Notch signaling is a key regulator of neural stem cell self-renewal. Song and Lu now dissect the molecular mechanism underlying this process. Using Drosophila as a model to recapitulate tumor formation from stem cells, they identify that a Notch signaling cascade, which includes a feedback loop between dMyc and eIF4E, is critical in maintaining neuroblast proliferation. Intriguingly, cancer stem cell-like cells exhibit greater dependence on eIF4E, and inhibition of eIF4E function preferentially down-regulates tumor-initiating stem cells. This work provides insights into cancer stem cell regulation that may have valuable therapeutic implications.
Keywords: neural stem cell, Notch, cancer stem cell, eIF4E, cell growth
Abstract
Cancer stem cells (CSCs) are postulated to be a small subset of tumor cells with tumor-initiating ability that shares features with normal tissue-specific stem cells. The origin of CSCs and the mechanisms underlying their genesis are poorly understood, and it is uncertain whether it is possible to obliterate CSCs without inadvertently damaging normal stem cells. Here we show that a functional reduction of eukaryotic translation initiation factor 4E (eIF4E) in Drosophila specifically eliminates CSC-like cells in the brain and ovary without having discernable effects on normal stem cells. Brain CSC-like cells can arise from dedifferentiation of transit-amplifying progenitors upon Notch hyperactivation. eIF4E is up-regulated in these dedifferentiating progenitors, where it forms a feedback regulatory loop with the growth regulator dMyc to promote cell growth, particularly nucleolar growth, and subsequent ectopic neural stem cell (NSC) formation. Cell growth regulation is also a critical component of the mechanism by which Notch signaling regulates the self-renewal of normal NSCs. Our findings highlight the importance of Notch-regulated cell growth in stem cell maintenance and reveal a stronger dependence on eIF4E function and cell growth by CSCs, which might be exploited therapeutically.
It is proposed that cancer arises from a small population of tumor-initiating stem cells called cancer stem cells (CSCs), which share many functional properties with normal stem cells (Reya et al. 2001; Pardal et al. 2003; Passegue et al. 2003; Lobo et al. 2007). In human brain or ovarian tumors, CSCs and their corresponding normal stem cells express similar cell surface markers or stem cell-specific factors and rely on similar signaling pathways for their self-renewal and differentiation (Singh et al. 2004; Curley et al. 2009; Liu et al. 2010). The ability to molecularly or mechanistically distinguish between normal and tumor-initiating stem cells is thus a prerequisite for new cancer therapies aimed at selectively targeting those malignant stem cells.
The type II neural stem cells (NSCs), known as neuroblasts (NBs), in Drosophila larval brain provide a powerful model for studying CSC-initiated tumorigenesis (Wodarz and Gonzalez 2006; Bello et al. 2008; Boone and Doe 2008; Bowman et al. 2008; Wirtz-Peitz et al. 2008; Izergina et al. 2009; Weng et al. 2010). These NBs—marked by the presence of Deadpan (Dpn), a transcriptional target of Notch involved in NB self-renewal (San-Juan and Baonza 2011), and the absence of differentiation-promoting Prospero (Pros) transcription factor—divide asymmetrically to self-renew and give rise to immature intermediate progenitors (IPs; Dpn−Pros−), which are of smaller cell sizes and readily proceed to become mature IPs (Dpn+; cytoplasmic Pros). Mature IPs undergo multiple rounds of self-renewing transit-amplifying divisions to produce ganglion mother cells (GMCs; Dpn−; nuclear Pros) and, eventually, differentiated neurons (marked by the expression of Pros and neuronal marker Elav) (Fig. 1A,B). Such cellular hierarchy within the lineage provides a valuable system for tracing the cellular origin of CSCs, including the possibility that they may arise from more differentiated progenitor cells within the lineage (Clarke and Fuller 2006; Lobo et al. 2007).
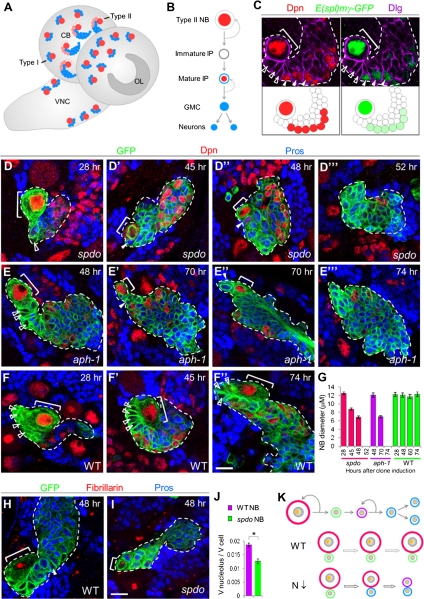
Figure 1.
N-dependent cell growth is required for type II NB maintenance. (A) A schematic drawing of Drosophila late larval CNS showing type I and type II NB lineages within the central brain area. (CB) Central brain; (OL) optical lobe; (VNC) ventral nerve cord. (B) A diagram of a type II NB lineage. Distinct cell types within the hierarchy can be identified by using combinations of cell fate markers: type II NB: Dpn (Deadpan, red)+, Pros (Prospero, blue)−; immature IPs: Dpn−, Pros−; mature IPs: Dpn+, cytoplasmic Pros; GMC or neurons: Dpn−, nuclear Pros. (C) N reporter E(spl)mγ-GFP (green) expression in type II NB lineage (mature IPs, closed arrowheads) showing differential N activity in different cell types. Dlg staining (purple) outlines the cell cortex. (White closed arrowheads) Mature IPs. From this panel on, NBs are marked with brackets and immature IPs are marked with white open arrowheads. (D–G) Clonal analysis of type II NBs of spdo (D) or aph-1 (E) mutants at various times ACI. Newly born daughter cells are marked with closed arrowheads. (F) Wild-type NBs served as control. (H,I) Analysis of the nucleoli (red; anti-fibrillarin) of the NBs and their daughter cells within spdo mutant (I) or wild-type (H) clones. (J) Quantification of nucleolar/cellular volume ratio of wild-type or spdo mutant NBs. (*) P < 0.002 versus control in Student's t-test; n = 6–8. (K) A working model depicting N regulation of NB fate through control of their cellular and nucleolar sizes. (Orange dot) Nucleolus; (red) NB; (green) immature IP; (purple) mature IP; (blue) GMCs or neurons. Bar, 10 μm.
When Notch (N) signaling is overactivated, transit-amplifying IPs could revert their cell fate back to a stem cell-like state, and their uncontrolled production leads to a brain tumor phenotype (Bowman et al. 2008; Wirtz-Peitz et al. 2008; Weng et al. 2010). N signaling is required for the proper maintenance of normal type II NBs (Wang et al. 2006; Bowman et al. 2008), a feature shared by stem cells from diverse tissues and species (Varnum-Finney et al. 2000; Luo et al. 2005; Mizutani et al. 2007; Ohlstein and Spradling 2007; Yu et al. 2008; Harrison et al. 2010). The molecular and cellular mechanisms by which N signaling promotes NSC maintenance are poorly understood. Parallel to N signaling modulation, the proliferation potential of IP cells is nonredundantly restricted by the asymmetrically segregated determinant Brain tumor (Brat) (Bello et al. 2006; Betschinger et al. 2006). Loss of Brat also leads to IP dedifferentiation and ectopic NB formation. Whether the N and Brat pathways impinge on similar downstream effectors is not known, and it remains unclear whether it is possible at all to selectively eliminate the ectopic NBs induced by either N overactivation or Brat inactivation without affecting normal NBs.
Here we show that the maintenance of both normal NBs and the CSC-like ectopic NBs in the Drosophila brain depend on N pathway-regulated cell growth involving the growth regulators eukaryotic translation initiation factor 4E (eIF4E) and dMyc. We provide evidence that eIF4E expression is up-regulated in CSC-like ectopic NBs and that eIF4E and dMyc form a regulatory loop to promote cell growth and stem cell fate. Importantly, normal NBs and CSC-like ectopic NBs can be distinguished based on their differential dependence on eIF4E function. Moreover, differential eIF4E dependency also differentiates normal germline stem cells (GSCs) from tumor-initiating stem cells in the ovary.
Results
Type II NBs exhibit gradually reduced cell growth and cell size when N signaling is inhibited
To search for distinguishing features between normal and tumor-initiating NSCs, we first investigated the mechanisms underlying the maintenance of normal type II NBs. Overactivation of N signaling is sufficient to induce ectopic type II NBs, and physiological N signaling is necessary for maintaining type II NB identity (Wang et al. 2006; Bowman et al. 2008). Thus, when N signaling is inhibited by RNAi-mediated N knockdown or by Numb overexpression, all type II NBs are lost (Bowman et al. 2008). However, the molecular mechanisms and downstream effectors of this signaling event remain undefined. We observed that E(spl)mγ-GFP, a reporter for N activity (Almeida and Bray 2005), was expressed highly in the NB and weakly in mature IPs, but was absent in immature IPs (Fig. 1C), suggesting that differential N activity within this lineage might confer distinct cell fates.
To investigate how N signaling helps maintain NB cell fate, we induced GFP-marked NB MARCM clones (Lee and Luo 2001) mutant for Sanpodo (Spdo) or Aph-1, two essential components of the N pathway (Skeath and Doe 1998; Hu and Fortini 2003), and examined the status of the mutant NBs at different time points after clone induction (ACI) (Fig. 1D,E). If the NB underwent symmetric division to yield two differentiating daughter cells when N signaling was inhibited, the loss of NB within the clone would be expected to occur after one mitotic division, which usually takes 2–3 h (Cabernard and Doe 2009). However, at ∼28 h ACI, NBs (bracket in Fig. 1D,F) were still present in spdo mutant clones and were of normal cell sizes (10–12 μm) (Fig. 1D,F). At 45–48 h ACI, mutant NBs became significantly smaller (6–8 μm), while other cells within the clone were unchanged in size (Fig. 1D′, D″). At ∼52 h ACI, cells with type II NB characters were no longer present (Fig. 1D′″). Similar observations were made in aph-1 mutant clones, although the phenotypes occurred with a delayed onset compared with the spdo mutant (Fig. 1E-E′″), presumably due to a longer perdurance of the Aph-1 protein (Hu and Fortini 2003). Consistently, gradual knockdown of Notch receptor itself, through the expression of Notch RNAi (N-IR) induced by the conditional 1407ts (1407-GAL4 [NB-specific GAL4]; tub-GAL80ts) system, revealed a gradual cell size reduction of type II NBs before their NB fate loss (Supplemental Fig. S1A,B). In comparison, the cell size of wild-type NBs remained constant during similar time courses (10–12 μm) (Fig. 1F–F″). Thus, the tight correlation between reduced cell size and stem cell fate loss is a common feature of type II NBs defective in Notch signaling.
In wild-type NB clones of the type II lineage, the newly born immature IPs (open arrowhead in Fig. 1D′,E′) and primary NBs are in direct contact and both are Pros−, whereas Pros+ cells are several cell diameters away from the NBs. In aged spdo or aph-1 clones, however, cells with nuclear Pros were found directly contacting the primary NBs (Fig. 1D′,E′). Consistently, low levels of cytoplasmic Pros not seen in wild-type type II NBs gradually appeared in spdo mutant NBs (Fig. 1D′; Supplemental Fig. S2A). However, NB loss still occurred in spdo pros double mutants at 52 h ACI (Supplemental Fig. S2B), arguing against the mild up-regulation of Pros being the primary cause of NB fate loss in spdo mutants. Following the reduction in cell sizes, Asense (Ase), normally present in mature IPs, was also detected in aged aph-1 NBs (Supplemental Fig. S2C), suggesting that the gradual reduction in cell size led to a progressive transition of the mutant NBs from a stem cell state to a differentiated one. In addition, similar-sized Dpn− Pros+ (white arrowhead in Fig. 1E″) and Dpn+ Pros− (bracket in Fig. 1E″) daughter cells were observed right after NB cytokinesis in aph-1 mutant clones (four out of four aph-1 mutant clones containing NB at this mitotic stage) (Fig. 1E″), indicating that mutant NBs might have undergone a division that is asymmetric in marker expression but symmetric in cell size. In contrast, the size or cell fate maintenance of type I NBs was unaffected by N inhibition (Supplemental Fig. S1C–E). Type I NBs, which divide asymmetrically to self-renew and produce a GMC that divides one more time to generate differentiated neurons (Supplemental Fig. S1C), do express the N reporter (Supplemental Fig. S1D). Thus, either N signaling is not involved in type I NB maintenance or there exists other redundant pathways.
To further investigate how N signaling might regulate NB size, we examined the nucleolus, which is important for ribosome biogenesis and whose size correlates with cell growth (Arabi et al. 2005; Grandori et al. 2005; Grewal et al. 2005). Wild-type NBs of the type II lineage exhibited larger nucleoli than their daughter cells (Fig. 1H), consistent with the notion that stem cells may grow at a faster rate than their differentiating daughter cells (Kohlmaier and Edgar 2008; Neumuller et al. 2008; Fichelson et al. 2009; Neumuller and Knoblich 2009). Importantly, the NB nucleolar sizes and, more significantly, the ratios between nucleolar and cellular volume were greatly reduced in spdo mutant NBs (Fig. 1H–J), suggesting that N signaling maintains type II NB fate at least in part through promoting cell growth (Fig. 1K).
Ectopic NBs show a faster growth rate than normal NBs within the type II lineages
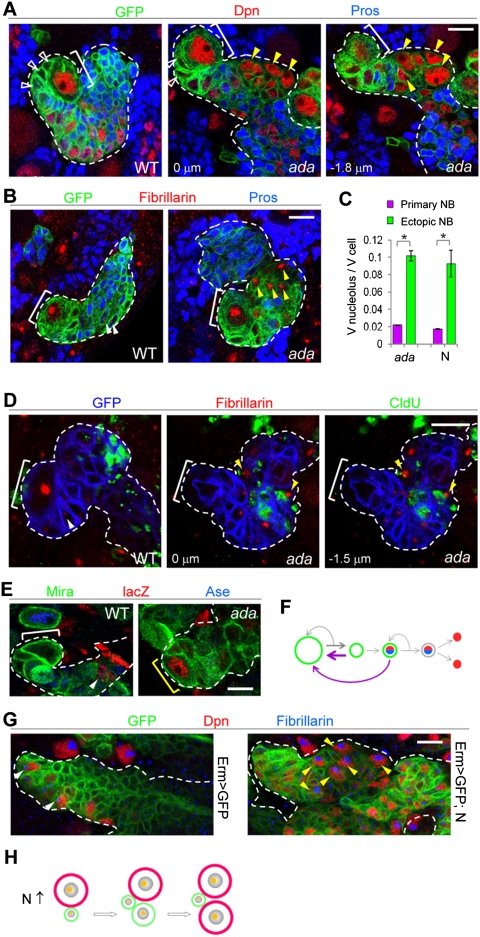
We next tested whether induction of ectopic NBs by N activation also involves regulated cell growth. We found that the endocytic protein α-Adaptin (Ada), which forms a protein complex with Numb to regulate the trafficking of cargo proteins such as N and Spdo in the sensory organ precursor (SOP) cells (Berdnik et al. 2002; Hutterer and Knoblich 2005), regulated type II NB homeostasis by down-regulating N signaling (Supplemental Fig. S3). In a time-course study, we found that at 30 h ACI, ada mutant MARCM clones contained a single NB in direct contact with immature IPs, followed by mature IPs, GMCs, and neurons (Supplemental Fig. S4A). No ectopic NBs were found in ada clones at this time point (Supplemental Fig. S4C). At 48 h ACI, ada mutant clones started to show ectopic NBs (yellow arrowhead in Fig. 2A), which were several cells away from the primary NB. Given that the newly born cells adjacent to the primary NB in ada clones were immature IPs, as in control clones (Fig. 2A; Supplemental Fig. S4A), and that the ectopic NBs were noticeably larger in cell size than the IPs, ada mutant NBs likely underwent asymmetric divisions to initially give rise to IP cells, which gradually increased their cell size while moving away from the primary NB and progressively dedifferentiated into ectopic NBs. Remarkably, the nucleolar to cellular volume ratios of the ectopic NBs within ada mutants or N-overexpressing clones were approximately fivefold higher than the primary NB (Fig. 2B,C), suggesting that cell growth is faster in ectopic NBs. Supporting the cell growth and dedifferentiation model, in ada mutant clones at 70 h ACI (Supplemental Fig. S4A,C), the cellular sizes of the ectopic NBs correlated with their distances from the primary NB (Supplemental Fig. S4A,C, white bracket), with the largest full-sized NBs (≥10 μm) (Supplemental Fig. S4A,C, yellow bracket) consistently being farthest away from the primary NB. A time-dependent increase of full-sized ectopic NBs was also observed in N overexpression clones (Supplemental Fig. S4B,C). These results reinforced the notion that, in response to N overactivation, an IP may dedifferentiate into a NB-like state in a process involving accelerated cell growth.
Figure 2.
N signaling promotes cell growth and dedifferentiation of IPs into ectopic NBs. (A) Ectopic NBs (Dpn+, Pros−; yellow arrowheads) in ada mutant clones. From this panel on, yellow arrowheads mark ectopic NBs. (B) Ectopic NBs in ada mutant clones contain enlarged nucleoli compared with IPs in wild-type clones (white arrowheads). (C) Quantification of nucleolar/cellular volume ratio in normal or ectopic NBs within ada mutant or N overactivation clones. (*) P < 0.0001; n = 8–10. (D) CldU pulse and chase revealed a cell cycle delay of ectopic NBs in ada mutant clones. After a 20-h chase, CldU was undetectable in the primary NBs (bracket) within wild-type or ada mutant clones. The only cells retaining the CldU label (white arrowheads) in wild-type clones were terminally differentiated neurons furthest from the primary NB. In contrast, CldU was detectable in some ectopic NBs within ada mutant clones (bigger nucleoli; yellow arrowheads). (Green) CldU; (red) fibrillarin; (blue) GFP. (E) Cell lineage tracing showing that mature IPs of ada mutants could dedifferentiate back into type II NBs (yellow bracket; identified by the expression of NB marker Mira and the absence of mature IP and GMC marker Ase), while wild-type mature IPs only generate GMCs or neurons (closed arrowhead; Mira−). Note that Ase is specifically expressed in type I but not type II NBs. A type I NB (Ase+Mira+) is shown in the left panel. (F) A schematic model summarizing cell lineage tracing data shown in E. (Green) Mira labeling NBs, immature IPs, and mature IPs; (red) lacZ; (blue) Ase. In ada mutants, immature and mature IPs can dedifferentiate into type II NBs (purple arrows). (G) Ectopic NBs induced by N overactivation specifically in mature IPs (driven by Erm-GAL4) contain larger nucleoli (yellow arrowheads) than control IPs (white arrowhead). (H) A working model proposing that when N signaling is overactivated (N↑), immature IPs (or mature IPs, not shown) gradually increase cellular and nucleolar sizes and dedifferentiate into a stem cell. Bar, 10 μm.
Using a chlorodeoxyuridine (CldU) pulse-and-chase strategy, we further examined the cell cycle of normal and ectopic NBs. After a 20-h chase, while CldU was undetectable in wild-type or ada mutant primary NBs due to dilution of label after multiple divisions, some ectopic NBs in ada mutant clones (containing an enlarged nucleolus) (yellow arrowhead in Fig. 2D) still retained the CldU label (Fig. 2D), indicating that the dedifferentiation process leading to ectopic NBs involved a transient cell cycle delay. Moreover, FLP-FRT-based lineage tracing by transiently inducing lacZ+ clones exclusively in mature IPs resulted in labeling of a few mature IPs, GMCs, and neurons (closed arrowhead in Fig. 2E), but never NBs (white bracket in Fig. 2E) in wild-type brains (five to six mature IPs; zero NBs/brain lobe are lacZ+; n = 10). However, in ada mutants, lacZ-labeled ectopic type II NBs (yellow bracket in Fig. 2E) were found after similar lineage tracing (two to three ectopic type II NBs/brain lobe are lacZ+; n = 12), indicating that mature IPs could indeed dedifferentiate back to NBs when N is overactivated (Fig. 2F). Consistently, expression of constitutively active N (NΔECD) specifically in mature IPs with Erm-Gal4 (Pfeiffer et al. 2008; Weng et al. 2010) was able to induce ectopic NBs, which contained enlarged nucleoli (Fig. 2G,H).
Ectopic NBs exhibit stronger dependence on eIF4E function than normal NBs
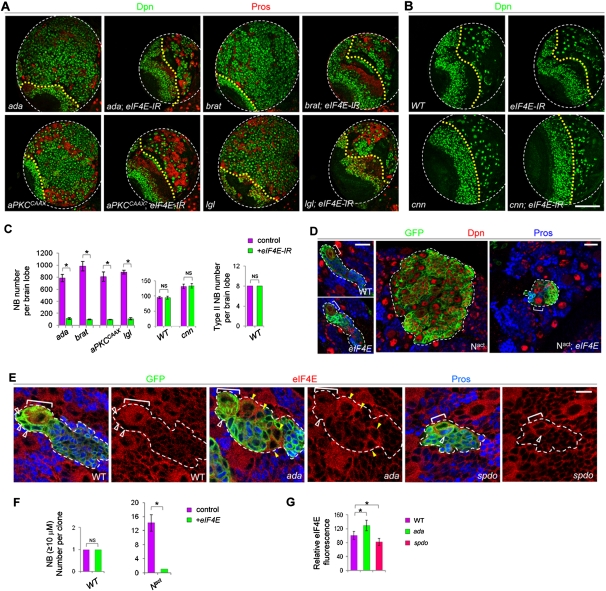
The differential cell growth rates observed between ectopic NBs and normal or primary NBs and the correlation between cell growth defects and NB fate loss prompted us to test whether slowing down cell growth might selectively affect the formation of ectopic NBs. Attenuation of TOR signaling, a primary mechanism of cell growth regulation, through NB-specific overexpression of TSC1/2 (Gao and Pan 2001; Potter et al. 2001; Tapon et al. 2001), a strong allele of eIF4E antagonist 4EBP [4EBP(LL)s] (Miron et al. 2001), or a dominant-negative form of TOR (TOR.TED) all partially suppressed ectopic NB formation in ada mutants without affecting normal or primary NBs (Supplemental Fig. S3). Interestingly, RNAi-mediated knockdown of eIF4E, a stimulator of oncogenic transformation (Lazaris-Karatzas et al. 1990) and a downstream effector of TOR signaling (Mamane et al. 2004), showed a better suppression than manipulating other TOR pathway components (Supplemental Fig. S5), suggesting that eIF4E might play a more important role in ectopic NB formation. Strikingly, the brain tumor phenotypes caused by overactivation of N signaling—as in lethal giant larvae (lgl) mutant (Betschinger et al. 2006; Lee et al. 2006a), aPKCCAAX overexpression (Lee et al. 2006a), or N overexpression conditions—were also fully suppressed by eIF4E knockdown (Fig. 3A,C; Supplemental Figs. S6A, S7A). Furthermore, the brain tumor phenotypes of brat mutants (Bello et al. 2006; Betschinger et al. 2006; Lee et al. 2006b) were also completely rescued by eIF4E RNAi (Fig. 3A,C; Supplemental Fig. S6A).
Figure 3.
eIF4E knockdown efficiently and specifically inhibits brain tumor formation. (A) Effects of NB-specific knockdown of eIF4E (driven by 1407-GAL4) on ectopic NB formation in ada, brat, or lgl mutants or aPKCCAAX overexpression backgrounds. (Green) NBs marked by Dpn; (red) neurons marked by Pros. Posterior views of a single brain lobe are shown. (B) eIF4E knockdown has no discernable effects on normal NB development or on ectopic NB formation resulting from symmetric division of type I NBs in cnn mutants. From this panel on, the yellow dotted line marks the boundary between the optic lobe (left) and the central brain (right) areas. Central brain NBs can be distinguished from optic lobe NBs based on their medial/superficial location in the brain and larger size. (C) Quantification of data from A and B. (*) P < 0.0001; n = 15–20. (D) Clonal analysis of type II NBs in wild-type, eIF4E mutant, Nact, or Nact; eIF4E backgrounds. (E) eIF4E expression (red) in wild-type, ada, or spdo mutant type II NBs. (F,G) Quantification of data from D and E. Bars: A,B, 100 μm; D, 20 μm; E, 10 μm.
In contrast, normal NB formation or maintenance was not affected by eIF4E knockdown (Fig. 3B,C; Supplemental Fig. S6B,C). NBs with eIF4E knockdown remained highly proliferative, as evidenced by the mitotic figures, and displayed relatively normal apical basal cell polarity (Supplemental Fig. S8). There are several other eIF4E-like genes in the fly genome (Hernandez et al. 2005), which may play partially redundant roles in normal NB maintenance. eIF4E knockdown appeared to specifically block ectopic NB formation caused by the dedifferentiation of IPs in type II NB lineages, since it did not affect ectopic type I NB formation in cnn (Cabernard and Doe 2009) or polo mutants (Wang et al. 2007) that are presumably caused by symmetric divisions of type I NBs (Fig. 3B,C; Supplemental Fig. S6D,E). In addition, cell fate transformation induced by N overactivation in the SOP lineage was not affected by eIF4E RNAi (Supplemental Fig. S6F), supporting that eIF4E is particularly required for type II NB homeostasis (Supplemental Fig. S6G). Supporting the specificity of the observed eIF4E RNAi effect, another eIF4E RNAi transgene (eIF4E-RNAi-s) also prevented ectopic NB formation (Supplemental Fig. S9). Moreover, a strong loss-of-function mutation of eIF4E also selectively eliminated ectopic NBs induced by N overactivation (Fig. 3D,F) without affecting normal NBs (Fig. 3D,F), reinforcing our hypothesis that ectopic NBs exhibit higher dependence on eIF4E.
To further support the notion that the ectopic NBs are particularly vulnerable to eIF4E depletion, we carried out a conditional expression experiment in which eIF4E-RNAi-s was turned on in brat mutants using the 1407ts system, after ectopic NBs had been generated. Whereas the brain tumor phenotype exacerbated over time in the brat mutants, 1407-GAL4-driven eIF4E-RNAi-s expression in brat mutants effectively eliminated ectopic NBs, leaving normal NBs largely unaffected (Supplemental Figs. S9, S10).
In normal type II NB lineage, eIF4E protein was enriched in the NBs (Fig. 3E). Ectopic NBs induced by N overactivation in ada mutants also expressed eIF4E at high levels (Fig. 3E, yellow arrowhead), whereas spdo mutant NBs exhibited reduced eIF4E expression (Fig. 3E,G). Thus, eIF4E up-regulation correlates with N-induced ectopic NB formation in a dedifferentiation process that likely involves elevated cell growth.
A Notch-dMyc–eIF4E molecular circuitry is crucial for NSC growth control
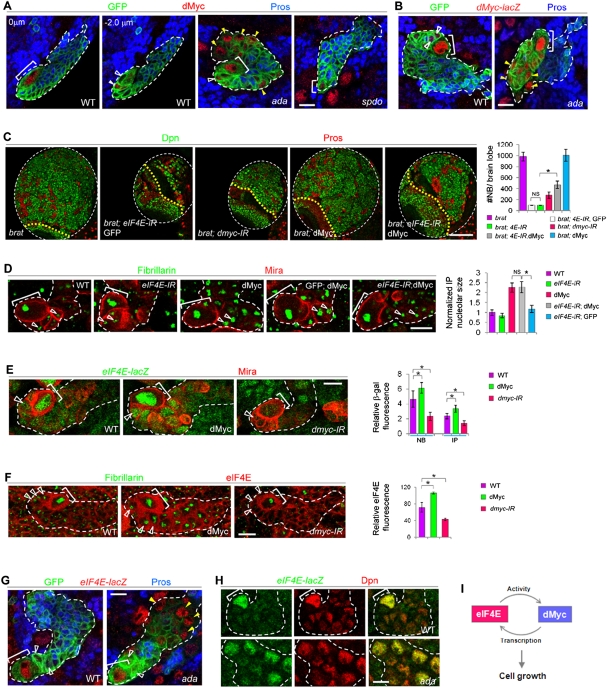
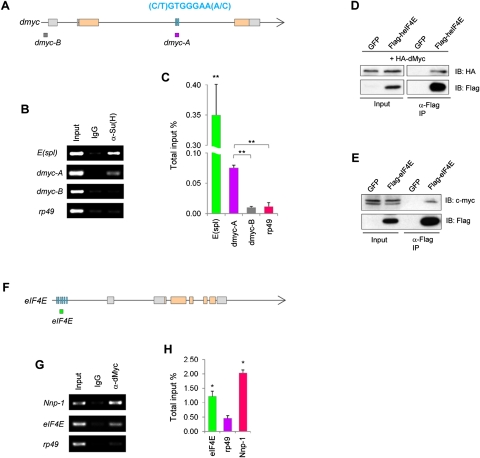
Given the coincidence of nucleolar size change with ectopic NB formation, we tested the involvement of the growth regulator dMyc (Arabi et al. 2005; Grandori et al. 2005; Grewal et al. 2005). dMyc protein levels were up-regulated in normal or N overactivation-induced ectopic NBs, but were down-regulated in spdo mutant NBs (Fig. 4A). Furthermore, dMyc transcription, as detected with a dMyc-lacZ transcriptional fusion reporter (Mitchell et al. 2010), was also up-regulated in both normal and ectopic NBs in ada mutants (Fig. 4B). A previous study in Drosophila S2 cells identified dMyc as a putative N target (Krejci et al. 2009). We carried out in vivo chromatin immunoprecipitation (ChIP) experiments to assess whether dmyc transcription is directly regulated by N signaling in NBs. Using chromatin isolated from wild-type larval brains and a ChIP-quality antibody against the N coactivator Suppressor of Hairless [Su(H)] (Krejci and Bray 2007), we could demonstrate specific binding of Su(H) to its putative binding sites within the second intron of dmyc (dmyc-A). No binding to an internal negative control region proximal to the first exon of dmyc (dmyc-B) or to the promoter region of the rp49 gene was detected (Fig. 5A–C). N signaling thus directly activates dMyc transcription in the NBs. Similar to eIF4E RNAi, knockdown of dMyc strongly suppressed ectopic NB formation induced by Brat or Ada inactivation or N overactivation (Fig. 4C; Supplemental Figs. S7, S11). Intriguingly, the strong tumor suppression effect of eIF4E knockdown was partially abolished by dMyc overexpression (Fig. 4C; Supplemental Fig. S7). Furthermore, dMyc function, as reflected by its promotion of nucleolar growth in IPs, was attenuated by eIF4E RNAi, although eIF4E RNAi alone had no obvious effect (Fig. 4D). Different from the reported eIF4E regulation of Myc expression in mammalian cells (Lin et al. 2008), dMyc promoter activity or protein levels remained unaltered under eIF4E RNAi conditions (Supplemental Fig. S12A,B), suggesting that eIF4E may modulate dMyc activity without altering its expression. One possibility is that eIF4E may enter the nucleus to interact with Myc and promote its transcriptional activity. To test this hypothesis, HEK293T cells were transfected with Flag-tagged human eIF4E alone or in combination with HA-tagged dMyc. Indeed, both Drosophila dMyc and endogenous human c-Myc specifically coimmunoprecipitated with human eIF4E from nuclear extracts (Fig. 5D,E), indicating a conserved interaction between eIF4E and Myc within the nuclei of proliferating cells. Consistent with these biochemical data, dMyc transcriptional activity within NBs, which could be monitored with an eIF4E-lacZ reporter (see below), was drastically reduced upon eIF4E knockdown (Supplemental Fig. S12C).
Figure 4.
dMyc and eIF4E constitute a regulatory loop in NB regulation. (A) dMyc (red) expression in wild-type, ada, or spdo mutant type II NBs. The MARCM clones were analyzed at 48 h ACI. (B) dMyc-lacZ is highly expressed in the NB but not its daughter cells in a wild-type type II NB clone. In ada mutant clones, however, ectopic NBs (yellow arrowheads) also show up-regulated dMyc transcription. (Green) GFP; (red) dMyc-lacZ; (blue) Pros; (brackets) NBs. (C) Ectopic NB formation in brat mutants is suppressed by dMyc RNAi, and the tumor suppression effect of eIF4E knockdown is partially relieved when dMyc is overexpressed. (*) P < 0.0001; n = 15–20. (D) An increase in IP nucleolar size (green, open arrowheads) promoted by dMyc overexpression is strongly suppressed by eIF4E RNAi. (*) P < 0.0001; n = 30–40. (E) Up-regulation of eIF4E-lacZ expression in NBs and IPs by dMyc. (*) P < 0.015; n = 20–30. (F) Positive regulation of eIF4E protein levels by dMyc in IPs (open arrowheads). (*) P < 0.01; n = 20–30. (G) High eIF4E-lacZ expression in both wild-type NBs and ectopic NBs of ada mutant. (Green) eIF4E-lacZ; (red) Dpn. (H) eIF4E-lacZ expression in wild-type or ada mutant type II NB clones. (Green) GFP; (red) eIF4E-lacZ; (blue) Pros; (brackets) NBs. (I) A working model depicting the eIF4E–dMyc feedback regulatory loop in promoting cell growth within NBs. Bars: A,B,D–H, 10 μm; C, 100 μm.
Figure 5.
Biochemical characterization of the N-dMyc–eIF4E molecular circuitry. (A–C) ChIP with the anti-Su(H) antibody or a control IgG on wild-type third instar larval brain chromatin. (A) Schematic representation of the dmyc locus. (Orange rectangles) Coding regions; (gray rectangles) noncoding regions; (lines) introns; (blue bars) two putative Su(H)-binding sites matching the consensus sequence (C/T)GTGGGAA(A/C). Enrichment of Su(H) at the dmyc-A but not dmyc-B amplicon is determined by both standard PCR (B) and real-time quantitative PCR (C). A region in E(spl) containing Su(H)-binding sites (Bailey and Posakony 1995) and a region in the rp49 promoter without such sites serve as positive and negative controls, respectively. (**) P < 0.0001. (D,E) Coimmunoprecipitation between transfected human eIF4E and Drosophila dMyc (D) or human eIF4E and endogenous c-Myc (E) in HEK293T nuclear extracts. GFP serves as a negative control. (IP) Immunoprecipitation; (IB) immunoblotting. Input represents 4% of total. (F–H) ChIP with anti-dMyc antibody or a control IgG on wild-type third instar larval brain chromatin. (F) Schematic representation of the eIF4E locus. Blue bars represent a cluster of adjacent noncanonical E boxes. Enrichment of dMyc at the eIF4E locus is shown by standard PCR (G) and real-time quantitative PCR (H). A region in Nnp-1 harboring canonical E boxes CACGTG and a region within the rp49 promoter serve as positive and negative controls, respectively. (*) P < 0.01. Error bars indicate standard deviation of three independent experiments.
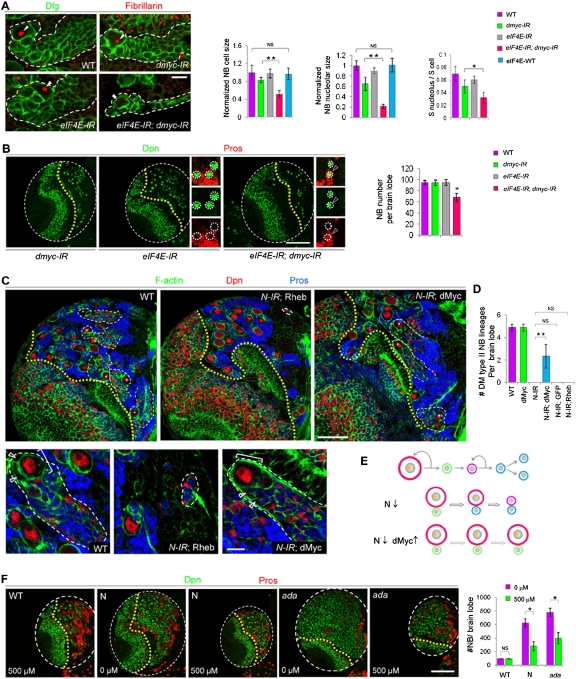
On the other hand, eIF4E transcription, as detected with an eIF4E-lacZ transcriptional fusion reporter, as well as eIF4E protein levels detected by immunostaining were up-regulated upon dMyc overexpression and down-regulated by dMyc RNAi (Fig. 4E,F). It is unlikely that the changes in eIF4E-lacZ activity were due to global increases or decreases in β-galactosidase (β-gal) translation caused by altered dMyc levels, since lacZ expression from a dMyc-lacZ reporter was unaffected under similar conditions (Supplemental Fig. S12B). Furthermore, like dMyc protein, eIF4E-lacZ reporter expression was up-regulated in normal NBs or ectopic NBs in ada mutants (Fig. 4G,H), further supporting the notion that dMyc may up-regulate eIF4E transcription. Moreover, ChIP experiments using chromatins isolated from wild-type larval brains and a ChIP-quality antibody against dMyc (Maines et al. 2004; Teleman et al. 2008) demonstrated specific binding of dMyc to an eIF4E promoter region harboring a cluster of adjacent noncanonical E boxes (Fig. 5F–H; Arabi et al. 2005), supporting a direct regulation of eIF4E transcription by dMyc. dMyc and eIF4E thus appeared to form a regulatory feedback loop that promoted NB growth and renewal (Fig. 4I). Consistent with this model, while knocking down either dMyc or eIF4E had no noticeable effect on type II NB maintenance and only a mild effect on NB nucleolar size in the case of dMyc RNAi, their simultaneous knockdown led to a significant reduction in nucleolar size, premature neuronal differentiation, and loss of NBs (Fig. 6A,B).
Figure 6.
The N-dMyc–eIF4E module operates in normal or ectopic NBs. (A) dMyc or eIF4E knockdown alone has a mild or no effect on NB cell growth, respectively, as assayed by their cell size, nucleolar size, or ratio of nucleolar/cell size, whereas knocking down both leads to a drastic reduction in NB cell growth. Overexpression of a wild-type form of the eIF4E transgene shows no discernible effect on NB cellular or nucleolar sizes. NB nucleoli are indicated by arrowheads. (Green) Dlg; (red) fibrillarin. (**) P < 0.0001; (*) P < 0.05; n = 30–40. (B) Knocking down either dMyc or eIF4E alone has no noticeable effect on type II NB maintenance, but knocking down both leads to premature differentiation and NB loss. (Green) Dpn; (red) Pros. (*) P < 0.001; n = 15–20. Magnified images are shown on the right, with individual NBs encircled with white dotted lines. (Arrowheads) Pros+ Dpn+ cell. (C) Type II NB lineages in wild-type, 1407>N RNAi; Rheb, or1407>N-IR; dMyc backgrounds are delineated by white dashed lines, and the NB in each lineage is indicated by a star. The yellow dotted line indicates the boundary between the optical lobe (left) and the central brain (right) region. Amplified images of a representative type II NB lineage in each genotype are shown at the bottom. Larvae were processed at 96 h ALH. (D) Quantification of type II NB number in wild-type, 1407>dMyc, 1407>N-IR, 1407>N-IR; dMyc, 1407>N-IR; GFP, or 1407>N-IR; Rheb brain lobes. (**) P < 0.0005; n = 15–20. Only dorsal–medial (DM) type II NBs generating surface layer progeny were counted. (E) A working model of dMyc overexpression preventing the stem cell fate loss in N signaling-defective NBs by boosting cell growth. (Orange dot) Nucleolus; (red) NB; (green) immature IP; (purple) mature IP; (blue) GMCs or neurons. (F) Effects of eIF4E inhibitor treatment on wild-type NB maintenance and ectopic NB formation induced by N overactivation or Ada inactivation. (*) P < 0.0001; n = 15–20. Bars: A,C, bottom panel, 10 μm; C, top panel, 50 μm; B,F, 100 μm.
Enhancing cell growth through dMyc overexpression prevents NB loss caused by N inhibition
If the dMyc–eIF4E axis of cell growth control is a crucial downstream effector of N signaling in regulating NB maintenance, its up-regulation might be able to rescue the type II NB depletion phenotype resulting from reduced N signaling. Indeed, the loss of NBs associated with reduced Notch signaling was preventable when cell growth was boosted by dMyc overexpression (Fig. 6C–E). Thus, while N-IR directed by 1407-GAL4 led to complete elimination of type II NBs (Supplemental Fig. S13A), the coexpression of dMyc, but not CD8-GFP or Rheb, an upstream component of the TOR pathway (Stocker et al. 2003), resulted in the preservation of approximately half of type II NBs with apparently normal cell sizes, cell fate marker expression, and lineage composition (Fig. 6C–E; Supplemental Fig. S13A). A similar effect was observed when dMyc was coexpressed with N-IR using the conditional 1407ts system, with transgene expression induced at the larval stage (Supplemental Fig. S13B,C). While both dMyc and Rheb promote cell growth, they do so through distinct mechanisms, with the former increasing nucleolar size and the latter expanding cytoplasmic volume (Saucedo and Edgar 2002). These results thus provide compelling evidence that control of cell growth, particularly nucleolar growth, is a critical component in the maintenance of NB identity by N signaling.
The differential responses of normal and tumor-initiating stem cells to functional reduction of eIF4E prompted us to test whether chemicals that specifically inhibit eIF4E function might have therapeutic potential in preventing CSC-induced tumorigenesis. Indeed, the brain tumor phenotypes induced by N overactivation or ada loss of function were effectively suppressed by feeding animals with fly food containing Ribavirin, an eIF4E inhibitor that interferes with eIF4E binding to mRNA 5′ caps and promotes the relocalization of eIF4E from the nucleus to the cytoplasm (Fig. 6F; Supplemental Fig. S14; Kentsis et al. 2004; Assouline et al. 2009).
CSC-like cells in the Drosophila ovary also particularly rely on the eIF4E–dMyc regulatory loop
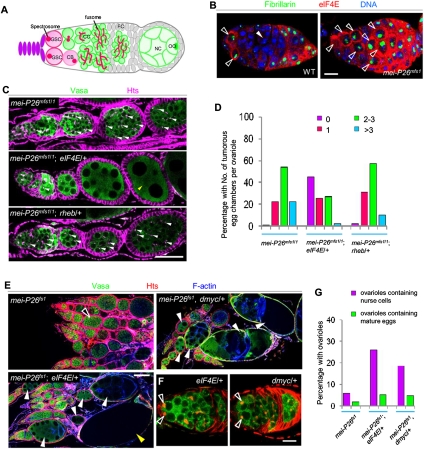
To test whether the higher dependence on eIF4E function may also apply to tumor-initiating stem cells in other tissues, we investigated the function of the dMyc–eIF4E axis in Drosophila ovarian GSCs (Gilboa and Lehmann 2004; Wong et al. 2005; Fuller and Spradling 2007). After mitotic division, each of these GSCs self-renews and produces a differentiating daughter cell, the cystoblast (CB), which subsequently undergoes four rounds of transit-amplifying divisions to generate a cyst of 16 interconnected cystocytes of smaller sizes. The cysts become surrounded by epithelial follicle cells before they bud off the germarium to become individual egg chambers (Fig. 7A; Lin 1997; Fuller and Spradling 2007; Kirilly and Xie 2007). It has been proposed that in mei-P26 mutant ovaries, cystocytes increase cellular and nucleolar sizes and gain self-renewing capacities to become ectopic CSC-like stem cells (Page et al. 2000; Neumuller et al. 2008; Liu et al. 2009), likely through a dedifferentiation mechanism (Kai and Spradling 2004).
Figure 7.
The dMyc–eIF4E regulatory loop also operates in Drosophila GSCs. (A) A schematic drawing of Drosophila early oogenesis. GSCs (pink) reside at the anterior tip of the germarium, directly contacting the stem cell niche (purple). The GSCs continuously self-renew and produce a daughter cell that moves away from the niche and becomes the differentiating CB (light pink), which gives rise to 16 post-mitotic cystocytes (CC; green). The GSCs and CBs can be identified by the presence of the membranous spectrosome (red spot), which develops into a branched fusome in differentiating cysts (Lin et al. 1994). (OO) Oocyte; (NC) nurse cells. (B) eIF4E (red) expression is enriched in the GSCs (open arrowhead, containing large nucleoli) compared with post-mitotic cysts (closed arrowhead, containing small nucleoli) in wild-type ovaries. In mei-P26mfs1 mutants, eIF4E is highly expressed in all CSCs that contained enlarged nucleoli. (C) mei-P26mfs1/1 mutant egg chambers often carry ectopic GSC-like cells (white arrowheads) marked by the spectrosomes. Such intermediate ovarian tumor phenotypes of mei-P26 can be effectively suppressed by removing one copy of eIF4E but not rheb. Note that the defect of irregular nurse cell number in mei-P26mfs1/1 mutants is not suppressed by reduction of eIF4E function (yellow arrowhead). (Green) Vasa (germ cell-specific marker); (red) Hts (marker for the spectrosome and the fusome). (D) Statistical analysis of data shown in C; n = 450–500 ovarioles examined for each genotype. (E,F) mei-P26fs1 mutants show a strong ovarian tumor phenotype (88.3% tumorous ovarioles [open arrowhead], 6.25% ovarioles containing nurse cells, 1.25% ovarioles containing eggs, and 4.2% empty ovarioles), which can be significantly suppressed by removing one copy of eIF4E (63.9% tumorous ovarioles, 26.0% ovarioles containing nurse cells [white arrowheads], 5.3% ovarioles containing eggs [yellow arrowhead], and 4.8% empty ovarioles) or one copy of dMyc (72.4% tumorous ovarioles, 18.6% ovarioles containing nurse cells [white arrowheads], 4.8% ovarioles containing eggs, and 4.2% empty ovarioles). Images in these panels are assembled from individual images acquired with a 40× objective. (F) eIF4E or dMyc heterozygosity has no effect on the maintenance of GSCs (open arrowheads) in an otherwise wild-type background (open arrowheads). (G) Statistical analysis of data shown in E; n = 480–520 ovarioles examined for each genotype. Bars: B,F, 10 μm; C, 50 μm; E, 100 μm.
dMyc protein expression and ribosomal biogenesis, as indicated by nucleolar size, are differentially regulated between GSCs and post-mitotic cystocytes, and their up-regulation coincides with the transformation of cystocytes into CSCs (Neumuller et al. 2008; Rhiner et al. 2009), although the physiological significance of such cell growth regulation is yet to be assessed. We found that, similar to dMyc, eIF4E protein levels were high in GSCs (open arrowheads in Fig. 7B) but low in cystocytes (closed arrowhead in Fig. 7B) of wild-type ovaries, and differential eIF4E expression correlated with the distinct nucleolar sizes of GSCs and their differentiated daughter cells (Fig. 7B). eIF4E distribution within the GSC lineages thus resembled its distribution in the type II NB lineages (Fig. 3E). In ovarian tumor tissues derived from mei-P26 mutants (Page et al. 2000; Neumuller et al. 2008), high levels of eIF4E expression could be detected in all ectopic GSCs (open arrowheads in Fig. 7B), similar to the expression of dMyc. This suggests that the dMyc–eIF4E regulatory loop may also operate in the GSCs to promote stem cell growth and cell fate (Supplemental Fig. S15B).
We examined the functional significance of up-regulated eIF4E and dMyc expression in ovarian tumor formation. Strikingly, removing one copy of eIF4E or dMyc effectively suppressed the formation of ectopic GSCs in mei-P26 mutants (Fig. 7C–E,G) without affecting normal GSCs (Fig. 7F). In contrast, removing one copy of rheb did not have a discernable effect (Fig. 7C,D). Together, these results indicate that CSCs having a stronger dependence on eIF4E function represents a common theme in cancer biology that might be exploited to specifically target these malignant stem cells.
Discussion
The CSC hypothesis was initially developed based on studies in mammalian systems. Various studies have supported the notion that CSCs share many functional features with normal stem cells, such as signaling molecules, pathways, and mechanisms governing their self-renewal versus differentiation choice. However, the cellular origin of CSCs and the molecular and cellular mechanisms underlying their development or genesis remain poorly understood. It has been proposed that CSCs could arise from (1) an expansion of normal stem cell niches, (2) normal stem cells adapting to different niches, (3) normal stem cells becoming niche-independent, or (4) differentiated progenitor cells gaining stem cell properties (Clarke and Fuller 2006). Here we showed that in the Drosophila larval brain, CSCs can arise from the dedifferentiation of transit-amplifying progenitor cells back to a stem cell-like state. Importantly, we identified eIF4E as a critical factor involved in this dedifferentiation process. More significantly, we showed that reduction of eIF4E function can effectively prevent the formation of CSCs without affecting the development or maintenance of normal stem cells. This particular dependence on eIF4E function by CSCs appears to be a general theme, as reduction of eIF4E function also effectively prevented the formation of CSCs, but not normal GSCs, in the fly ovary. These findings may have important implications for stem cell biology and cancer biology, in terms of both mechanistic understanding and therapeutic intervention.
Our study also offers mechanistic insights into the cellular processes leading to the dedifferentiation of progenitors back to stem cells. In Drosophila type II NB clones with overactivated N signaling, ribosome biogenesis within ectopic NBs appears to be faster than in normal NBs, as shown by the fact that the ratio of nucleolar to cellular volume of the ectopic NBs is approximately fivefold higher than that of normal NBs. The faster growth rate is accompanied by the up-regulation of dMyc and eIF4E and appears to be essential for transit-amplifying progenitors to undergo complete dedifferentiation back to a stem cell-like state. When the function of cell growth-promoting factors such as eIF4E is attenuated, the faster cell growth of ectopic NBs can no longer be sustained and the dedifferentiation process stalls. As a result, brain tumor formation caused by uncontrolled production of ectopic NBs is suppressed. In contrast, normal NBs, which presumably have relatively lower requirements for cell growth and hence eIF4E function, maintain their stem cell fate and development under similar conditions (Supplemental Fig. S6G). Therefore, a potential key to a successful elimination of CSC-induced tumors would be to find the right level of functional reduction in eIF4E, which causes minimal effects on normal stem cells but effectively obliterates CSCs. An ongoing clinical trial with Ribavirin in treating acute myeloid leukemia (AML) (Assouline et al. 2009), a well-characterized CSC-based cancer, demonstrated exciting proof of principle that such a strategy is feasible. The current version of Ribavirin, however, has certain limitations, such as its poor specificity and the high dosage (micromolar range) required for effective treatment (Sulkowski et al. 2011). Thus, more specific and effective eIF4E inhibitors are urgently needed. Our drug treatment experiments with Ribavirin validated Drosophila NBs as an excellent CSC model for searching further improved drugs. More importantly, the nuclear interaction between eIF4E and Myc unraveled by our biochemical analysis not only provides a new mechanistic explanation for the synergistic effects of eIF4E and Myc in tumorigenesis (Ruggero et al. 2004; Wendel et al. 2007), but also sheds new light on how to rationally optimize drug design and therapy for treating CSC-based cancer.
Our results offer new information on how N signaling helps specify and maintain NSC fate. N signaling regulates stem cell behavior in various tissues of diverse species (Varnum-Finney et al. 2000; Luo et al. 2005; Mizutani et al. 2007; Ohlstein and Spradling 2007; Yu et al. 2008; Harrison et al. 2010). However, it remains unclear how differential N signaling determines distinct cell fate within the stem cell hierarchy. Here we demonstrate that N signaling maintains Drosophila NSC fate at least in part through promoting cell growth. The following evidence supports that cell growth, but not cell fate, change is the early and primary effect of N signaling inhibition in type II NBs: (1) Pros expression is not immediately turned on in spdo mutant NBs with reduced cell sizes. Instead, it gradually increases during the course of spdo mutant NB divisions. (2) Up-regulation of Pros is not the cause of stem cell fate loss in spdo mutant NBs, as shown by spdo pros double-mutant analysis. (3) Cell growth defects precede the up-regulation of Ase expression in aph-1 mutant NBs. (4) Promotion of cell growth, and particularly nucleolar growth, by dMyc is sufficient to prevent NB loss caused by N inhibition. At the molecular level, N signaling appears to regulate the transcription of dMyc, which in turn up-regulates the transcription of eIF4E. Such a transcriptional cascade and feedback regulation of dMyc activity by eIF4E may help to sustain and amplify the activity of the Notch-dMyc–eIF4E molecular circuitry. Hence, differential N signaling within the lineage can lead to different cell growth rates, which partially determine differential cell fates. Consistent with this notion, knockdown of both eIF4E and dMyc results in defects of NB cell growth and loss of stem cell fate.
While many signaling pathways and molecules have been implicated in the maintenance of stem cell identity, the question of how a stem cell loses its “stemness” at the cellular level remains poorly understood. A stem cell may lose its stem cell fate by undergoing a symmetric division to yield two daughter cells that are both committed to differentiation or through cell death. Earlier studies provided intriguing hints that cell growth and translational regulation could influence stem cell maintenance in the Drosophila ovary (Kai et al. 2005; Fichelson et al. 2009; Shen et al. 2009). Here, our detailed clonal analyses of NSCs over multiple time points provide direct evidence that a NSC with impaired N signaling will gradually lose its identity due to a gradual slowing down of cell growth and loss of cell mass. Remarkably, such loss of stem cell fate can be prevented when cell growth is restored by dMyc, but not Rheb, overexpression, demonstrating the functional significance of regulated cell growth, particularly nucleolar growth, in stem cell maintenance. More importantly, this information offers clues on how to specifically eliminate tumor-initiating stem cells. Our studies suggest that a stem cell, normal or malignant, has to reach a certain growth rate in order to acquire and maintain its stemness, presumably because when the stem cell grows below such a threshold, its proliferative capacity becomes too low, whereas the concentration of differentiation-promoting factors becomes too high to be compatible with the maintenance of stem cell fate. Consistent with this notion are the strong correlation between the expression of ribosomal proteins and cellular proliferation (van Riggelen et al. 2010) as well as the correlation between the reduction of NB sizes and the up-regulation of differentiation-promoting factor Pros or Ase in different developmental contexts (Fig. 6B; Supplemental Fig. S2; Maurange et al. 2008; Siegrist et al. 2010).
Our results also provide new insights into how the evolutionarily conserved tripartite motif and Ncl-1, HT2A, and Lin-41 (TRIM-NHL) domain proteins regulate stem cell homeostasis. The TRIM-NHL protein family, to which Brat and Mei-P26 belong, include evolutionarily conserved stem cell regulators that prevent ectopic stem cell self-renewal by inhibiting Myc (Bello et al. 2006; Betschinger et al. 2006; Lee et al. 2006b; Neumuller et al. 2008; Schwamborn et al. 2009). However, the downstream effectors of the TRIM-NHL proteins remain largely unknown. Here we identify eIF4E as such a factor. NB-specific knockdown of eIF4E completely suppresses the drastic brain tumor phenotype caused by loss of Brat. Interestingly, eIF4E knockdown is even more effective than dMyc knockdown in this regard. N signaling and Brat have been proposed to act in parallel in regulating Drosophila type II NB homeostasis (Bowman et al. 2008; Weng et al. 2010). However, at the molecular level, how deregulation of these two rather distinct pathways causes similar brain tumor phenotypes remain largely unknown. Our results suggest that these two pathways eventually converge on the dMyc–eIF4E regulatory loop to promote cell growth and stem cell fate (Supplemental Fig. S15). N overactivation and loss of Brat both result in up-regulation of eIF4E and dMyc in transit-amplifying progenitors, accelerating their growth rates and helping them acquire stem cell fate. Consistent with a general role of eIF4E and dMyc in stem cell regulation, we show that partial reduction of eIF4E or dMyc function in the Drosophila ovary effectively rescues the ovarian tumor phenotype due to the loss of Mei-P26. The vertebrate member of the TRIM-NHL family, TRIM32, is shown to suppress the stem cell fate of mouse neural progenitor cells, partially through degrading Myc (Schwamborn et al. 2009). Whether eIF4E acts as a downstream effector of TRIM32 in balancing stem cell self-renewal versus differentiation in mammalian tissues awaits future investigation.
Materials and methods
Clonal analysis
To generate NB MARCM clones, larvae at 42 h after larvae hatching (ALH) were heat-shocked for 90 min at 37°C and further aged at 25°C before dissection. For inducing eIF4E mutant or knockdown clones, newly hatched larvae were heat-shocked for 90 min at 37°C and further aged for 4 d at 25°C before dissection.
For lineage tracing experiments, ada, UAS-Flp/Bc-Gla; actin-FRT-stop-FRT-lacZ flies were crossed to UAS-GAL80ts; Erm-GAL4 flies or ada, UAS-GAL80ts/Bc-Gla; Erm-GAL4 flies at 22°C. Larvae at 24 h ALH were shifted for 30 min to 30°C to induce clones and further aged for 4 d at 22°C before dissection.
Immunohistochemistry
For larval brain immunostaining, larvae were dissected in Shields and Sang M3 insect medium (Sigma-Aldrich) and fixed with 4% formaldehyde in PEM buffer (100 mM PIPES at pH 6.9, 1 mM EGTA, 1 mM MgCl2) for 21 min at room temperature. Brains were washed several times with 1× PBS plus 0.1% Triton X-100 (PBST) and were incubated with appropriate primary antibody overnight at 4°C or for 2 h at room temperature, labeled with secondary antibodies according to standard procedures, and mounted in Vectashield (Vector Laboratories). Ovaries were dissected, fixed, and stained as described (Palacios and St Johnston 2002). For DNA staining, TO-PRO-3 (Molecular Probes) was added in the wash step at a dilution of 1:3000. Images were obtained on a Leica TCS SP5 AOBS confocal microscope and were processed with LAS AF (Leica) and Adobe Photoshop.
The primary antibodies used were chicken anti-GFP (1:2000; Abcam), mouse anti-Pros (1:100; Developmental Studies Hybridoma Bank [DSHB]), rabbit anti-Miranda (1:1000; F. Matsuzaki), mouse anti-Miranda (1:10; F. Matsuzaki), guinea pig anti-Dpn (1:1000; J. Skeath), guinea pig anti-dMyc (1:200; M Milán), rabbit anti-eIF4E (1:1000; A Nakamura), mouse anti-β-galactosidase (1:100; Promega), rabbit anti-β-galactosidase (1:1000; Cappel), rat anti-BrdU (1:500; Novus Biologicals, for CldU detection), guinea pig anti-Ase (1:400; YN. Jan), rat anti-Vasa (1:2; DSHB), mouse anti-Hts (1:2; 1B1; DSHB), rabbit anti-fibrillarin (1:20; Abcam), and mouse anti-fibrillarin (1:20; Abcam).
Data quantification
For NB quantification, embryos of various genotypes were collected for 4-6 h at 25°C and allowed to develop to the third instar larval stage (96 h or 120 h ALH). Ten to 20 larvae of each genotype were dissected and stained with anti-Mira or anti-Dpn antibodies to identify NBs and anti-Pros to identify neurons. Central brain NBs can be distinguished from optic lobe NBs based on their medial/superficial location in the brain, larger size, and dispersed pattern. For NB size measurement, the means of the longest and shortest cell diameters were used in the statistics. To determine the volume of a NB or its nucleolus, semiaxes (a, b, and c) of a NB or its nucleolus were measured from stacked images collected at 0.2-μm intervals, and its volume as an ellipsoid [(4/3)πabc] was calculated. For quantification of the intensity of antibody staining, images were taken with the same confocal settings, and the mean fluorescence intensity was measured with NIH ImageJ or the Histogram function of Adobe Photoshop.
CldU pulse and chase in MACRM clones
Larvae at 42 h ALH were heat-shocked for 90 min at 37°C to induce MARCM clones, and further aged for 28 h at 25°C before being fed with 1 mg/mL CldU (Sigma-Aldrich) mixed with freshly made yeast paste for 4 h at room temperature. After CldU pulse, larvae were rinsed and transferred to standard fly food for a 20-h CldU-free chase at 25°C before dissection.
Statistical analysis
Unpaired Student's t-tests were used for statistical analysis between two groups.
Acknowledgments
We are grateful to Drs. S.J. Bray, W. Chia, T. Cline, P.J. DiMario, C.Q. Doe, F. Demontis, B. Edgar, M. Fortini, N.J. Gay, Y.-N. Jan, C.Y. Lee, T. Lee, J. Lipsick, L. Luo, F. Matsuzaki, H. McNeill, M. Milán, A. Nakamura, J. Ng, P. Pereira, G. Rubin, J.B. Skeath, N. Sonenberg, D. Stein, and T. Xu; University of Iowa DSHB; VDRC; Bloomington Drosophila Stock Center; Szeged Drosophila Stock Centre; and the TRiP at Harvard Medical School for fly stocks and reagents. We thank Dr. S. Guo for reading the manuscript, Dr. S. Gehrke for the pcDNA3-Flag-heIF4E construct, J. Gaunce and G. Silverio for technical assistance, and members of the Lu laboratory for discussions and help. This work was supported by NIH R01NS043167 grant (to B.L.) and a Stanford University School of Medicine Dean's Post-doctoral Fellowship (to Y.S.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.171959.111.
References
- Almeida MS, Bray SJ 2005. Regulation of post-embryonic neuroblasts by Drosophila Grainyhead. Mech Dev 122: 1282–1293 [DOI] [PubMed] [Google Scholar]
- Arabi A, Wu S, Ridderstrale K, Bierhoff H, Shiue C, Fatyol K, Fahlen S, Hydbring P, Soderberg O, Grummt I, et al. 2005. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol 7: 303–310 [DOI] [PubMed] [Google Scholar]
- Assouline S, Culjkovic B, Cocolakis E, Rousseau C, Beslu N, Amri A, Caplan S, Leber B, Roy DC, Miller WH Jr, et al. 2009. Molecular targeting of the oncogene eIF4E in acute myeloid leukemia (AML): A proof-of-principle clinical trial with ribavirin. Blood 114: 257–260 [DOI] [PubMed] [Google Scholar]
- Bailey AM, Posakony JW 1995. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev 9: 2609–2622 [DOI] [PubMed] [Google Scholar]
- Bello B, Reichert H, Hirth F 2006. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development 133: 2639–2648 [DOI] [PubMed] [Google Scholar]
- Bello BC, Izergina N, Caussinus E, Reichert H 2008. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev 3: 5 doi: 1186/1749-8104-3-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdnik D, Torok T, Gonzalez-Gaitan M, Knoblich JA 2002. The endocytic protein α-Adaptin is required for numb-mediated asymmetric cell division in Drosophila. Dev Cell 3: 221–231 [DOI] [PubMed] [Google Scholar]
- Betschinger J, Mechtler K, Knoblich JA 2006. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell 124: 1241–1253 [DOI] [PubMed] [Google Scholar]
- Boone JQ, Doe CQ 2008. Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Dev Neurobiol 68: 1185–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman SK, Rolland V, Betschinger J, Kinsey KA, Emery G, Knoblich JA 2008. The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev Cell 14: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabernard C, Doe CQ 2009. Apical/basal spindle orientation is required for neuroblast homeostasis and neuronal differentiation in Drosophila. Dev Cell 17: 134–141 [DOI] [PubMed] [Google Scholar]
- Clarke MF, Fuller M 2006. Stem cells and cancer: Two faces of eve. Cell 124: 1111–1115 [DOI] [PubMed] [Google Scholar]
- Curley MD, Therrien VA, Cummings CL, Sergent PA, Koulouris CR, Friel AM, Roberts DJ, Seiden MV, Scadden DT, Rueda BR, et al. 2009. CD133 expression defines a tumor initiating cell population in primary human ovarian cancer. Stem Cells 27: 2875–2883 [DOI] [PubMed] [Google Scholar]
- Fichelson P, Moch C, Ivanovitch K, Martin C, Sidor CM, Lepesant JA, Bellaiche Y, Huynh JR 2009. Live-imaging of single stem cells within their niche reveals that a U3snoRNP component segregates asymmetrically and is required for self-renewal in Drosophila. Nat Cell Biol 11: 685–693 [DOI] [PubMed] [Google Scholar]
- Fuller MT, Spradling AC 2007. Male and female Drosophila germline stem cells: Two versions of immortality. Science 316: 402–404 [DOI] [PubMed] [Google Scholar]
- Gao X, Pan D 2001. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev 15: 1383–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa L, Lehmann R 2004. How different is Venus from Mars? The genetics of germ-line stem cells in Drosophila females and males. Development 131: 4895–4905 [DOI] [PubMed] [Google Scholar]
- Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ 2005. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol 7: 311–318 [DOI] [PubMed] [Google Scholar]
- Grewal SS, Li L, Orian A, Eisenman RN, Edgar BA 2005. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat Cell Biol 7: 295–302 [DOI] [PubMed] [Google Scholar]
- Harrison H, Farnie G, Howell SJ, Rock RE, Stylianou S, Brennan KR, Bundred NJ, Clarke RB 2010. Regulation of breast cancer stem cell activity by signaling through the Notch4 receptor. Cancer Res 70: 709–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez G, Altmann M, Sierra JM, Urlaub H, Diez del Corral R, Schwartz P, Rivera-Pomar R 2005. Functional analysis of seven genes encoding eight translation initiation factor 4E (eIF4E) isoforms in Drosophila. Mech Dev 122: 529–543 [DOI] [PubMed] [Google Scholar]
- Hu Y, Fortini ME 2003. Different cofactor activities in γ-secretase assembly: Evidence for a nicastrin-Aph-1 subcomplex. J Cell Biol 161: 685–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutterer A, Knoblich JA 2005. Numb and α-Adaptin regulate Sanpodo endocytosis to specify cell fate in Drosophila external sensory organs. EMBO Rep 6: 836–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izergina N, Balmer J, Bello B, Reichert H 2009. Postembryonic development of transit amplifying neuroblast lineages in the Drosophila brain. Neural Dev 4: 44 doi: 10.1186/1749-8104-4-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai T, Spradling A 2004. Differentiating germ cells can revert into functional stem cells in Drosophila melanogaster ovaries. Nature 428: 564–569 [DOI] [PubMed] [Google Scholar]
- Kai T, Williams D, Spradling AC 2005. The expression profile of purified Drosophila germline stem cells. Dev Biol 283: 486–502 [DOI] [PubMed] [Google Scholar]
- Kentsis A, Topisirovic I, Culjkovic B, Shao L, Borden KL 2004. Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap. Proc Natl Acad Sci 101: 18105–18110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirilly D, Xie T 2007. The Drosophila ovary: An active stem cell community. Cell Res 17: 15–25 [DOI] [PubMed] [Google Scholar]
- Kohlmaier A, Edgar BA 2008. Proliferative control in Drosophila stem cells. Curr Opin Cell Biol 20: 699–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci A, Bray S 2007. Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev 21: 1322–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci A, Bernard F, Housden BE, Collins S, Bray SJ 2009. Direct response to Notch activation: Signaling crosstalk and incoherent logic. Sci Signal 2: ra1 doi: 10.1126/scisignal.2000140 [DOI] [PubMed] [Google Scholar]
- Lazaris-Karatzas A, Montine KS, Sonenberg N 1990. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature 345: 544–547 [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L 2001. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci 24: 251–254 [DOI] [PubMed] [Google Scholar]
- Lee CY, Robinson KJ, Doe CQ 2006a. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature 439: 594–598 [DOI] [PubMed] [Google Scholar]
- Lee CY, Wilkinson BD, Siegrist SE, Wharton RP, Doe CQ 2006b. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev Cell 10: 441–449 [DOI] [PubMed] [Google Scholar]
- Lin H 1997. The tao of stem cells in the germline. Annu Rev Genet 31: 455–491 [DOI] [PubMed] [Google Scholar]
- Lin H, Yue L, Spradling AC 1994. The Drosophila fusome, a germline-specific organelle, contains membrane skeletal proteins and functions in cyst formation. Development 120: 947–956 [DOI] [PubMed] [Google Scholar]
- Lin CJ, Cencic R, Mills JR, Robert F, Pelletier J 2008. c-Myc and eIF4F are components of a feedforward loop that links transcription and translation. Cancer Res 68: 5326–5334 [DOI] [PubMed] [Google Scholar]
- Liu N, Han H, Lasko P 2009. Vasa promotes Drosophila germline stem cell differentiation by activating mei-P26 translation by directly interacting with a (U)-rich motif in its 3′ UTR. Genes Dev 23: 2742–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HK, Wang Y, Belz T, Bock D, Takacs A, Radlwimmer B, Barbus S, Reifenberger G, Lichter P, Schutz G 2010. The nuclear receptor tailless induces long-term neural stem cell expansion and brain tumor initiation. Genes Dev 24: 683–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo NA, Shimono Y, Qian D, Clarke MF 2007. The biology of cancer stem cells. Annu Rev Cell Dev Biol 23: 675–699 [DOI] [PubMed] [Google Scholar]
- Luo D, Renault VM, Rando TA 2005. The regulation of Notch signaling in muscle stem cell activation and postnatal myogenesis. Semin Cell Dev Biol 16: 612–622 [DOI] [PubMed] [Google Scholar]
- Maines JZ, Stevens LM, Tong X, Stein D 2004. Drosophila dMyc is required for ovary cell growth and endoreplication. Development 131: 775–786 [DOI] [PubMed] [Google Scholar]
- Mamane Y, Petroulakis E, Rong L, Yoshida K, Ler LW, Sonenberg N 2004. eIF4E–from translation to transformation. Oncogene 23: 3172–3179 [DOI] [PubMed] [Google Scholar]
- Maurange C, Cheng L, Gould AP 2008. Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cell 133: 891–902 [DOI] [PubMed] [Google Scholar]
- Miron M, Verdu J, Lachance PE, Birnbaum MJ, Lasko PF, Sonenberg N 2001. The translational inhibitor 4E-BP is an effector of PI(3)K/Akt signalling and cell growth in Drosophila. Nat Cell Biol 3: 596–601 [DOI] [PubMed] [Google Scholar]
- Mitchell NC, Johanson TM, Cranna NJ, Er AL, Richardson HE, Hannan RD, Quinn LM 2010. Hfp inhibits Drosophila myc transcription and cell growth in a TFIIH/Hay-dependent manner. Development 137: 2875–2884 [DOI] [PubMed] [Google Scholar]
- Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N 2007. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature 449: 351–355 [DOI] [PubMed] [Google Scholar]
- Neumuller RA, Knoblich JA 2009. Wicked views on stem cell news. Nat Cell Biol 11: 678–679 [DOI] [PubMed] [Google Scholar]
- Neumuller RA, Betschinger J, Fischer A, Bushati N, Poernbacher I, Mechtler K, Cohen SM, Knoblich JA 2008. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature 454: 241–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A 2007. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science 315: 988–992 [DOI] [PubMed] [Google Scholar]
- Page SL, McKim KS, Deneen B, Van Hook TL, Hawley RS 2000. Genetic studies of mei-P26 reveal a link between the processes that control germ cell proliferation in both sexes and those that control meiotic exchange in Drosophila. Genetics 155: 1757–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios IM, St Johnston D 2002. Kinesin light chain-independent function of the Kinesin heavy chain in cytoplasmic streaming and posterior localisation in the Drosophila oocyte. Development 129: 5473–5485 [DOI] [PubMed] [Google Scholar]
- Pardal R, Clarke MF, Morrison SJ 2003. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer 3: 895–902 [DOI] [PubMed] [Google Scholar]
- Passegue E, Jamieson CH, Ailles LE, Weissman IL 2003. Normal and leukemic hematopoiesis: Are leukemias a stem cell disorder or a reacquisition of stem cell characteristics? Proc Natl Acad Sc 100: 11842–11849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, et al. 2008. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci 105: 9715–9720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CJ, Huang H, Xu T 2001. Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size. Cell 105: 357–368 [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL 2001. Stem cells, cancer, and cancer stem cells. Nature 414: 105–111 [DOI] [PubMed] [Google Scholar]
- Rhiner C, Diaz B, Portela M, Poyatos JF, Fernandez-Ruiz I, Lopez-Gay JM, Gerlitz O, Moreno E 2009. Persistent competition among stem cells and their daughters in the Drosophila ovary germline niche. Development 136: 995–1006 [DOI] [PubMed] [Google Scholar]
- Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, Pandolfi PP 2004. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med 10: 484–486 [DOI] [PubMed] [Google Scholar]
- San-Juan BP, Baonza A 2011. The bHLH factor deadpan is a direct target of Notch signaling and regulates neuroblast self-renewal in Drosophila. Dev Biol 352: 70–82 [DOI] [PubMed] [Google Scholar]
- Saucedo LJ, Edgar BA 2002. Why size matters: Altering cell size. Curr Opin Genet Dev 12: 565–571 [DOI] [PubMed] [Google Scholar]
- Schwamborn JC, Berezikov E, Knoblich JA 2009. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 136: 913–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R, Weng C, Yu J, Xie T 2009. eIF4A controls germline stem cell self-renewal by directly inhibiting BAM function in the Drosophila ovary. Proc Natl Acad Sci 106: 11623–11628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist SE, Haque NS, Chen CH, Hay BA, Hariharan IK 2010. Inactivation of both Foxo and reaper promotes long-term adult neurogenesis in Drosophila. Curr Biol 20: 643–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Hide T, Dirks PB 2004. Cancer stem cells in nervous system tumors. Oncogene 23: 7267–7273 [DOI] [PubMed] [Google Scholar]
- Skeath JB, Doe CQ 1998. Sanpodo and Notch act in opposition to Numb to distinguish sibling neuron fates in the Drosophila CNS. Development 125: 1857–1865 [DOI] [PubMed] [Google Scholar]
- Stocker H, Radimerski T, Schindelholz B, Wittwer F, Belawat P, Daram P, Breuer S, Thomas G, Hafen E 2003. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat Cell Biol 5: 559–565 [DOI] [PubMed] [Google Scholar]
- Sulkowski MS, Cooper C, Hunyady B, Jia J, Ogurtsov P, Peck-Radosavljevic M, Shiffman ML, Yurdaydin C, Dalgard O 2011. Management of adverse effects of Peg-IFN and ribavirin therapy for hepatitis C. Nat Rev Gastroenterol Hepatol 8: 212–223 [DOI] [PubMed] [Google Scholar]
- Tapon N, Ito N, Dickson BJ, Treisman JE, Hariharan IK 2001. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell 105: 345–355 [DOI] [PubMed] [Google Scholar]
- Teleman AA, Hietakangas V, Sayadian AC, Cohen SM 2008. Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab 7: 21–32 [DOI] [PubMed] [Google Scholar]
- van Riggelen J, Yetil A, Felsher DW 2010. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer 10: 301–309 [DOI] [PubMed] [Google Scholar]
- Varnum-Finney B, Xu L, Brashem-Stein C, Nourigat C, Flowers D, Bakkour S, Pear WS, Bernstein ID 2000. Pluripotent, cytokine-dependent, hematopoietic stem cells are immortalized by constitutive Notch1 signaling. Nat Med 6: 1278–1281 [DOI] [PubMed] [Google Scholar]
- Wang H, Somers GW, Bashirullah A, Heberlein U, Yu F, Chia W 2006. Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts. Genes Dev 20: 3453–3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ouyang Y, Somers WG, Chia W, Lu B 2007. Polo inhibits progenitor self-renewal and regulates Numb asymmetry by phosphorylating Pon. Nature 449: 96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel HG, Silva RL, Malina A, Mills JR, Zhu H, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Teruya-Feldstein J, Pelletier J, et al. 2007. Dissecting eIF4E action in tumorigenesis. Genes Dev 21: 3232–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M, Golden KL, Lee CY 2010. dFezf/Earmuff maintains the restricted developmental potential of intermediate neural progenitors in Drosophila. Dev Cell 18: 126–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz-Peitz F, Nishimura T, Knoblich JA 2008. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell 135: 161–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A, Gonzalez C 2006. Connecting cancer to the asymmetric division of stem cells. Cell 124: 1121–1123 [DOI] [PubMed] [Google Scholar]
- Wong MD, Jin Z, Xie T 2005. Molecular mechanisms of germline stem cell regulation. Annu Rev Genet 39: 173–195 [DOI] [PubMed] [Google Scholar]
- Yu X, Zou J, Ye Z, Hammond H, Chen G, Tokunaga A, Mali P, Li YM, Civin C, Gaiano N, et al. 2008. Notch signaling activation in human embryonic stem cells is required for embryonic, but not trophoblastic, lineage commitment. Cell Stem Cell 2: 461–471 [DOI] [PMC free article] [PubMed] [Google Scholar]