β-Catenin performs two varieties of cellular functions: It is a pivotal component of the canonical Wnt signaling pathway, and it has a structural role in regulating cell–cell adhesion. Due to these dual functions, obtaining insight into specific arms of β-catenin activity in a whole-organism, especially mammalian, context has been problematic. Balser and colleagues now provide a breakthrough by creating a β-catenin mutant form that maintains its adhesion function but specifically blocks its Wnt signaling activity. Performed in both mice and Drosophila, this study provides an invaluable tool in specifically dissecting Wnt/β-catenin signaling activity in a tissue- and developmental stage-specific manner.
Keywords: canonical Wnt signaling, signaling versus structural function of β-catenin, mouse strains expressing transcriptionally inactive β-catenin, cell fate determination in the dorsal neural tube
Abstract
β-Catenin, apart from playing a cell-adhesive role, is a key nuclear effector of Wnt signaling. Based on activity assays in Drosophila, we generated mouse strains where the endogenous β-catenin protein is replaced by mutant forms, which retain the cell adhesion function but lack either or both of the N- and the C-terminal transcriptional outputs. The C-terminal activity is essential for mesoderm formation and proper gastrulation, whereas N-terminal outputs are required later during embryonic development. By combining the double-mutant β-catenin with a conditional null allele and a Wnt1-Cre driver, we probed the role of Wnt/β-catenin signaling in dorsal neural tube development. While loss of β-catenin protein in the neural tube results in severe cell adhesion defects, the morphology of cells and tissues expressing the double-mutant form is normal. Surprisingly, Wnt/β-catenin signaling activity only moderately regulates cell proliferation, but is crucial for maintaining neural progenitor identity and for neuronal differentiation in the dorsal spinal cord. Our model animals thus allow dissecting signaling and structural functions of β-catenin in vivo and provide the first genetic tool to generate cells and tissues that entirely and exclusively lack canonical Wnt pathway activity.
β-Catenin fulfills two key functions in metazoan organisms. First, it serves as an important component in the cadherin-based cell–cell adhesion system that is required for the structural integrity and functional polarization of epithelia. β-Catenin binds the transmembrane protein E-cadherin and regulates actin filament assembly via α-catenin (Heuberger and Birchmeier 2010). Experimental removal of β-catenin in the mouse embryo results in progressive body disintegration caused by the collapse of cadherin-based cell adhesiveness (Hierholzer and Kemler 2010).
Second, β-catenin is the central regulator of gene expression in the canonical Wnt signal transduction pathway. In the absence of a Wnt signal, β-catenin is phosphorylated by a multiprotein degradation complex. Phosphorylation of β-catenin results in its ubiquitylation and subsequent degradation by the proteasome. Binding of Wnts to their cell surface receptors triggers a complex signaling cascade that leads to the inhibition of β-catenin degradation and hence increased levels of cytoplasmic β-catenin. Some of this β-catenin enters the nucleus and associates with transcription factors of the TCF/LEF family (Cadigan and Liu 2006). On their own, these factors possess only a limited ability to regulate transcription. Indeed, in the absence of Wnt signals, TCF/LEF proteins mediate transcriptional repression by forming a complex with Groucho/TLE repressors. The binding of β-catenin displaces Groucho and converts TCF/LEF factors into activators, thereby translating the Wnt signal into a specific transcriptional program (Mosimann et al. 2009).
The β-catenin protein consists of a central region that is made up of 12 imperfect Armadillo (Arm) repeats flanked by specific N- and C-terminal tails (Huber et al. 1997; Xing et al. 2008). Some mutations in the gene encoding the Drosophila ortholog of β-catenin, Armadillo, interfere with the adhesion function but not its role in Wnt/Wingless (Wg) signaling and vice versa, indicating that these two functions are independent and separable (Orsulic and Peifer 1996). The signaling activity of Arm/β-catenin depends on different regions within the polypeptide sequence. The N-terminal domain of the first Arm repeat region is necessary and sufficient for recruitment of Legless (Lgs)/B-cell lymphoma 9 (BCL9), which in turn recruits Pygopus (Pygo). The latter appears to act as a transcriptional coactivator (Hoffmans and Basler 2004; Hoffmans et al. 2005). The central Arm repeats mediate the interaction with TCF/LEF transcription factors. This region is also required for the binding to components of the adherens junctions; notably, E-cadherin (Graham et al. 2000). Finally, a region comprising Arm repeats R11–R12 and the C-terminal tail recruits a multitude of proteins involved in the transcription process, such as TATA-binding protein (TBP), Brahma/Brahma-related gene-1 (Brg-1), CREB-binding protein (CBP)/p300, Mediator subunit 12 (MED12), and Hyrax/Parafibromin. Many of these proteins are components of chromatin remodelling complexes or allow β-catenin to connect with the basic transcription initiation and elongation machinery (for a detailed review, see Mosimann et al. 2009).
Despite the immense interest in deepening our understanding of canonical Wnt signaling, its roles in development and disease, as well as the in vivo activities of this pathway, are only poorly defined. Genetic investigations in vertebrates are confounded by the large size of the Wnt ligand and receptor families, the multiple roles played by the transduction components, and the varied signs of action of the TCF/LEF transcription factors. Despite potential complications due to its dual role, major attention has been focused on studying the in vivo requirements of β-catenin, the nonredundant pivot of the canonical pathway. In mice, a large body of experiments has been generated using conditional ablation of β-catenin by using tissue-specific Cre drivers (for review, see Grigoryan et al. 2008). However, this approach does not reveal the contribution of defective cell adhesion to the observed phenotypes. For example, the loss of β-catenin in Wnt1-Cre-expressing tissues severely affects midbrain and hindbrain development and also causes defects in the dorsal neural tube, but to what extent the signaling phenotypes are responsible for the observed defects is obscured by impaired epithelial integrity (Brault et al. 2001; Ille et al. 2007).
Here we exploit the molecular knowledge of β-catenin and assays for monitoring Wg and Wnt pathway activity to develop and validate a tool for the in vivo analysis specifically of canonical Wnt signaling in mice. By depriving β-catenin of only its transcriptional activity, leaving its adhesive functions intact, we can generate cells and tissues that entirely lack canonical pathway activity. Moreover, the tools described here allow us to compare the contributions of BCL9/Legless and the C-terminal coactivators to Wnt/β-catenin-mediated transcription in different tissues at different times of development.
Results
The N- and C-terminal coactivator branches both contribute to the transcriptional output of β-catenin (Armadillo)
To discriminate between the transcriptional output of β-catenin and its structural role, we generated mutant forms of Arm and β-catenin, which lack the ability to bind N-terminal and/or C-terminal coactivators. We introduced a single amino acid change into the first Arm repeat of β-catenin (D164A), blocking the interaction between β-catenin and its N-terminal coactivator, BCL9/BCL9L. A C-terminal truncation (ΔC) in β-catenin abrogated the interaction with the multitude of coactivators that bind there. Finally, β-catenin-D164A-ΔC contains both the D164A mutation and the C-terminal deletion and is referred to as β-catenin-dm (for double mutant). These mutations were validated biochemically (Supplemental Fig. 1A,B). Equivalent forms of Arm were also generated (Arm D172 corresponds to D164 of mammalian β-catenin).
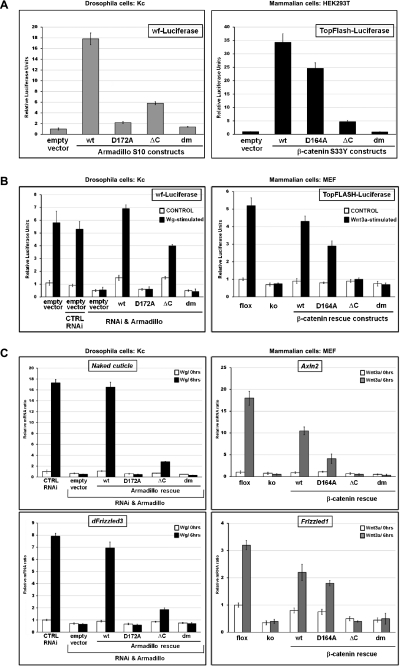
As an initial step, we compared the functional consequences of the D172A/D164A mutation and the C-terminal truncation on Arm and β-catenin function in transactivation reporter assays. The effect of the mutations on the signaling capacities of constitutively active forms of Arm (ArmS10) and β-catenin (β-cateninS33Y) was assayed in Drosophila Kc cells and clone-8 cells (data not shown) and mammalian HEK293 cells, respectively. ArmS10 strongly activates transcription of the Wg pathway-specific reporter wf-Luciferase (Fig. 1A). Similarly, β-cateninS33Y strongly activates expression of the mammalian Wnt reporter TOPFlash. Only double-mutant forms of either ArmS10 or β-cateninS33Y lacked any detectable transcription-inducing activity; single mutants retained transcription potential, especially in mammalian cells (Fig. 1A).
Figure 1.
Requirements for N- and C-terminal coactivators for Arm- and β-catenin-mediated transcription. (A–C) The following mutant forms of Arm/β-catenin were tested: wild type (wt), compromised in binding to Lgs/BCL9 (D172A for Arm and D164A for β-catenin), lacking the ability to interact with C-terminal coactivators (ΔC), and the composite of these two individual mutations (dm). (A,B) Luciferase reporter assays in Drosophila Kc and mammalian HEK293T or MEF cells. The activity of Wnt/β-catenin transcription was determined in Kc cells by a wingful-Luciferase (wf-Luciferase) reporter and in mammalian cells by TopFlash. The Y-axes show relative luciferase units normalized to the levels of constantly expressed Renilla. Error bars represent standard deviations. (A) Expression vectors are based on constructs encoding a constitutive active form of Arm (ArmS10) or β-catenin (β-cateninS33Y); the different mutant plasmids were transfected into Kc or HEK293T cells as indicated. Luciferase units shown were normalized relative to those from cells that were transfected in parallel with empty vector (and set to 1). (B) Drosophila Kc cells were treated with dsRNA directed against UTRs of arm mRNA or with control dsRNAi. Cells treated with dsRNAi were transfected with tubulinα1 promoter-driven mutant arm constructs to rescue Arm protein expression. All cells were cultivated with Wg-conditioned medium or control medium. Mammalian MEFs containing a conditional knockout allele of β-catenin (flox) were used for the generation of β-catenin-null MEFs (ko). ko cells were infected with retroviral particles encoding different β-catenin rescue constructs as indicated. Cells were stimulated by Wnt3a-conditioned medium or control medium. Luciferase units shown were normalized relative to those from cells that expressed endogenous Arm or β-catenin in the Wnt-unstimulated situation. (C) Relative expression of the Wg targets Naked cuticle and dFrizzled3 in Drosophila Kc cells (which expressed different mutant forms of Arm instead of endogenous wild-type Arm) and relative expression levels of the Wnt/β-catenin targets Axin2 and Frizzled1 upon Wnt3a treatment in MEFs whose endogenous β-catenin expression was substituted with mutant forms. Cells were generated as in B. mRNA levels were normalized to those of the housekeeping genes SDHA and GAPDH. Y-axes show normalized relative mRNA levels; Wnt-unstimulated cells expressing endogenous Arm and β-catenin, respectively, were set to 1. Error bars show standard deviations.
The above reporter assays were based on constitutively active, overexpressed forms of Arm and β-catenin. We also examined the effect of the mutations in otherwise wild-type proteins that were expressed at normal levels and that constitute the only source of Arm and β-catenin, respectively (Supplemental Fig. 1C). For mammalian β-catenin, we started with mouse embryonic fibroblasts (MEFs) derived from conditional knockout animals (β-cateninflox/flox), eliminated endogenous β-catenin by infection with self-excising lentiviral Cre, and then reinstalled expression of different β-catenin mutants by retroviral infection. In Drosophila cells, we used a highly effective dsRNA targeting the untranslated regions (UTRs) of arm, followed by transfection of expression constructs under control of the moderately active tubulinα1 promoter. Expression of wild-type constructs rescued the responsiveness of both mammalian and Drosophila cells to exogenously provided Wnt3a and Wg, respectively (Fig. 1B). The transcriptional activation of endogenous target genes was also restored: We analyzed Axin2 and Frizzled1 in MEFs and Naked cuticle and dFrizzled3 in Drosophila cells (Fig. 1C). The behavior of the mutant forms of β-catenin and Arm in this more refined system confirmed that both the N- and C-terminal coactivators are important for proper transcriptional activity of β-catenin/Armadillo. Consistent with earlier results, the individual contribution of the two branches seems to differ in Drosophila and mammals (Fig. 1C). While in Drosophila Arm output is critically dependent on its interaction with Legless (via residue D172; i.e., D164 in mammals), β-catenin output relies more heavily on the presence of the C-terminal tail. Whatever the case, the double-mutant proteins are transcriptionally inert in both Drosophila and mammals and hence provide a means to totally abrogate canonical Wnt signaling activity in vivo.
Double-mutant Arm (Arm-D172A-ΔC) sustains adherens junctions but fully blocks Wg-mediated transcription in vivo
The most stringent assay to check whether the double-mutant form of either Arm or β-catenin lost its activity in signaling, but not its structural function in adherens junctions, is to test the mutant proteins in vivo. Using the attP/ΦC31 integration system, we generated a series of Drosophila transgenic lines in which the arm transgenes were driven by the tubulinα1 promoter. All transgenes were integrated at the same predefined landing site and expressed equal levels of the Arm protein, comparable with those of the endogenous arm locus (Supplemental Fig. 2). We first analyzed to what extent the Arm variants can substitute for the wild-type form in a rescue assay with the arm2a9-null allele. Hemizygous arm2a9 mutants die as embryos, but can be fully rescued to adulthood by a transgene encoding wild-type Arm (Supplemental Table 1). In contrast, the N-terminally mutant form of Arm (Arm-D172A), as well as the double-mutant Arm-D172A-ΔC, failed to rescue the lethality of arm2a9 males. Only Arm-ΔC retains some modest rescuing function (1%). This assay indicates that in Drosophila, the C-terminal coactivators are important for proper Arm function, while the N-terminal branch is essential.
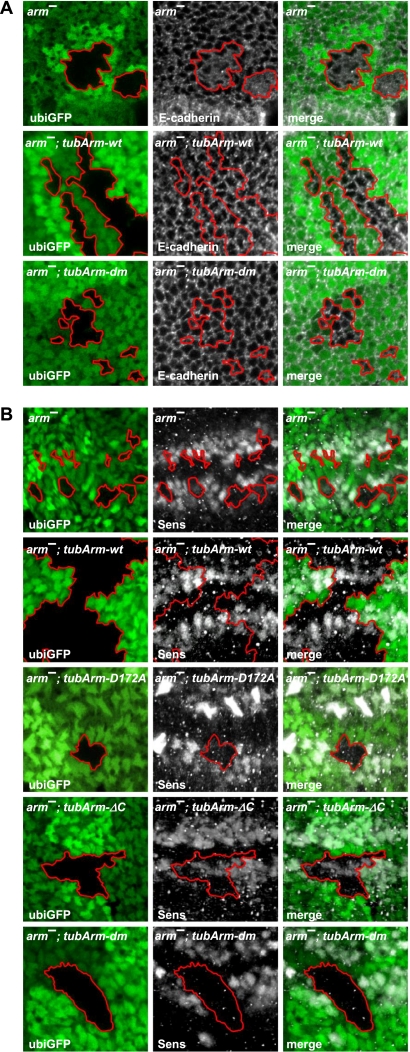
To assess whether the cell–cell adhesive function is intact in our Arm mutants, we induced arm-null (arm2a9) clones in the wing imaginal discs of second instar larvae. arm2a9 clones remain small and often become eliminated ∼48 h after induction. In addition to being small, cells within the clone exhibit an irregular apical morphology. This disrupted epithelial shape is not seen in clones lacking other Wg pathway components and thus appears to be a consequence of the loss of Arm's structural function. In order to visualize the cell–cell adhesive junctions, we stained wing discs with an antibody recognizing E-cadherin, which is the main binding partner of β-catenin in adherens junctions. In arm2a9 clones, the pattern of E-cadherin is collapsed, and E-cadherin is lost from the cell membranes, where it is normally localized, and exhibits a diffuse distribution (Fig. 2A). In the presence of the tub-arm-wt transgene, the expression levels of Arm are restored, leading to a rescue of cell shapes and the E-cadherin expression pattern (Fig. 2A; Supplemental Fig. 2). The double-mutant form of Arm (Arm-D172A-ΔC) also rescued the change in cell shape and the E-cadherin pattern in arm2a9 clones, indicating that Arm-D172A-ΔC retains its function as a building stone of epithelial adherent junctions (Fig. 2A). In these assays, the single-mutant transgenes did not significantly differ from the double mutant in terms of rescuing the clonal morphology and pattern of cell–cell contacts (data not shown).
Figure 2.
The double-mutant form of Arm restores irregular adherens junctions of arm-null clones, but fails to rescue Wg-mediated transcription in vivo. (A,B) Confocal sections of wing discs are shown that contain arm-null (arm−) clones marked by the loss of ubi-GFP expression (borders of clones are highlighted). The particular form of ubiquitously expressed transgenic Arm is indicated in the top corner. Panels to the left show the shape of the clones and the expression of GFP, the middle panels show immunostainings of proteins indicated in the bottom left corner, and the panels to the right show the merged images. (A) Loss of Arm in arm− clones results in defective cell shapes and improper adherens junctions as revealed by the adherens junction component E-cadherin. Ubiquitous expression of wild-type Arm fully restores the normal cell adhesion pattern within arm-null clones (arm−, tubArm-wt). Expression of Arm lacking the ability to bind both Lgs and C-terminal coactivators sustains normal adhesivity, indistinguishable from the wild-type situation. (B) Arm− clones fail to express the Wg target gene senseless (sens). Ubiquitous expression of wild-type Arm (tubArm-wt) fully restores the Sens expression pattern. Arm mutants lacking the ability to bind Lgs (tubArm-D172A) or missing the interaction domain with C-terminal coactivators (tubArm-ΔC) only partially restore Sens expression levels. The double-mutant form of Arm (tubArm-dm) completely lacks the ability to restore Sens expression.
Using the same clonal system, we analyzed Wg target gene expression to determine the Wg-transducing capacity of Arm mutants. Senseless (Sens) is a high-threshold target expressed in two well-confined stripes of cells along the dorsoventral compartment boundary. Sens expression was completely lost in arm2a9 clones that overlap the Sens domain, but was restored in clones from tub-arm-wt animals (Fig. 2B). Transgenes encoding single-mutant forms of Arm showed different rescuing activity. Whereas tub-arm-D172A did not restore Sens expression, tub-arm-ΔC partially rescued Wg signaling. Double-mutant Arm (dm; i.e., Arm-D172A-ΔC) totally failed to rescue Sens expression. Thus, this latter genetic setup equals the Arm-null situation in terms of lack of Wg transduction; however, as concluded above, it differs in its ability to restore epithelial integrity.
Generation of mouse strains with mutant β-catenin alleles
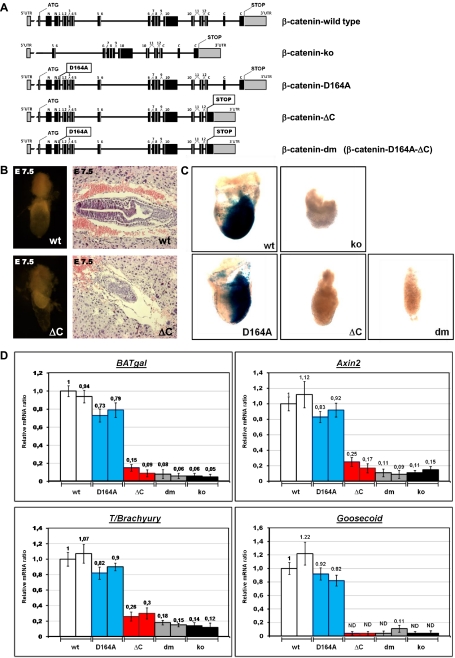
The successful separation of signaling and adhesive functions described above served as a rationale for achieving the same in mammalian β-catenin. By homologous recombination in TC-1 (129Sv) mouse embryonic stem (mES) cells, we replaced the endogenous β-catenin allele with one of the three mutant variants that correspond to the mutants we tested in our tissue culture system (Fig. 3A; Supplemental Fig. 3A; for further details, see the Materials and Methods): (1) A point mutation was introduced in the fourth exon of the β-catenin locus, resulting in a D164A mutation in the first Arm repeat that prevents binding to the N-terminal coactivator BCL9/BCL9L. (2) A stop codon was engineered yielding a C-terminally truncated β-catenin. In this allele, codon 673 in exon 13 was changed to a premature stop followed by a frameshift mutation. Moreover, in order to eliminate possible nonsense-mediated RNA decay, we fused exon 15, encompassing the β-catenin 3′ UTR, directly to exon 13 (see Fig. 3A; Supplemental Fig. 6). (3) Finally, we also generated an ES cell line carrying the double-mutant form of the β-catenin gene, which was created by two subsequent rounds of homologous recombinations. These cells were then used to reconstitute mice of the same genotype. Upon successful elimination of the various selection markers, we obtained three heterozygous strains harboring the mutant β-catenin alleles: β-cateninD164A/β-cateninwt, β-cateninΔC/β-cateninwt, and β-catenindm/β-cateninwt.
Figure 3.
Contribution of the C-terminal β-catenin coactivators is essential for gastrulation. (A) Schematic representation of the β-catenin loci of the mutant strains generated. β-Cateninko was generated by crossing a CMV-Cre line with a conditional β-catenin strain (β-cateninflox/flox; i.e., B6.129-Catnbtm2Kem/J); the resulting allele does not contain an in-frame ATG, and exons 2–6, which encode domains essential for binding to E-cadherin and/or TCF/LEF, are eliminated. β-Catenin-D164A harbors a single-amino-acid exchange (D164A) in exon 4, preventing the interaction between the resulting β-catenin and BCL9/BCL9L. In the β-catenin-ΔC locus, a preliminary stop codon is followed by a frameshift introduced into exon 13; moreover, exon 15 was directly fused to exon 13, thus eliminating exon 14. The β-Catenin-dm strain carries both individual mutations. Boxes represent exons, with black indicating coding and gray indicating noncoding; numbers denominate the Arm repeats. (B) Morphology of wild-type (wt) and β-cateninΔC/ΔC (ΔC) embryos at E7.5, including extraembryonic tissues. (Right panels) Embryos dissected from decidual tissues. (Left panels) Sagittal sections of E7.5 embryos within the decidua stained by H&E. (C) Developmental failure during gastrulation caused by C-terminal truncation of β-catenin is associated with absence of TCF/β-catenin-mediated transcription, as monitored by the Wnt-specific reporter BAT-gal. Dissected embryos at E7.5; each individual embryo inherited one allele of the BAT-gal transcriptional reporter and is homozygous for the indicated mutant allele of β-catenin. (ko) Total loss of β-catenin. LacZ expression from the BAT-gal reporter was determined using enzymatic staining based on X-gal (blue). (D) Transcription of Wnt/β-catenin target genes (transgenic reporter BAT-gal and endogenous genes Axin2 and T/Brachyury) is strongly reduced in E6.5 embryos homozygous for the β-catenin allele that prevents the binding of β-catenin to C-terminal transcription coactivators (ΔC). Expression of factors regulating mesoderm formation (T/Brachyury and Goosecoid) is also strongly affected in β-cateninΔC/ΔC embryos. Levels of mRNA were determined by quantitative real-time PCR and normalized to the housekeeping genes SDHA and GAPDH. The levels of a β-cateninwt/wt (wild-type) embryo are set as 1. Embryos from two independent litters were tested for each homozygous β-catenin mutant as indicated. (dm) Double-mutant form; (ko) β-catenin-null embryos. Each embryo carries one allele of the BAT-gal reporter. Error bars show standard deviation. (ND) Nondetectable levels.
Transcriptional output via β-catenin's C terminus is essential for gastrulation
None of the three mutant β-catenin strains gave birth to homozygous animals. To determine the time point and cause of embryonic lethality, we analyzed litters of such crosses. β-CateninΔC/ΔC and β-catenindm/dm embryos showed strong defects during gastrulation before embryonic day 7.5 (E7.5), while β-cateninD164A/D164A animals developed normally until E10.0, when they exhibited the first signs of developmental delay. The defects of β-cateninΔC/ΔC and β-catenindm/dm embryos resemble those described for the β-cateninko/ko embryos (Haegel et al. 1995; Huelsken et al. 2000). While wild-type and β-cateninD164A/D164A animals form mesoderm at the posterior side at E7.5 and are in a dynamic process of gastrulation, in β-cateninΔC/ΔC embryos, the majority of cells detached from the ectodermal cell layer and compact uniform cell masses formed without any signs of mesoderm or other defined germ layers (Fig. 3B.C). Extraembryonic tissues do not seem to be affected, presumably because β-catenin is normally not expressed in these tissues (Supplemental Fig. 3B). The total failure of embryonic development at E7.5 in β-cateninΔC/ΔC as well as β-catenindm/dm and β-cateninko/ko animals mirrors the loss of Wnt/β-catenin-mediated transcription as monitored by the β-galactosidase activity of the TCF/β-catenin reporter BAT-gal (Maretto et al. 2003). Wild-type or β-cateninD164A/D164A embryos have strong BAT-gal activity at the posterior side of the embryo in the primitive streak, the area where the mesoderm is formed. No such signal could be detected in embryos homozygous for a C-terminally truncated β-catenin allele (alone or as double mutant together with the D164A mutant), or in embryos missing β-catenin entirely (Fig. 3C). The dramatic block in TCF/β-catenin transcriptional activity in β-cateninΔC/ΔC, β-catenindm/dm, and β-cateninko/ko embryos could also be confirmed at E6.5 using quantitative real-time PCR for the BAT-gal reporter or for the endogenous TCF/β-catenin target genes Axin2 and LEF-1 (Fig. 3D; data not shown). The disappearance of TCF/β-catenin-mediated transcription directly correlates with the failure in developing the mesodermal layer, as indicated by the loss of expression of the mesodermal marker T/Brachyury (Fig. 3D). The transcription factor T/Brachyury is not only encoded by a direct target of TCF/β-catenin-mediated transcription, but is also one of the key elements defining mesoderm formation in the primitive streak (Yamaguchi et al. 1999; Arnold et al. 2000). The lack of mesoderm was further confirmed by the absence of mRNA of the early mesodermal gene Goosecoid (Fig. 3D). We interpret our findings as an indication that the interactions of β-catenin with its C-terminal coactivators are essential for proper mesodermal formation and thus for gastrulation.
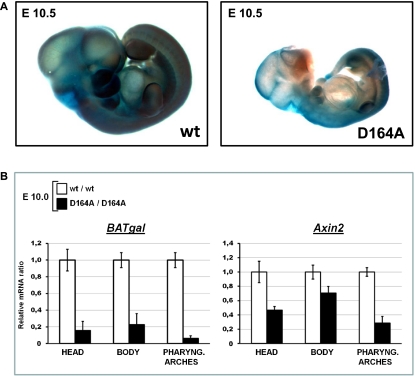
In contrast to embryos expressing C-terminally truncated β-catenin, the development of embryos expressing the D164A form of β-catenin, which cannot bind BCL9/BCL9L, is indistinguishable from wild type until E10.0 (Supplemental Fig. 3C). After this stage, β-cateninD164A/D164A embryos are developmentally delayed and smaller than wild-type control embryos. At E10.5, their forebrain is not properly developed, pharyngeal arches are rudimentary, and, in 43% of the embryos (18 out of 42 analyzed embryos), defects can be observed in craniofacial development (Fig. 4A). These developmental defects are associated with reduced Wnt signaling activity, since β-cateninD164A/D164A embryos have strongly reduced BAT-gal expression in their pharyngeal arches and reduced reporter gene activity in the developing brain and throughout the body (Fig. 4A; Supplemental Fig. 3C). The deficit in Wnt signaling activity is the most likely reason for the developmental delay and the subsequent defects, since at E10.0, when no obvious phenotypic defects are detectable, β-cateninD164A/D164A embryos already exhibit a strong reduction of Wnt/β-catenin transcriptional activity, as assessed by quantitative real-time PCR for the BAT-gal reporter (Fig. 4B). Endogenous Axin-2 expression is also reduced, especially in pharyngeal arches (Fig. 4B). Thus, it appears that N-terminal coactivators, represented by BCL9/BCL9L, are important regulators of the Wnt/β-catenin output during embryonic development independently of the C-terminal coactivator branch.
Figure 4.
Developmental defects and reduction in β-catenin-mediated transcription in embryos expressing the D164A mutant form. (A) Wild-type (wt) and homozygous β-cateninD164A/D164A (D164A) embryos at E10.5 carrying one allele of the Wnt/β-catenin transcriptional reporter BAT-gal and stained for enzymatic activity with X-gal (blue). The mutant embryos show severe defects in size and in the development of brain, craniofacial structures, and pharyngeal arches. These defects are associated with reduced BAT-gal activity. Representative embryos are shown. (B) Wnt/β-catenin transcription is reduced in such mutant embryos as determined by quantitative real-time PCR in the head and the pharyngeal arches at E10.0. Expression of the BAT-gal reporter and of the endogenous Wnt/β-catenin target gene Axin2 in the head, pharyngeal arches, and the rest of the body was monitored. Transcript levels are normalized to those of the housekeeping genes SDHA and GAPDH. Levels for wild-type (wt) embryos are set to 1. Error bars show standard deviations. Representative results are shown.
Replacing β-catenin with its signaling-defective variant in Wnt1-expressing tissues
To overcome the early embryonic lethality, we placed the mutant β-catenin alleles over a conditional knockout allele (β-cateninflox) in combination with a Cre driver line. In Cre-expressing cells that undergo mitotic recombination, our mutant alleles are the only source for β-catenin, essentially replacing the wild-type protein with the mutant form of interest (for a detailed crossing scheme, see Supplemental Fig. 5). As a model Cre driver, we used Wnt1-Cre, which is expressed in the early developing neural tube, resulting in recombination in the midbrain, in the dorsal spinal cord, and in neural crest cells (Danielian et al. 1998). Previous reports showed that complete loss of β-catenin mediated by Wnt1-Cre dramatically affects the development of midbrain–hindbrain structures, the craniofacial apparatus, and neural crest cell derivatives. However, for most of these phenotypes, it is difficult to decipher what is due to the loss of Wnt signaling activity as opposed to loss of adhesion (Brault et al. 2001; Hari et al. 2002).
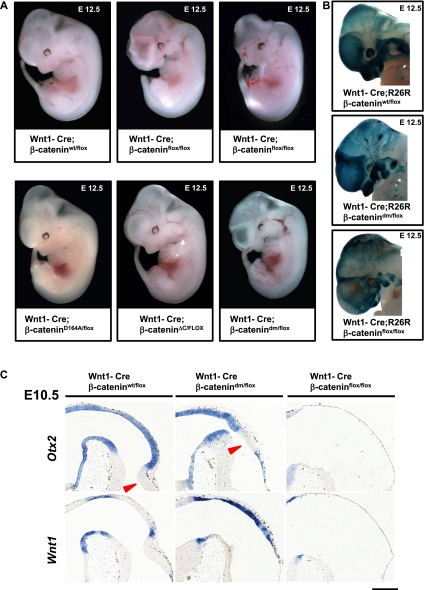
To resolve this, we used Wnt1-Cre in combination with our mutant β-catenin alleles. Upon Wnt1-Cre-mediated recombination, Wnt1-Cre; β-cateninflox/flox embryos exhibit severe phenotypes at E12.5: defective craniofacial structures, malformed telencephalic lobes, and a missing midbrain–hindbrain boundary (MHB). The phenotypes of Wnt1-Cre; β-catenindm/flox embryos are less drastic and comprise milder craniofacial and mesencephalic defects than the complete loss of β-catenin. The defects in Wnt1-Cre; β-catenindm/flox are unvarying, while the Wnt1-Cre; β-cateninflox/flox embryos exhibit considerable phenotypic variability.
Embryos of the genotypes Wnt1-Cre; β-cateninΔC/flox and Wnt1-Cre; β-cateninD164A/flox resemble wild-type embryos and display less severe phenotypes (Fig. 5A) compared with those expressing double-mutant β-catenin. This indicates that both the N- and C-terminal coactivators contribute to the transcriptional output of β-catenin in Wnt1-expressing tissues, validating the approach of using the double-mutant form to fully block the transcriptional output of β-catenin.
Figure 5.
Total loss of β-catenin in Wnt1-Cre-expressing tissues leads to more severe phenotypes than blocking the Wnt signaling output of β-catenin. (A) E12.5 embryos expressing Wnt1-Cre, one conditional allele of β-catenin (β-cateninflox), and a second allele of β-catenin as indicated. In Wnt1-Cre; β-cateninflox/flox embryos, Cre activity results in the absence of β-catenin in Wnt1-expressing tissues. Such embryos show strong defects in craniofacial development, midbrain, and hindbrain. The phenotype of Wnt1-Cre; β-catenindm/flox embryos is milder compared with that of Wnt1-Cre; β-cateninflox/flox, especially in the case of telencephalic lobes and midbrain and hindbrain areas. Representative embryos are shown. (B) X-gal staining visualizing Wnt1-Cre-mediated recombination using R26R in the heads of embryos at E12.5. The strong dark-blue signal in forming craniofacial structures (jaw/maxilla and meninges) and pharyngeal arches in wild type represents neural crest derivatives. Similarly strong signals could be partially observed in Wnt1-Cre/R26R; β-catenindm/flox, while in Wnt1-Cre/R26R; β-cateninflox/flox, there are fewer, irregularly scattered X-gal-positive cells. (C) In situ hybridization with early midbrain–hindbrain junction markers on sagittal sections of embryonic heads at E10.5. (Top left panel) In wild-type embryos, the transcription factor Otx2 is expressed in the midbrain with a sharp boundary at the midbrain–hindbrain junction. (Top middle panel) In Wnt1-Cre; β-catenindm/flox, the midbrain–hindbrain junction marked by Otx2 expression is shifted, and the prospective cerebellum is not developed. (Bottom left panel) Wnt1 is expressed in wild type in the posterior midbrain and along the dorsal midline. (Bottom middle panel) Wnt1-Cre; β-catenindm/flox animals show increased expression of Wnt1 along the dorsal midline. (Right panels) In contrast, Wnt1-Cre; β-cateninflox/flox (ko) brains lack expression of both Otx2 (top) and Wnt1 (bottom), indicating absence of any midbrain and cerebellar structures. The arrowheads point to hindbrain structures (cerebellum). Bar, 200 μm.
The significant differences between β-catenin double-mutant and null phenotypes prompted us to examine the Wnt1-Cre recombined cells using the ROSA26 reporter (R26R) (Soriano 1999). Wnt1-Cre activates R26R expression in the entire neural crest population, including the migrating pool of cells (Danielian et al. 1998). At E12.5, our control embryos exhibit strong R26R activity in the developing craniofacial structures, which are constituted from the pharyngeal arches into which neural crest cells migrate. In Wnt1-Cre; R26R; β-catenindm/flox embryos, the strong R26R expression in the craniofacial structures indicates that neural crest stem cells (NCSCs) have migrated to form craniofacial skeletal structures, although these were hypoplastic and malformed. In contrast, Wnt1-Cre; R26R; β-cateninflox/flox embryos entirely lack maxillae and mandibles (Fig. 5B), and residual neural crest cells reside in rudimentary pharyngeal arches (Fig. 5B).
Closer examination of midbrain sagittal sections of mutant embryos at E10.5 revealed further differences between Wnt1-Cre; β-cateninflox/flox and Wnt1-Cre; β-catenindm/flox animals. In wild-type embryos, the expression domain of the transcription factor Otx2 spreads throughout the forebrain and midbrain with a sharp border at the MHB (Millet et al. 1996). In null embryos, the midbrain and hindbrain structures are entirely absent, while in Wnt1-Cre; β-catenindm/flox animals, the midbrain is present as a thin layer of Otx2-positive cells (Fig. 5C), but the prospective cerebellum is not developed. In both genotypes, the MHB, demarcated by the expression of FGF8, is absent (data not shown). We also checked the distribution of Wnt1, which is expressed in a stripe along the dorsal neural tube with a gap at rhombomere 1 and as a transverse band dorsal to the MHB (Wilkinson et al. 1987). Upon total loss of β-catenin, we did not detect any expression of Wnt1 at the dorsal midline, where recombined cells are apoptotic (Brault et al. 2001), but increased expression laterally, where the recombination rate decreases. Similarly, embryos expressing the double-mutant β-catenin exhibit enhanced dorsal Wnt1 expression (Fig. 5C), underscoring the importance of Wnt signaling for midbrain development and of distinguishing loss of signaling function versus lack of β-catenin.
Epithelial integrity in tissue with double-mutant β-catenin (D164A-ΔC)
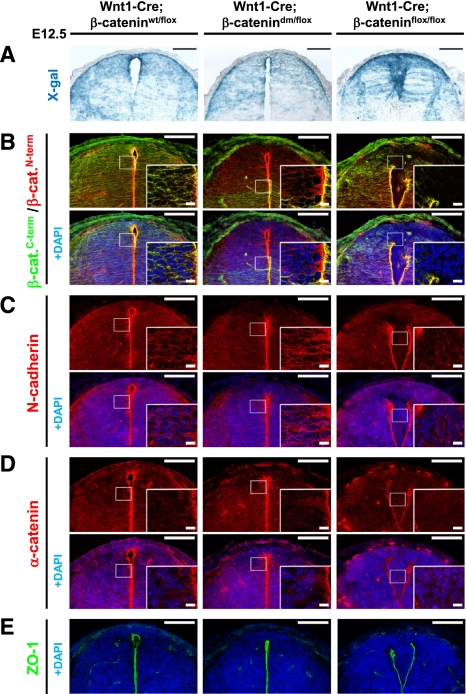
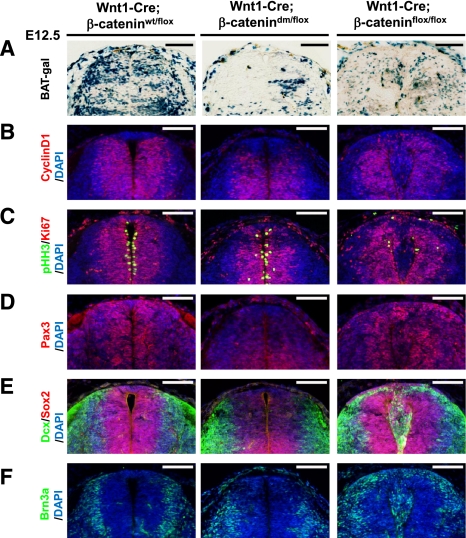
As described above, the Wnt1-Cre driver is active in the dorsal neural tube (Fig. 6A; Ille et al. 2007), an epithelial tissue well suited to analyze the capacity of different β-catenin forms to sustain cell–cell adhesion. Antibodies specific for either the N or C terminus of β-catenin allowed us to detect the presence of both termini in control spinal cord expressing wild-type β-catenin, a C-terminally truncated protein in Wnt1-Cre; β-catenindm/flox, and lack of β-catenin in Wnt1-Cre; β-cateninflox/flox (Fig. 6B). As expected, Wnt1-Cre-mediated loss of β-catenin in β-cateninflox/flox embryos caused the disruption of apical neural tube morphology and led to the migration of cells into the neural canal, as shown by immunostaining against the adherens junction components N-cadherin, α-catenin, and the tight junction marker ZO-1 (Fig. 6C–E). In contrast to the Wnt1-Cre; β-cateninflox/flox embryos, the structure of Wnt1-Cre; β-catenindm/flox neural tubes is intact, and adherens junction components are distributed like in controls. Importantly, however, just like in the case of Arm, the double-mutant form of β-catenin cannot provide transcriptional output for the canonical Wnt pathway, as the expression of the Wnt/β-catenin reporter BAT-gal and the direct target CyclinD1 is severely reduced in the dorsal region of Wnt1-Cre; β-catenindm/flox neural tubes (Fig. 7A,B). Taken together, we conclude that the mutations introduced into the β-catenin protein indeed ablate specifically the signaling roles of β-catenin independently of its adhesion function.
Figure 6.
β-Catenin-dm can fully restore the corrupted dorsal spinal cord morphology caused by defective adherens junctions in tissue lacking β-catenin. (A) X-gal staining visualizing Wnt1-Cre-mediated expression of β-galactosidase from R26R in the dorsal spinal cord of E12.5 embryos. Wild-type embryos (Wnt1-Cre/R26R; β-cateninwt/flox) and embryos expressing double-mutant β-catenin (Wnt1-Cre/R26R; β-catenindm/flox) display normal morphology and shape of the dorsal spinal cord. On the other hand, absence of β-catenin (Wnt1-Cre/R26R; β-cateninflox/flox) results in breakage of medial apical contacts and severe morphological defects. (B) Double-mutant β-catenin effectively restores the adhesion defects caused by the loss of β-catenin in Wnt1-Cre-positive tissues. Immunostaining with antibody recognizing either the N terminus or C terminus of β-catenin. The C terminus is missing in the double-mutant form, but protein expression is recognized by the antibody against the N terminus. No β-catenin is detected in the recombination-prone region of Wnt1-Cre; β-cateninflox/flox neural tubes. (C–E) N-cadherin (C), α-catenin (D), and ZO-1 (E) immunostainings for adherens and tight junctions do not show a significant difference between wild-type (Wnt1-Cre; β-cateninwt/flox) and mutant (Wnt1-Cre; β-catenindm/flox) spinal cord. Lack of β-catenin (Wnt1-Cre; β-cateninflox/flox) leads to adhesion defects associated with the breakdown of apical junctional complexes. Bar, 100 μm; bar in inset, 10 μm.
Figure 7.
Block of Wnt signaling output by double-mutant β-catenin affects cell fate determination in the dorsal spinal cord. (A–F) Immunostaining of transversal sections of dorsal spinal cord isolated from E12.5 embryos. The double-mutant form of β-catenin fully blocks expression of the Wnt reporter BAT-gal (A) and of the endogenous Wnt target gene CyclinD1 (B) in the dorsal spinal cord, where Wnt1-Cre is active. (C) Cells expressing the double-mutant β-catenin are still proliferating, indicated by Ki67 and pHH3 expression. In contrast, cell proliferation was strongly reduced in Wnt1-Cre; β-cateninflox/flox animals. The spinal cord of Wnt1-Cre; β-catenindm/flox animals is characterized by strongly reduced expression of Pax3 (D) and Sox2 (E), indicating a loss of undifferentiated precursor cells. Neuronal differentiation in the dorsal neural tube of these animals is affected as indicated by the absence of early neuronal markers Dcx (E) and Brn3 (F). Bar, 100 μm.
Block of transcriptional β-catenin output affects cell fate determination in the dorsal spinal cord
The rescued morphology of the dorsal neural tube in E12.5 Wnt1-Cre; β-catenindm/flox embryos enabled the analysis of progenitors in the absence of Wnt transduction. Blocking β-catenin-mediated transcription in the dorsal neural tube does not noticeably affect cell proliferation, as determined by the expression of the proliferation marker Ki67 and the distribution of the mitotic marker phosphorylated histone H3 (pHH3) in Wnt1-Cre; β-catenindm/flox animals (Fig. 7C). Moreover, we did not observe any sign of increased apoptosis determined by caspase-3 staining in the dorsal neural tube of such embryos (Supplemental Fig. 4A). However, when monitoring the differentiation of sensory neurons by pSmad1/5/8 activity, we found this process severely affected (Supplemental Fig. 4B). Furthermore, the expression of Pax3, a key transcription factor regulating the fate of neuronal precursor cells (Christova et al. 2010), as well as Sox2, normally expressed in spinal cord precursors (Wakamatsu et al. 2004), was strongly reduced (Fig. 7D,E). The severe decrease in expression of these transcription factors was accompanied by the disappearance of the neuronal marker Dcx and by a partial loss of dorsal interneurons marked with Brn3a (Fig. 7E,F; Fedtsova and Turner 1995). Thus, Wnt/β-catenin transcription is essential for the maintenance and proper development of neuronal precursors in the dorsal neural tube.
In contrast to this phenotype, we found that β-catenin-null cells underwent premature differentiation and often dispersed from the roof plate to the outer dorsal rim of the tube (Fig. 7E,F, right panels). These defects must be caused by the concurrent loss of β-catenin-mediated transcription and disruption of dorsal neural tube morphology (Fig. 7 A–F, right panels). The loss of medial apical junctions in the dorsal spinal cord thus severely limits the possibility of determining the spatial and temporal fates of neurons in this tissue.
Together, our observations uncover the requirement of β-catenin-mediated transcription for maintenance of a neural progenitor state and for neuronal differentiation in the neural tube. They illustrate the importance of discriminating between the nuclear and adhesive roles of β-catenin when studying canonical Wnt signaling.
Discussion
The first specific model to study lack of canonical Wnt/β-catenin signaling
The importance of canonical Wnt/β-catenin signaling during development is only rivaled by the difficulty of studying it in vivo. The main obstacles for a genetic analysis of the transcriptional output of the canonical Wnt pathway are the following: First, mammalian genomes encode almost 20 different Wnt proteins. In many cases, their expression patterns are overlapping, and the range of action is unknown. The elimination of canonical Wnt signaling at the level of the ligand thus not only faces the technical difficulty of removing the function of numerous genes at once, it is further complicated by the problem that each of these Wnts may additionally act in a noncanonical fashion. A similar complexity exists at the level of Wnt receptors (van Amerongen et al. 2008). The nuclear mediators of the canonical Wnt pathway, the TCF/LEF proteins, are somewhat better suited for the analysis of canonical Wnt output, as they are less numerous and have a better-defined pathway specificity. However, there are still four family members existing as differently spliced variants that are all expressed in different combinations and at different stages of development (Najdi et al. 2011). Moreover, some of these transcription factors have been shown to act—in addition to their role as activators—also as repressors of target genes in the absence of Wnt signaling. Their removal, therefore, does not necessarily reflect the lack of canonical pathway activity, but rather a mixture of derepression and loss-of-activation states. Overexpressing a dominant-negative form of TCF/LEF would also be problematic, as it might also elicit transcriptional repression in genomic regions that are not relevant for canonical Wnt signaling. Understandably, therefore, much hope and attention have been focused on β-catenin, which is a nonredundant and essential component of the pathway. Indeed, loss-of-function studies with the β-catenin gene form the largest foundation for our current understanding of the canonical Wnt pathway.
A complication for using β-catenin to study Wnt signaling, however, is that it also acts as an organizer of polarized epithelia—an essential function that is independent of its signaling role. Accentuating this situation is the uncertainty about the extent to which the loss of β-catenin's adhesive function contributes to a particular phenotype; estimates of this contribution range from negligible to substantial at different stages in different tissues. Here we describe a genetic tool that resolves these issues. We generated a β-catenin allele that is devoid of canonical Wnt signaling activity, yet retains cadherin-dependent cellular adhesion function. Using it, we can now disentangle the consequences of loss of Wnt signaling from those of loss of cell adhesion. This relieves β-catenin of its sole significant drawback and enables us to experimentally exploit its key position in the canonical Wnt pathway.
From Arm to β-catenin
The development of the signaling-defective form of β-catenin built on our understanding of Arm. The epithelium of the Drosophila wing imaginal disc served as a validation system for the separation of the signaling and the adhesive function. Clones of cells null mutant for Arm are eliminated 20–24 h earlier than clones expressing the double-mutant form of Arm. The loss of Arm's adhesive activity led to the destabilization of cadherin-based adherens junctions, as indicated by the delocalization of E-cadherin from the membrane. Clones with affected cadherin-based adhesion are rapidly extruded from the wing disc epithelium and eliminated (Widmann and Dahmann 2009). Cells expressing the double-mutant form of Arm, which preserves the adhesion function, survive longer than null clones, but are also smaller than clones rescued by the wild-type form of Arm, supporting previous observations that Wg/Arm signaling activity is essential for proper cell proliferation and survival (for review, see Gonsalves and DasGupta 2008; Widmann and Dahmann 2009). Hence, in the wing disc, loss of Arm affects the behavior of mutant clones on two levels: adhesion and signaling.
After validating mutant mammalian β-catenin in biochemical and cell-based signaling assays, we introduced the mutations into the mouse germline. Using the resulting alleles in combination with tissue-specific knockout of the β-catenin gene, cell populations could be generated that correspond to the arm clones in the Drosophila wing disc. With the Wnt1-Cre driver, we observed in the dorsal neural tube corrupted adherens junctions associated with morphological changes that would prevent studying the fate of cells lacking canonical Wnt output. Double-mutant β-catenin, however, restores the cell and tissue structures and serves now as an adequate basis to assess the role of Wnt/β-catenin signaling as well as its cross-talk with other signaling pathways.
Early embryonic role of β-catenin
Embryos with transcriptionally inactive β-catenin die at E7.5, just like embryos with total loss of β-catenin. Therefore, β-catenin's role of mediating cadherin-dependent cellular adhesion does not seem to play an important function during gastrulation, although it is a dynamic process dominated by epithelial-to-mesenchymal transitions (EMT) and cell migrations (for review, see Arnold and Robertson 2009). Moreover, at these early stages, β-catenin expression is more pronounced in areas where no E-cadherin is detected (E-cadherin is prominent in extraembryonic tissues and β-catenin in the developing embryo). It appears, therefore, that mutant embryos are not significantly affected by the lack of β-catenin's adhesive role; rather, their phenotype results from the inability to form mesoderm. This, in turn, can be attributed to the loss of Wnt signaling-dependent expression of transcription factors that are important for mesoderm formation. Indeed, embryos lacking MED12 die at E7.5, also from a failure to develop mesoderm (Rocha et al. 2010), and MED12 is an important coactivator of β-catenin (Kim et al. 2006).
Dual role of β-catenin in craniofacial and CNS development
Using Wnt1-Cre to remove wild-type β-catenin revealed situations in which the consequences of total lack of β-catenin do not correspond to those of substituting it with the adhesion-competent form. Striking differences were observed in craniofacial and mesencephalic structures. Lack of β-catenin results in the absence of the midbrain and anterior hindbrain structures, yet when the signaling function is specifically ablated, the dorsal part of the midbrain develops. More posteriorly in the CNS, the specific removal of Wnt/β-catenin signaling seems to affect the ground state and the differentiation potential of precursors in the dorsal neural tube. As expected when Wnt signaling is blocked, expression of its downstream target, CyclinD1, is lost. However, the cell cycle of dorsal neural tube progenitors does not seem to be halted, as assessed by Ki67/pHH3 staining. Previous studies using the same Wnt1-Cre driver in conjunction with a constitutively active form of β-catenin indicated that Wnt signaling promotes proliferation, while BMP signaling induces differentiation (Ille et al. 2007). Indeed, we also observed decreased differentiation of spinal cord precursors to sensory neurons, based on reduced pSmad1/5/8 activity in those cells. Thus, Wnt signaling not only plays an essential role for maintaining dorsal neural tube precursors in a proliferative state, it also affects their differentiation.
Our analysis of the developing neural tube demonstrates how the double-mutant β-catenin allele can be used to specifically block canonical Wnt signaling and that it represents a powerful new tool to discriminate between the structural and signaling function of β-catenin. Additionally, our single-mutant alleles will help to analyze the needs for individual β-catenin transcriptional coactivators during normal development and in disease and could thus be invaluable to validate therapeutic strategies targeting the interaction of β-catenin with its coactivators.
Materials and methods
Vectors and constructs
Drosophila Arm expression constructs were based on the pT2-attB(+) backbone driving protein expression under the control of tubulinα1 promoter (Basler and Struhl 1994). For mammalian expression, constructs derived from pcDNA3.1 (Invitrogen) were used. Single-amino-acid change variant Armadillo-D172A or β-catenin-D164A was introduced using site-directed mutagenesis (QuickChange, Stratagene). C-terminally truncated fragments of Arm (amino acids 1–691) or β-catenin (amino acids 1–673) were generated by PCR and cloned into particular vectors. Double-mutant forms were created by combinations of individual mutations using the internal restriction site StuI conserved in arm or β-catenin. For testing constitutively active forms of Arm, constructs arising from armS10 in the pT2-attB(+) backbone with the same mutations as in the case of wild-type Arm were generated (i.e., ArmadilloS10-wild type, ArmadilloS10-D172A, ArmadilloS10-ΔC, and ArmadilloS10-dm). For the mammalian system, corresponding constructs were prepared based on the constitutively active β-cateninS33Y (β-cateninS33Y-wt, β-cateninS33Y-D164A, β-cateninS33Y-ΔC, and β-cateninS33Y-dm).
Drosophila cell culture experiments
RNAi against the 5′ and 3′ UTRs of arm was performed as described previously by Mosimann et al. (2006) and Hoffmans and Basler (2007), including dsRNA generation and application to Kc cells (for primer sequences, see Supplemental Table 2). Four hours after RNAi application, cells were transfected using CellFectin (Invitrogen) with expression constructs encoding different Arm variants (0.5 μg of vector per 106 cells), eventually together with the luciferase reporter plasmids. Cells were incubated for 24 h to allow turnover of endogenous Arm and replacement by exogenous mutant forms. After this, cells were stimulated for an additional 24 h by Wg-conditioned medium (or control one) prepared from tubulin-Wg-S2 cells (Drosophila Genomic Resource Center, stock no. 165).
Mammalian cell culture experiments
HEK293T cells and MEFs were cultured at 37°C in 5% CO2 in DMEM, 10% FCS, and 1% P/S. MEFs were isolated from E13.5 β-cateninflox/flox embryos according to standard protocols. MEFs lacking β-catenin (MEF-ko) were generated from MEF-β-cateninflox/flox cells by infection with lentiviral particles encoding self-excising Cre recombinase as published by Cudré-Mauroux et al. (2003). Expression of β-catenin was reconstituted in MEF-ko cells through retroviral infection. Retroviral vectors based on the pBABE-Puro backbone were transfected into Phoenix-Eco (Orbigen) retrovirus producer cells via the Lipofectamine transfection reagent (Invitrogen), and subsequent retroviral infection was performed as previously described (Valenta et al. 2003). Two days after infection, MEF-ko cells were treated with puromycin (5 μg/mL) for 5 d to establish four different polyclonal cell lines: MEF-wt (expressing wild-type β-catenin), MEF-D164A (expressing β-catenin-D164A), MEF-ΔC (expressing β-catenin-ΔC), and MEF-dm (expressing β-catenin-dm; i.e., D164A-ΔC). To activate Wnt/β-catenin-mediated transcription, MEFs with reconstituted β-catenin expression (plus MEF-ko and parental MEFs) were stimulated by Wnt3a-conditioned medium acquired from Wnt3a-producing L cells (L Wnt-3A; American Type Culture Collection no. CRL-2647) as described by the supplier.
Luciferase assay
Drosophila Kc (or clone 8) cells were transfected in 96-well plates by CellFectin (Invitrogen) with a total amount of 300 ng of plasmid mixture per three wells as follows: 50 ng of wf-Luciferase reporter (Hoffmans and Basler 2007), 30 ng of tubulin-α1-Renilla, and 220 ng of empty vector, or, alternatively, 100 ng of ArmS10-based construct with 120 ng of empty vector. Luciferase reporter gene assays in mammalian cells were performed as described previously (Valenta et al. 2006). To assay Wnt/β-catenin-mediated transcription, the firefly luciferase reporter pTOPFLASH was used. In both cases, Luciferase activity was measured by the Promega Dual-Luciferase reporter system, and firefly luciferase levels were normalized to Renilla values to standardize the transfection efficiency.
Quantitative real-time PCR
Total RNA was extracted from Drosophila Kc and mouse MEF cells and from embryos (whole E6.5 embryos or parts of E10.0 embryos) using Nucleospin RNAII kit (Marchery-Nagel). Quantitative real-time PCR reactions were performed in triplicate and monitored by the Applied Biosystems SYBR Green kit and the ABI Prism 7900HT system (Applied Biosystem). All experiments were repeated in parallel at least twice; in the case of mouse embryos, RNA isolated from different litters at the same developmental stage was tested. Results were normalized to Actin5C, β-tubulin, and GAPDH in the case of Drosophila Kc cells. SDHA, GAPDH, and β-actin were used for normalization of mouse samples (MEFs and embryos). The ΔΔCt method (Applied Biosystem user bulletin) was used for result calculations. For details concerning primers, see Supplemental Table 2.
Fly stocks
Four transgenic fly lines expressing Arm under the control of the tubulinα1 promoter were generated using the ΦC31 integration system developed previously in our laboratory (Bischof et al. 2007): tub-Arm-wt; tub-Arm-D172A, tub-Arm-ΔC, and tub-Arm-dm. Constructs based on the pT2-attB(+) vector backbone were injected into the ZP-attP-86Fb fly line harboring a landing site on the third chromosome.
Disc clones
Mutant imaginal disc clones were generated by crossing arm2a9 FRT19/FM7 females with hs-flp ubiGFP FRT19; tub-arm-R/TM6b males, where R represents an individual variant of an arm rescue construct. Seventy-two hours after egg laying, larvae were heat-shocked for 45 min at 38°C. Female third instar larvae that did not carry TM6b were dissected 48 h after the heat shock. Imaginal discs were fixed and stained by standard techniques. Pictures were captured by a Zeiss LSM710 confocal microscope.
Generation of β-catenin knock-in mouse strains
To replace the endogenous β-catenin locus with mutant forms, two independent targeting vectors for homologous recombination were cloned. The single-point amino-acid change D164A, blocking the interaction between β-catenin and BCL9/BCL9L, was introduced using pL-PGK#12(loxP-Hyg-loxPrev) (a gift from D. Shmerling) encoding the D164A mutation in exon 4 cloned in the 5′ arm of the vector. For the C-terminal truncation, the vector pTargetVectorpBS-FRT-neo-TK was generated, which encodes in its 3′ arm a preliminary stop codon at the position of amino acid 673 followed by a frameshift mutation. Moreover, exon 13 harboring the premature stop codon was directly fused to exon 15 encoding the 3′ UTR of β-catenin (exon 14 and the introns between exons 13, 14, and 15 were thus eliminated). Electroporation of targeting vectors into TC-1 (129Sv) mouse embryonic cells, blastocyst injection, and further generation of knock-in animals, including verification, were performed by Polygene. Double-mutant animals were generated by two subsequent steps of homologous recombination. mES cells harboring C-terminal truncation were electroporated with targeting vectors encoding the D164A mutation, and double-mutant mES cells were further used for blastocyst injection. For detailed information concerning targeting vectors and genotyping, see the Supplemental Material and Supplemental Figure 6.
Mouse experiments
Mouse experiments were performed in accordance with Swiss guidelines and approved by the Veterinarian Office of the Kanton of Zürich, Switzerland. All animals were kept on a C57BL/6 background. Strains harboring the β-catenin-null allele (β-cateninko) were generated by crossing a CMV-Cre line with the conditional β-catenin strain β-cateninflox/flox (B6.129-Catnbtm2Kem/J, Jackson Laboratory). To monitor Wnt/β-catenin transcription, compound animals heterozygous for the mutated forms of β-catenin were crossed with the BAT-gal reporter line (Maretto et al. 2003), and offspring were further crossed to get β-cateninmutant/mutant; BAT-gal+/− embryos, where “mutant” represents D164A, ΔC, or dm (D164A-ΔC). The fate of neuroepithelial cells was followed in compound transgenic animals (β-cateninmutant/flox) expressing Cre (Wnt1-Cre) and bred to the mouse R26R line (Soriano 1999). Activity of β-galactosidase expressed in BAT-gal or ROSA26 was determined by X-gal staining as described previously (Maretto et al. 2003; Ille et al. 2007).
Coimmunoprecipitations and staining of tissue sections
Coimmunoprecipitation experiments and staining of mouse tissue sections were preformed according to standard protocols. For details, see the Supplemental Material.
Acknowledgments
We thank H. Bellen, E. Wieschaus, T. Uemura, and the Hybridoma Bank for antibodies; S. Piccolo for providing the BAT-gal strain; L. Hari, I. Miescher, and D. Shmerling for help with mouse experiments; and G. Hausmann for critical reading of the manuscript. This work was supported by the European Research Council, the Swiss National Science Foundation, and the Kanton of Zürich.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.181289.111.
References
- Arnold SJ, Robertson EJ 2009. Making a commitment: Cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol 10: 91–103 [DOI] [PubMed] [Google Scholar]
- Arnold SJ, Stappert J, Bauer A, Kispert A, Herrmann BG, Kemler R 2000. Brachyury is a target gene of the Wnt/β-catenin signaling pathway. Mech Dev 91: 249–258 [DOI] [PubMed] [Google Scholar]
- Basler K, Struhl G 1994. Compartment boundaries and the control of Drosophila limb pattern by hedgehog protein. Nature 368: 208–214 [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K 2007. An optimized transgenesis system for Drosophila using germ-line-specific ΦC31 integrases. Proc Natl Acad Sci 104: 3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R 2001. Inactivation of the β-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development 128: 1253–1264 [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Liu YI 2006. Wnt signaling: Complexity at the surface. J Cell Sci 119: 395–402 [DOI] [PubMed] [Google Scholar]
- Christova T, Mojtahedi G, Hamel PA 2010. Lymphoid enhancer factor-1 mediates loading of Pax3 to a promoter harbouring lymphoid enhancer factor-1 binding sites resulting in enhancement of transcription. Int J Biochem Cell Biol 42: 630–640 [DOI] [PubMed] [Google Scholar]
- Cudré-Mauroux C, Occhiodoro T, König S, Salmon P, Bernheim L, Trono D 2003. Lentivector-mediated transfer of Bmi-1 and telomerase in muscle satellite cells yields a duchenne myoblast cell line with long-term genotypic and phenotypic stability. Hum Gene Ther 14: 1525–1533 [DOI] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP 1998. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol 8: 1323–1326 [DOI] [PubMed] [Google Scholar]
- Fedtsova NG, Turner EE 1995. Brn-3.0 expression identifies early post-mitotic CNS neurons and sensory neural precursors. Mech Dev 53: 291–304 [DOI] [PubMed] [Google Scholar]
- Gonsalves FC, DasGupta R 2008. Function of the wingless signaling pathway in Drosophila. Methods Mol Biol 469: 115–125 [DOI] [PubMed] [Google Scholar]
- Graham TA, Weaver C, Mao F, Kimelman D, Xu W 2000. Crystal structure of a β-catenin/Tcf complex. Cell 103: 885–896 [DOI] [PubMed] [Google Scholar]
- Grigoryan T, Wend P, Klaus A, Birchmeier W 2008. Deciphering the function of canonical Wnt signals in development and disease: Conditional loss- and gain-of-function mutations of β-catenin in mice. Genes Dev 22: 2308–2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegel H, Larue L, Ohsugi M, Fedorov L, Herrenknecht K, Kemler R 1995. Lack of β-catenin affects mouse development at gastrulation. Development 121: 3529–3537 [DOI] [PubMed] [Google Scholar]
- Hari L, Brault V, Kléber M, Lee HY, Ille F, Leimeroth R, Paratore C, Suter U, Kemler R, Sommer L 2002. Lineage-specific requirements of β-catenin in neural crest development. J Cell Biol 159: 867–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuberger J, Birchmeier W 2010. Interplay of cadherin-mediated cell adhesion and canonical Wnt signaling. Cold Spring Harb Perspect Biol 2: a002915 doi: 10.1101/cshperspect.a002915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierholzer A, Kemler R 2010. β-Catenin-mediated signaling and cell adhesion in postgastrulation mouse embryos. Dev Dyn 239: 191–199 [DOI] [PubMed] [Google Scholar]
- Hoffmans R, Basler K 2004. Identification and in vivo role of the Armadillo–Legless interaction. Development 131: 4393–4400 [DOI] [PubMed] [Google Scholar]
- Hoffmans R, Basler K 2007. BCL9-2 binds Arm/β-catenin in a Tyr142-independent manner and requires Pygopus for its function in Wg/Wnt signaling. Mech Dev 124: 59–67 [DOI] [PubMed] [Google Scholar]
- Hoffmans R, Städeli R, Basler K 2005. Pygopus and legless provide essential transcriptional coactivator functions to armadillo/β-catenin. Curr Biol 15: 1207–1211 [DOI] [PubMed] [Google Scholar]
- Huber O, Krohn M, Kemler R 1997. A specific domain in α-catenin mediates binding to β-catenin or plakoglobin. J Cell Sci 110: 1759–1765 [DOI] [PubMed] [Google Scholar]
- Huelsken J, Vogel R, Brinkmann V, Erdmann B, Birchmeier C, Birchmeier W 2000. Requirement for β-catenin in anterior–posterior axis formation in mice. J Cell Biol 148: 567–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ille F, Atanasoski S, Falk S, Ittner LM, Märki D, Büchmann-Møller S, Wurdak H, Suter U, Taketo MM, Sommer L 2007. Wnt/BMP signal integration regulates the balance between proliferation and differentiation of neuroepithelial cells in the dorsal spinal cord. Dev Biol 304: 394–408 [DOI] [PubMed] [Google Scholar]
- Kim S, Xu X, Hecht A, Boyer TG 2006. Mediator is a transducer of Wnt/β-catenin signaling. J Biol Chem 281: 14066–14075 [DOI] [PubMed] [Google Scholar]
- Maretto S, Cordenonsi M, Dupont S, Braghetta P, Broccoli V, Hassan AB, Volpin D, Bressan GM, Piccolo S 2003. Mapping Wnt/β-catenin signaling during mouse development and in colorectal tumors. Proc Natl Acad Sci 100: 3299–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet S, Bloch-Gallego E, Simeone A, Alvarado-Mallart RM 1996. The caudal limit of Otx2 gene expression as a marker of the midbrain/hindbrain boundary: A study using in situ hybridisation and chick/quail homotopic grafts. Development 122: 3785–3797 [DOI] [PubMed] [Google Scholar]
- Mosimann C, Hausmann G, Basler K 2006. Parafibromin/Hyrax activates Wnt/Wg target gene transcription by direct association with β-catenin/Armadillo. Cell 125: 327–341 [DOI] [PubMed] [Google Scholar]
- Mosimann C, Hausmann G, Basler K 2009. β-Catenin hits chromatin: Regulation of Wnt target gene activation. Nat Rev Mol Cell Biol 10: 276–278 [DOI] [PubMed] [Google Scholar]
- Najdi R, Holcombe RF, Waterman ML 2011. Wnt signaling and colon carcinogenesis: Beyond APC. J Carcinog 10: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsulic S, Peifer M 1996. An in vivo structure–function study of armadillo, the β-catenin homologue, reveals both separate and overlapping regions of the protein required for cell adhesion and for wingless signaling. J Cell Biol 134: 1283–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha PP, Scholze M, Bleiss W, Schrewe H 2010. Med12 is essential for early mouse development and for canonical Wnt and Wnt/PCP signaling. Development 137: 2723–2731 [DOI] [PubMed] [Google Scholar]
- Soriano P 1999. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 21: 70–71 [DOI] [PubMed] [Google Scholar]
- Valenta T, Lukas J, Korinek V 2003. HMG box transcription factor TCF-4’s interaction with CtBP1 controls the expression of the Wnt target Axin2/Conductin in human embryonic kidney cells. Nucleic Acids Res 31: 2369–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenta T, Lukas J, Doubravska L, Fafilek B, Korinek V 2006. HIC1 attenuates Wnt signaling by recruitment of TCF-4 and β-catenin to the nuclear bodies. EMBO J 25: 2326–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Amerongen R, Mikels A, Nusse R 2008. Alternative wnt signaling is initiated by distinct receptors. Sci Signal 1: re9 doi: 10.1126/scisignal.135re9 [DOI] [PubMed] [Google Scholar]
- Wakamatsu Y, Endo Y, Osumi N, Weston JA 2004. Multiple roles of Sox2, an HMG-box transcription factor in avian neural crest development. Dev Dyn 229: 74–86 [DOI] [PubMed] [Google Scholar]
- Widmann TJ, Dahmann C 2009. Wingless signaling and the control of cell shape in Drosophila wing imaginal discs. Dev Biol 334: 161–173 [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, Bailes JA, McMahon AP 1987. Expression of the proto-oncogene int-1 is restricted to specific neural cells in the developing mouse embryo. Cell 50: 79–88 [DOI] [PubMed] [Google Scholar]
- Xing Y, Takemaru K, Liu J, Berndt JD, Zheng JJ, Moon RT, Xu W 2008. Crystal structure of a full-length β-catenin. Structure 16: 478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi TP, Takada S, Yoshikawa Y, Wu N, McMahon AP 1999. T (Brachyury) is a direct target of Wnt3a during paraxial mesoderm specification. Genes Dev 13: 3185–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]