Abstract
Explorations of the vaginal microbiota (VMB) began over 150 years ago. Using light microscopy and bacterial cultures the concept of normal versus abnormal microbiotain women began to emerge. The latter became known by the term “bacterial vaginosis” or BV. BV microbiota is dominated by Gardnerella vaginalis and includes a number of anaerobic organisms. In contrast normal flora is dominated various Lactobacilli. BV microbiota is associated with vaginal discharge, poor pregnancy outcomes, pelvic inflammatory disease, post-operative wound infections, and endometritis following elective abortions. Additionally, BV flora predisposes women to infection by HIV as well as other STDs. Application of molecular techniques over the last decade has significantly advanced our understanding of the VMB. It is far more complex than previously recognized and is comprised of many previously unknown organisms in addition to those already identified by culture. Analyses using high-throughput sequencing techniques have revealed unique microbial communities not previously recognized within the older, established vaginal flora categories. These new findings will inform the design of future clinical investigations of the role of the VMB in health and disease.
Introduction
It has long been recognized that humans coexist with complex bacterial communities that are relatively unique to specific niches such as the gastrointestinal tract and the oral cavity. However, the scope of these relationships has only recently been recognized through the application of high throughput sequencing modalities. It is now clear that the microbes closely associated with humans outnumber the total number of human cells by a ratio of more than 10:1 and that, in fact, the total number of genes in the human genome is outnumbered by those in the bacterial microbiome by a factor greater than 1000:1.1 (Note: In this paper the term “microbiome” will be used to refer primarily to the sum total of all bacteria that live in close association with humans and all of their genes. The term “microbiota” will be used primarily to refer to human site-specific bacterial communities as determined by 16s RNA sequencing studies.) These organisms and the communities they form have evolved with us over thousands of years and are critical to our survival. For example, we rely on bacteria for absorption of key nutrients and for conditioning the innate immune response; however, we are just now beginning to understand the complex mechanisms of these mutually beneficial interactions. On the other hand, specific disturbances in our microbiome may result in disease as well, and thus improved knowledge of these relationships is critical to a better understanding of both human health and illness.
Anton Van Leeuwenhoek, the “Father of Microbiology” and the inventor of the microscope, was the first observer of a human bacterial community as he used biofilm scraped from his own teeth for his first experiments. Beginning in the late nineteenth century, his work was further advanced by numerous investigators through the use of ever more sophisticated aerobic and anaerobic culture techniques to explore the human microbiome. Despite this work, scientists recognized relatively early that there likely were members of these communities that could not be cultured ex vivo, and thus the true nature of the human microbiome remained obscure. The discovery that it was possible to amplify bacterial 16S RNA genes directly from natural samples (reviewed in Marrazzo et al2) and use the sequences thus obtained to identify all bacterial species present in such samples resulted in the first accurate descriptions of the human microbiome. Such molecular analyses highlighted the inadequacies of the methods of classic bacteriology, important as they were, to describe the true complexity of the human microbiome accurately. In fact, it is now recognized that, depending on the anatomical location, less than 10% of the organisms comprising the human microbiome are non-cultivatable.3
Recent discoveries have demonstrated the potential of molecular microbiology to reveal heretofore hidden characteristics of the human microbiome that are of great importance to human health. For example, Ley and colleagues discovered significant differences in the colonic microbiota of obese compared to lean study individuals.4 This work was extended to a germ-free mouse model in which it was demonstrated that experimentally induced changes in the gut flora induced weight gain.5 These and similar results demonstrate the insights gained in part through the NIH-funded Human Microbiome Project that began in 2007. As stated in the “marker paper” describing the project, “The goals of the HMP are to demonstrate the feasibility of characterizing the human microbiome well enough to enable study of the variation in the human microbiome (with population, geno-type, disease, age, nutrition, medication, and environment) and its influence on disease, while providing both a standardized data resource and technological developments to enable such studies to be undertaken broadly in the scientific community. The HMP is a limited effort per se, but has the ultimate objective of creating broad opportunities to improve human health through monitoring or manipulating the human microbiome.”6
One of the original HMP-targeted sites for characterization of the microbiota was the vagina. This site was selected in large part because of the extensive body of data developed over the last 40 years using the techniques of classical microbiology, which clearly established the effect of differing vaginal microbial communities on women’s health. The goal of this review is to provide a brief summary of the early work based on the classic microbiologic techniques that provide a context for the molecular work published over the last 10 years, which will then be summarized in the following section of the review.
Early studies of vaginal microbiota based on culture and microscopy
In 1892 Doderlein described an organism that he isolated from a vaginal specimen of normal pregnant women. This organism, which he originally named Doderlein’s bacillus after himself, was later renamed Lactobacillus.7 At the end of the nineteenth century, Menge and Kronig first described the isolation of anaerobic organisms in addition to Lactobacillus from the vagina.8 In 1923 Curtis described a vaginal discharge syndrome in women that he termed the “white discharge” syndrome.9 Using culture techniques, he associated this syndrome with black-pigmented anaerobes, curved anaerobic motile rods, anaerobic cocci, and Gram-variable diphtheroidal rods. He also noted a relative dearth of Doderlein’s bacillus in women with this syndrome. Schroeder confirmed and extended this work in the 1920s.10 Thus, over 80 years ago it was known that significant shifts in the vaginal microbiota were associated with a symptomatic vaginal discharge syndrome in women. Despite this work, many investigators interested in women’s health continued to believe that this syndrome must in some way be caused by a single organism. These beliefs provided fertile ground for the work of Gardner and Duke, who first described the strong association of a Gram-variable coccobacillus initially named Haemophilus vaginalis (later renamed Gardnerella vaginalis) with nonspecific vaginitis (the name then used for Curtis’s white-discharge syndrome).11 They believed that this organism was the cause of the syndrome despite the fact that direct inoculation of cultivated G. vaginalis did not reproduce the disease. Clinicians and investigators over the next 25 years nearly completely forgot about the earlier work, and in fact, the term “Gardnerella vaginitis” was frequently used to describe the syndrome, reflecting the general belief that this single organism was the cause.7
It was the seminal work of the University of Washington group of investigators that redirected attention to the fact that nonspecific vaginitis was associated with dramatic shifts in the vaginal microbiota, including not only increased abundance of G. vaginalis but, by striking increases, a number of anaerobic species and genital mycoplasmas.12–15 Based on this work, the syndrome associated with these changes in the composition of the vaginal microbiota was renamed “bacterial vaginitis” and later changed to “bacterial vaginosis” (BV) due to the fact that few inflammatory cells were observed microscopically in the vaginal fluid. Additionally, this group of investigators identified clinical markers of BV that could be used to differentiate the majority of symptomatic women who harbored this diverse microbiota from aymptomatic women with Lactobacillus crisptatus- and Lactobacillus jensenii-dominated microbiota.16 These markers consisted of the following: 1. vaginal secretion with a pH level of >4.5; 2. a fishy odor that was best elicited by mixing vaginal secretions with 10% KOH solution; 3. a rate of at least 20 percent of microscopically observed vaginal epithelial cells coated with bacteria (these cells were named “clue cells”); and 4. a typically white, skim-milk-like vaginal discharge. In order for a clinical diagnosis of BV to be made, at least three of the four markers needed to be present. Soon thereafter, Nugent et al. revised the vaginal Gram-stain criteria for BV first described by Spiegel by developing a numerical score based on semi-quantization of Gram-positive rods, Gram-negative coccobacilli forms and curved Gram-negative rods.17 These morphotypes were thought to represent Lactobacillus spp., G. vaginalis and Mobiluncus spp., respectively. Scores of 0–3 were considered normal (lactobacillus dominant), 4–6 were labeled as intermediate (mixed morphotypes), and 7–10 were indicative of BV (absence of lactobacilli and predominance of the other two morphotypes). This score became known as the “Nugent score.” Though the microscopic work is exacting and time-consuming, in the hands of trained research technicians, it is highly reproducible. 17–19 Over the years the Nugent score has been used widely as the standard for diagnosing BV for the purposes of clinical research. It is not applied as a part of routine clinical care very often because of the time it takes to read the slides and the requirement for specially trained microscopists.
Clinically, women with symptomatic BV present with complaints of vaginal discharge; vaginal and/or perineal pruritus; foul, fishy odor; and dyspareunia. Symptoms may occur alone or in any combination of the above though BV diagnosed by the Amsel criteria in the absence of Gram-stain data requires the presence of three of the four markers. In 1 study the sensitivity and specificity of the clinical diagnosis relative to the Nugent score were 70% and 94%, respectively.20 Treatment of symptomatic BV is with metronidazole 500 mg BID for 7 days, metronidazole containing vaginal gel, or clindamycin containing vaginal cream. Unfortunately, the clinical response rate one month post treatment is only 71% to 89%,7 and the relapse rate of clinical disease after 1 to 6 months is 50% to 75%.21,22
Clinical studies have associated BV as determined by the Nugent score with abnormal pregnancy outcomes including preterm birth, premature rupture of the membranes, early labor, and postpartum endometritis.23–25 However, efforts to prevent these complications by treatment have not uniformly met with success. There is evidence that treatment of BV in women who have had a previous preterm delivery decreases the probability of a subsequent preterm birth.25,26 Treatment of asymptomatic BV in women at low risk for preterm delivery has not proven successful.27 Whether this is related to the fact that the Nugent score does not differentiate microbiota subpopulations that have differential effects on pregnancy outcome or is related to the poor efficacy of currently available therapies or both is not clear. As discussed below, this is an instance in which a more targeted preventive treatment approach based on better knowledge of the vaginal microbiota might yield better results. Additional complications of Amsel criteria and/or a Nugent score defined as BV include post-hysterectomy wound infections and endometritis following elective abortion.28–30 Pelvic inflammatory disease’s relationship to BV is less well established, but the indirect evidence strongly indicates an association with endometritis, which, in turn, is associated with pelvic inflammatory disease.31–33 Of greatest concern is that BV appears to increase the risk of acquiring sexually transmitted infections (STIs), including HIV.
While several studies have suggested that BV by Nugent score increases the risk of chlamydial, gonococcal, and trichomonal infections, the most compelling is that by Brotmen et al.34 These investigators followed women recruited from primary-care clinics over a period of up to three years with quarterly assessments of vaginal flora by Nugent score and detection of STIs by nucleic acid amplification. After adjusting for sexual behavior risk factors, investigators found that women with BV at a prior visit were twice as likely as women with normal Nugent scores to acquire chlamydial infection and trichomoniasis. The odds ratio for the acquisition of gonorrhea was lower (1.4) but still significant. Four papers demonstrate a significant association between herpes simplex virus infection and BV,35–38 and one shows a weaker but still significant association with human papillomavirus infection.39 Of most concern is the association of HIV infection and BV.40,41 Myer et al.42 performed a prospective nested-case control study of women in South Africa. Women with BV as determined by Nugent score at enrollment had a 2.01-fold (95% CI, 1.1–3.6) increased risk of an incident HIV infection than women who were found not to have BV after adjustment for prevalent STIs, sexual behaviors, and demographics. Thus, BV defined by Nugent score appears to be a significant risk factor for acquisition of a number of STIs including HIV. If true, then treatment of BV has the potential to decrease the incidence of STIs significantly and could have a major impact in the HIV epidemic, especially in Africa and other locations where prevalent rates of BV are very high. However, it should be kept in mind that even though many of the studies referenced above attempted to control for the role of sexual behavior as the major STI risk factor, undetected residual confounding is always a possibility. There are two reasons that seem to point to the increased likelihood of this possibility. First, it is difficult to envision host physiological and/or immunobiological factors induced by a disturbed vaginal microbiota such as BV that would increase susceptibility to infection to such a wide array of organisms that differ dramatically from each other in their biology and mechanisms of pathogenesis. Second, as well summarized by Schwebke in a 2009 editorial,43 there is strong evidence that though Nugent-score BV is not the result of infection by a single organism, it is sexually transmitted. Women with incident BV are more likely to have had a recent new sex partner, to have had more partners in the last few months, to have had partners who did not use condoms, and to have had partners with another STI. The importance of this issue is that if BV is primarily a fellow traveler with other STIs, treating BV—even with optimal therapeutic options—would not likely result in the prevention of HIV or any other STI.
In summary, the pre-molecular microbiota research over the last 40 years based on classical bacterial culture methods and the use of a relatively simple microscopic assessment of the vaginal flora has resulted in a great deal of new knowledge concerning vaginal microbiota and its role in reproductive health. Complications of abnormal vaginal flora reflected by Nugent-score BV are well established, and much has been learned about the epidemiology of the disease, including its strong correlation with recognized STIs. However, there many issues that are not understood. Why have preventive treatment trials failed in pregnant women? Why do some women respond to treatment of symptomatic disease while others do not? Why are some women symptomatic while others are not? To answer these questions and others, better definitions of the vaginal microbiota are clearly needed as are investigations of the pathophysiology of differing vaginal microbiota and the immunobiologic responses of the human host.
Molecular microbiologic investigations of vaginal microbiota
Molecular studies of vaginal microbiota are based on nucleic acid extraction, amplification, and sequencing of variable regions of the 16S rRNA gene using oligonucleotide primer sequences that complement regions of this gene conserved across a broad phylogenetic spectrum of bacteria. The PCR amplicons represent a sample of all the bacteria present in a specimen. The resulting individual 16S rDNA amplicons are separated by various means and then sequenced. Web-based data bases such as the Ribosomal Database Project (RDP) are queried via computer for sequences of bacteria that match those retrieved from clinical specimens. Sequences for which close matches are not found are likely to be previously unknown organisms. The methods of separating amplicons for sequencing vary widely. Many early studies were carried out using denaturing gradient gel electrophoresis (DGGE) to separate amplicons. Amplicons forming distinct bands in the gels are sampled, re-amplified, and then directly sequenced. DGGE-based studies led to the discovery of several organisms not previously recognized as belonging to the vaginal microbiota, including Lactobacillus iners and Atopobium vaginae.44,45 In subsequent studies, amplicons were ligated into plasmids and transferred into E. coli for expansion, and then the purified plasmid with 16S rDNA inserts were sequenced 46–49 or studied by terminal restriction fragment polymorphism analysis.50 The latter approach avoids the cost of sequencing but has limited ability to identify and classify unknown bacteria. Both provide better resolution of bacterial community structure than DGGE but are more labor intensive and therefore more expensive to perform. Using the cloning and sequencing methodology, Fredericks et al.48 confirmed the association of A. vaginae with BV and, in addition, described three organisms strongly associated with BV with unique 16s RNA sequences never before described and only distantly related to other organisms in the sequence databases. These were named “BV-associated bacteria” (BVAB) 1, 2, and 3. These studies were extended by more detailed work, demonstrating that among the organisms commonly found in the vagina, there were many related at the genus level to well-known species but with unique 16s RNA sequences never before described.51,52 If these new bacterial species can be shown in future studies to be exclusively associated with the genitourinary tract, it would constitute evidence that they have evolved to survive in this specialized human ecologic niche and must play key roles in the function of the vaginal microbiome.
While these studies were groundbreaking in terms of demonstrating the potential of molecular techniques for expanding our understanding of the vaginal microbiota, the methods used were limited in that the number of sequences that could be reasonably analyzed by traditional Sanger sequencing is far too low to enable a complete understanding of microbial community structure, and the cost of increasing that number was beyond what the usual NIH grant would support. What microbiologists needed was a “high-throughput” sequencing technique that could provide thousands of sequences per specimen at a manageable cost. The method first adapted to this purpose was pyrosequencing using the 454 sequencing platform.53
Over the past 4 or 5 years, competing technologies have become available, and the cost of high-throughput sequencing has plummeted. Additionally, computational analysis programs to analyze large numbers of sequences have become increasingly available.54–56 The combination of increased availability of high-throughput sequencing and new computational analyses has “democratized” the field by facilitating the entry of many more investigators. Pyrosequencing produces a shorter sequence length than the amplification and cloning approaches described above, but these shorter sequences are sufficient for identifying bacteria at least at the genus level in most cases. The number of reads available enables characterization of individual patient vaginal microbiota to the extent that there is reasonable confidence that the true structure of each individual community can be analyzed and compared to the microbiota of other individuals.
Using this approach, Ravel et al57 recently reported the results of a pyrosequencing analysis of vaginal specimens from a sample of 400 normal women. The sample was balanced by ethnicity. Five main clusters of women were identified by Unifrac analysis. Four of these were dominated by lactobacilli, including L. crispatus, L. iners, L. jensenii, and L. gasserii. The former 2 groups were by far the largest. A fifth cluster (termed the “diverse” group) was not dominated by any one species and contained high abundances of the organisms traditionally associated with BV, including G. vaginalis and Prevotella spp. Additionally, many of the newly described organisms found to be associated with Nugent scores indicative of BV as described above were present. One of the most important findings of this study was the relative non-specificity of the Nugent score. While L. crispatus- and L. jensenii-dominated vaginal specimens almost uniformly had normal Nugent scores, those dominated by L. iners and L. gasserii frequently had intermediate and BV Nugent scores. Though the diverse cluster of cases had BV Nugent scores in the majority of cases, this group also contained many samples with normal and intermediate scores. In other words, the microbiotas of many women with normal Nugent scores often were indistinguishable from those with BV Nugent scores. In an as-yet-unpublished study, we have confirmed these findings in women studied in the New Orleans STD Clinic using pyrosequencing. The significance of these findings is that past research based on the Nugent score misclassified a number of cases. Of particular note is that pyrosequencing analysis showed that the microbial community composition of women with intermediate Nugent scores in these studies was about evenly divided between those whose microbiotas were most closely related to the larger groups of women with normal Nugent scores and those with BV Nugent scores. This may be significant as clinical research in the past has often divided study subjects into 2 groups, those with BV by Gram stain and those without (i.e., both normal and intermediate Nugent-score groups combined). Overall, the pyrosequencing data indicate that at least half of the intermediate cases really should have been included in the BV Nugent-score group.
The implications for interpretation of past research based on the Nugent score are as follows: 1. Several studies utilizing only the Gram stain to assess vaginal flora have suggested that microbiota as determined by Nugent score is unstable and that this is particularly so for intermediate Nugent scores.58–60 In 1 study of serially obtained vaginal specimens, after an interval of as few as 7 days, women with intermediate Nugent scores had converted to either normal or BV Nugent scores in about a third of cases.60 These observations have led to the conclusion that the microbiota may shift significantly in composition over relatively short periods of time. However, as summarized above, pyrosequencing data suggest that in some cases there is little difference between the microbiota of a woman with a normal Nugent score and 1 with a BV Nugent score. Thus, it is possible that despite Nugent score shifts over time, the microbiota itself may not be experiencing major changes. Longitudinal studies incorporating both Gram stains and sequence-based analyses will be required to test this hypothesis. 2. Use of Nugent scores to define the status of the vaginal microbiota for the purposes of clinical research introduces bias towards the null hypothesis in studies of BV as a risk factor for complications or acquisition of another STI. Therefore, though published studies that have shown disease associations with Nugent-score BV are valid and, in fact, the odds and/or risk ratios may be stronger than those published, it is clear that future studies of the relationship of vaginal microbiota and adverse women’s health events must be based on more specific definitions of the microbiota than the Nugent score. For example, poor treatment outcomes for Nugent-score BV may be restricted to certain microbiota subpopulations while others may respond much better. Another example is pelvic inflammatory disease, where it is possible that a virulent microbiota indistinguishable from others by Nugent score is primarily responsible for disease. Prospective treatment studies in the future should be analyzed based on the microbiota present, not exclusively on the Nugent score.
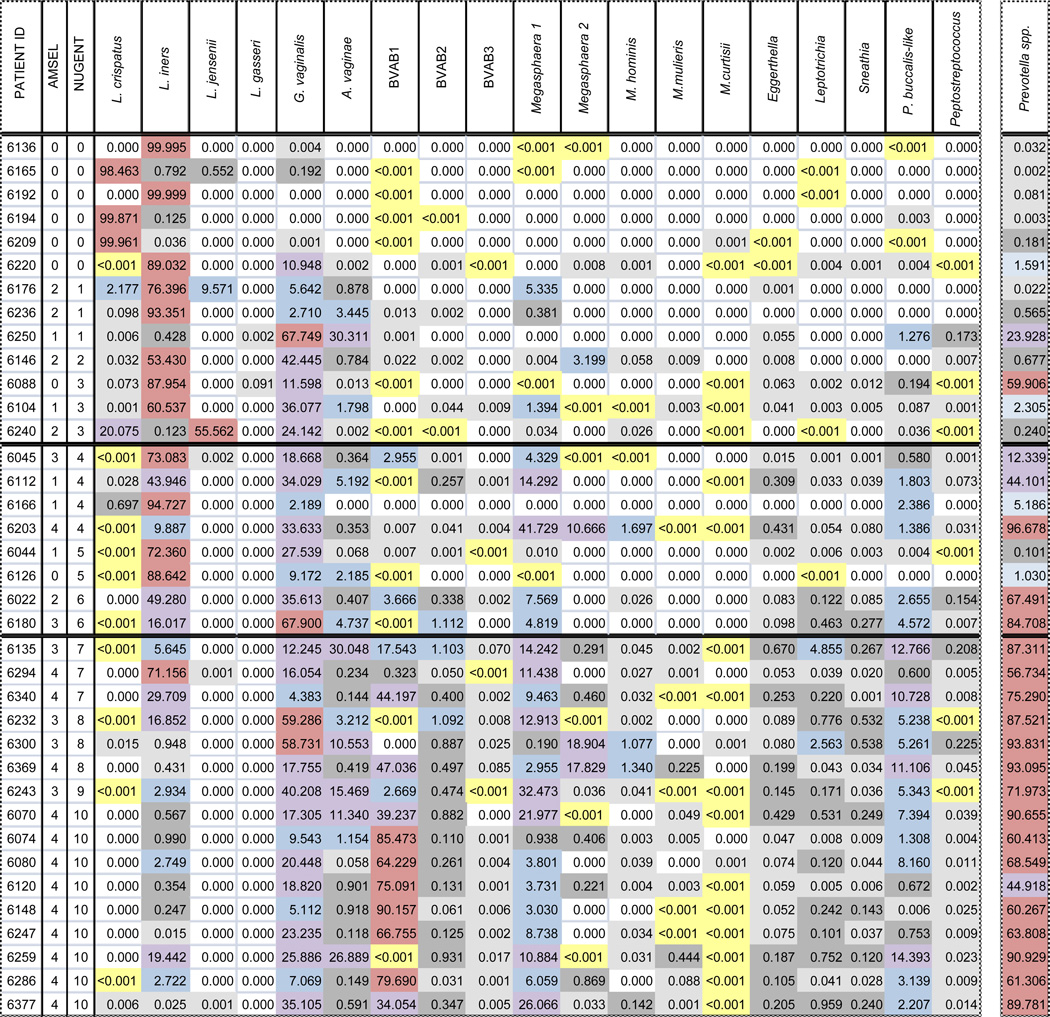
Based on the initial molecular work described above, the consensus among workers in the field has been that the most abundant members of the vaginal microbiota have been identified.2 Given that the cost of pyrosequencing was far too high for use in population-based studies, our group embarked on the development of a panel of 19 quantitative species and genus-specific quantitative PCR (qPCR) assays for assessing the concentrations of bacterial species in the vaginal microbiota.61 The results of a survey of women drawn from our STI clinic population are summarized in Table 1. The main findings were that L. iners is ubiquitously present in the vagina as are Prevotella spp. G. vaginalis was not universally present but was nearly so. These three organisms likely play important core roles in vaginal bacterial community structure. Organisms such as BVAB3 and Sneathia sanguinegens were present in more than 90% of Nugent-score BV cases but in only a few of those with normal scores. These organisms have potential as markers for the disturbed vaginal microbiota that is characteristic of BV as defined by the Nugent score. Another observation of potential importance was that in a subgroup of specimens with BV Nugent scores of 10, the dominant single species measured was BVAB1, the previously unknown organism first described by Fredricks et al.48 It is possible that the microbiota in these cases is distinctively different than that of other women with BV by Nugent score. Recently completed pyrosequencing studies in our laboratories on this same population of women appear to confirm this idea. The data of Hummelen et al, who used Illumina technology to perform very deep sequencing of variable region six of the 16S rRNA gene on vaginal specimens from HIV-infected African women, are suggestive of a similar clustering, but the short sequence reads prevented classification of many organisms beyond the family level.62 Such a subpopulation was not revealed in the pyrosequencing data of Ravel et al. described above.57 These disparate findings likely are reflective of differences in methodology. For example, the Ravel group use primers that amplify the V1–V3 region of the 16s RNA gene, whereas we amplify the V4–V6 region. The former primer set is biased against the detection of G. vaginalis, and ours, no doubt, has subtle biases of its own. A critical challenge in the field is to perform cross-comparisons of different high throughput sequencing techniques in order to define clearly the various vaginal microbiota subpopulations that are present in representative human populations.
Table 1. Relative bacterial species composition (%) based on DNA extracted from vaginal specimens clinically diagnosed as normal (Nuget score of 0 to 3), intermediate (4 to 6), or BV (7 to 10).
 0%;
0%;  <0.001%;
<0.001%;  0.001 to <0.1%;
0.001 to <0.1%;  0.1 to <1%;
0.1 to <1%;  1 to <10%;
1 to <10%;  10 to <50%;
10 to <50%;  50 to 100%
50 to 100%
 |
Once this is accomplished, it will be important to determine the limits of variation of bacterial communities over time. Relative stability would be required to support the hypothesis that one or another microbiota type or subtype is causative of adverse health outcomes.
Assuming that this is the case, future clinical research to address the role of newly defined “types” of vaginal microbiota in health outcomes will require the development of low-cost markers for each “type” of vaginal community as high throughput sequencing, may be too cumbersome for the performance of large epidemiologic studies and treatment trials. Developing relatively small sets of qPCR assays for the detection of abundance patterns specific for differing vaginal microbiota would seem to be a reasonable approach.
Despite the advances in understanding the composition of the vaginal microbiota brought about by pyrosequencing analyses of 16S rRNA sequences, the focus of research in this field may be shifting as, increasingly, experts are suggesting the need for moving away from simply describing the microbiota to determining what the organisms are actually doing. Metagenomic studies using shotgun sequencing approaches are being used to identify most of the bacterial genes present in the microbiota at a particular site at a specific point in time. Such data permit the determination of metabolic pathways most common in a chosen environment. Moreover, direct measurement of metabolites in samples is possible and is being done. An additional approach to discovering what the microbiota is doing is transcriptomics, which measures the specific mRNAs being produced by the microbiota. Ultimately, proteomic approaches can be used to identify and measure the gene products actually being produced by the microbiota. Further adding to the complexity is determining how the human host responds to differing microbiota. Of particular interest are the inflammatory response pathways. While all of these approaches will generate interesting information, the cost of generating the data will be high despite decreases in sequencing, transcriptome measurement, and protein identification costs, and there is a need for new mega-data analysis tools beyond what is currently available. Therefore, specimens for such analyses will need to be carefully selected in order to ensure that the most useful data are obtained. In my opinion, until it is clear which of the vaginal microbiota variants is associated with human disease, there is risk of wasting scarce research funding on detailed metabolomic, transcriptomic, and proteomic studies if the data may not have clinical relevance. On the other hand, it could be argued that the longitudinal risk-association studies required to establish the significance of newly defined vaginal microbiota categories will also be very expensive and will require many years. An alternative approach might be cross-sectional studies of the host inflammatory response to the differing vaginal microbiota, which would provide clues as to which research approaches will likely yield the most useful results.
In summary, over the last 10 years a clearer picture of the complexities of the vaginal microbiota has begun to emerge based on the techniques of molecular microbiology. We have learned that many previously unknown bacteria are important components of the vaginal microbiota. At the same time, it has become more and more apparent that none of these organisms alone is likely to be of great significance. High-throughput sequencing studies have provided a more complete picture of total bacterial diversity in the vaginal environment and also are revealing previously unrecognized categories of vaginal microbiota. A key challenge for the immediate future of research in this field is to tie together the findings from studies utilizing differing sequencing approaches in order to reach agreement as to the identity of clinically important vaginal microbiota categories. Only then will researchers be able to proceed with the clinical research necessary to identify which categories are associated with health and which are associated with disease. Once microbiotas that are clearly beneficial or clearly detrimental to women’s health have been identified, “omic” technology will be available to investigate mechanisms of pathogenesis and/or protection, which, in turn, may lead to interventions designed to improve women’s health.
Acknowledgments
Grant Support: NIAID - RO1 AI079071 and Louisiana Board of Regents - HEF (2001-2004)-04
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gordon JI, Klaenhammer TR. A rendezvous with our microbes. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4513–4515. doi: 10.1073/pnas.1101958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marrazzo JM, Martin DH, Watts DH, et al. Bacterial vaginosis: identifying research gaps proceedings of a workshop sponsored by DHHS/NIH/NIAID. Sexually transmitted diseases. 2010;37:732–744. doi: 10.1097/OLQ.0b013e3181fbbc95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 5.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 6.Peterson J, Garges S, Giovanni M, et al. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hillier S, Marrazzo J, Holmes KK. Holmes KK, Sparling FP, Stamm WA, et al. Sexually transmitted diseases. 4th ed. New York: McGraw Hill Medical; 2008. Bacterial Vaginosis; pp. 737–768. [Google Scholar]
- 8.Menge C, Kronig B. Bakteriologie des weiblichen genitalkanales. Monatschr Geburtsh. 1899;9 [Google Scholar]
- 9.Curtis AH. On the etiology and bacteriology of leucorrhoea. Surg Gynecol Obstet. 1914;18 [Google Scholar]
- 10.Schroder R. Zur pathogenese and klinik des vaginalen fluors. Zentralb Gynakol. 1921;38 [Google Scholar]
- 11.Gardner HL, Dukes CD. Haemophilus vaginalis vaginitis: a newly defined specific infection previously classified non-specific vaginitis. Amer J Obstet and Gyn. 1955;69:962–976. [PubMed] [Google Scholar]
- 12.Spiegel CA, Amsel R, Eschenbach D, Schoenknecht F, Holmes KK. Anaerobic bacteria in nonspecific vaginitis. New Eng J Med. 1980;303:601–607. doi: 10.1056/NEJM198009113031102. [DOI] [PubMed] [Google Scholar]
- 13.Spiegel CA, Amsel R, Holmes KK. Diagnosis of bacterial vaginosis by direct gram stain of vaginal fluid. J Clin Microbiol. 1983;18:170–177. doi: 10.1128/jcm.18.1.170-177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spiegel CA, Eschenbach DA, Amsel R, Holmes KK. Curved anaerobic bacteria in bacterial (nonspecific) vaginosis and their response to antimicrobial therapy. J Infect Dis. 1983;148:817–822. doi: 10.1093/infdis/148.5.817. [DOI] [PubMed] [Google Scholar]
- 15.Hillier SL, Krohn MA, Rabe LK, Klebanoff SJ, Eschenbach DA. The normal vaginal flora, H2O2-producing lactobacilli, and bacterial vaginosis in pregnant women. Clin Infect Dis. 1993;16 Suppl 4:S273–S281. doi: 10.1093/clinids/16.supplement_4.s273. [DOI] [PubMed] [Google Scholar]
- 16.Amsel R, Totten PA, Spiegel CA, Chen KC, Eschenbach D, Holmes KK. Nonspecific vaginitis. Diagnostic criteria and microbial and epidemiologic associations. Am J Med. 1983;74:14–22. doi: 10.1016/0002-9343(83)91112-9. [DOI] [PubMed] [Google Scholar]
- 17.Nugent RP, Krohn MA, Hillier SL. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of gram stain interpretation. J Clin Microb. 1991;29:297–301. doi: 10.1128/jcm.29.2.297-301.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forsum U, Jakobsson T, Larsson PG, et al. An international study of the interobserver variation between interpretations of vaginal smear criteria of bacterial vaginosis. Apmis. 2002;110:811–818. doi: 10.1034/j.1600-0463.2002.1101107.x. [DOI] [PubMed] [Google Scholar]
- 19.Hillier SL, Krohn MA, Nugent RP, Gibbs RS. Characteristics of three vaginal flora patterns assessed by gram stain among pregnant women. Vaginal Infections and Prematurity Study Group. Amer J Obstet and Gyn. 1992;166:938–944. doi: 10.1016/0002-9378(92)91368-k. [DOI] [PubMed] [Google Scholar]
- 20.Schwebke JR, Hillier SL, Sobel JD, McGregor JA, Sweet RL. Validity of the vaginal gram stain for the diagnosis of bacterial vaginosis. Obstet and Gyn. 1996;88:573–576. doi: 10.1016/0029-7844(96)00233-5. [DOI] [PubMed] [Google Scholar]
- 21.Sobel JD, Ferris D, Schwebke J, et al. Suppressive antibacterial therapy with 0.75% metronidazole vaginal gel to prevent recurrent bacterial vaginosis. Amer J Obstet and Gyn. 2006;194:1283–1289. doi: 10.1016/j.ajog.2005.11.041. [DOI] [PubMed] [Google Scholar]
- 22.Schwebke JR, Desmond RA. A randomized trial of the duration of therapy with metronidazole plus or minus azithromycin for treatment of symptomatic bacterial vaginosis. Clin Infect Dis. 2007;44:213–219. doi: 10.1086/509577. [DOI] [PubMed] [Google Scholar]
- 23.Hillier SL, Nugent RP, Eschenbach DA, et al. Association between bacterial vaginosis and preterm delivery of a low-birth-weight infant. The Vaginal Infections and Prematurity Study Group. New Eng J Med. 1995;333:1737–1742. doi: 10.1056/NEJM199512283332604. [DOI] [PubMed] [Google Scholar]
- 24.Jacobsson B, Pernevi P, Chidekel L, Jorgen Platz-Christensen J. Bacterial vaginosis in early pregnancy may predispose for preterm birth and postpartum endometritis. Acta obstetricia et gynecologica Scandinavica. 2002;81:1006–1010. doi: 10.1034/j.1600-0412.2002.811103.x. [DOI] [PubMed] [Google Scholar]
- 25.Leitich H, Bodner-Adler B, Brunbauer M, Kaider A, Egarter C, Husslein P. Bacterial vaginosis as a risk factor for preterm delivery: a meta-analysis. Amer J Obstet and Gyn. 2003;189:139–147. doi: 10.1067/mob.2003.339. [DOI] [PubMed] [Google Scholar]
- 26.Hauth JC, Goldenberg RL, Andrews WW, DuBard MB, Copper RL. Reduced incidence of preterm delivery with metronidazole and erythromycin in women with bacterial vaginosis. New Eng J Med. 1995;333:1732–1736. doi: 10.1056/NEJM199512283332603. [DOI] [PubMed] [Google Scholar]
- 27.Carey JC, Klebanoff MA, Hauth JC, et al. Metronidazole to prevent preterm delivery in pregnant women with asymptomatic bacterial vaginosis. National Institute of Child Health and Human Development Network of Maternal-Fetal Medicine Units. New Eng J Med. 2000;342:534–540. doi: 10.1056/NEJM200002243420802. [DOI] [PubMed] [Google Scholar]
- 28.Larsson PG, Bergman B, Forsum U, Platz-Christensen JJ, Pahlson C. Mobiluncus and clue cells as predictors of PID after first-trimester abortion. Acta obstetricia et gynecologica Scandinavica. 1989;68:217–220. doi: 10.3109/00016348909020992. [DOI] [PubMed] [Google Scholar]
- 29.Larsson PG, Platz-Christensen JJ, Thejls H, Forsum U, Pahlson C. Incidence of pelvic inflammatory disease after first-trimester legal abortion in women with bacterial vaginosis after treatment with metronidazole: a double-blind, randomized study. Amer J Obstet and Gyn. 1992;166:100–103. doi: 10.1016/0002-9378(92)91838-2. [DOI] [PubMed] [Google Scholar]
- 30.Soper DE, Bump RC, Hurt WG. Bacterial vaginosis and trichomoniasis vaginitis are risk factors for cuff cellulitis after abdominal hysterectomy. Amer J Obstet and Gyn. 1990;163:1016–1021. doi: 10.1016/0002-9378(90)91115-s. discussion 21-3. [DOI] [PubMed] [Google Scholar]
- 31.Hillier SL, Kiviat NB, Hawes SE, et al. Role of bacterial vaginosis-associated microorganisms in endometritis. Amer J Obstet and Gyn. 1996;175:435–441. doi: 10.1016/s0002-9378(96)70158-8. [DOI] [PubMed] [Google Scholar]
- 32.Wiesenfeld HC, Hillier SL, Krohn MA, et al. Lower genital tract infection and endometritis: insight into subclinical pelvic inflammatory disease. Obstet and Gyn. 2002;100:456–463. doi: 10.1016/s0029-7844(02)02118-x. [DOI] [PubMed] [Google Scholar]
- 33.Korn AP, Hessol NA, Padian NS, et al. Risk factors for plasma cell endometritis among women with cervical Neisseria gonorrhoeae, cervical Chlamydia trachomatis, or bacterial vaginosis. Amer J Obstet and Gyn. 1998;178:987–990. doi: 10.1016/s0002-9378(98)70536-8. [DOI] [PubMed] [Google Scholar]
- 34.Brotman RM, Klebanoff MA, Nansel TR, et al. Bacterial vaginosis assessed by gram stain and diminished colonization resistance to incident gonococcal, chlamydial, and trichomonal genital infection. J Infect Dis. 2010;202:1907–1915. doi: 10.1086/657320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cherpes TL, Melan MA, Kant JA, Cosentino LA, Meyn LA, Hillier SL. Genital tract shedding of herpes simplex virus type 2 in women: effects of hormonal contraception, bacterial vaginosis, and vaginal group B Streptococcus colonization. Clin Infect Dis. 2005;40:1422–1428. doi: 10.1086/429622. [DOI] [PubMed] [Google Scholar]
- 36.Cherpes TL, Meyn LA, Krohn MA, Hillier SL. Risk factors for infection with herpes simplex virus type 2: role of smoking, douching, uncircumcised males, and vaginal flora. Sex Trans Dis. 2003;30:405–410. doi: 10.1097/00007435-200305000-00006. [DOI] [PubMed] [Google Scholar]
- 37.Cherpes TL, Meyn LA, Krohn MA, Lurie JG, Hillier SL. Association between acquisition of herpes simplex virus type 2 in women and bacterial vaginosis. Clin Infect Dis. 2003;37:319–325. doi: 10.1086/375819. [DOI] [PubMed] [Google Scholar]
- 38.Gottlieb SL, Douglas JM, Jr, Foster M, et al. Incidence of herpes simplex virus type 2 infection in 5 sexually transmitted disease (STD) clinics and the effect of HIV/STD risk-reduction counseling. J Infect Dis. 2004;190:1059–1067. doi: 10.1086/423323. [DOI] [PubMed] [Google Scholar]
- 39.Watts DH, Fazzari M, Minkoff H, et al. Effects of bacterial vaginosis and other genital infections on the natural history of human papillomavirus infection in HIV-1-infected and high-risk HIV-1-uninfected women. J Infect Dis. 2005;191:1129–1139. doi: 10.1086/427777. [DOI] [PubMed] [Google Scholar]
- 40.Taha TE, Hoover DR, Dallabetta GA, et al. Bacterial vaginosis and disturbances of vaginal flora: association with increased acquisition of HIV. AIDS (London, England) 1998;12:1699–1706. doi: 10.1097/00002030-199813000-00019. [DOI] [PubMed] [Google Scholar]
- 41.Martin HL, Richardson BA, Nyange PM, et al. Vaginal lactobacilli, microbial flora, and risk of human immunodeficiency virus type 1 and sexually transmitted disease acquisition. J Infect Dis. 1999;180:1863–1868. doi: 10.1086/315127. [DOI] [PubMed] [Google Scholar]
- 42.Myer L, Denny L, Telerant R, Souza M, Wright TC, Jr, Kuhn L. Bacterial vaginosis and susceptibility to HIV infection in South African women: a nested case-control study. J Infect Dis. 2005;192:1372–1380. doi: 10.1086/462427. [DOI] [PubMed] [Google Scholar]
- 43.Schwebke JR. Bacterial vaginosis: are we coming full circle? J Infect Dis. 2009;200:1633–1635. doi: 10.1086/648093. [DOI] [PubMed] [Google Scholar]
- 44.Ferris MJ, Masztal A, Aldridge KE, Fortenberry JD, Fidel PL, Jr, Martin DH. Association of Atopobium vaginae, a recently described metronidazole resistant anaerobe, with bacterial vaginosis. BMC Infect Dis. 2004;4:5. doi: 10.1186/1471-2334-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burton JP, Reid G. Evaluation of the bacterial vaginal flora of 20 postmenopausal women by direct (Nugent score) and molecular (polymerase chain reaction and denaturing gradient gel electrophoresis) techniques. J Infect Dis. 2002;186:1770–1780. doi: 10.1086/345761. [DOI] [PubMed] [Google Scholar]
- 46.Burton JP, Devillard E, Cadieux PA, Hammond JA, Reid G. Detection of Atopobium vaginae in postmenopausal women by cultivation-independent methods warrants further investigation. J Clin Microbiol. 2004;42:1829–1831. doi: 10.1128/JCM.42.4.1829-1831.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verhelst R, Verstraelen H, Claeys G, et al. Cloning of 16S rRNA genes amplified from normal and disturbed vaginal microflora suggests a strong association between Atopobium vaginae, Gardnerella vaginalis and bacterial vaginosis. BMC Microbiol. 2004;4:16. doi: 10.1186/1471-2180-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fredricks DN, Fiedler TL, Marrazzo JM. Molecular identification of bacteria associated with bacterial vaginosis. New Eng J Med. 2005;353:1899–1911. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- 49.Hyman RW, Fukushima M, Diamond L, Kumm J, Giudice LC, Davis RW. Microbes on the human vaginal epithelium. Proc Natl Acad Sci U S A. 2005;102:7952–7957. doi: 10.1073/pnas.0503236102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou X, Bent SJ, Schneider MG, Davis CC, Islam MR, Forney LJ. Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiol. 2004;150:2565–2573. doi: 10.1099/mic.0.26905-0. [DOI] [PubMed] [Google Scholar]
- 51.Zozaya-Hinchliffe M, Martin DH, Ferris MJ. Prevalence and abundance of uncultivated Megasphaera-like bacteria in the human vaginal environment. Appl and Environ Microbiol. 2008;74:1656–1659. doi: 10.1128/AEM.02127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oakley BB, Fiedler TL, Marrazzo JM, Fredricks DN. Diversity of human vaginal bacterial communities and associations with clinically defined bacterial vaginosis. Appl and Environ Microbiol. 2008;74:4898–4909. doi: 10.1128/AEM.02884-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spear GT, Sikaroodi M, Zariffard MR, Landay AL, French AL, Gillevet PM. Comparison of the diversity of the vaginal microbiota in HIV-infected and HIV-uninfected women with or without bacterial vaginosis. J Infect Dis. 2008;198:1131–1140. doi: 10.1086/591942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl and Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lozupone C, Hamady M, Knight R. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ravel J, Gajer P, Abdo Z, et al. Microbes and Health Sackler Colloquium: Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwebke JR, Desmond R. Natural history of asymptomatic bacterial vaginosis in a high-risk group of women. Sex Trans Dis. 2007;34:876–877. doi: 10.1097/OLQ.0b013e318073bd82. [DOI] [PubMed] [Google Scholar]
- 59.Brotman RM, Ravel J, Cone RA, Zenilman JM. Rapid fluctuation of the vaginal microbiota measured by Gram stain analysis. Sex Trans Infect. 2010;86:297–302. doi: 10.1136/sti.2009.040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thoma ME, Gray RH, Kiwanuka N, et al. The short-term variability of bacterial vaginosis diagnosed by Nugent Gram stain criteria among sexually active women in Rakai, Uganda. Sex Trans Dis. 2011;38:111–116. doi: 10.1097/OLQ.0b013e3181f0bdd0. [DOI] [PubMed] [Google Scholar]
- 61.Zozaya-Hinchliffe M, Lillis R, Martin DH, Ferris MJ. Quantitative PCR assessments of bacterial species in women with and without bacterial vaginosis. J Clin Microbiol. 2010;48:1812–1819. doi: 10.1128/JCM.00851-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hummelen R, Fernandes AD, Macklaim JM, et al. Deep sequencing of the vaginal microbiota of women with HIV. PLoS One. 2010;5:e12078. doi: 10.1371/journal.pone.0012078. [DOI] [PMC free article] [PubMed] [Google Scholar]


