A Perspective on the study by Hu et al. discussing long noncoding RNAs and the implications of identifying an erythroid-specific lncRNA with an anti-apoptotic role in red blood cell formation.
Keywords: long noncoding RNA, erythroid terminal differentiation, apoptosis
Abstract
Long noncoding RNAs (lncRNAs) are a large and diverse class of functional RNAs that regulate important biological processes, including cell division, survival, and differentiation. In this issue of Genes & Development, Hu and colleagues (2573–2578) report the discovery of LincRNA erythroid prosurvival (LincRNA-EPS), a murine lncRNA that facilitates red blood cell formation (erythropoiesis) by suppressing apoptosis. This finding expands the repertoire of lncRNA functions and illustrates a novel genetic pathway that potentially can be exploited for treating anemias.
The central dogma, originally proposed by Crick in 1958 (Crick 1958, 1970) implies that most genetic information is transferred from DNA to RNA to protein. However, we now know that <2% of our DNA is accounted for by protein-coding genes (Lander et al. 2001). Indeed, the majority of transcribed DNA encodes noncoding RNAs (ncRNAs) that do not specify any protein. Moreover, the relative proportion of noncoding genomic DNA increases with developmental complexity, suggesting that ncRNAs have served increasingly important biological functions during eukaryotic evolution (Mattick 2004). Exciting new studies over the past decade to define and exploit the functions of ncRNAs have revolutionized biology and revised our view of the central dogma.
ncRNAs are broadly classified into housekeeping RNAs (rRNAs, tRNAs, and snoRNAs), small ncRNAs (<200 base pairs [bp] in size; microRNAs [miRNAs], siRNAs, and PIWI-interacting RNAs [piRNAs]), and long ncRNAs (lncRNAs; >200 bp in size) (Chen and Carmichael 2010). miRNAs are now universally recognized as an extensive and ubiquitous class of regulatory molecules (Bartel 2009). More recently, lncRNAs have risen to prominence with newly defined roles in a broad range of biological processes, including cell division, survival, and differentiation. The impact of lncRNAs on biology is likely massive. Thus far, ∼100 lncRNAs studied in detail have been shown to regulate multiple biological processes. However, recent genome-wide studies indicate that thousands of lncRNAs are encoded within mammalian genomes and expressed in unique combinations in virtually all tissues (Mercer et al. 2008; Cabili et al. 2011). While recent studies suggest that many lncRNAs are functional, the entire scope of their actions throughout mammalian biology is largely undefined. In this issue of Genes & Development, Hu et al. (2011) show for the first time a role for lncRNAs in red blood cell (RBC) development (erythropoiesis). Specifically, they identify a lncRNA that inhibits apoptosis of RBC precursors.
Life and death decisions during erythropoiesis
The major function of RBCs is to transport oxygen from the lungs to peripheral tissues. Healthy humans continually synthesize ∼2 million RBCs every second to replace those lost by senescence. This process must be controlled to maintain RBC numbers within a narrow physiological range, as too few or too many are deleterious. Moreover, homeostatic mechanisms are in place to rapidly increase erythropoiesis during blood loss or premature RBC destruction. The production of RBCs is largely regulated through mechanisms that influence the survival of lineage-committed precursors. Thus, erythropoietin (Epo), the major erythropoietic cytokine, engages the Epo receptor to initiate intracellular signaling pathways that inhibit apoptosis of erythroid precursors (Koury and Bondurant 1990). Proapoptotic pathways, including signal transduction through the Fas and tumor necrosis factor (TNF) receptors, also influence erythropoiesis, particularly during inflammatory stresses such as infection and cancer (Testa 2004). Engagement of these death receptors by their cognate ligands triggers apoptotic cascades by activating intercellular caspases. Thus, the competitive outcome between pro- and anti-apoptotic signaling determines erythropoietic output. The current study by Hu et al. (2011) indicates that this balance is regulated by at least one lncRNA.
LincRNA-EPS: an erythroid-specific lncRNA that inhibits apoptosis
Day 14.5 gestation murine fetal liver is largely a blood-forming tissue. Hu et al. (2011) sequenced polyadenylated RNAs from purified fetal liver erythroid precursors and identified 428 lncRNAs that are expressed at distinct developmental stages. From this group, they identified LincRNA erythroid prosurvival (LincRNA-EPS), a 2.5-kb lncRNA that is strongly induced when erythroid precursors begin to synthesize hemoglobin and other lineage-specific proteins. LincRNA-EPS is expressed minimally in other hematopoietic lineages, indicating erythroid specificity.
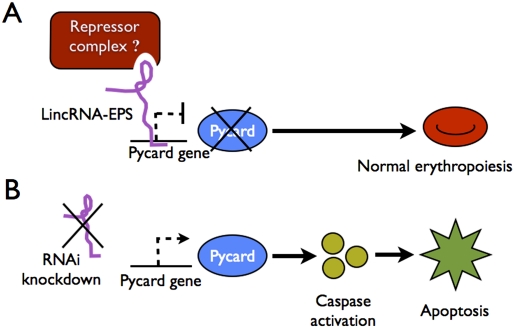
Hu et al. (2011) examined LincRNA-EPS further though gain- and loss-of-function studies in cultured mouse fetal liver erythroid precursors, which normally progress from an immature erythroblast stage to bone fide enucleate RBCs during 3 d of incubation with Epo. Remarkably, 40%–50% reduction of LincRNA-EPS by shRNAs significantly reduced the accumulation of erythroid cells by inducing massive apoptosis. Conversely, ectopic expression of LincRNA-EPS at physiological levels protected cultured erythroid precursors from cell death that normally occurs upon Epo starvation. Thus, LincRNA-EPS fosters erythropoiesis by preventing apoptosis. Careful follow-up studies demonstrated that LincRNA-EPS mediates this effect at least partly by suppressing the expression of the apoptosis-associated speck-like protein (ASC)/Pycard, a signaling molecule that promotes cell death by activating caspases (Ohtsuka et al. 2004). Of note, manipulation of LincRNA-EPS expression did not alter erythroid differentiation, suggesting that its biological activities may be restricted to influencing apoptotic pathways. This work illustrates an interesting new facet of erythroid development and extends the scope of known lncRNA actions (Fig. 1).
Figure 1.
A potential mechanism for LincRNA-EPS action in erythropoiesis. (A) The study by Hu et al. (2011) demonstrates that LincRNA-EPS represses Pycard to inhibit apoptosis of RBC progenitors. LincRNA-EPS may bind the Pycard gene and inhibit its transcription by recruiting transcriptional repressor complexes. (B) Knockdown of LincRNA-EPS permits transcription of Pycard, which activates intercellular caspases and induces apoptosis.
Mechanisms of lncRNA actions
Like all interesting studies, the work by Hu et al. (2011) raises many new questions and areas for future investigation. Foremost, it will be important to determine how LincRNA-EPS represses Pycard and perhaps other proapoptotic targets. LncRNAs act through remarkably diverse mechanisms using both digital (base-pairing) and analog (structural) features common to RNA molecules.
The best-characterized lncRNA function is the regulation of chromatin states and gene expression by facilitating histone modifications. The lncRNAs Xist, HOTAIR, HOTTIP, and Mistral act as scaffolds to recruit one or more histone-modifying enzymes to specific regions of the genome. Xist, a 17-kb lncRNA produced by one X chromosome in female (XX) cells, acts in cis to trigger inactivation of that chromosome by recruiting the polycomb-repressive complex 2 (PRC2) (Tattermusch and Brockdorff 2011). HOTAIR silences gene expression in trans by recruiting PRC2 and CoREST/REST-repressive complexes to multiple genes on different chromosomes (Rinn et al. 2007; Tsai et al. 2010). In HeLa cells and human fibroblasts, ∼20% of lncRNAs are bound by PRC2 (Khalil et al. 2009), and, conversely, PRC2 may bind as many as 9000 lncRNAs in mouse embryonic stem (ES) cells (Zhao et al. 2010). Thus, LincRNA-EPS may recruit repressive protein complexes, including PRC2, to the Pycard gene (Fig. 1). In support, this lncRNA localizes predominantly to the nucleus.
lncRNAs can also activate transcription. For example, HOTTIP and Mistral recruit chromatin-activating complexes (WDR5/MLL1) in cis to neighboring genes (Bertani et al. 2011; Wang et al. 2011). Thus, it is formally possible that LincRNA-EPS represses Pycard indirectly by inducing the synthesis of transcriptional repressors.
lncRNAs also regulate gene expression through additional mechanisms that could reflect actions of LincRNA-EPS. For example, MALAT1 is a 6.5-kb lncRNA that localizes to nuclear speckles, sequesters the serine/arginine-rich (SR) family of splicing factors, and regulates splicing (Tripathi et al. 2010). A class of lncRNAs termed 1/2-sbsRNAs contains Alu repeat elements that bind to complementary sequences in mRNAs and mediate their decay by recruiting the protein Staufen 1 (Gong and Maquat 2011). Heat-shock RNA 1 (HSR1) binds heat-shock transcription factor 1 (HSF1), facilitating its ability to activate genes encoding heat-shock proteins (Shamovsky et al. 2006). These examples illustrate how lncRNAs can influence the central dogma at many levels through remarkably diverse mechanisms.
At least three other lncRNAs are known to protect against apoptosis: PINC protects mammary gland cells against apoptosis (Ginger et al. 2006), TUG1 protects rat retina cells against apoptosis (Young et al. 2005), and CUDR down-regulates caspase 3 to protect cancer cell lines against chemotherapy-induced death (Tsang et al. 2007). The current study extends this list by identifying the LincRNA-EPS–Pycard axis in erythroid cells. Future studies should identify the mechanisms through which LincRNA-EPS inhibits Pycard and whether anti-apoptotic lncRNAs act through common mechanisms. Hu et al. (2011) propose that one reasonable starting point for future mechanistic studies will be to identify LincRNA-EPS-interacting proteins through pull-down experiments (Tsai et al. 2010).
Potential clinical relevance
In humans, increased apoptosis of RBC progenitors is associated with a wide range of anemias, including those associated with inflammation and cancer (Testa 2004). It is possible that pharmacologic inhibition of proapoptotic signaling pathways could be useful clinically, particularly since many of these anemias are resistant to Epo therapy. In this regard, the LincRNA-EPS–Pycard axis could represent a “druggable” pathway. To explore this possibility, it will be important to assess how manipulated LincRNA-EPS and Pycard impact erythropoiesis in whole-animal models and, eventually, in humans.
It will also be important to determine whether a human version of LincRNA-EPS exists, which was not possible using basic DNA homology searches. In general, the conservation of lncRNAs is challenging to quantify bioinformatically. Xist, a lncRNA that is critical for X-chromosome inactivation in mammals, exhibits poorly conserved primary sequence between species. However, evidence of conservation can be found by computation of nucleotide substitution rates and measuring conservation of promoters, splice junctions, or secondary structures (Pang et al. 2006; Guttman et al. 2009). Similar approaches could be applied to the study of LincRNA-EPS. Additionally, it may be possible to identify a human version of murine LincRNA-EPS by examining transcripts arising from syntenic genomic regions. Subsequently, it will be feasible to investigate whether misregulation of LincRNA-EPS occurs in various human anemias and whether this regulatory axis can be manipulated therapeutically.
Fathoming the scope of lncRNA actions
LincRNA-EPS is only one of the 428 lncRNAs that Hu et al. (2011) found to be dynamically regulated during erythropoiesis. Thus, this process is likely to be modulated by additional lncRNAs. Indeed, we speculate that specific lncRNAs function at distinct developmental stages as erythroid precursors mature into RBCs and at different times during embryonic development as the site of erythropoiesis shifts from yolk sac to fetal liver to bone marrow. Guttman et al. (2011) recently used RNAi to knock down ∼150 different lncRNAs present in mouse ES cells. They found that the majority of these lncRNAs regulate many protein-coding genes. Moreover, knockdown of ∼35% of these lncRNAs led to loss of ES cell pluripotency and/or activated expression of differentiation markers. Thus, in ES cells, many lncRNAs are expressed, and most of these exert important cellular functions (Guttman et al. 2011). The same probably holds true for lncRNAs in erythroid cells, and the findings of Hu et al. (2011) provide the means to test this hypothesis. Without a doubt, the tremendous knowledge of lncRNA biology gained over the past several years represents only the tip of the iceberg. It will be exciting and rewarding to explore what lies beneath.
Acknowledgments
We thank Vijay Sankaran for comments on the manuscript. Work in our laboratory is funded by The Roche Foundation for Anemia Research (RoFAR).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.183020.111.
References
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani S, Sauer S, Bolotin E, Sauer F 2011. The noncoding RNA mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell 43: 1040–1046 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL 2011. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Carmichael GG 2010. Long noncoding RNAs in mammalian cells: What, where and why? Wiley Interdiscipl Rev 1: 2–21 [DOI] [PubMed] [Google Scholar]
- Crick FH 1958. On protein synthesis. Symp Soc Exp Biol 12: 138–163 [PubMed] [Google Scholar]
- Crick F 1970. Central dogma of molecular biology. Nature 227: 561–563 [DOI] [PubMed] [Google Scholar]
- Ginger MR, Shore AN, Contreras A, Rijnkels M, Miller J, Gonzalez-Rimbau MF, Rosen JM 2006. A noncoding RNA is a potential marker of cell fate during mammary gland development. Proc Natl Acad Sci 103: 5781–5786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Maquat LE 2011. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470: 284–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. 2011. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Yuan B, Flygare F, Lodish HF 2011. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev (this issue) doi: 10.1101/gad.178780.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. 2009. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci 106: 11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koury MJ, Bondurant MC 1990. Erythropoietin retards DNA breakdown and prevents programmed death in erythroid progenitor cells. Science 248: 378–381 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Mattick JS 2004. RNA regulation: A new genetics? Nat Rev Genet 5: 316–323 [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS 2008. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci 105: 716–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka T, Ryu H, Minamishima YA, Macip S, Sagara J, Nakayama KI, Aaronson SA, Lee SW 2004. ASC is a Bax adaptor and regulates the p53–Bax mitochondrial apoptosis pathway. Nat Cell Biol 6: 121–128 [DOI] [PubMed] [Google Scholar]
- Pang KC, Frith MC, Mattick JS 2006. Rapid evolution of noncoding RNAs: Lack of conservation does not mean lack of function. Trends Genet 22: 1–5 [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E 2006. RNA-mediated response to heat shock in mammalian cells. Nature 440: 556–560 [DOI] [PubMed] [Google Scholar]
- Tattermusch A, Brockdorff N 2011. A scaffold for X chromosome inactivation. Hum Genet 130: 247–253 [DOI] [PubMed] [Google Scholar]
- Testa U 2004. Apoptotic mechanisms in the control of erythropoiesis. Leukemia 18: 1176–1199 [DOI] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. 2010. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39: 925–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast B, Wang JK, Lan F, Shi Y, Segal E, Chang HY 2010. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang WP, Wong TW, Cheung AH, Co CN, Kwok TT 2007. Induction of drug resistance and transformation in human cancer cells by the noncoding RNA CUDR. RNA 13: 890–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. 2011. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472: 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TL, Matsuda T, Cepko CL 2005. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol 15: 501–512 [DOI] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT 2010. Genome-wide identification of Polycomb-associated RNAs by RIP-seq. Mol Cell 40: 939–953 [DOI] [PMC free article] [PubMed] [Google Scholar]