Glioblastomas are responsible for nearly 60% of malignant primary brain tumors. Those with a mesenchymal (MES) gene expression signature present with worse survival and increased treatment resistance. Performing an extensive gene network analysis, Bhat et al. now identify the TAZ transcriptional coactivator as a master modulator of this severe MES signature. Validating their bioinformatic analysis in patient-derived glioma stem cells and murine neural stem cells, they further observe that TAZ expression is restrained in lower-grade gliomas due to promoter hypermethylation and that TAZ regulates MES genes by directly binding their promoters in complex with TEAD2. This study reveals a direct role for TAZ in driving MES differentiation in malignant gliomas.
Keywords: HIPPO, TAZ, TEAD, glioma, mesenchymal
Abstract
Recent molecular classification of glioblastoma (GBM) has shown that patients with a mesenchymal (MES) gene expression signature exhibit poor overall survival and treatment resistance. Using regulatory network analysis of available expression microarray data sets of GBM, including The Cancer Genome Atlas (TCGA), we identified the transcriptional coactivator with PDZ-binding motif (TAZ), to be highly associated with the MES network. TAZ expression was lower in proneural (PN) GBMs and lower-grade gliomas, which correlated with CpG island hypermethylation of the TAZ promoter compared with MES GBMs. Silencing of TAZ in MES glioma stem cells (GSCs) decreased expression of MES markers, invasion, self-renewal, and tumor formation. Conversely, overexpression of TAZ in PN GSCs as well as murine neural stem cells (NSCs) induced MES marker expression and aberrant osteoblastic and chondrocytic differentiation in a TEAD-dependent fashion. Using chromatin immunoprecipitation (ChIP), we show that TAZ is directly recruited to a majority of MES gene promoters in a complex with TEAD2. The coexpression of TAZ, but not a mutated form of TAZ that lacks TEAD binding, with platelet-derived growth factor-B (PDGF-B) resulted in high-grade tumors with MES features in a murine model of glioma. Our studies uncover a direct role for TAZ and TEAD in driving the MES differentiation of malignant glioma.
Glioblastoma (GBM) is the most common and aggressive form of glioma responsible for nearly 60% of malignant primary brain tumors (Furnari et al. 2007; Huse and Holland. 2010). Although all GBMs share similar histological features, such as microvascular proliferation and pseudopalisading necrosis, patients present with differential treatment response and survival rates that can be predicted based on molecular determinants (Burton et al. 2002; Freije et al. 2004; Hegi et al. 2005; Nigro et al. 2005; Phillips et al. 2006; Verhaak et al. 2010). In particular, recent work has shown that patients whose tumors have a signature enriched in genes associated with neural development (proneural [PN]) have better survival compared with those that have signatures resembling the mesenchyme (mesenchymal [MES]) (Phillips et al. 2006; Colman et al. 2010). Although the genetic abnormalities associated with these signatures are not fully understood, studies by The Cancer Genome Atlas (TCGA) consortium point to alterations of Neurofibromatosis-1 (NF1) and platelet-derived growth factor receptor-A (PDGFRA)/isocitrate dehydrogenase 1 (IDH1) as defining features of MES and PN subtypes, respectively (Verhaak et al. 2010). Gene regulatory network analyses identified the transcription factors (TFs) signal transducer and activator of transcription 3 (STAT3) and CCAAT enhancer-binding protein-β (C/EBP-β) as synergistic initiators and master regulators of the MES signature in glioma (Carro et al. 2010). Silencing these TFs collapsed the MES network and led to improved survival in mice implanted with tumor cells lacking these TFs. While this study represents the first comprehensive mapping of the transcriptional control of the MES signature, additional TFs could be uncovered by subjecting broader data sets such as TCGA to master regulatory analysis.
MES differentiation is a well-studied phenomenon in a variety of solid tumors, including breast carcinoma (Kalluri and Weinberg 2009; Thiery et al. 2009). Epithelial-to-MES transition (EMT) is a dynamic and reversible cellular event in which cells lose epithelial features, such as polarity and intercellular junctions, and acquire MES characteristics, leading to increased migration and invasion (Kalluri and Weinberg 2009; Thiery et al. 2009). Cancer cells that have undergone EMT exhibit enhanced metastatic potential and acquire stem cell-like properties (Mani et al. 2008; Gupta et al. 2009). These events are predominantly orchestrated by TFs (snail homolog 1 [SNAI1], SNAI2, twist homolog 1 [TWIST1] forkhead box C2 [FOXC2], zinc finger E-box-binding homeobox-1 [ZEB-1], and ZEB-2) or microRNAs (miR-141, the miR-200 family, and miR-205), many of which are integral components of early embryonic development and pathological conditions that also require EMT (Gregory et al. 2008; Park et al. 2008; Thiery et al. 2009). Until recently, however, the presence of a MES component in tumors such as GBM had not been demonstrated (Phillips et al. 2006; de Groot et al. 2008; Verhaak et al. 2010). If the proposed cell of origin of these tumors is a neural stem cell (NSC) or oligodendrocyte precursor cell (OPC) (Alcantara Llaguno et al. 2009; Ward et al. 2009; Liu et al. 2011; Sugiarto et al. 2011), it is intriguing how most GBMs exhibit MES characteristics. Furthermore, it has been shown that PN GBMs tend to shift to a MES subclass upon recurrence (Phillips et al. 2006). One hypothesis is that aberrant activation of TFs occurring during GBM progression and/or recurrence can trigger a global MES shift. This is a well-established phenomenon in breast cancer progression, where overexpression of EMT inducers, SNAI1, TWIST1, or FOXC2 is sufficient to cause metastasis to distant organs (Yang et al. 2004; Mani et al. 2007; Kudo-Saito et al. 2009). Thus, TFs playing a role in EMT in other tumor types could be candidate master regulators of MES differentiation in GBM.
Transcriptional coactivator with PDZ-binding motif (TAZ), also known by gene name WW domain-containing transcription regulator 1 (WWTR1), is a transcription cofactor that plays pivotal roles in EMT, cell growth, and organ development (Hong and Yaffe 2006; Zhao et al. 2010). TAZ functions by transactivating numerous TFs, including runt-related transcription factor 2 (RUNX2), paired box-3 (PAX3), PAX8, transcription termination factor-1 (TTF-1), T-box 5 (TBX5), mothers against decapentaplegic homologs (SMADs), and TEA domain family members (TEAD) (Hong et al. 2005; Murakami et al. 2005, 2006; Varelas et al. 2008; Di Palma et al. 2009; Zhang et al. 2009). The regulation of TAZ and its paralog, Yes-associated protein (YAP), occurs primarily via HIPPO tumor suppressor pathway, many components of which are conserved from flies to humans (Zhao et al. 2008a; Pan 2010; Halder and Johnson 2011). Autophosphorylation of mammalian sterile 20-like-1 (MST-1) and MST-2 (the vertebrate homolog for HIPPO) in complex with its regulatory protein, salvador-1 (SAV1), triggers phosphorylation and activation of large tumor suppressor homolog-1 (LATS-1) and LATS-2, which in turn phosphorylate TAZ on four serine residues (S66, S89, S117, and S311) (Zhao et al. 2008a; Pan 2010; Halder and Johnson 2011). This kinase cascade causes cytoplasmic sequestration of phospho-TAZ by 14-3-3-ɛ. Phosphorylation of TAZ by LATS on Ser 311 also primes subsequent phosphorylation by casein kinase I (CKIδ/ɛ), which induces recruitment of SCFβ-TRCP E3 ligase, leading to the ubiquitination and degradation of TAZ (Liu et al. 2010). Recently discovered membrane sequestration of TAZ/YAP by PDZ domain-containing tight junction proteins adds another complex layer of control of these crucial transcription coactivators (Duning et al. 2010; Remue et al. 2010; Varelas et al. 2010b; Chan et al. 2011; Zhao et al. 2011). Upon translocation to the nucleus, TAZ induces cell proliferation, migration, invasion, and EMT (Lei et al. 2008; Zhang et al. 2009). Silencing TEAD or even preventing the TAZ–TEAD interaction blocks the ability of TAZ to promote cell proliferation and EMT, implying TEAD as a major facilitator of TAZ functions. Thus, restraining TAZ interaction with TEAD may be an important step in preventing aberrant events central to cancer progression.
The current study was specifically initiated to identify candidate TFs/cofactors that are causally associated with the MES signature. We employ gene network analyses of TCGA data sets, mimic glioma-associated alterations in patient-derived glioma stem cells (GSCs) and murine models of glioma, and show that reprogramming of otherwise PN GSCs to a MES state can be achieved by overexpression of a single transcriptional coactivator.
Results
Identification of TAZ association with the MES network, grade IV gliomas, and its epigenetic control
We subjected microarray expression data sets from TCGA (n = 385) to the previously described Algorithm for the Reconstruction of Accurate Cellular Networks (ARACNE) to identify novel TFs/cofactors associated with the MES gene signature (Margolin et al. 2006). For the initial screen, we used an expanded list of transcriptional regulators that included cofactors associated with transcription (Supplemental Table S1A) that were not part of a previous study (Carro et al. 2010). We next generated a subnetwork containing known GBM MES genes (Supplemental Table S1B) as defined by two independent groups (Phillips et al. 2006; Verhaak et al. 2010) and the associated TFs/cofactors as predicted by ARACNE. Henceforth, we refer to the two data sets simply as Phillips and Verhaak. We subsequently filtered for the higher-order “hub” (i.e., most connected) TFs/cofactors, which represent critical regulators in a scale-free network model. Our analyses revealed ∼70% overlap with previously identified candidates, including STAT3, C/EBP-β, and other TFs that had strong correlation with the MES network as either activators or repressors (Supplemental Table S1C; Carro et al. 2010). The unique transcriptional regulators that were identified to be positively correlated with the MES signature in our study included v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (MAFB), hematopoietic cell-specific Lyn substrate 1 (HCLS1), and the HIPPO pathway transcription cofactors TAZ and YAP (Supplemental Table S1C). While a role for MAFB and HCLS1 in inducing MES differentiation cannot be ruled out, TAZ and YAP have previously been demonstrated to induce EMT in other cancer types (Lei et al. 2008; Zhao et al. 2008b). Furthermore, for reasons that will become obvious, we chose to characterize TAZ over YAP. TAZ inferred MES network targets were relatively nonoverlapping with those of STAT3 and C/EBP-β (Fig. 1A,B; Supplemental Fig. S1A,B; Supplemental Table S1D). Intrigued by this, we were curious what other types of functions were predicted to be regulated by TAZ in the overall ARACNE-generated GBM regulatory network. Thus, we generated a list of genes predicted to be regulated by TAZ and analyzed this using the Database for Annotation, Visualization, and Integrated Discovery tool (DAVID), which distills a list of genes to biologically meaningful activities (Dennis et al. 2003; Huang da et al. 2009). This analysis further supported TAZ as playing a role in MES activities (e.g., wound response) and immunologic functions (Fig. 1C). In concordance with this, the expression of TAZ in the TCGA data set was strongly correlated with a MES metagene score generated from the union of the Phillips and Verhaak MES genes (R2 = 0.48) (Supplemental Fig. S1C). Thus, in silico analysis uncovered a positive association of TAZ with the MES signature.
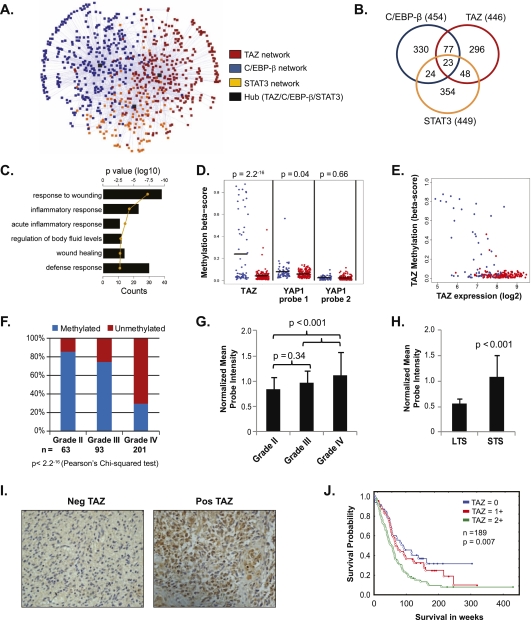
Figure 1.
Association of TAZ to the MES network, the MES subclass of gliomas, and its epigenetic control. (A) Subnetwork generated from the ARACNE analysis limited to the union of Phillips and Verhaak MES target genes (n = 281). Nodes are color-coded to show the membership of a given gene to a regulatory network. (B) Venn diagram of gene overlap of the TAZ, C/EBP-β, and STAT3 networks. The number in parentheses is the total number of genes in the ARACNE network for the given TF. (C) Bar graph depicting enrichment of genes possessing MES properties in the initial ARACNE network. Functional categories showing significant enrichment based on log10 of the P-value (line) and counts (bars) are shown. (D) Methylation status of TAZ and YAP in 62 PN (blue) or 147 MES (red) GBMs from TCGA data set (Illumina Infinium platform). The black bar is the mean of the methylation β-score. Two-sample t-test between the groups was performed to assess statistical significance. (E) Correlation of TAZ expression with TAZ methylation status. Two-hundred-nine GBMs from TCGA data set with both expression data (Affymetrix platform) and methylation data (Illumina Infinium platform) are plotted. GBMs were color-coded as either MES (red) or PN (blue) based on the composite metagenes, as defined by Phillips et al. (2006) and Verhaak et al. (2010). (F) Bar graphs showing the frequency of methylation on TAZ CpG sites across various grades of glioma. Red bars indicate unmethylation and blue bars indicate methylation. Pearson's χ2 test was used to estimate significance. (G) TAZ expression in multi-institutional microarray data sets (∼800). Bar graphs indicate normalized mean probe intensity of TAZ across grades. (H) Bar graphs indicate normalized mean probe intensity of TAZ across survivors. Long-term survivors (LTS) were defined as those who lived longer than 2 years, and those who survived less than that were short-term survivors (STS). Student's t-test values are shown for significance. (I) Representative IHC images (40×) of TAZ expression. (J) Kaplan-Meier survival analysis of TAZ expression based on the IHC staining pattern. Tumors lacking TAZ expression were scored 0, and staining in the nucleus or cytoplasm was scored 1, whereas tumors showing both nuclear and cytoplasmic staining were scored 2. Log rank test values are shown.
Since methylation of CpG islands of gene promoters can influence their expression (Deaton and Bird 2011), we analyzed the methylation status of TAZ and other components of the HIPPO pathway (Supplemental Fig. S1D) from TCGA data sets. The CpG island of TAZ (Fig. 1D,E; Supplemental Fig. S1E) was dramatically hypermethylated in the PN subgroup compared with MES tumors. YAP, LATS2, and MST1 also appeared methylated in PN tumors, albeit to a less significant extent (Fig. 1D; Supplemental Fig. S1F). If the TCGA tumors are stratified by Verhaak subtypes, the TAZ promoter is hypermethylated only in the PN group, but no obvious differences were seen in the YAP promoter methylation across all four subtypes (Supplemental Fig. S1G,H). We next compared TAZ methylation across grades and found higher methylation frequencies in grade II and grade III gliomas (86% and 75%, respectively) compared with only 30% in grade IV tumors (Pearson's χ2 test, P = 2.2−16) (Fig. 1F). Since lower-grade gliomas are typically PN in nature (Li et al. 2009; Cooper et al. 2010), whereas GBMs tend to be both PN and MES, we view the association of TAZ methylation with lower grade as reflective of the gene expression signature rather than differences in grade. Consequently, TAZ expression was lower in grades II/III when compared with grade IV gliomas (P < 0.001) (Fig. 1G), as well as in long-term (>1 year) versus short-term survivors (P < 0.001) (Fig. 1H). Consistent with gene expression patterns, TAZ protein was higher in grade IV when compared with grade II/III tumors (Supplemental Fig. S1I). A similar pattern emerged for MES marker fibronectin1 (FN1), YAP, and TEAD4, but not other TEAD family members (Supplemental Fig. 1I). Interestingly, LATS2 and MST1, whose promoters were methylated in the PN subgroup, showed higher expression in grade IV gliomas, but activation of these kinases was not seen, as evidenced by weak induction of p-MST1 in all grades of glioma and p-LATS1/2 in only one out of the eight GBMs tested (Supplemental Fig. 1I). This, in turn, could facilitate TAZ localization to the nucleus. All other HIPPO pathway components failed to exhibit notable differences between grades (Supplemental Fig. 1I). Because FN1 is also associated with endothelial cells (Martinez et al. 1994), and to delineate whether the increased FN1 expression was a consequence of MES shift or increased vasculature, we performed immunohistochemistry (IHC) on paraffin-embedded sections. A membranous pattern of staining on tumor cells was observed in grade IV tumors, but not in grade II, where only endothelial cells expressed FN1 (Supplemental Fig. S1J).
We next analyzed TAZ mRNA expression and its correlation with survival in the four subtypes of GBM in the TCGA data sets using a Cox proportional hazards model. In the Verhaak-called subtypes (see the Supplemental Material), overall survival was predicted by TAZ expression only in the PN group (Supplemental Table S2). The reason for this is unclear, although this could be partially attributed to the PN group having the widest range of TAZ expression, and so its influence is more readily seen. On the contrary, when using calculated Verhaak subtypes (see the Supplemental Material), TAZ expression predicted overall survival in both the PN and MES groups (Supplemental Table S2). Like the PN group as determined by either method, the calculated MES group had a wider spread of TAZ expression.
To assess the clinical impact of subcellular localization of TAZ, we tested 187 gliomas for the expression of TAZ by IHC. TAZ was readily detected in the nucleus in a large subset of the cases that were predominantly grade IV, whereas most grade II gliomas failed to express TAZ (representative images shown in Fig. 1I). In cases where TAZ was expressed in both nucleus and cytoplasm (score = 2), patients showed significantly reduced survival (P = 0.007) (Fig. 1J) compared with cases that had either one or the other (score = 1) or neither (score = 0). This is not entirely surprising, since TAZ has been shown to promote Wnt/β-catenin signaling in the cytoplasm (Varelas et al. 2010a), raising the possibility that cytoplasmic TAZ could still promote gliomagenesis by alternate mechanisms. However, nuclear TAZ expression strictly correlated with high CD44 cases, arguing that nuclear TAZ aligns with MES subtypes of tumors (Supplemental Fig. S1K). To rule out TAZ expression by nontumorigenic cell types that are part of the microenvironment, we costained TAZ with a monoclonal antibody (J1-31) that specifically detects reactive astrocytes (Ridet et al. 1997). Tumor areas that showed nuclear TAZ expression lacked J1-31 staining and vice versa (Supplemental Fig. S1L). Taken together, these data imply that TAZ is intricately connected to the MES network and is regulated by promoter methylation and that its nuclear expression correlates with the MES signature, higher grade, and worse overall survival in gliomas.
TAZ is required for MES transition and aggressive gliomagenesis in GSCs
Studies in our laboratory have shown a dichotomous nature of patient-derived GSCs—based on their gene expression signatures—that mirror the GBM subtypes of PN and MES (KPL Bhat and KD Aldape, unpubl.). We tested the expression of TAZ in these GSCs and compared them against TAZ promoter methylation status. TAZ expression was undetectable or significantly lower in PN GSCs (GSC7–11, GSC8–11, GSC11, and GSC23) when compared with those deemed MES (GSC6–27 and GSC20) (Fig. 2A). A similar pattern was seen for MES marker CD44 (Fig. 2A; Schieker et al. 2004; Schieker et al. 2007). YAP expression did not necessarily correlate with the PN or MES status of the GSCs (Fig. 2A). For example, a PN line (GSC23) had levels of YAP expression equal to GSC20, a MES line (Fig. 2A). Thus, TAZ, but not YAP, expression parallels the MES nature of GSCs, similar to clinical observations. Moreover, using cell fractionation, we show that nuclear TAZ was undetectable in PN GSCs compared with MES GSCs that exhibited dramatically higher levels of TAZ in the nucleus (Supplemental Fig. S2A). Cytosolic TAZ was also higher in the MES GSCs, but to only modest levels compared with PN GSCs. This could be attributed to increased amounts of active MST1 (phosphorylated form) in MES GSCs. Based on these considerations, and given that YAP is not a predictor of survival in the TCGA data set (Supplemental Fig. S2B), we chose TAZ for further characterization. Bisulfite sequencing of the TAZ promoter revealed hypermethylation in GSC7–11 and GSC8–11 and a converse hypomethylation in GSC6–27 and GSC20 (Fig. 2B). Treatment with the demethylation agent 5-aza-2′-deoxycytidine (DAC) induced TAZ expression by greater than twofold in GSC8–11 (Fig. 2C), whereas it failed to have an effect in GSC20 (data not shown). Interestingly, expression of CD44 and CTGF was also increased upon DAC treatment (Fig. 2C), implying that induction of TAZ parallels expression of MES genes. Our findings uncover a previously unknown epigenetic regulation of TAZ in gliomas.
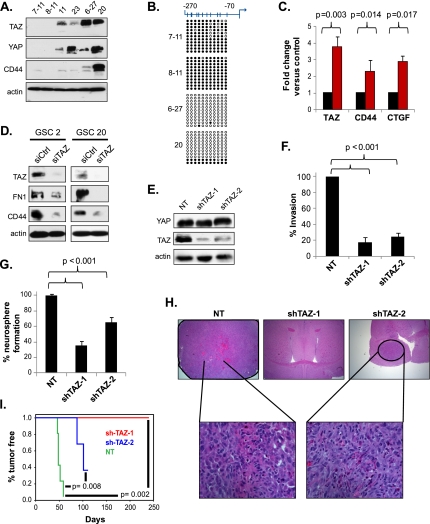
Figure 2.
TAZ is required for expression of MES markers, invasion, self-renewal, and tumor initiation in GSC20. (A) Western analyses of basal YAP, TAZ, and CD44 levels in GSCs. (B) DNA methylation analysis by bisulfite conversion followed by sequencing of cloned PCR products. (Filled circle) Methylated CG pair; (white circle) unmethylated CG pair. Each column corresponds to 11 CpG sites proximal to the transcription start site of TAZ (shown on top). Each row represents the methylation status of a clone. (C) Real-time qPCR of TAZ, CD44, and CTGF expression in GSC8–11 after treatment with 5 μM DAC for 72 h. Fold change before and after treatment was used for comparison of transcript levels and assessed using a t-test. (D) Western analysis of TAZ, FN1, and CD44 after transient knockdown of TAZ in GSC2 and GSC20. Cells were cultured in laminin and poly-L-ornithine-coated plates and transfected with siRNA for 48 h prior to immunoblotting. (E) Western analysis of stable TAZ knockdown clones showing reduction of TAZ but not YAP levels. (F) Invasion assay of TAZ stable knockdown clones in GSC20. The invasion efficiency of the nontargeting controls was set to 100% for comparison. P-values were generated using a t-test for significance. (G) Neurosphere assay of TAZ knockdown clones. Bar graphs indicate percentage of neurosphere formation, and P-values are shown for significance. (H) Representative hematoxylin and eosin-stained brain images from SCID mice injected intracranially with the TAZ stable knockdown clones. The bottom row shows higher magnification (40×) of tumors on the top row (10×). (I) Kaplan-Meier analysis of tumor-free progression for the TAZ knockdown clones compared with the nontargeting control.
To directly test whether TAZ is required for the expression of MES markers, we performed transient knockdown of TAZ using siRNA. Silencing TAZ for 72 h led to dramatic reduction of its protein levels with concomitant decreases in CD44 and FN1 (Fig. 2D). Encouraged by this finding, we sought to analyze the phenotypic consequence of stable silencing of TAZ using shRNA. Western analysis of two independent GSC20 clones targeting nonoverlapping regions of the TAZ transcript showed a significant reduction of TAZ expression compared with a nontargeting control, but YAP levels were unaltered (Fig. 2E). In line with previous reports, silencing TAZ caused reduced invasion across a chemotactic gradient compared with control GSC20 cells (Fig. 2F). Furthermore, self-renewal of GSC20 as measured by neurosophere formation was also reduced in both knockdown clones when compared with the nontargeting control (Fig. 2G). These data imply that TAZ is required for the invasive ability and self-renewal of GSC20. We then compared the tumor-initiating capacity of these clones using an orthotopic intracranial model using severe combined immunodeficiency (SCID) mice. All of the mice injected with control GSC20 (n = 5) developed tumors characterized by pseudopalisading necrosis and microvascular proliferation (Fig. 2H). In contrast, none of the mice injected with shTAZ-1 and only two out of five of the mice injected with shTAZ-2 formed tumors, with a consequent improved survival in these animals (Fig. 2H,I). Histologically, the tumors that did form in shTAZ-2 were well circumscribed and lacked expression of TAZ and CD44 as detected by IHC (Supplemental Fig. S2C). In sum, TAZ expression is regulated by CpG island methylation in GSCs and is required for expression of MES markers, invasion, self-renewal, and tumor initiation in GSCs.
The TAZ–TEAD complex mediates MES transition in GSCs and murine primary NSCs
Next, we asked whether overexpressing TAZ is sufficient to induce MES transition in otherwise PN GSCs. TAZ interacts with numerous TFs, but TEAD and RUNX2 play prominent roles in TAZ-mediated EMT and osteogenic differentiation, respectively (Hong et al. 2005; Zhang et al. 2009), and thus represent candidate TFs that could mediate TAZ-induced MES differentiation. To test this hypothesis, we used previously characterized mutants of TAZ (Lei et al. 2008; Zhang et al. 2009). Four serine residues (S66, S89, S117, and S311) substituted to alanine results in unphosphorylated TAZ that is constitutively nuclear (termed 4SA). An additional point mutation in the TEAD-binding domain (S51 to alanine) results in TAZ that lacks TEAD binding (4SA-S51A). We transduced a retrovirus-expressing Flag-tagged vector, 4SA, or 4SA-S51A, into GSC11 and generated stable clones (Fig. 3A). Consistent with previous reports, immunofluorescence (IF) analysis revealed predominant nuclear localization of 4SA, whereas 4SA–S51A showed both cytoplasmic and nuclear expression (data not shown). Immunoprecipitation using Flag antibody showed specific interaction of 4SA with both TEAD4 and RUNX2 (Fig. 3B). 4SA was also found to interact with TEAD2, suggesting that TAZ can bind to other TEAD family TFs with equal efficiency in GSCs (data not shown). On the contrary, the 4SA-S51A mutant retained binding only to RUNX2, but not TEAD4. Immunoblotting showed dramatic induction of MES markers—namely, connective tissue growth factor (CTGF), CD44, caveolin 2 (CAV2), and FN1—in 4SA, but not 4SA-S51A (Fig. 3C). The increase in MES markers was also observed in GSC7–11 and GSC8–11 that lack TAZ expression (Supplemental Fig. 3). Functionally, GSC11 overexpressing 4SA showed increased invasion, whereas 4SA-S51A showed efficiency similar to vector controls (Fig. 3D). Previous studies have shown that Yorkie (Yki), the Drosophila homolog of TAZ/YAP, promotes cell proliferation via activation of cyclin E (Huang et al. 2005). To test whether this effect was conserved in glial tumors, we assessed cell proliferation rates using EdU incorporation assay, but found no significant differences between the GSC11 clones (Fig. 3E). In addition, the expression of cell cycle regulatory proteins p-CDK1, p-CDK4, Cyclin A, Cyclin B1, and Cyclin E did not significantly differ between the groups (Fig. 3F). Thus, at least in GSCs, the primary event in response to TAZ activation and signaling via TEAD appears to be MES differentiation, rather than enhanced proliferation as observed in other cell types (Lei et al. 2008). Stable knockdown of TEAD2 and TEAD4 in 4SA also resulted in decreased MES protein expression, providing further evidence that TEAD is an important player in TAZ functions (Fig. 3G).
Figure 3.
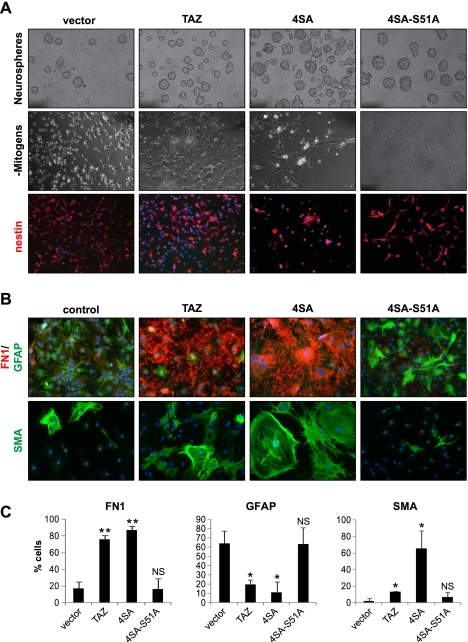
The TAZ–TEAD interaction mediates MES transition in GSCs. (A) Western analysis of GSC11 clones overexpressing vector, 4SA, or 4SA-S51A showing Flag expression. Retrovirus encoding pBabe vector or TAZ mutants was used for transduction of GSC11. Stable clones were generated by culturing transduced cells in puromycin. (B) Immunoprecipitation of TAZ using Flag antibody followed by Western analysis of TAZ, TEAD4, and RUNX2 in 4SA- and 4SA-S51A-expressing cells. (C) Western analyses of MES proteins in GSC11 clones expressing vector, 4SA, and 4SA-S51A. (D) Invasion assay of GSC11 clones across Matrigel-coated transwell plates. Bar graphs indicate percentage of invasion. A t-test was used to compare statistical differences. (E) GSC11 clones were subjected to EdU incorporation assay; cells were plated on laminin/poly-L-ornithine-coated chamber slides, fixed, and counterstained with DAPI; and Alexa 488-positive cells were quantified using a fluorescent microscope in 10 independent fields. Bar graphs indicate mean values. A t-test was used to assess statistical significance. (NS) Not significant. (F) Western analyses of cell cycle proteins in GSC11 clones. (G) Western analyses of MES proteins after stable knockdown of TEADs. Two independent stable knockdown pools (shTEAD-a and shTEAD-b) were generated using pGipZ lentivirus. Both clones appeared to silence TEAD2 and TEAD4 effectively. (H) Osteoblast induction of GSC11 clones. Cells were cultured in differentiation medium for 3–4 wk, fixed in 70% ethanol, stained with Alizarin Red for assessing intercellular calcium deposition, and photographed. (I) Chondrocyte induction of vector control and 4SA. Cells were grown as pellets in chondrocyte medium for 4 wk, fixed in buffered 10% formalin, and embedded in paraffin. Five-micron were slide-mounted and stained for glycosaminoglycans with Safranin O.
Previous studies have shown that TAZ is highly expressed in MES stem cells (MSCs) and promotes osteogenic differentiation via interaction with RUNX2 (Hong et al. 2005). Because GSC11 expressing 4SA-S51A retains binding to RUNX2, but lacks MES properties, we asked whether TAZ–TEAD interaction mediates osteogenic differentiation of GSCs. GSC11 clones were cultured in osteogenesis induction medium for 30 d and stained for alizarin red to assess calcium chelation (Meloan et al. 1972). No staining was observed in vector control or 4SA-S51A, whereas 4SA-overexpressing cells showed dramatic red coloration (Fig. 3H). Cartilage-specific proteoglycan expression, as determined by Safranin O staining (Rosenberg 1971), was also induced in 4SA when compared with vector transfected cells (Fig. 3I). 4SA-S51A-expressing cells failed to form a detectable pellet in chondrogenic medium and hence could not be assessed in this assay. The induction of bone and cartilage marker expression by TAZ in GSCs is consistent with a robust MES transition occurring via a TAZ–TEAD interaction in these cells.
Because GSCs contain numerous genetic alterations that could cooperate with TAZ to induce MES differentiation, we asked whether TAZ can reprogram primary cells of neural origin into MES lineages. To address this, we isolated NSCs from embryonic day 14 (E14) mouse embryonic brain telencephalon and cultured them as neurospheres. Immunoblotting revealed very low levels of basal TAZ expression, but TEAD2 and TEAD4 where highly expressed in NSCs at levels comparable with GSC20 (Supplemental Fig. S4A). We generated wild-type and mutant TAZ-overexpressing stable clones of NSCs by retroviral transduction. 4SA-expressing NSCs appeared larger, showed fibroblast-like morphology upon attachment (Fig. 4A), and grew at slower rates (data not shown) than the other NSC clones, suggesting a growth inhibitory effect of constitutive TAZ activation. However, basal levels of nestin expression were not altered significantly among the NSC clones under self-renewing conditions (Fig. 4A). In response to environmental cues such as withdrawal of growth factors or presence of serum, NSCs differentiate into neurons, astrocytes, and oligodendrocytes (Gage et al. 1995). To test whether TAZ interferes with these processes, we cultured NSC clones in the presence of serum for 6 d and analyzed expression of various proteins by IF. The astrocytic marker glial fibrillary acidic protein (GFAP) (Takizawa et al. 2001) was expressed in >60% of the cells transduced with a vector, whereas TAZ- or 4SA-expressing cells showed significantly lower percentages (10%–20%) (Fig. 4B,C). Conversely, TAZ and 4SA clones showed dramatic induction of MES markers FN1 and smooth muscle-specific α-actin (SMA) (Nakajima et al. 1997)—in some cases, up to an 80% increase compared with vector control (Fig. 4B,C). The gain of MES markers was abolished and astrocytic differentiation was restored in clones expressing 4SA-S51A. Similar results were obtained in independent differentiation experiments upon withdrawal of growth factors (Supplemental Fig. S4B). These experiments suggest that the TAZ–TEAD interaction causes aberrant MES reprogramming of primary NSCs at the expense of normal differentiation toward neural lineages.
Figure 4.
The TAZ–TEAD interaction mediates MES differentiation in murine NSCs. (A) NSCs were isolated from E14 mouse brains and transduced with vector, wild-type, or mutant TAZ, and stable pools were generated using puromycin. (Top panel) Phase-contrast images of neurospheres of corresponding NSC stable pools. (Middle panel) Phase-contrast images of NSCs after 1 d of differentiation in mitogen-free medium. (Bottom panel) Dissociated NSC clones were stained for NSC marker nestin (red), and the nuclei were stained with Hoeschst dye. Bars, 100 μm. (B) Differentiation of TAZ-overexpressing NSCs in 5% serum-containing medium. The top panel shows dual staining for GFAP (green) and FN1 (red). The bottom panel shows SMA (green) staining in these clones. Bars, 100 μm. (C) Quantification of expression of various markers in NSCs. For each experiment, FN1-, GFAP-, or SMA-positive cells were counted in 10 randomized microscopic fields. Bars indicate the mean value (±SD) for at least two to three independent experiments. A t-test was used to assess statistical significance. (*) P < 0.05; (**) P < 0.005; (NS) not significant.
Global MES differentiation is induced by TAZ–TEAD via direct recruitment to target promoters
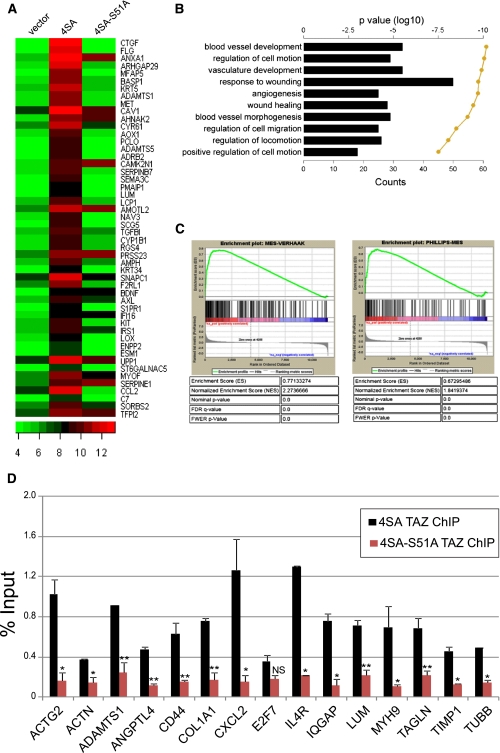
We next asked whether the TAZ/TEAD complex directly activates a global MES transcriptional program by target promoter occupancy. First, to rule out a plausible hierarchical transcription module between TAZ, STAT3, and C/EBP-β, and therefore indirect activation of MES genes by TFs (such as STAT3 and C/EBP-β) that could signal downstream from TAZ, we silenced these TFs individually in GSC20 and tested their protein levels by Western analysis. Despite significant reduction of all three TFs, none of them individually altered the expression of the other two TFs, indicating a lack of transcriptional interdependency between TAZ, STAT3, and CEBP-β (Supplemental Fig. S5A). Hypothetically, TAZ may also cause MES differentiation via CTGF, a direct downstream target of TAZ (Zhang et al. 2009) that by itself has been proven to induce EMT in other cell types (Gore-Hyer et al. 2002; Burns et al. 2006). To rule out this possibility, we silenced CTGF in GSC11 overexpressing 4SA, but found no alteration in the expression of MES markers (Supplemental Fig. S5B). These data are consistent with the hypothesis that TAZ is a direct inducer of MES differentiation in GSCs. To test this, we subjected GSC11 clones to microarray analyses and found 975 genes that were significantly (>1.5 fold) up-regulated in 4SA versus vector control (Fig. 5A; Supplemental Fig. S5C; Supplemental Table S3A). Of these, expression returned to vector control levels in 61% of the genes in the 4SA-S51A group. Quantitative real-time PCR (qRT–PCR) confirmed the induction of several microarray-inferred targets (CTGF, CD44, FN1, ADAMTS1, and IL8) in 4SA, but not control or 4SA-S51A, cells (Supplemental Fig. S5D). Functional pathway analyses using DAVID revealed that the genes induced by 4SA showed significant similarities to the MES subclass (Fig. 5B), while gene set enrichment analyses (GSEA) (Mootha et al. 2003; Subramanian et al. 2005) against Phillips and Verhaak MES genes showed significant enrichment with the two independent groups (4SA vs. Verhaak normalized enrichement score = 2.27; 4SA vs. Phillips normalized enrichment score = 1.84) (Fig. 5C). Interestingly, genes that were significantly down-regulated by overexpression of 4SA showed a PN characteristic (Supplemental Fig. S5E; Supplemental Table S3B). These data point to the important role of TAZ as master modulator of the PN-to-MES transition in GBM.
Figure 5.
Global MES differentiation induced by TAZ–TEAD via direct recruitment to target promoters. (A) Heat map showing ranked top 50 induced genes in 4SA-expressing GSC11 compared with control. Plots are the log2 Affymetrix expression values. (B) Bar graph showing the top 10 gene ontology (GO) terms associated with the twofold or greater 4SA-induced genes, as ranked by P-value. Shown is the number of genes that are common between the GO term's gene set and the 4SA gene set. The line is the log10 of the P-value as determined by DAVID functional analysis. (C) GSEA analysis of 4SA-mediated global gene expression. A ranked list of all of the 4SA-induced genes on the HGU133a2 microarray chip was compared against Phillips (left) or Verhaak (right) MES genes. (D) ChIP-qPCR analysis of TAZ recruitment at selected target gene promoters in GSC11 overexpressing 4SA or S51A. Bar graphs represent mean percentage of input occupancy by TAZ. A t-test was used to determine statistical significance. (*) P < 0.05; (**) P < 0.005; (NS) not significant.
Next, to address whether target genes inferred by microarray were indeed direct targets of TAZ–TEAD, we performed chromatin immunoprecipitation (ChIP) coupled with qPCR (ChIP-qPCR). We designed primers around putative TEAD consensus sequences 3 kb proximal to the transcriptional start site. All genes had at least one TEAD consensus sequence, and a 100- to 200-base-pair (bp) region surrounding this region was chosen for amplification. We tested 15 microarray-inferred targets, including CTGF (Supplemental Fig. S5F), which has been previously annotated as a direct target of TAZ and YAP (Zhao et al. 2008b; Zhang et al. 2009). E2F7, a gene in the proliferative subclass of GBM (de Bruin et al. 2003; Phillips et al. 2006), was chosen for comparison against occupancy of the MES targets by TAZ/TEAD. Strikingly, all 15 of the MES gene promoters were bound by TAZ in 4SA cells, whereas S51A showed significantly lower enrichment, implying that TAZ is recruited to MES gene promoters via interaction with TEAD (Fig. 5D; Supplemental Fig. S5F). Of note, no differential enrichment of TAZ on the E2F7 promoter was seen between 4SA and 4SA-S51A. On the contrary, TEAD2 was recruited to MES promoters in both 4SA- and 4SA-S51A-expressing cells (data not shown), implying that the binding of TAZ to TEAD—and in turn to target promoters—is a critical determinant of MES gene induction. Thus, biochemical studies prove that TAZ and TEAD2 are recruited directly to a majority of MES promoters.
The TAZ–TEAD pathway decreases survival and increases tumor grade in the RCAS/N-tva mouse model
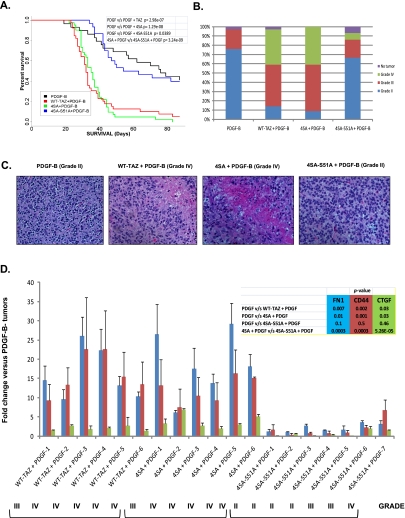
Although GSCs isolated from patients are valuable tools to study pathway alterations, they present two major technical limitations. First, by using GSCs, one cannot address the transforming potential of candidate oncogenes; that is, if TAZ-mediated MES transition is an oncogenic event, it cannot be addressed using GSCs. Second, tumor-initiating capacity in GSCs is assessed in immunocompromised recipient mice that form tumors, lacking immunological features of clinical tumors. To address these limitations and determine whether TAZ behaves as an oncogene, we used the well-characterized RCAS/N-tva model, in which the expression of retrovirus-encoded genes can be directed to express in neural progenitor cells (NPCs) (Uhrbom and Holland 2001; Begemann et al. 2002). Chicken fibroblast (DF1) cells transfected with the RCAS vector were directly injected into the brains of N-tva mice as previously described. Because this model allows for testing of specific gene alteration combinations in an otherwise normal background, PDGF-B was overexpressed with WT-TAZ, 4SA, and 4SA-S51A to test the effects on gliomagenesis. Moreover, PDGF-driven tumors exhibit predominantly PN characteristics (Lei et al. 2011), thus providing us with an ideal model to test TAZ effects in vivo. Consistent with prior studies, PDGF-B expression in Nestin+ NPCs resulted in grade II gliomas with a median survival of ∼11 wk (Fig. 6A; Dai et al.2001). Overexpression of TAZ or 4SA alone had no impact, since mice lived longer than 90 d post-implantation with no detectable tumors, suggesting that aberrant expression or activation of TAZ is insufficient by itself to cause transformation in NPCs (data not shown). On the other hand, when WT-TAZ or 4SA was coexpressed with PDGF-B, survival was dramatically reduced to <5 wk (Fig. 6A). A majority of the tumors from WT-TAZ or 4SA+PDGF-B were grade III (45%–50%) or grade IV (38%–41%), while those from PDGF-B alone were predominantly grade II (76%) (Fig. 6B,C). Although 4SA mice exhibit survival rates similar to the wild-type TAZ, 4SA+PDGF-B tumors were highly angiogenic and showed increased vascular proliferation (data not shown). Tumors resulting from 4SA-S51A coexpression with PDGF-B were predominantly grade II (67%) and were indistinguishable from those of PDGF-B alone (Fig. 6B,C). Last, we analyzed the expression of FN1, CD44, CTGF, and ACTG2 in tumors that arose from these mice to understand the spectrum of MES differentiation induced by TAZ. The expression of FN1, CD44, and CTGF was dramatically induced in both TAZ- and 4SA-driven tumors (Fig. 6D). ACTG2 was not induced in any of the groups compared with PDGF (data not shown). We next analyzed four grade II, two grade III, and one grade IV gliomas from the 4SA-S51A+PDGF-B tumors. Interestingly, expression levels of FN1, CD44, and CTGF were modestly increased with higher grade (PDGF+4SA-S51A-6 and 4SA-S51A-7). Thus, in rare cases, it appears that TAZ can induce high-grade MES tumors in a TEAD-independent fashion. However, the overall expression of MES markers in 4SA-S51A+PDGF tumors was significantly lower compared with that stemming from 4SA+PDGF overexpression (Fig. 6D). Thus, in line with our clinical observations and in vitro findings, overexpression of TAZ in conjunction with PDGF-B increases tumor grade and induces predominant MES differentiation in murine NPCs via interaction with TEAD.
Figure 6.
TAZ enhances tumor grade, reduces survival, and promotes MES differentiation in the RCAS/N-tva mouse model. (A) Kaplan-Meier survival analysis of PDGF-B-, WT-TAZ+PDGF-B-, 4SA+PDGF-B-, or 4SA-S51A+PDGF-B-injected mice. (B) Stacked bar graph showing WHO glioma grades within each group. (C) Representative images of hematoxylin and eosin-stained slides of brains isolated from mice injected with PDGF-B, WT-TAZ+PDGF-B, 4SA+PDGF-B, or 4SA-S51A+PDGF-B. Note that necrosis is observed only in WT-TAZ+PDGF-B and 4SA+PDGF-B mouse tumors. (D) Real-time qPCR analyses of gene expression in RCAS mouse tumors. The gene expression value of PDGF control was normalized to 1, and the relative expression of CD44, CTGF, and FN1 is shown.
Discussion
Our study demonstrates that the HIPPO pathway transcriptional coactivator TAZ is integral to the MES phenotype in glioma. First, we show that TAZ is epigenetically silenced in lower-grade gliomas as well as PN GBMs when compared with MES tumors. Second, manipulating TAZ expression in GSCs as well as murine NSCs affects expression of MES genes. Third, we demonstrate direct promoter co-occupancy of TAZ and TEAD2 in a majority of MES gene targets. Finally, we show that TAZ cooperates with PDGF-B to induce high-grade MES gliomas in the RCAS/N-tva model and that TEAD interaction is required for all of these processes. The results presented here implicate TAZ as a third key modulator of MES transition in glioma in addition to STAT3 and C/EBP-β.
Identification of TAZ as a key regulator of the MES network
Previous expression profiling efforts in GBM had failed to identify distinct patterns of gene expression owing primarily to fewer sample numbers. Large-scale studies by multiple groups eventually identified three (PN, MES, and proliferative) or four (PN, MES, neural, and classical) subtypes of tumors (Phillips et al. 2006; Verhaak et al. 2010). Of these, it is becoming evident that two robust, mutually exclusive gene expression patterns can readily be found across multiple data sets (PN and MES) (Huse et al. 2011). In the study by Phillips et al. (2006), a Proliferative subtype was identified in addition to the PN and MES groups. However, the Proliferative group may simply peel off tumors from the other two groups that have increased cell cycling, as suggested by the analysis of Huse et al. (2011). It may be reasonable to think of proliferation as an independent tumor axis, along with the mutually exclusive PN/MES axis. Likewise, the additional Neural and Classical groups that Verhaak et al. (2010) described seem to reclassify tumors that would be distributed equally between the PN and MES groups if these were the only classifications. Even though the active signaling pathways are different among the four Verhaak groups, the clinical relevance of adding the additional two Verhaak groups is not readily apparent. The MES subgroup, however, is of high interest to us, since these patients exhibit worse survival and treatment resistance compared with the other subtypes (Pelloski et al. 2005; Phillips et al. 2006; Colman et al. 2010). Identifying molecular drivers of the MES signature is a first step in effectively treating patients with this subclass of tumors, and a beginning for personalized medicine for GBM. Although various methods of genome-wide analyses of regulatory networks have been described, ARACNE is unique in that it identifies direct transcriptional interactions without a priori assumptions (Margolin et al. 2006). Using this method, the MES gene expression program was shown to be controlled in part by STAT3 and C/EBP-β in a recent report (Carro et al. 2010). However, the Carro et al. (2010) study and ours have several key differences that may account for us finding additional master regulators of the MES network. First, we used a larger data set (386 vs. 176 expression profiles) with a different microarray platform (HU-133A vs. HT-HU133). Second, our network analyses were limited to GBM only, whereas Carro et al. (2010) used a combination of grade III and grade IV astrocytomas. Third, we used a more comprehensive list of hub markers, which included transcriptional coregulators, TAZ being one of those absent from the prior work's TF list.
Epigenetic regulation of TAZ is important for its activity
Out of all of the HIPPO pathway components that had annotated CpG islands, only the TAZ promoter showed dramatic methylation differences between PN and MES subclasses of GBMs in the TCGA data sets. We further confirmed that TAZ is epigenetically silenced in lower-grade gliomas, PN GBMs, and PN GSCs. The paradoxical methylation of a tumor-enhancing gene like TAZ in gliomas is unconventional, since frequent epigenetic inactivation is typically associated with tumor suppressors (Palii and Robertson 2007). However, TAZ CpG methylation is concordant with a clinically distinct subclass of gliomas that is constitutively methylated in a cassette of genes (termed glioma CpG island methylator phenotype [G-CIMP]) (Noushmehr et al. 2010). A closer analysis of a few G-CIMP methylated genes from this study (CHI3L1, MMP9, LGALS3, PDPN, etc.) reveals that they are predominantly MES in nature (Hiratsuka et al. 2002; Wicki et al. 2006; Zhao et al. 2009). Perhaps methylation of TAZ and other G-CIMP genes restrains their expression in PN GBMs and lower-grade gliomas, which in turn results in favorable clinical response. In fact, the improved outcome in the PN subclass of tumors could be accounted for by a G-CIMP-positive subset that included methylated TAZ. As an aside, we noted that three of the 16 ARACNE-identified TFs in the MES network are part of the G-CIMP signature (Supplemental Table S4). Additionally, 12% of the ARACNE-identified TAZ target genes were G-CIMP (Supplemental Table S4).
The epigenetic control of TAZ further underlines the need for tight regulation of its expression and, in turn, its activity. Taken together with previous studies, TAZ now appears to be regulated at multiple levels, including promoter methylation (this study), subcellular localization, and phosphorylation. Indeed, our studies affirm that epigenetic regulation and cellular partitioning of TAZ play a major role in regulating its functions in high-grade gliomas and GSCs. For example, treating GSC8–11 with DAC not only induced TAZ expression, but also that of CTGF and CD44. We note, however, that DAC did not induce TAZ levels to those seen in GSC20, a MES GSC. Therefore, additional mechanisms may act to induce TAZ in MES GSCs.
MES transition is induced by the TAZ/TEAD complex
An important finding of our study is that TAZ alone can cause MES reprogramming in murine NSCs and human GBM-derived GSCs and that the TAZ–TEAD interaction is required for this process. Previous studies have shown that TAZ and its paralog, YAP, act as coactivators of numerous TFs (Yagi et al. 1999; Vassilev et al. 2001; Basu et al. 2003; Komuro et al. 2003; Hong et al. 2005; Murakami et al. 2005, 2006; Varelas et al. 2008; Di Palma et al. 2009; Zhang et al. 2009). Of particular interest is the association of TAZ/YAP with the TEAD family of TFs, as elucidated by the Guan group (Zhao et al. 2008b, Zhang et al. 2009). The TAZ/YAP interaction with TEAD promotes cell proliferation, EMT, invasion, and cellular transformation. In addition, YAP has been shown to regulate epidermal stem cell proliferation and expansion in mice via TEAD (Schlegelmilch et al. 2011). Other recent studies implicate TAZ/YAP as coregulators of SMAD activation induced by transforming growth factor-β (TGF-β) (Varelas et al. 2008). Although TGF-β has been shown to promote GSC self-renewal and tumor-initiating properties (Penuelas et al. 2009; Anido et al. 2010), we did not observe induction of MES genes by TGF-β in our GSC studies (data not shown). This observation, coupled with previous reports highlighting the importance of TEAD in mediating TAZ-induced EMT, led us to pursue TEAD as a candidate TF for TAZ-induced MES transition in GSCs. Indeed, TEAD interaction was sufficient and required for MES transition induced by TAZ, while RUNX2 was dispensable for these functions.
We aimed at delineating TAZ and YAP functions in this study in the context of glial tumors. While a role for YAP in inducing MES differentiation cannot be formally ruled out, we found YAP to be highly expressed in PN GSCs at levels comparable with MES. Thus, YAP may not be required for maintenance of the MES phenotype. Previous studies point to a more predominant role for YAP in embryonic stem cell self-renewal and proliferation (Lian et al. 2010), and in the chicken neural tube, YAP overexpression caused expansion of neural progenitor numbers, whereas loss of function promotes apoptosis and premature neuronal differentiation (Cao et al. 2008). On the contrary, we found that TAZ induces MES markers in murine NSCs, with a parallel loss of glial/neuronal differentiation, indicating that the predominant effect of TAZ in NSCs is to promote MES differentiation. In fact, NSCs overexpressing 4SA invariably exited the cell cycle after few rounds of proliferation (data not shown), arguing against a role for TAZ in inducing proliferation in these cells. Similarly, in mice, deletion of YAP or TEAD1/2 induces embryonic lethality (Morin-Kensicki et al. 2006; Sawada et al. 2008), whereas mice lacking TAZ are viable and develop renal cysts, polycystic kidney disease, and minor skeletal abnormalities (Hossain et al. 2007; Makita et al. 2008). Thus, although some properties of TAZ and YAP, including TEAD binding, appear to be shared, gain and loss of function of these cofactors elicit differential responses in a spatiotemporal manner.
While we show that TAZ induces MES high-grade gliomas in nestin-positive NPCs using the PDGF-driven RCAS mouse model, it is currently unclear whether TAZ can reprogram other cell types. The exact cell of origin for GBM is debatable, and the concept is still evolving. Early studies showed that Ink4a/Arf−/− NSCs or astrocytes with constitutively active EGFR induce high-grade gliomas (Bachoo et al. 2002). Subsequently, Parada and colleagues showed that specific ablation of key tumor suppressor genes in NSCs, but not in the adult nonneurogenic brain, caused gliomagenesis (Alcantara Llaguno et al. 2009). On the contrary, recent studies point to OPCs, but not other NSC-derived lineages or NSCs themselves, as the cell of origin of gliomas (Liu et al. 2011; Sugiarto et al. 2011). It remains unknown whether TAZ can reprogram every cell type that is proposed to be the cell of origin of glioma. Future studies wherein TAZ is expressed in the Ctv-a mouse, in which tumor formation can be examined in OPCs (Lindberg et al. 2009). could help address this important issue.
Clinical implications
Our in silico, in vitro, and in vivo studies of TAZ in glioma have important therapeutic implications. Patients whose tumors express MES genes present a considerable clinical challenge, since they show worse survival and radiation resistance (Phillips et al. 2006). We identified TAZ as an additional key transcriptional activator of the MES phenotype, which therefore presents another molecular target. Furthermore, we developed a mouse model using the RCAS/N-tva system and showed that TAZ in combination with PDGFB can drive formation of high-grade MES gliomas and therefore could be a useful preclinical model. This, to our knowledge, remains the first demonstration of TAZ as a tumor promoter in an in vivo setting. Identifying inhibitors of TAZ expression or TAZ–TEAD interaction will be the next challenge. With numerous clinical trials using STAT3 inhibitors for solid tumors currently ongoing, combination therapy with a TAZ inhibitor could be a viable strategy for treating the aggressive MES subclass of GBMs.
Materials and methods
Bioinformatic analyses
The ARACNE algorithm, as implemented in the geWorkBench suite (https://cabig.nci.nih.gov/tools/geWorkbench), was performed on the TCGA Affymetrix expression data set (downloaded April 28, 2011; n = 385), which was processed using a custom CDF and RMA normalization using R and Bioconductor (http://www.R-project.org). The resulting matrix of data was processed with the following ARACNE settings: P-value of 0.01, adaptive partitioning, and data processing inequality (DPI) tolerance of 0.1. For hub markers and the DPI target list, a composite set of TFs was generated from the TRANSFAC Web site and genes with the term “transcription factor” in their parent gene ontology (GO) term, which includes transcription cofactors. The initial resulting ARACNE network was limited to GBM MES genes by combining the gene lists from Phillips et al. (2006) and Verhaak et al. (2010) and then selecting the immediately adjacent hub genes. Target genes that were identified to be associated with TAZ by ARACNE analysis were processed using the DAVID Web tool (http://david.abcc.ncifcrf.gov) to obtain association of these genes with specific GO functional categories. Default processing was done, except the analysis was limited to DAVID's GO biological process FAT (GOTERM_BP_FAT) category. The resulting GO terms were ranked from smallest to largest P-values after removing GO terms that had 10 or fewer genes overlapping with the query gene list. To determine whether a TCGA GBM was PN or MES, first a metagene score for MES or PN was generated using a union of the respective Phillips and Verhaak gene sets. The two metagene scores were then compared, with class being assigned based on the greater metagene score. Tumors that had both Illumina Infinium methylation data and Affymetrix gene expression data were subsequently analyzed for correlation of TAZ methlation with TAZ expression and GBM subtype.
Cell culture and transfection
GSCs were isolated from patients undergoing surgery at M.D. Anderson Cancer Center (MDACC) and grown in neural basal medium (Dulbecco's Modified Eagle Medium [DMEM]/F12 50/50; Cellgro) supplemented with B27 (Invitrogen), 20 ng/mL EGF (Chemicon), and 20 ng/mL FGF (Akron-Biotech). Phoenix Ampho (for GSCs) or Phoenix Eco (for mouse NSCs) was obtained from a commercial source (Orbigen), grown in DMEM–F12 plus 10% fetal bovine serum (FBS), and transfected with 16 μg of DNA (pBABE vector containing wild-type TAZ, 4SA, 4SA-S51A) using Lipofectamine 2000 (Invitrogen) or Fugene HD (Roche) according to the manufacturer's instructions. Viral supernatants were collected 48 h after transfection, filtered, and immediately used for transduction of GSCs. For lentivial transduction, pGIPZ vectors expressing shRNA against two independent mRNA regions of TAZ were used. Viral particles were generated using the Trans-lentiviral packing system (Open Biosystems). Cells were plated on laminin (BD Biosciences)-coated plates, and then transduced with viral supernatant plus polybrene (Chemicon). The pBABE-4SA-S51A plasmid was made using the QuikChange II XL Site-Directed Mutatgenesis kit (Stratagene 200521) and primers designed to introduce serine-to-alanine mutation (5′-CGGAAGAAGATCCTGCCGGAGGCCTTCTTTAAGGAGCCTG-3′; 5′-CAGGCTCCTTAAAGAAGGCCTCCGGCAGGATCTTCTTCCG-3′). Both pBABE and pGIPZ transduced GSCs were selected in neurobasal medium containing puromycin (4 μg/mL) for generation of stable clones.
Intracranial mouse injections
SCID mice were bolted as previously described (Lal et al. 2000) and injected with cells (10 K/μL) in serum-free medium. Five mice were injected for each group. Mice were sacrificed once they showed neurological symptoms or appeared moribund. Brains were immediately fixed in 4% paraformaldehyde and processed for hematoxylin and eosin staining or IHC.
NSC isolation
NSCs were isolated from the telecephalon of C57/BL6 mouse embryos at E14 and cultured as described (Sher et al. 2008). Briefly, the telecephalon were dissociated, incubated with Accutase (Sigma-Aldrich), and cultured in neurobasal medium supplemented with 2% B27, 20 ng/mL EGF, 20 ng/mL bFGF, 1% GlutaMAX (GIBCO), 100 mg/mL primocin, and 5 mg/mL heparin (Sigma-Aldrich). After two passages, NSCs were used for viral transduction as described previously. For differentiation, stable pools of NSCs were dissociated and plated in poly-d-lysine and laminin-coated 12-mm glass coverslips in NBM supplemented with 5% FBS or after withdrawal of bFGF and EGF from the proliferation medium for 6 d and subsequently processed for IF. The following antibodies were used to identify undifferentiated NSCs and differentiated cell types: nestin, FN1 (Millipore), GFAP (DAKO), and SMA (Sigma-Aldrich).
Microarray analyses
Fifteen micrograms of cRNA was used in the hybridizations to U133A 2.0 human GeneChip expression arrays done according to the specifications of the manufacturer (Affymetrix). Intensity data were obtained from array images, and data were analyzed using R Suite as previously described.
ChIP assay
ChIP assay was performed after cross-linking cells using formaldehyde. DNA was sonicated using an Ultrasonic Processor (GE130, Sorvall) at two cycles of six pulses each at 50% power with a 1-min interval between cycles. Sonicated DNA was then centrifuged at 13,5000 rpm at 4°C. Supernatant from 100,000 cells was used for each ChIP assay using MAGnify ChIP system (Invitrogen). Two micrograms of mouse IgG, TAZ (BD Biosciences), or TEAD2 (Novus) was used per ChIP. Immunoprecipitated DNA was analyzed by Syber Green PCR, and Ct values were used to calculate the percentage of input enrichment. Primer sequences are shown in Supplemental Table S5.
RCAS mice
The RCAS model for somatic gene transfer has been previously described in detail (Uhrbom and Holland 2001; Begemann et al. 2002). Briefly, DF1 transfected cells were directly injected intracranially on both sides (10,000 cells per microliter) into 2-d-old pups. Mice were sacrificed when neurological symptoms were present (i.e., hydrocephalus, seizures, inactivity, and/or ataxia). Mice were sacrificed after 90-d post-injection if they did not show symptoms. Brains were fixed in formalin and embedded in paraffin blocks for further analyses.
Acknowledgments
We thank the generosity of Dr. Kun-Lian Guan (University of California at San Diego) for donating the pBABE plasmids. This research was supported by funding from the American Brain Tumor Association basic research fellowship, Odyssey Special Fellowship, Caroline Ross Endowment Fellowship, and MDACC Brain Tumor SPORE developmental research project grant (to K.P.L.B.); the Center for Clinical and Translational Sciences T32 grant (to K.S.); the NINDS grant (NS070928 to G.R.); the Brain Tumor Funders' Collaborative, the V Foundation, Rose Foundation, National Brain Tumor Society basic research award, and SPORE grant P50CA127001 from NIH/NCI (to K.A.); Ben and Cathy Ivy Foundation Research Award (to F.F.L., K.A., and E.P.S); and SPORE Animal Core grant (to F.F.L.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.176800.111.
References
- Alcantara Llaguno S, Chen J, Kwon CH, Jackson EL, Li Y, Burns DK, Alvarez-Buylla A, Parada LF 2009. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell 15: 45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anido J, Saez-Borderias A, Gonzalez-Junca A, Rodon L, Folch G, Carmona MA, Prieto-Sanchez RM, Barba I, Martinez-Saez E, Prudkin L, et al. 2010. TGF-β receptor inhibitors target the CD44(high)/Id1(high) glioma-initiating cell population in human glioblastoma. Cancer Cell 18: 655–668 [DOI] [PubMed] [Google Scholar]
- Bachoo RM, Maher EA, Ligon KL, Sharpless NE, Chan SS, You MJ, Tang Y, DeFrances J, Stover E, Weissleder R, et al. 2002. Epidermal growth factor receptor and Ink4a/Arf: Convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell 1: 269–277 [DOI] [PubMed] [Google Scholar]
- Basu S, Totty NF, Irwin MS, Sudol M, Downward J 2003. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell 11: 11–23 [DOI] [PubMed] [Google Scholar]
- Begemann M, Fuller GN, Holland EC 2002. Genetic modeling of glioma formation in mice. Brain Pathol 12: 117–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns WC, Twigg SM, Forbes JM, Pete J, Tikellis C, Thallas-Bonke V, Thomas MC, Cooper ME, Kantharidis P 2006. Connective tissue growth factor plays an important role in advanced glycation end product-induced tubular epithelial-to-mesenchymal transition: Implications for diabetic renal disease. J Am Soc Nephrol 17: 2484–2494 [DOI] [PubMed] [Google Scholar]
- Burton EC, Lamborn KR, Feuerstein BG, Prados M, Scott J, Forsyth P, Passe S, Jenkins RB, Aldape KD 2002. Genetic aberrations defined by comparative genomic hybridization distinguish long-term from typical survivors of glioblastoma. Cancer Res 62: 6205–6210 [PubMed] [Google Scholar]
- Cao X, Pfaff SL, Gage FH 2008. YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes Dev 22: 3320–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, et al. 2010. The transcriptional network for mesenchymal transformation of brain tumours. Nature 463: 318–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Chong YF, Pobbati AV, Huang C, Hong W 2011. Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J Biol Chem 286: 7018–7026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman H, Zhang L, Sulman EP, McDonald JM, Shooshtari NL, Rivera A, Popoff S, Nutt CL, Louis DN, Cairncross JG, et al. 2010. A multigene predictor of outcome in glioblastoma. Neuro Ooncol 12: 49–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper LA, Gutman DA, Long Q, Johnson BA, Cholleti SR, Kurc T, Saltz JH, Brat DJ, Moreno CS 2010. The proneural molecular signature is enriched in oligodendrogliomas and predicts improved survival among diffuse gliomas. PLoS ONE 5: e12548 doi: 10.1371/journal.pone.002548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Celestino JC, Okada Y, Louis DN, Fuller GN, Holland EC 2001. PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes Dev 15: 1913–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, Bird A 2011. CpG islands and the regulation of transcription. Genes Dev 25: 1010–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin A, Maiti B, Jakoi L, Timmers C, Buerki R, Leone G 2003. Identification and characterization of E2F7, a novel mammalian E2F family member capable of blocking cellular proliferation. J Biol Chem 278: 42041–42049 [DOI] [PubMed] [Google Scholar]
- de Groot JF, Gilbert MR, Aldape K, Hess KR, Hanna TA, Ictech S, Groves MD, Conrad C, Colman H, Puduvalli VK, et al. 2008. Phase II study of carboplatin and erlotinib (Tarceva, OSI-774) in patients with recurrent glioblastoma. J Neurooncol 90: 89–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4: 3 doi: 10.1186/gb-2003-4-5-p3 [PubMed] [Google Scholar]
- Di Palma T, D'Andrea B, Liguori GL, Liguoro A, de Cristofaro T, Del Prete D, Pappalardo A, Mascia A, Zannini M 2009. TAZ is a coactivator for Pax8 and TTF-1, two transcription factors involved in thyroid differentiation. Exp Cell Res 315: 162–175 [DOI] [PubMed] [Google Scholar]
- Duning K, Rosenbusch D, Schluter MA, Tian Y, Kunzelmann K, Meyer N, Schulze U, Markoff A, Pavenstadt H, Weide T 2010. Polycystin-2 activity is controlled by transcriptional coactivator with PDZ binding motif and PALS1-associated tight junction protein. J Biol Chem 285: 33584–33588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freije WA, Castro-Vargas FE, Fang Z, Horvath S, Cloughesy T, Liau LM, Mischel PS, Nelson SF 2004. Gene expression profiling of gliomas strongly predicts survival. Cancer Res 64: 6503–6510 [DOI] [PubMed] [Google Scholar]
- Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al. 2007. Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Genes Dev 21: 2683–2710 [DOI] [PubMed] [Google Scholar]
- Gage FH, Ray J, Fisher LJ 1995. Isolation, characterization, and use of stem cells from the CNS. Annu Rev Neurosci 18: 159–192 [DOI] [PubMed] [Google Scholar]
- Gore-Hyer E, Shegogue D, Markiewicz M, Lo S, Hazen-Martin D, Greene EL, Grotendorst G, Trojanowska M 2002. TGF-β and CTGF have overlapping and distinct fibrogenic effects on human renal cells. Am J Physiol Renal Physiol 283: F707–F716 doi: 10.1152/ajprenal.00007.2002 [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ 2008. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10: 593–601 [DOI] [PubMed] [Google Scholar]
- Gupta PB, Chaffer CL, Weinberg RA 2009. Cancer stem cells: Mirage or reality? Nat Med 15: 1010–1012 [DOI] [PubMed] [Google Scholar]
- Halder G, Johnson RL 2011. Hippo signaling: Growth control and beyond. Development 138: 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, et al. 2005. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 352: 997–1003 [DOI] [PubMed] [Google Scholar]
- Hiratsuka S, Nakamura K, Iwai S, Murakami M, Itoh T, Kijima H, Shipley JM, Senior RM, Shibuya M 2002. MMP9 induction by vascular endothelial growth factor receptor-1 is involved in lung-specific metastasis. Cancer Cell 2: 289–300 [DOI] [PubMed] [Google Scholar]
- Hong JH, Yaffe MB 2006. TAZ: A β-catenin-like molecule that regulates mesenchymal stem cell differentiation. Cell Cycle 5: 176–179 [DOI] [PubMed] [Google Scholar]
- Hong JH, Hwang ES, McManus MT, Amsterdam A, Tian Y, Kalmukova R, Mueller E, Benjamin T, Spiegelman BM, Sharp PA, et al. 2005. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science 309: 1074–1078 [DOI] [PubMed] [Google Scholar]
- Hossain Z, Ali SM, Ko HL, Xu J, Ng CP, Guo K, Qi Z, Ponniah S, Hong W, Hunziker W 2007. Glomerulocystic kidney disease in mice with a targeted inactivation of Wwtr1. Proc Natl Acad Sci 104: 1631–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D 2005. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122: 421–434 [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Huse JT, Holland EC 2010. Targeting brain cancer: Advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer 10: 319–331 [DOI] [PubMed] [Google Scholar]
- Huse JT, Phillips HS, Brennan CW 2011. Molecular subclassification of diffuse gliomas: Seeing order in the chaos. Glia 59: 1190–1199 [DOI] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA 2009. The basics of epithelial–mesenchymal transition. J Clin Invest 119: 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro A, Nagai M, Navin NE, Sudol M 2003. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem 278: 33334–33341 [DOI] [PubMed] [Google Scholar]
- Kudo-Saito C, Shirako H, Takeuchi T, Kawakami Y 2009. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell 15: 195–206 [DOI] [PubMed] [Google Scholar]
- Lal S, Lacroix M, Tofilon P, Fuller GN, Sawaya R, Lang FF 2000. An implantable guide-screw system for brain tumor studies in small animals. J Neurosurg 92: 326–333 [DOI] [PubMed] [Google Scholar]
- Lei QY, Zhang H, Zhao B, Zha ZY, Bai F, Pei XH, Zhao S, Xiong Y, Guan KL 2008. TAZ promotes cell proliferation and epithelial–mesenchymal transition and is inhibited by the hippo pathway. Mol Cell Biol 28: 2426–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, Sonabend AM, Guarnieri P, Soderquist C, Ludwig T, Rosenfeld S, Bruce JN, Canoll P 2011. Glioblastoma models reveal the connection between adult glial progenitors and the proneural phenotype. PLoS ONE 6: e20041 doi: 10.1371/journal.pone.0020041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Walling J, Ahn S, Kotliarov Y, Su Q, Quezado M, Oberholtzer JC, Park J, Zenklusen JC, Fine HA 2009. Unsupervised analysis of transcriptomic profiles reveals six glioma subtypes. Cancer Res 69: 2091–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, Chinnaiyan A, Israel MA, Goldstein LS, Abujarour R, et al. 2010. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev 24: 1106–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg N, Kastemar M, Olofsson T, Smits A, Uhrbom L 2009. Oligodendrocyte progenitor cells can act as cell of origin for experimental glioma. Oncogene 28: 2266–2275 [DOI] [PubMed] [Google Scholar]
- Liu CY, Zha ZY, Zhou X, Zhang H, Huang W, Zhao D, Li T, Chan SW, Lim CJ, Hong W, et al. 2010. The hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCFβ-TrCP E3 ligase. J Biol Chem 285: 37159–37169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Sage JC, Miller MR, Verhaak RG, Hippenmeyer S, Vogel H, Foreman O, Bronson RT, Nishiyama A, Luo L, et al. 2011. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell 146: 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makita R, Uchijima Y, Nishiyama K, Amano T, Chen Q, Takeuchi T, Mitani A, Nagase T, Yatomi Y, Aburatani H, et al. 2008. Multiple renal cysts, urinary concentration defects, and pulmonary emphysematous changes in mice lacking TAZ. Am J Physiol Renal Physiol 294: F542–F553 doi: 10.1152/ajprenal.00201.2007 [DOI] [PubMed] [Google Scholar]
- Mani SA, Yang J, Brooks M, Schwaninger G, Zhou A, Miura N, Kutok JL, Hartwell K, Richardson AL, Weinberg RA 2007. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci 104: 10069–10074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. 2008. The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell 133: 704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin AA, Wang K, Lim WK, Kustagi M, Nemenman I, Califano A 2006. Reverse engineering cellular networks. Nat Protoc 1: 662–671 [DOI] [PubMed] [Google Scholar]
- Martinez J, Chalupowicz DG, Roush RK, Sheth A, Barsigian C 1994. Transglutaminase-mediated processing of fibronectin by endothelial cell monolayers. Biochemistry 33: 2538–2545 [DOI] [PubMed] [Google Scholar]
- Meloan SN, Puchtler H, Valentine LS 1972. Alkaline and acid alizarin red S stains for alkali-soluble and alkali-insoluble calcium deposits. Arch Pathol 93: 190–197 [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. 2003. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34: 267–273 [DOI] [PubMed] [Google Scholar]
- Morin-Kensicki EM, Boone BN, Howell M, Stonebraker JR, Teed J, Alb JG, Magnuson TR, O'Neal W, Milgram SL 2006. Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol Cell Biol 26: 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Nakagawa M, Olson EN, Nakagawa O 2005. A WW domain protein TAZ is a critical coactivator for TBX5, a transcription factor implicated in Holt-Oram syndrome. Proc Natl Acad Sci 102: 18034–18039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Tominaga J, Makita R, Uchijima Y, Kurihara Y, Nakagawa O, Asano T, Kurihara H 2006. Transcriptional activity of Pax3 is co-activated by TAZ. Biochem Biophys Res Commun 339: 533–539 [DOI] [PubMed] [Google Scholar]
- Nakajima Y, Mironov V, Yamagishi T, Nakamura H, Markwald RR 1997. Expression of smooth muscle α-actin in mesenchymal cells during formation of avian endocardial cushion tissue: A role for transforming growth factor β3. Dev Dyn 209: 296–309 [DOI] [PubMed] [Google Scholar]
- Nigro JM, Misra A, Zhang L, Smirnov I, Colman H, Griffin C, Ozburn N, Chen M, Pan E, Koul D, et al. 2005. Integrated array-comparative genomic hybridization and expression array profiles identify clinically relevant molecular subtypes of glioblastoma. Cancer Res 65: 1678–1686 [DOI] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. 2010. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17: 510–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palii SS, Robertson KD 2007. Epigenetic control of tumor suppression. Crit Rev Eukaryot Gene Expr 17: 295–316 [DOI] [PubMed] [Google Scholar]
- Pan D 2010. The hippo signaling pathway in development and cancer. Dev Cell 19: 491–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SM, Gaur AB, Lengyel E, Peter ME 2008. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 22: 894–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelloski CE, Mahajan A, Maor M, Chang EL, Woo S, Gilbert M, Colman H, Yang H, Ledoux A, Blair H, et al. 2005. YKL-40 expression is associated with poorer response to radiation and shorter overall survival in glioblastoma. Clin Cancer Res 11: 3326–3334 [DOI] [PubMed] [Google Scholar]
- Penuelas S, Anido J, Prieto-Sanchez RM, Folch G, Barba I, Cuartas I, Garcia-Dorado D, Poca MA, Sahuquillo J, Baselga J, et al. 2009. TGF-β increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell 15: 315–327 [DOI] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, et al. 2006. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9: 157–173 [DOI] [PubMed] [Google Scholar]
- Remue E, Meerschaert K, Oka T, Boucherie C, Vandekerckhove J, Sudol M, Gettemans J 2010. TAZ interacts with zonula occludens-1 and -2 proteins in a PDZ-1 dependent manner. FEBS Lett 584: 4175–4180 [DOI] [PubMed] [Google Scholar]
- Ridet JL, Malhotra SK, Privat A, Gage FH 1997. Reactive astrocytes: Cellular and molecular cues to biological function. Trends Neurosci 20: 570–577 [DOI] [PubMed] [Google Scholar]
- Rosenberg L 1971. Chemical basis for the histological use of safranin O in the study of articular cartilage. J Bone Joint Surg Am 53: 69–82 [PubMed] [Google Scholar]
- Sawada A, Kiyonari H, Ukita K, Nishioka N, Imuta Y, Sasaki H 2008. Redundant roles of Tead1 and Tead2 in notochord development and the regulation of cell proliferation and survival. Mol Cell Biol 28: 3177–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieker M, Pautke C, Reitz K, Hemraj I, Neth P, Mutschler W, Milz S 2004. The use of four-colour immunofluorescence techniques to identify mesenchymal stem cells. J Anat 204: 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieker M, Pautke C, Haasters F, Schieker J, Docheva D, Bocker W, Guelkan H, Neth P, Jochum M, Mutschler W 2007. Human mesenchymal stem cells at the single-cell level: Simultaneous seven-colour immunofluorescence. J Anat 210: 592–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, Kreger BT, Vasioukhin V, Avruch J, Brummelkamp TR, et al. 2011. Yap1 acts downstream of α-catenin to control epidermal proliferation. Cell 144: 782–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher F, Rossler R, Brouwer N, Balasubramaniyan V, Boddeke E, Copray S 2008. Differentiation of neural stem cells into oligodendrocytes: Involvement of the polycomb group protein Ezh2. Stem Cells 26: 2875–2883 [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiarto S, Persson AI, Munoz EG, Waldhuber M, Lamagna C, Andor N, Hanecker P, Ayers-Ringler J, Phillips J, Siu J, et al. 2011. Asymmetry-defective oligodendrocyte progenitors are glioma precursors. Cancer Cell 20: 328–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa T, Nakashima K, Namihira M, Ochiai W, Uemura A, Yanagisawa M, Fujita N, Nakao M, Taga T 2001. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell 1: 749–758 [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA 2009. Epithelial–mesenchymal transitions in development and disease. Cell 139: 871–890 [DOI] [PubMed] [Google Scholar]
- Uhrbom L, Holland EC 2001. Modeling gliomagenesis with somatic cell gene transfer using retroviral vectors. J Neurooncol 53: 297–305 [DOI] [PubMed] [Google Scholar]
- Varelas X, Sakuma R, Samavarchi-Tehrani P, Peerani R, Rao BM, Dembowy J, Yaffe MB, Zandstra PW, Wrana JL 2008. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat Cell Biol 10: 837–848 [DOI] [PubMed] [Google Scholar]
- Varelas X, Miller BW, Sopko R, Song S, Gregorieff A, Fellouse FA, Sakuma R, Pawson T, Hunziker W, McNeill H, et al. 2010a. The Hippo pathway regulates Wnt/β-catenin signaling. Dev Cell 18: 579–591 [DOI] [PubMed] [Google Scholar]
- Varelas X, Samavarchi-Tehrani P, Narimatsu M, Weiss A, Cockburn K, Larsen BG, Rossant J, Wrana JL 2010b. The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-β–SMAD pathway. Dev Cell 19: 831–844 [DOI] [PubMed] [Google Scholar]
- Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML 2001. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev 15: 1229–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. 2010. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17: 98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward RJ, Lee L, Graham K, Satkunendran T, Yoshikawa K, Ling E, Harper L, Austin R, Nieuwenhuis E, Clarke ID, et al. 2009. Multipotent CD15+ cancer stem cells in patched-1-deficient mouse medulloblastoma. Cancer Res 69: 4682–4690 [DOI] [PubMed] [Google Scholar]
- Wicki A, Lehembre F, Wick N, Hantusch B, Kerjaschki D, Christofori G 2006. Tumor invasion in the absence of epithelial–mesenchymal transition: Podoplanin-mediated remodeling of the actin cytoskeleton. Cancer Cell 9: 261–272 [DOI] [PubMed] [Google Scholar]
- Yagi R, Chen LF, Shigesada K, Murakami Y, Ito Y 1999. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J 18: 2551–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA 2004. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117: 927–939 [DOI] [PubMed] [Google Scholar]
- Zhang H, Liu CY, Zha ZY, Zhao B, Yao J, Zhao S, Xiong Y, Lei QY, Guan KL 2009. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial–mesenchymal transition. J Biol Chem 284: 13355–13362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Lei QY, Guan KL 2008a. The Hippo-YAP pathway: New connections between regulation of organ size and cancer. Curr Opin Cell Biol 20: 638–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, et al. 2008b. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 22: 1962–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Guo X, Nash GB, Stone PC, Hilkens J, Rhodes JM, Yu LG 2009. Circulating galectin-3 promotes metastasis by modifying MUC1 localization on cancer cell surface. Cancer Res 69: 6799–6806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Lei Q, Guan KL 2010. The Hippo-YAP pathway in organ size control and tumorigenesis: An updated version. Genes Dev 24: 862–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q, Guan KL 2011. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev 25: 51–63 [DOI] [PMC free article] [PubMed] [Google Scholar]