analyzed individually. These experiments had similar Nt values. Therefore, the raw fluctuation data were combined and analyzed as one large data set (right column, Supplementary Table 3) to obtain the best estimate of mutation rate (bottom right in bold, Supplementary Table 3). Per-base-pair mutation rates were calculated from the phenotypic ouabain-resistance rates assuming an effective target size (τ) of 30 base pairs. This τ value was estimated as follows. Base substitution mutations in any one of sixteen codons in the Na,K-ATPase α1 gene (Atp1a1) are known to confer genetically dominant resistance to μM concentrations of ouabain in human cells22. Mouse cells, however, are naturally resistant to μM concentrations of ouabain due to differences at 2 of these 16 codons (Q111R and N122D)23,24. Our fluctuation assays were conducted with 2 mM ouabain, conditions expected to only detect mutations that confer exceptionally high ouabain resistance. We estimate the target size to be ~5 base pairs per allele, corresponding to 2 Atp1a1 codons (D121 and T797) known to effect >50-fold ouabain resistance when mutated22,25. The mouse fibroblast cell line used in our experiments exhibited a near-hexaploid karyotype (data not shown). Therefore, τ = 5 base pairs per allele × 6 alleles = 30 base pairs. The combined fluctuation data yielded a mutation rate of 4.2×10−5 ouabain-resistant mutants per cell division. This phenotypic rate corresponds to a per-base-pair rate of 4.2 x 10−5/30 base pairs = 1.4×10−6.
Primer Design
Arbitrary sequences of length 8, 9, or 10 were generated randomly, with the exception that the 3′ base was always either G or C (with equal probability). 23,000 sequences were screened against the mouse NCBI37/mm9 build (http://www.ncbi.nlm.nih.gov/mapview/stats/BuildStats.cgi?taxid=10090&build=37&ver=1) derived from the C57BL/6J strain, using 2 Perl programs (Supplementary Information). The software calculated the frequency of primer binding, the distribution of lengths for predicted amplicons, and the expected number of amplified nucleotides contained within a specified range of amplified product lengths. Potential primer candidates were picked based on a goal of amplifying 5×10−5 to 3×10−4 fraction of the mouse genome in amplicons ≤2 Kb in length. Next, the potential candidates were screened based on stability, G·C basepair content, predicted melting temperature, and the possibility of dimer, hairpin, and repetitive sequence run formation using Clone Manager Professional Suite 6.0 (Sci-Ed Software). Based on those data, a total 105 sequences were selected for experimental testing by PCR using C57BL/6J mouse genomic DNA as the template and visualization on a 1.5% TBE agarose gel with ethidium bromide for evaluation. Of the 105 sequences tested, 7 were 8-mers, 65 were 9-mers, and 33 were 10-mers. Amplicons were seen in 14% of 8-mers, 48% of 9-mers, and 73% of 10-mers. The 5 most promising candidates (3 9-mers and 2 10-mers) plus a combination of 2 promising candidates were tested by sequencing the resulting amplicons on the Roche 454 genomic sequencing platform in duplicate using 12 multiplex identifier adapters. Our amplification goal was ~500,000 nucleotides at ≥30× coverage. The best primer sequence was the 10-mer: 5′-GGGGGGGGAG-3′ which mapped 501 Kb in common between the duplicates at ≥1× coverage but only 23.8 Kb in common at ≥30× coverage. We switched sequencing platforms to the ABI SOLiD platform given higher throughput and improved accuracy which allowed us to reduce our fold-coverage goal to ≥15×. The 4 most promising primer candidates were then tested on the SOLiD system version 2.0 in duplicates on one flow cell divided into 8 segments. The best primer sequence again was the same 10-mer: 5′-GGGGGGGGAG-3′ which mapped 7.11 Gb in common between the duplicates at ≥1× coverage and 621 Kb in common at ≥15× coverage, achieving the design specification.
PCR
50 ng of DNA from each sample was mixed with 2 μM of primer, 250 μM of each dNTP, and 5 units of Agilent PfuUltra High-Fidelity DNA Polymerase AD in a final volume of 50 μl of manufacturer-supplied buffer. Following initial denaturation at 95° C for 2 min., samples underwent 50 cycles of denaturation at 95 °C for 30 sec., annealing at 25° C for 30 sec., extension at 72° C for 3 min., and final extension at 72° C for 15 min., followed by purification using QIAGEN QIAquick Nucleotide Removal Kit. In order to diminish contributions from PCR-related errors, 8 independent PCR reactions were performed per sample and then combined (~10 μg DNA total) for sequencing.
DNA Sequencing
Massively parallel sequencing was performed using the Applied Biosystems SOLiD (version 3+, read length = 50 nucleotides) platform utilizing the manufacturer-supplied reagents and protocols for fragment analysis, with 5 μg DNA starting material per sample. Outputted. csfasta and .QV.qual files were mapped to mouse reference genome build NCBI37/mm9 using manufacturer’s supplied Bioscope 1.0.1 software, with default parameters. SAMtools software26 was used to prepare .pileup files. Data files are available at http://horwitz.genetics.washington.edu/trees/. Mapped sequences for all 15 samples can be viewed as custom tracks in the UCSC Genome Browser. Chromosomes 1–10 are viewable at http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=Horwitz%20Lab&hgS_otherUserSessionName=CH1-10 Chromosomes 11–19 and the X chromosome are viewable at http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=Horwitz%20Lab&hgS_otherUserSessionName=CH11-19X. Custom tracks can also be accessed at http://horwitz.genetics.washington.edu/trees/tracks/.
Mutation Detection
We wrote a Perl program (Supplementary Information) to compute the most likely genotypes at each locus covered by all of the samples, given a collection of sequencing reads from multiple samples of a common population. The inputs to the program are the reads, read quality values at each locus, and the mapping quality values for each sample. The program assumes that at each locus, some of the samples are homozygous and others are heterozygous, but that all samples have at least one allele in common. Then the program computes the probability that each subset is the set of heterozygotes. The subset which has the largest probability P, is chosen as the heterozygote, and the Phred score Q=10log(1-P) is the quality value of this consensus call. Following Bayes’ formula, the a posteriori probabilities are proportional to the products of the a priori probabilities and the probability of the data, given the subset of heterozygotes. The a priori probability is based on the (unknown) coalescent tree of the samples. All trees are assumed to be equally likely, and the a priori probability depends only on the cardinality of the heterozygote set. We calculate the fraction C(n,k) of clades of size k among all clades of all trees on n nodes. This can be calculated recursively, based on a well-known recursive construction of all tree shapes27. The a priori probability for the empty set (all samples are homozygous) is: 1-het-mut, where het is the heterozygosity of the mouse (assumed 10−3) and mut is an approximation to the combined mutation rate over all branches of the coalescent tree. The a priori probability for the full set (all samples are heterozygous) is het. For any other set, S, the a priori probability is: mut×C(n,k)/nchoosek, where k=#(S). The calculation of the probability of the data, given the set S, is calculated in a manner similar to that of the genotype calling algorithm of Maq software15. Briefly, we form the product of the probabilities for each sample. For samples not in S, this is the product of the error probabilities of all variant reads, while for samples in S, we use the probability nchoosek/2n, where n is the total number of reads and k is the number of variant reads. We additionally performed receiver operating characteristic analysis (Supplementary Fig. 6), employing a read depth cutoff ≥15×, in order to evaluate optimal Phred quality scores.
Phylogenetic Analysis
Bayesian phylogenetic trees were inferred using MrBayes 3.2.128. As previously described in detail6,7, a generalized time reversible DNA substitution model was selected with gamma-distributed rate variation across sites. For both reconstructions, 106 generations were produced and consensus trees were calculated using a burn-in of 2,500 trees (about twice the number required for convergence of the runs). Phylogenies were edited for clarity using FigTree v1.3.1 (A. Rambaut, University of Edinburgh, http://tree.bio.ed.ac.uk/software/figtree/).
Supplementary Material
Acknowledgments
We thank D. Anderson, J. Salk, and L. Loeb for discussion and comments. Supported by NIH grants DP1OD003278 and R01DK078340 (to M.S.H.); R01CA111582 and R01CA098243 (to B.D.P.); T32HL007093 (for C.A.C.); F30AG030316 (to S.J.S.) and T32GM007266 and ARCS Fellowship grants to the University of Washington Medical Scientist Training Program (for S.J.S.).
Footnotes
Author Contributions C.A.C., B.D.P., L.E.H., S.J.S. and M.S.H. designed the experiments. C.A.C., S.J.S, and L.E.H. carried out the experiments. C.A.C., A.K., R.K., and M.S.H. contributed to analyzing the data. M.S.H., with input from other authors, wrote the paper.
Competing Financial Interests The authors declare no competing financial interests.
References
- 1.Frumkin D, Wasserstrom A, Kaplan S, Feige U, Shapiro E. Genomic variability within an organism exposes its cell lineage tree. PLOS Computational Biology. 2005;1:e50. doi: 10.1371/journal.pcbi.0010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salipante SJ, Horwitz MS. Phylogenetic fate mapping. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:5448–5453. doi: 10.1073/pnas.0601265103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frumkin D, et al. Cell lineage analysis of a mouse tumor. Cancer Res. 2008;68:5924–5931. doi: 10.1158/0008-5472.CAN-07-6216. [DOI] [PubMed] [Google Scholar]
- 4.Wasserstrom A, et al. Reconstruction of cell lineage trees in mice. PLoS ONE. 2008;3:e1939. doi: 10.1371/journal.pone.0001939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wasserstrom A, et al. Estimating cell depth from somatic mutations. PLoS Comput Biol. 2008;4:e1000058. doi: 10.1371/journal.pcbi.1000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salipante SJ, Thompson JM, Horwitz MS. Phylogenetic fate mapping: theoretical and experimental studies applied to the development of mouse fibroblasts. Genetics. 2008;178:967–977. doi: 10.1534/genetics.107.081018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salipante SJ, Kas A, McMonagle E, Horwitz MS. Phylogenetic analysis of developmental and postnatal mouse cell lineages. Evol Dev. 2010;12:84–94. doi: 10.1111/j.1525-142X.2009.00393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shibata D, Tavare S. Stem cell chronicles: autobiographies within genomes. Stem Cell Rev. 2007;3:94–103. doi: 10.1007/s12015-007-0022-6. [DOI] [PubMed] [Google Scholar]
- 9.Dasgupta S, et al. Following mitochondrial footprints through a long mucosal path to lung cancer. PLoS One. 2009;4:e6533. doi: 10.1371/journal.pone.0006533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fellous TG, et al. A methodological approach to tracing cell lineage in human epithelial tissues. Stem Cells. 2009;27:1410–1420. doi: 10.1002/stem.67. [DOI] [PubMed] [Google Scholar]
- 11.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sulston JE. Caenorhabditis elegans: the cell lineage and beyond (Nobel lecture) Chembiochem. 2003;4:688–696. doi: 10.1002/cbic.200300577. [DOI] [PubMed] [Google Scholar]
- 13.Preston BD, Albertson TM, Herr AJ. DNA replication fidelity and cancer. Semin Cancer Biol. 2010;20:281–293. doi: 10.1016/j.semcancer.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McClelland M, Welsh J. DNA fingerprinting by arbitrarily primed PCR. PCR Methods Appl. 1994;4:S59–65. doi: 10.1101/gr.4.1.s59. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nye TM, Lio P, Gilks WR. A novel algorithm and web-based tool for comparing two alternative phylogenetic trees. Bioinformatics. 2006;22:117–119. doi: 10.1093/bioinformatics/bti720. [DOI] [PubMed] [Google Scholar]
- 17.Petit AC, Legue E, Nicolas JF. Methods in clonal analysis and applications. Reprod Nutr Dev. 2005;45:321–339. doi: 10.1051/rnd:2005024. [DOI] [PubMed] [Google Scholar]
- 18.Baker SM, et al. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet. 1996;13:336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- 19.Albertson TM, et al. DNA polymerase epsilon and delta proofreading suppress discrete mutator and cancer phenotypes in mice. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:17101–17104. doi: 10.1073/pnas.0907147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng Q. New algorithms for Luria-Delbruck fluctuation analysis. Mathematical biosciences. 2005;196:198–214. doi: 10.1016/j.mbs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 21.Zheng Q. A note on plating efficiency in fluctuation experiments. Mathematical biosciences. 2008;216:150–153. doi: 10.1016/j.mbs.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Croyle ML, Woo AL, Lingrel JB. Extensive random mutagenesis analysis of the Na+/K+-ATPase alpha subunit identifies known and previously unidentified amino acid residues that alter ouabain sensitivity--implications for ouabain binding. European journal of biochemistry/FEBS. 1997;248:488–495. doi: 10.1111/j.1432-1033.1997.00488.x. [DOI] [PubMed] [Google Scholar]
- 23.Fallows D, et al. Chromosome-mediated transfer of the murine Na,K-ATPase alpha subunit confers ouabain resistance. Molecular and cellular biology. 1987;7:2985–2987. doi: 10.1128/mcb.7.8.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price EM, Lingrel JB. Structure-function relationships in the Na,K-ATPase alpha subunit: site-directed mutagenesis of glutamine-111 to arginine and asparagine-122 to aspartic acid generates a ouabain-resistant enzyme. Biochemistry. 1988;27:8400–8408. doi: 10.1021/bi00422a016. [DOI] [PubMed] [Google Scholar]
- 25.Cantley LG, Cunha MJ, Zhou XM. Ouabain-resistant OR6 cells express the murine alpha 1-subunit of the Na,K-ATPase with a T797-I797 substitution. The Journal of biological chemistry. 1994;269:15358–15361. [PubMed] [Google Scholar]
- 26.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Felsenstein J. Inferring Phylogenies. Sinauer Associates; 2004. [Google Scholar]
- 28.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.