Abstract
Purpose
Identify and explain morphologic changes of the extraocular muscles (EOMs) in anophthalmic patients.
Methods
Retrospective chart review of patients with congenital anophthalmia, using MRI and intraoperative findings to characterize EOM morphology. We then employ molecular biology techniques in zebrafish and chick embryos to determine the relationships among the developing eye, periocular neural crest, and EOMs.
Results
In three human patients with bilateral congenital anophthalmia and preoperative orbital imaging, we observed a spectrum of EOM morphologies ranging from indiscernible muscle tissue to well-formed, organized EOMs. Timing of eye loss in zebrafish and chick embryos correlated with the morphology of EOM organization in the orbit (“eye socket”). In congenitally eyeless Rx3 zebrafish mutants, or following genetic ablation of the cranial neural crest cells, EOMs failed to organize, which was independent of other craniofacial muscle development.
Conclusions
Orbital development is dependent on interactions between the eye, neural crest, and developing EOMs. Timing of the ocular insult, in relation to neural crest migration and EOM development, is a key determinant of aberrant EOM organization. Additional research will be required to study patients with unilateral and syndromic anophthalmia, and assess for possible differences in clinical outcomes among patients with varied EOM morphology.
Clinical relevance
The presence and organization of EOMs in anophthalmic sockets may serve as a marker for the timing of genetic or teratogenic insults, improving genetic counseling, and assisting with surgical reconstruction and family counseling efforts.
INTRODUCTION
The volume of the adult human orbit (“eye socket”) is 30 mL and contains the ocular globe and a surrounding complex set of neuronal, vascular, muscular and glandular structures. These include six extraocular muscles (EOMs), which differentiate contemporaneously with the eye vesicle, form precise anatomic attachments to sclera and bone, and are innervated by cranial nerves 3, 4 and 6. Vertebrate EOMs are required for the precise control of eye movements to achieve directionality of gaze. In binocular animals with overlapping visual fields (e.g. primates, birds), eye movement control is required for stereopsis and fusion. EOMs are skeletal muscles with unique properties, including patterns of gene expression very different from skeletal muscle and altered susceptibility to systemic myopathies.1-5
Vertebrate EOMs are derived from two populations of mesenchymal cells: the unsegmented paraxial head mesoderm, which likely contributes to the lateral rectus (LR) and superior oblique (SO) muscles (innervated by the abducens and trochlear nerves, respectively), and the prechordal head mesoderm, which likely gives rise to the superior (SR), inferior (IR) and medial rectus (MR) and inferior oblique (IO) muscles (innervated by the oculomotor nerve). These presumptive origins of the EOMs are based on ablation, transplantation and genetic data using mostly chick and rodent models.6-11 EOM development appears to initiate caudal to the eye field, followed by anterior migration of mesodermal cells and subsequent muscle morphogenesis.
The mesodermal mesenchyme that forms the EOMs is intermingled with cells of the cranial neural crest that will form sclera and part of the choroid layer of the eye. Experiments that removed the neural crest have lead to the suggestion that interactions between mesoderm and neural crest may be involved in EOM formation. However, due to differences in animal models, the involvement of periocular neural crest in EOM development and organization is somewhat controversial. This may be due to the use of different techniques in the ablation of the neural crest. For example, surgical ablation may fail to completely and reproducibly ablate the entire cranial neural crest, whereas genetic ablation may cause unanticipated collateral damage to surrounding tissues, including the developing EOMs directly. Still, given that cranial neural crest cells contribute to craniofacial bone, connective tissue, nerves and the developing eye, it is likely that cranial neural crest plays an important role at least in the organization of the EOMs, if not the induction of differentiation.8, 9, 11-14
Anophthalmia and microphthalmia are rare conditions, with a combined birth prevalence of up to 30 per 100,000.15 These are caused by genetic mutations or teratogenic insults that interfere with differentiation of the neuroectodermal optic tissues and surrounding structures, resulting in variable attenuation of eye development (unilateral or bilateral). The genetic or teratogenic insult may primarily disrupt eye development and secondarily affect surrounding structures. Alternatively, the primary insult may disrupt orbitofacial mesenchymal development, secondarily affecting eye development. Regardless of their etiologies, studies of anophthalmia and microphthalmia have been very fruitful in elucidating the genetics and complex tissue interactions that are required for eye formation.16 These conditions likely represent a phenotypic continuum, and are commonly associated with systemic phenotypes as part of a syndrome. As a consequence, it is often difficult to determine the precise timing of the ocular insult retrospectively.
Because the bony orbits of congenitally anophthalmic patients are abnormally small, socket reconstruction includes bony expansion (non-surgically using conformers, or surgically using implants, expanders or osteotomies) before placing a permanent orbital implant and fashioning a custom ocular prosthesis.17 The organization and functionality of EOMs in congenitally anophthalmic patients is critical to the reconstructive surgical efforts - functional EOMs can be attached to orbital implants, providing the ocular prosthesis with movement characteristics that create a more natural appearance. Even non-functional EOMs can be helpful in securing the implant to prevent post-operative migration and reduce the risk of implant exposure or extrusion.
Based on our clinical work with anophthalmic patients, we investigated the temporal dependence of EOM development on the presence of the ocular globe in zebrafish and chick embryos. Our findings reveal a strong correlation between the extent and organization of remnant EOMs and the timing of the ocular insult. This correlation merits further study, because it could serve as a longitudinal marker for anophthalmia genetic research and aid in clinical care and the development of new therapies.
MATERIALS AND METHODS
Human patients
A retrospective chart review of 22 patients with unilateral or bilateral congenital anophthlamia treated at the University of Michigan Kellogg Eye Center was performed with the approval of the University of Michigan Institutional Review Board (# HUM00040783). We identified 3 patients with congenital bilateral anophthalmia that were well matched by age (1-2 years old) and clinical exam and who underwent orbital magnetic resonance imaging prior to any surgical intervention. Their MRIs were carefully reviewed for the presence and organization of EOMs, and correlated with intraoperative findings. In one case, a unilateral orbital cyst was excised, and the paraffin-embedded tissue block was re-sectioned and stained for microscopic evaluation. Mosaics of microscope images were obtained using Photoshop CS4 (Adobe Systems Inc., San Jose, CA).
Zebrafish
Zebrafish (Danio rerio) were raised in a laboratory breeding colony on a 14 hour light/10 hour dark cycle at 28.5 degrees Celsius and staged as described18 using hours post fertilization (hpf). The Tg(α-actin::EGFP) strain was a generous gift of Dr. Simon Hughes, King’s College London, United Kingdom.19 The Rx3/chokh mutant strain20 was the generous gift of Dr. Herwig Baier, University of California, San Francisco. When noted, embryos were treated with 0.003% (200 μM) propylthiouracil at 16-20 hpf to inhibit pigmentation. The protocols have met guidelines established by the University of Michigan Committee on the Use and Care of Animals.
Zebrafish enucleation experiments
Embryos from wild type and Tg(α-actin::EGFP) zebrafish were dechorionated and enucleated at 16-18 hpf or 26hpf using sharp tungsten needles using a dissecting microscope. Care was taken not to injure the cranium or the surrounding orbit during the procedure. Embryos were then returned to zebrafish growth medium and allowed to recover at 28.5 degrees Celsius. Live Tg(α-actin::EGFP) embryos were imaged at 72 and 96 hpf and harvested in 4% paraformaldehyde (PFA). Additional embryos were raised to adulthood using standard aquarium, light cycle and feeding protocols. At 4-5 months, they were sacrificed by deep anesthesia and immersion in 10% buffered formalin. Adults were processed for histology as described below.
Zebrafish microinjections
Morpholino oligonucleotide (MO; Gene Tools, Cowallis, OR) were reconstituted in de-ionized water to concentrations of 0.1 to 0.3 mM. MO sequences were previous published.21-24 and sequences available upon request.
Embryos obtained from timed-matings were injected with 0.2-1pmol MO at the 1-2 cell stage. MO against a control sequence (β-globin, Gene Tools) was also injected for each experiment. Microinjections were done at multiple doses to establish concentrations at which consistent and reproducible phenotypes were yielded. Live embryos were imaged and harvested at 72 and 96 hpf for further analysis as described below.
In situ hybridization
Whole-mount in situ hybridization was performed as described25 using digoxigenin (DIG)-labeled RNA antisense probes. RNA probes were generated by transcribing a PCR product of a cDNA fragment using a 3′ primer that included a T3 or T7 promoter. The only exception was the crestin probe, which was obtained from a plasmid digested with EcoRI and transcribed with T7 RNA Polymerase (the crestin plasmid was a generous gift from Mary Halloran, Madison, WI). Tissue was fixed in 4% buffered PFA, treated with proteinase K, acetylated, and then prehybridized. Tissue was incubated with probe in hybridization buffer overnight at 65 degrees Celsius. Probes were detected using anti-DIG alkaline phosphatase-conjugated antibody and visualized with 4-nitroblue tetrazolium/5-bromo- 4-chloro-3-indolyl phosphate (NBT/BCIP; Roche Molecular Biochemicals, Indianapolis, IN). Embryos were cryoprotected and embedded in OCT for sectioning.
Zebrafish histology and microscopy
Zebrafish embryos were fixed in 10% buffered formalin overnight at room temperature and embedded in paraffin. Adult zebrafish specimens were fixed in 10% formalin for at least 48 hours at room temperature, then decalcified for 1 hour in 5% nitric acid solution, neutralized, washed, and embedded in paraffin.
All specimens were sectioned at 5 micrometers and mounted on slides. Sections were stained with hematoxylin and eosin or Masson Trichrome using standard techniques26 Permanent cover slips were placed using CytoSeal (Richard-Allan Scientific).
Live embryos were embedded in methylcellulose and imaged using a Leica M205FA Combi microscope using Leica DFC290 (brightfield) and Hamamatsu ORCA-ER (fluorescence) cameras. Animals processed for in situ hybridizations were imaged with a Leica DM6000B microscope using a Leica DFC500 camera (Leica Microsystems CMS GmbH). Images were processed using Photoshop (Adobe Systems) and Leica LAS AF software.
Chick enucleation and analysis
Chick research protocols followed Royal Veterinary College guidelines. Fertile white leghorn chicken eggs (Henry Stewart and Co. Ltd, Lincolnshire, UK) were incubated at 37C. Embryos were staged according to Hamburger and Hamilton.27 Enucleations were performed as described,28 collected at E14 (HH stage 40-42) and fixed in 4% PFA in PBS. Embryos were embedded in paraffin and sectioned at 8 μm and mounted on slides. Staining with hematoxylin and eosin or Masson’s Trichrome was performed using standard techniques. For immunostaining, sections were dewaxed,rehydrated, and treated with 0.05% hydrogen peroxide before incubating overnight in anti-mouse myosin (1:10; DSHB, University of Iowa) at 4 degrees Celsius. Slides were then incubated in goat anti-mouse HRP secondary antibody (1:250; Invitrogen, Paisley, UK) and the peroxidase reaction product was generated using DAB. Labeled muscles were imaged using a Leica DM4000B microscope. In situ hybridizations for RXR-γ were performed as previously described.28
RESULTS
Human anophthalmia and extraocular muscles
In the course of providing surgical care to patients with congenital anophthalmia and microphthalmia, we noted that EOM anatomy could vary widely. Variations included the presence or absence of EOMs, variable numbers of EOMs and alterations in the spatial organization of the muscles within the orbit. Twenty-two patient charts were reviewed, which included patients with bilateral and unilateral clinical anophthalmia, often associated with hemifacial microsomia and other craniofacial abnormalities. We identified 3 patients with congenital bilateral anophthalmia that were well matched by age and clinical exam and who underwent orbital magnetic resonance imaging prior to any surgical intervention. A careful assessment of the pre-operative MRI scans and intraoperative findings suggested that pre-operative MRI accurately represented the presence and organization of EOMs (patient 1; Fig. 1). The MRI of patient 2 (Fig. 2) and patient 3 (Fig. 3) showed less muscle mass and reduced organization, respectively. Patient 2 had a 6mm orbital cyst removed from the right orbit, and histopathology (Fig. 2B) revealed an extremely microphthalmic remnant with minimal retinal development, small and malformed lens (large arrow), fibrovascular scar, and irregular muscle insertions (small arrow). Of note, the EOMs of patient 2 were identified intraoperatively. The EOMs were attached to the orbital implant during socket reconstruction in order to stabilize the implant and facilitate prosthesis movement. Patient 3 underwent reconstructive socket surgery, during which EOMs were barely identifiable on the right, and could not be identified on the left. The intraoperative report noted the presence of fibrovascular connective tissue where the EOMs would normally be found, consistent with the pre-operative MRI findings that lacked identifiable EOM structures (Fig. 3).
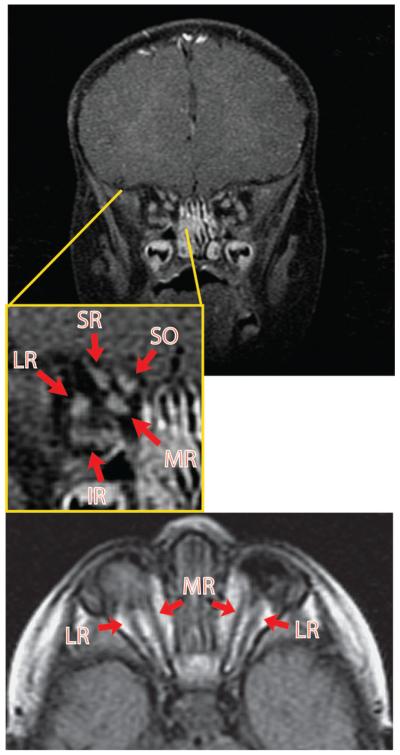
Figure 1. Pre-operative MRI of patient 1 with congenital bilateral anophthalmia but well-defined EOMs.
MRI of patient 1 reveals the presence of relatively organized EOM groups. Arrows point to the EOMs and EOMs are labeled according to position in the anophthalmic socket. This patient wore small custom socket conformers fitted by an ocularist. Note the absence of an optic nerve. MR, medial rectus; LR, lateral rectus; SR, superior rectus; IR, inferior rectus; SO, superior oblique.
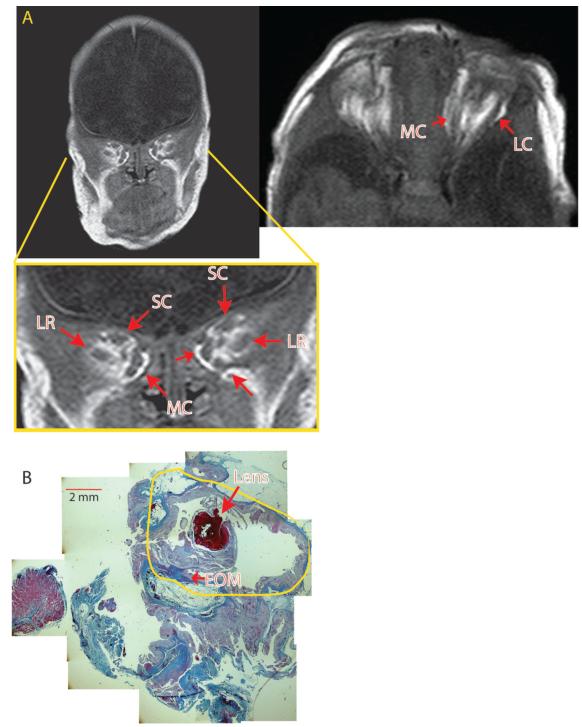
Figure 2. Pre-operative MRI of anophthalmic patient 2 with less well defined XEOMs.
A) MRI of patient 2 reveals EOMs that are less defined, with EOM clusters. The coronal image is taken from a more posterior section closer to the muscle origin compared to the MRI in Figure 1. The EOMs of the right orbit are less defined than on the left. B) Section of a cystic eye remnant that was removed from the left socket of patient 2, revealing a small lens (large arrow) within a malformed eye remnant (yellow tracing) surrounded by connective tissue, with irregular EOM insertions onto thickened, poorly formed scleral tissues (small arrow). Intraoperatively, thin EOM remnants were identified. (scale bar = 2 mm.) MC, medial complex; SC, superior complex; LR, lateral rectus.
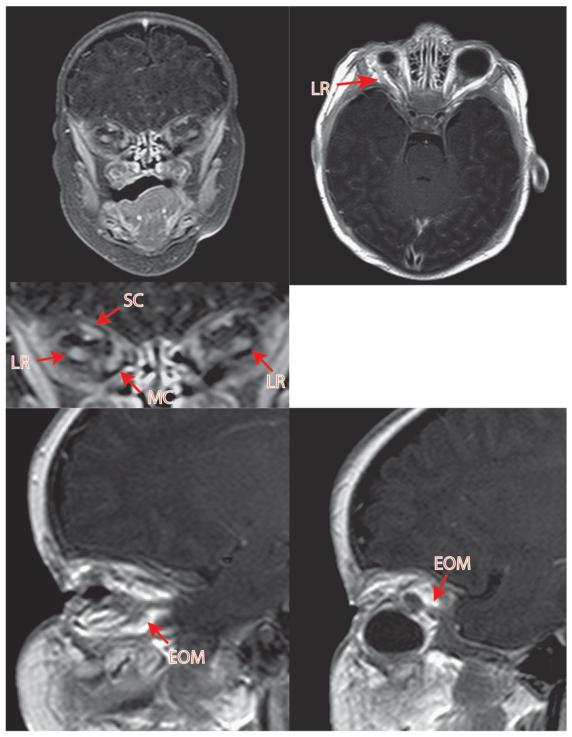
Figure 3. Pre-operative MRI of patient 3 with congenital bilateral anophthlamia and almost complete lack of EOMs.
MRI reveals near total lack of EOMs. A very posterior coronal section is required to identify an EOM cluster, worse on the left, without significant organization. Intraoperatively, EOMs could not be identified on the left side, although fibrovascular connective tissue strands were noted. The hypo-intense round objects in both orbits represent custom external socket conformers fitted by an ocularist.
Zebrafish rx3/chokh eyeless mutants reveal modest EOM development with poor organization
To explore the correlation between anophthalmia and EOM organization, we turned to a zebrafish experimental model. In situ hybridization for myhz2, the zebrafish gene encoding myosin heavy chain, labeled the six EOMS at 72hpf with anatomic positions similar to that found in humans (Fig. 4G-I; Schematic Fig. 4J-L). We next utilized a zebrafish strain homozygous for the rx3/chokh mutation, which prevents optic vesicle evagination.20, 29 In humans, rx mutations are associated with anophthalmia and sclerocornea.30 In situ hybridization for myhz2 at 72 hpf in the rx3/chokh mutants (Fig. 4A-C; Schematic Fig. 4D-F) reveal that the EOMs are differentiated, but appeared to cluster as ventral and dorsal muscle groups (Fig 5A, B) that did not further organize into rectus and oblique muscles. In contrast, the jaw and pharyngeal arch muscles differentiate and form normally (Fig. 4A-C, purple arrows) in the rx3/chokh mutants.
Figure 4. Genetic ablation of the optic vesicle in Rx3/chokh −/− mutant embryos results in poorly formed EOMs.
Wholemount in situ hybridization for myhz2, the gene encoding myosin heavy chain, in Rx3 −/− mutants at 72hpf (A-C) demonstrates the presence of differentiated muscle in the anophthalmic sockets (red arrows). However, the muscles are not properly organized into 6 EOMs, (A-C) as seen in wildtype embryos (G-I).In the Rx3/chokh mutants, muscles of the jaw and pharyngeal arch differentiate and form normally (purple arrows). Schematics of the rx3/chokh mutant (D-F) and wildtype (J-L) show the relation of the EOMs (red) to the eye (blue) and jaw musculature (purple). The dotted line in the lateral view corresponds to the coronal cut shown on the sections shown in Fig 5. SR, superior rectus; MR, medial rectus; LR, lateral rectus; IR, inferior rectus; SO, superior oblique; IO, inferior oblique.
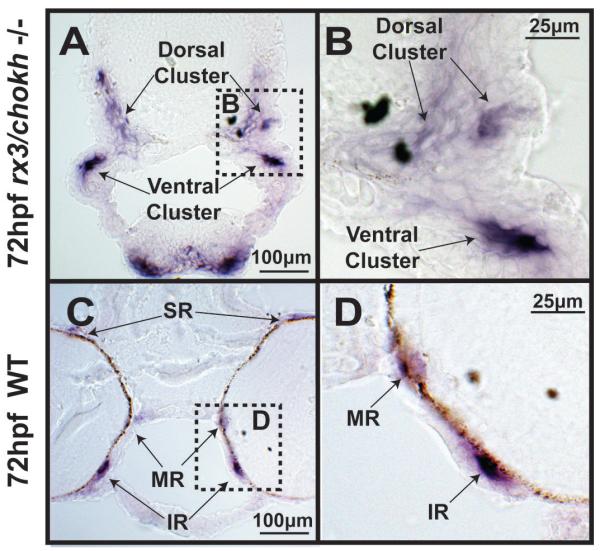
Figure 5. Rx3/chokh −/− mutant embryos express myosin but fail to form distinct EOM structures.
Cryosection in situ hybridization for the myosin gene myhz2 of 72hpf Rx3/chokh mutants (A, B) and wildtype (C, D) embryos demonstrates that the EOMs in the Rx3/chokh −/− mutant form dorsal and ventral clusters (double arrows), but do not organize into distinct rectus and oblique muscles. SR, superior rectus; MR, medial rectus; LR, lateral rectus; IR, inferior rectus; SO, superior oblique; IO, inferior oblique.
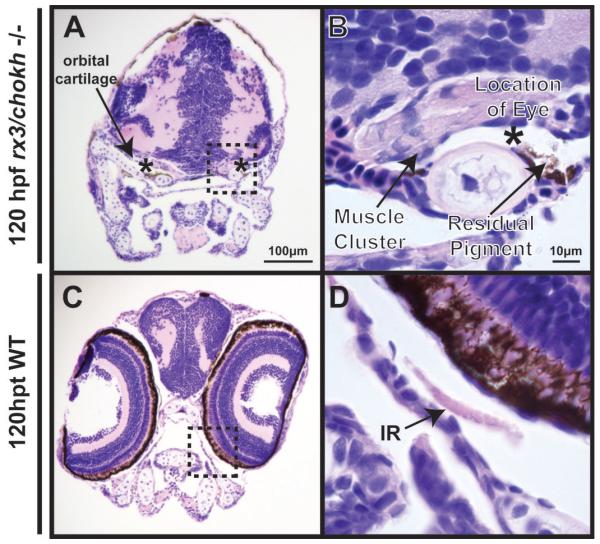
Histologic sections taken at 120 hpf from the rx3/chokh mutants (Fig. 6A, B) revealed that there was cartilage demarcating the anophthalmic socket which contained some pigmented epithelium. In addition, there were small clusters of muscle tissue in the erstwhile orbits (asterisks). However, no discrete muscles could be identified as in the wildtype (Fig. 6C, D). Thus, genetic ablation of the developing eye results in EOM differentiation, but impaired EOM organization.
Figure 6. Rx3/chokh −/− mutant embryos lack orbits with distinct EOMs.
H&E staining of coronal sections in Rx3−/− (left) embryos at 120 hpf demonstrate bilateral anophthalmia compared to wildtype controls (right). Rx3 −/− mutants showed cartilage (double arrow) surrounding the shrunken anophthalmic socket (asterik) which contained a small amount of remnant pigment (arrowhead), but no appreciable ocular structures. EOMs (arrows) are present in Rx3 −/− mutants. However, they are thickened and disorganized compared to wildtype control. SR, superior rectus; MR, medial rectus; LR, lateral rectus; IR, inferior rectus; SO, superior oblique; IO, inferior oblique.
Surgical removal of zebrafish optic vesicle reveals a time-dependent interaction between the developing eye and the developing EOMs
The rx3 gene mutation may directly cause EOM maldevelopment. Alternatively, it may prevent EOM development through its effects on the interactions between the eye and periocular mesenchyme. Failure of eye development, including in the Rx3/chokh mutants, was found to cause abnormal neural crest cell migration to the orbit as evidenced by crestin expression31 supporting an eye-mediated effect on the surrounding mesenchyme. So we next tested whether surgical removal of the optic vesicle would interfere with EOM development in a similar manner to genetic ablation. In order to visualize differentiated muscles with fluorescence microscopy, we unilaterally enucleated Tg(actin::EGFP) embryos, which express green fluorescent protein (GFP) under the control of the α-actin-promotor. Enucleations were performed at 16 hpf, prior to cranial neural crest migration to the orbit (see Fig. 7A left, and Ref 32). Removal of the optic vesicle at 16-18 hpf resulted in minimal EOM development at 96 hpf (data not shown). Wildtype embryos that were unilaterally enucleated at 16 hpf were also raised to adulthood. Transverse sections stained with H&E (Fig. 7B) confirmed the presence of a small rectus muscle remnant and the complete absence of oblique muscles in the anophthalmic socket. In contrast, enucleation of embryos at 26 hpf, which is after migration of the cranial neural crest (Fig. 7A) but prior to the earliest signs of EOM differentiation, revealed well developed EOMs at 72 hpf (data not shown) and 96 hpf (Fig. 8C,E,G,H). Despite, the absence of the eye, and the relatively late development of the EOMs,33, 34 individual rectus and oblique muscles could be identified (Fig. 8C,E,G,H) with normal anatomic positions. Together, these studies reveal a critical period for the interaction between the developing eye and the periocular mesenchyme that is required for EOM development and morphogenesis.
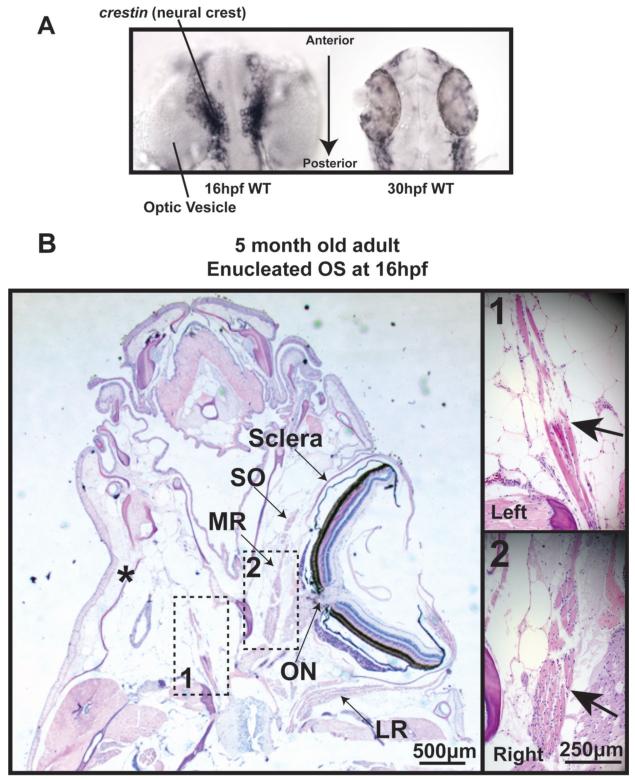
Figure 7. Surgical removal of the developing eye alters EOM formation.
A) In situ hybridization for crestin expression demonstrates that neural crest is adjacent to the neural tube at 16 hpf (left) and has migrated to peripheral locations by 30 hpf (right). B) H&E staining of a transverse section of 5 month old zebrafish that was enucleated at 16 hpf reveals one rectus muscle close to the origin, but a complete lack of other rectus and oblique muscles in the anophthalmic orbit. (scale bar 250μm)
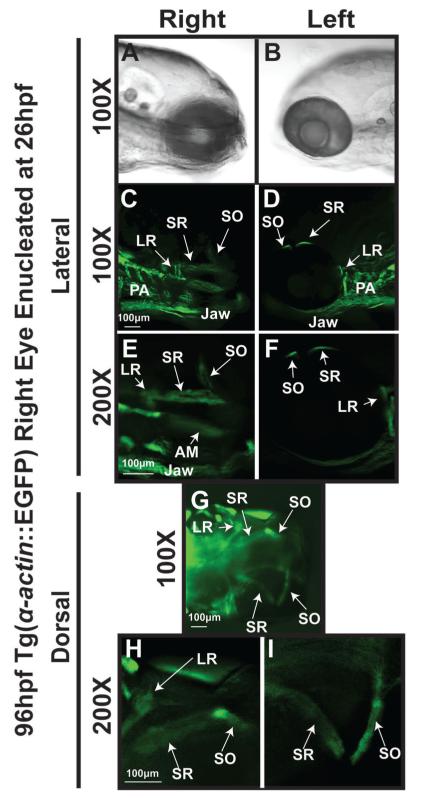
Figure 8. EOM properly form in orbits that were enucleated following neural crest migration.
Tg(α-actin::EGFP) embryo that was enucleated at 26 hpf and imaged at 96hpf (top) demonstrates well formed and distinct EOMs including superior rectus and superior oblique. Contralateral control side (bottom) shows 6 distinct EOMs. SR, superior rectus; SO, superior oblique; LR, lateral rectus; MR, medial rectus.
Zebrafish cranial neural crest is required for proper EOM morphogenesis
The interaction between the eye vesicle and the EOMs occurs in a developmental window that overlaps with cranial neural crest migration into the orbit.35 The role of the cranial neural crest in EOM development is somewhat controversial, but there is significant data to suggest an important relationship between the cranial neural crest and the mesodermal mesenchyme that forms the EOMs.7, 9 In zebrafish, the eye is required for proper migration of neural crest cells to the orbit,31 and therefore we investigated whether the interactions between the developing eye and EOMs are mediated by the cranial neural crest.
As previously described,21, 22 microinjection of antisense morpholino oligonucleotide (MO) directed against the transcription factor Sox10 at the 1-2 cell stage (within 30 minutes of fertilization) effectively knocks down the expression of sox10 and results in fish that survive to the early larval stage, but with neural crest-derived craniofacial cartilages and hindbrain structures that fail to develop. Non-specific effects of the Sox10 MO, such as heart edema, improve with co-injection of p53 MO.23 Using Sox10 gene knockdown, we tested whether EOM differentiation and organization were affected by inhibition of neural crest development. Inhibition of sox10 expression in the Tg(α-actin::EGFP) strain showed that EOMs failed to organize around the eye by 72 hpf (Fig 9A-C) and were severely maldeveloped by 96 hpf (Fig 9G-I). Further, we determined by in situ hybridization that the EOMs as well as the pharyngeal muscles expressed myosin heavy chain in the sox10 morphants (data not shown), and confirmed that the EOMs did not form proper attachments to the eye. The overall α-actin signal was decreased, revealing a smaller muscle mass (Fig 9A-C, G-I). Similar results were obtained with MO knockdown of FoxD3,24 another transcription factor required for neural crest development (data not shown). Thus, in the zebrafish embryo the cranial neural crest is required for proper EOM development and organization, and lack of orbital neural crest cells mimicked the effect of early anophthalmia on EOM development.
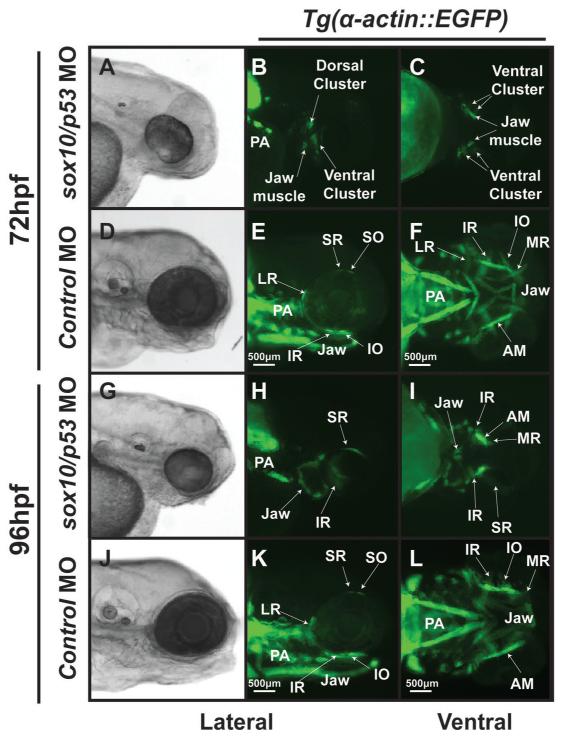
Figure 9. Inhibition of cranial neural crest development by knockdown of sox10 expression by antisense oligonucleotide morpholinos is required for EOM development.
Microinjection of antisense oligonucleotide morpholinos at the 1-2 cell stage (within 30 minutes of fertilization) effectively knocked down the expression of sox10 and inhibited neural crest development.. Knockdown of sox10 in the Tg(α-actin::EGFP) embryos demonstrates poor development of EOMs at 72 (A-C) and 96 hpf (G-I) when compared to control animals (D-F, J-L). MO against p53 was coinjected with the sox10 MO to decrease non-specific (off target) effects, but knockdown of p53 did not alter the EOM phenotype. At 72 hpf, the sox10/p53 morphants showed significant delay in EOM development and overall decreased α-actin expression. By 96 hpf, the sox10/p53 morphants had rudimentary EOM which were severely malformed and not organized around the eye compared to control MO.
Surgical removal of the chick optic vesicle confirms that EOMs can develop in the absence of an eye
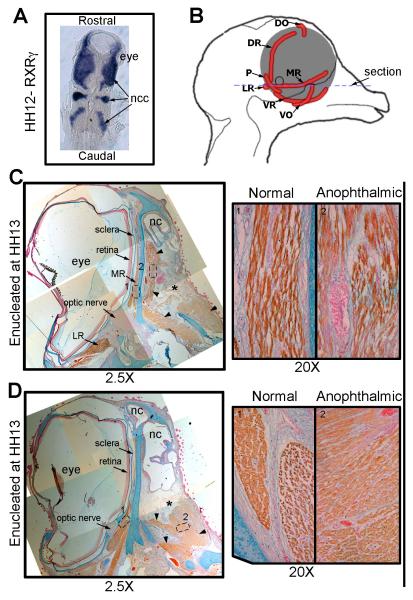
We next tested whether our experimental observations using zebrafish can be replicated in another vertebrate model. We utilized the chick embryo, a well-established model for ocular and craniofacial development, to test whether EOM organization is dependent on the presence of the cranial neural crest and the optic vesicle. The cranial neural crest in the chick embryo migrates into the craniofacial region at HH stage 11/12, which is at an earlier stage (early optic cup) than in zebrafish. In situ hybridization for RXR-γ (Fig 10A), which is expressed in the neural crest in chick embryos,28 demonstratesthe close relationship between the cranial neural crest and the developing eye at HH 12. The right eye of chick embryos was enucleated at HH stage 13 and EOM development was examined at HH stage 40 (E14; Fig 10B-D). Immunohistochemistry for myosin heavy chain (brown) demonstrates that the EOMs formed and organized properly on the normal control side (Fig 10C, D). On the anophthalmic side, large clusters of orbital muscle were identifiable in the area normally occupied by the medial rectus muscle (Fig 10C, right), and caudally in the lateral rectus field (Fig 10D, right). These clusters, however, lacked discernible anatomic organization within the orbit. Hence, early enucleation of a chick eye immediately after periocular neural crest cell migration resulted in the development of EOMs with very limited spatial organization.
Figure 10. Surgical removal of the developing eye in the chick disrupts EOMs.
A) Dorsal view in situ hybridization for RXRγ on a HH stage 12 (~E2) chick embryo, showing the close contact between the developing eye and the neural crest cells (blue). B) Schematic representing the chick head of an E14 embryo seen from the midline showing the positioning of the EOMs (red) of the left eye. The blue dotted line suggests the position of the sections in C and D. (C and D) HH stage 40 (E14) chick embryo that was enucleated at HH stage 13 labeled for Myosin (brown), Alcian blue (cartilage- blue) and nuclear fast red (nuclei-red). The position where the eye would have developed is denoted by the *. No retina, optic nerve or sclera has developed in this region. However, extraocular muscles are seen (arrow heads). C) The medial rectus muscle on the anophthalmic side of the embryo (right) has become fully differentiated muscle but is misshapen and has a looser morphology compared to those seen on the control (left) side. D) The other EOMs show reduced organization (right) compared to the control side (left)and individual muscles are no longer discrete and identifiable. Ncc, neural crest cell; MR, medial rectus; VR, ventral rectus; LR, lateral rectus; VO, ventral oblique; P, pyramidalis; DR, dorsal rectus; DO, dorsal oblique; nc, nasal cavity.
DISCUSSION
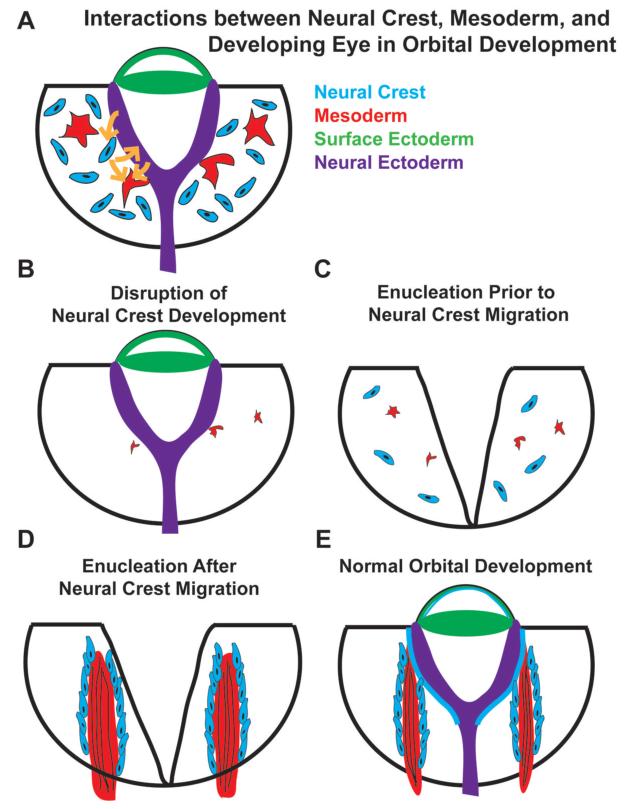
Our evaluations of human patients with clinical congenital anophthalmia revealed significant variability in their EOM anatomy. In zebrafish, microsurgical removal of the developing eye at 16 hpf, prior to optic cup evagination or orbital neural crest migration, resulted in poorly developed and disorganized EOMs (Fig 11C). Using a genetic eyeless mutant (rx3), in which ocular development aborts at 18-20 hpf, resulted in differentiated EOMs that were small and disorganized. Microsurgical removal of the developing eye in zebrafish at 26 hpf, after orbital neural crest migration, resulted in discernible EOM organization (Fig. 11D), even though expression of early muscle transcription factors such as Myf5 and MyoD in EOMs only begins at 26-32 hpf.33, 34 In the chick model, the eye was enucleated shortly after the neural crest migrated to the developing eye. Analysis of the resulting chick orbit revealed muscle clusters that did not develop into properly organized EOMs. Our results, based on observations made with human patients, as well as experimental data using zebrafish and chick, suggest that the observed clinical variability in EOM development may reflect the onset and extent of ocular maldevelopment.
Figure 11. Model.
The developing eye, EOMs, and migratory cranial neural crest cells form interdependent relationships that are necessary for the proper development of one another (A, E). Disrupting the development of either the eye or the neural crest during early stages of orbital development (B, C) impact one another and also leave a permanent mark on the structural organization of EOMs, whose development requires input signals from both the developing eye and surrounding cranial neural crest cells. Removal of the eye after the migration of the neural crest into the orbit has less effect on EOM development (D).
Previous studies of the relationship between the eye and cranial neural crest show that the developing eye is required for proper cranial neural crest migration.31. Similarly, cells of the cranial neural crest and EOMs show a complex interdependence that can vary depending on the tools and experimental models.8, 9, 11-14 Our results suggest that the developing eye and the cranial neural crest provide early signals that are required to drive the morphogenic process of EOM development.
Based on our results along with published data, we suggest that the developing eye, migrating cells of the cranial neural crest, and head paraxial and pre-chordal mesoderm (Fig. 11A) interact to create the functional orbit. Absence or abnormality in one group of cells has a lasting impact on the other groups. For example, patients with neurocristopathies and craniofacial defects will often have ocular manifestations, including microphthalmia/anophthalmia (e.g. Goldenhar Syndrome and hemifacial microsomia) and strabismus (e.g. Saethre-Chotzen syndrome).
Based on these results, and in order to build a more complete biological picture, we propose that the timing of abnormal eye development induces lasting alterations in the development and organization of the EOMs. In turn, clinically observable alterations in EOMs can provide important clues regarding the timing and mechanisms of anophthalmia and microphthalmia. We further suggest that the relationship between the developing eye and surrounding EOMs reflects, in part, the timing of cranial neural crest migration. Early loss of the optic vesicle, prior to cranial neural crest migration, results in poorly differentiated muscle that fails to organize into discernible EOMs. Later loss of the developing eye, after cranial neural crest cells had migrated into the orbit, leads to well-defined EOMs with somewhat variable levels of organization likely related to the lack of actual eye-muscle attachments.
Whereas results in different animal models may be somewhat divergent, an understanding of this temporal window of EOM development, and the relative difference in timing between species, will help guide future studies on the causes and treatment of anophthalmia, microphthalmia, and associated syndromes. We suggest that part of the evaluation of clinical anophthalmia, and in the context of genetic evaluations, an MRI of the orbits be obtained prior to any medical or surgical interventions. The analysis of EOM development in these patients, and correlation with any identified genetic alleles, may help reveal the timing of ocular insult, and suggest placement of that allele within a timeline of ocular and orbital development. In addition, preoperative evaluation of EOM development may help with surgical reconstructive and family counseling efforts. Further studies will be necessary to elaborate on these findings.
Acknowledgements
The authors wish to thank Herwig Baier, Mary Halloran and Simon Hughes for sharing plasmids and strains, Hemant Parmar and Nancy Dudek for their assistance with radiology images, Peter Hitchcock and Thomas Glaser for helpful comments on the manuscript, and Mary Halloran and Daniel Goldman for their support and mentorship. BLB and AK acknowledge Fight For Sight, Inc. for generous research support. BLB is supported in part by a post-doctoral training grant from the National Eye Institute of theNational Institutes of Health (NIH; T32 EY013934). AK is a recipient of a Research to Prevent Blindness Career Development Award. The research described in this manuscript was funded by NIH grant K08EY018689 to AK, and a Vision Research Core Award P30EY007003 to the Department of Ophthalmology and Visual Sciences, Kellogg Eye Center, University of Michigan. IM and HT are funded by the Wellcome Trust, London, UK.
References
- 1.Zhou Y, Liu D, Kaminski HJ. Myosin heavy chain expression in mouse extraocular muscle: More complex than expected. Invest Ophthalmol Vis Sci. 2010;51(12):6355–6363. doi: 10.1167/iovs.10-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kusner LL, Young A, Tjoe S, Leahy P, Kaminski HJ. Perimysial fibroblasts of extraocular muscle, as unique as the muscle fibers. Invest Ophthalmol Vis Sci. 2010;51(1):192–200. doi: 10.1167/iovs.08-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeiger U, Khurana TS. Distinctive patterns of microRNA expression in extraocular muscles. Physiol Genomics. 2010;41(3):289–296. doi: 10.1152/physiolgenomics.00169.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porter JD, Israel S, Gong B, et al. Distinctive morphological and gene/protein expression signatures during myogenesis in novel cell lines from extraocular and hindlimb muscle. Physiol Genomics. 2006;24(3):264–275. doi: 10.1152/physiolgenomics.00234.2004. [DOI] [PubMed] [Google Scholar]
- 5.Yu Wai, Man CY, Chinnery PF, Griffiths PG. Extraocular muscles have fundamentally distinct properties that make them selectively vulnerable to certain disorders. Neuromuscul Disord. 2005;1(1):17–23. doi: 10.1016/j.nmd.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Couly CF, Coltey PM, Le Douarin NM. The developmental fate of the cephalic mesoderm in quail-chick chimeras. Development. 1992;114(1):1–15. doi: 10.1242/dev.114.1.1. [DOI] [PubMed] [Google Scholar]
- 7.Noden DM, Trainor PA. Relations and interactions between cranial mesoderm and neural crest populations. J Anat. 2005;207(5):575–601. doi: 10.1111/j.1469-7580.2005.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noden DM. The embryonic origins of avian cephalic and cervical muscles and associated connective tissues. Am J Anat. 1983;168(3):257–276. doi: 10.1002/aja.1001680302. [DOI] [PubMed] [Google Scholar]
- 9.Tzahor E, Kempf H, Mootoosamy RC, et al. Antagonists of Wnt and BMP signaling promote the formation of vertebrate head muscle. Genes Dev. 2003;17:3087–3099. doi: 10.1101/gad.1154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wachtler F, Jacob M. Origin and development of the cranial skeletal muscles. Bibl Anat. 1986;29:24–46. [PubMed] [Google Scholar]
- 11.von Scheven G, Alvares LE, Mootoosamy RC, Dietrich S. Neural tube derived signals and Fgf8 act antagonistically to specify eye versus mandibular arch muscles. Development. 2006;133(14):2731–2745. doi: 10.1242/dev.02426. [DOI] [PubMed] [Google Scholar]
- 12.Gage PJ, Suh H, Camper SA. Dosage requirement of Pitx2 for development of multiple organs. Development. 1999;126(20):4643–4651. doi: 10.1242/dev.126.20.4643. [DOI] [PubMed] [Google Scholar]
- 13.Schilling TF, Walker C, Kimmel CB. The chinless mutation and neural crest cell interactions in zebrafish jaw development. Development. 1996;122(5):1417–1426. doi: 10.1242/dev.122.5.1417. [DOI] [PubMed] [Google Scholar]
- 14.Rinon A, Lazar S, Marshall H, et al. Cranial neural crest cells regulate head muscle patterning and differentiation during vertebrate embryogenesis. Development. 2007;134(17):3065–3075. doi: 10.1242/dev.002501. [DOI] [PubMed] [Google Scholar]
- 15.Verma AS, Fitzpatrick DR. Anophthalmia and microphthalmia. Orphanet J Rare Dis. 2007;26(2):47. doi: 10.1186/1750-1172-2-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chow RL, Lang RA. Early eye development in vertebrates. Annu Rev Cell Dev Biol. 2001;17:255–296. doi: 10.1146/annurev.cellbio.17.1.255. [DOI] [PubMed] [Google Scholar]
- 17.Chen D, Heher K. Management of the anophthalmic socket in pediatric patients. Curr Opin Ophthalmol. 2004;15(5):449–453. doi: 10.1097/01.icu.0000137855.45573.02. [DOI] [PubMed] [Google Scholar]
- 18.Kimmel CB, Ballard WW, Kimmel SR, Ullmann BB, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 19.Higashijima S, Okamoto H, Ueno N, Hotta Y, Eguchi G. High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin. Dev Biol. 1997;192(2):289–299. doi: 10.1006/dbio.1997.8779. [DOI] [PubMed] [Google Scholar]
- 20.Loosli F, Staub W, Finger-Baier KC, et al. Loss of eyes in zebrafish caused by mutation of chokh/rx3. EMBO Rep. 2003;4(9):894–899. doi: 10.1038/sj.embor.embor919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dutton KA, Pauliny A, Lopes SS, et al. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development. 2001 Nov;128(21):4113–4125. doi: 10.1242/dev.128.21.4113. [DOI] [PubMed] [Google Scholar]
- 22.Kelsh RN, Eisen JS. The zebrafish colourless gene regulates development of non- ectomesenchymal neural crest derivatives. Development. 2000 Feb;127(3):515–525. doi: 10.1242/dev.127.3.515. [DOI] [PubMed] [Google Scholar]
- 23.Langheinrich U, Hennen E, Stott G, Vacun G. Zebrafish as a model organism for identification and characterization of drugs and genes affecting p53 signaling. Current Biology. 2002;12(23):2023–2028. doi: 10.1016/s0960-9822(02)01319-2. [DOI] [PubMed] [Google Scholar]
- 24.Lister JA, Cooper C, Nguyen K, Modrell M, Grant K, Raible DW. Zebrafish Foxd3 is required for development of a subset of neural crest derivatives. Dev Biol. 2006 Feb 1;290(1):92–104. doi: 10.1016/j.ydbio.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 25.Barthel LK, Raymond PA. In situ hybridization studies of retinal neurons. Methods Enzymol. 2000;316:579–590. doi: 10.1016/s0076-6879(00)16751-5. [DOI] [PubMed] [Google Scholar]
- 26.Armed forces institute of pathology . In: Laboratory methods in histotechnology. Prophet EB, Mills B, Arrington JB, Sobin LH, editors. American Registry of Pathology; Washington, D.C.: 1992. [Google Scholar]
- 27.Hamburger V, Hamilyton HL. A series of normal stages in the development of the chick embryo. J Morph. 1951;88:49–92. [PubMed] [Google Scholar]
- 28.Thompson H, Griffiths JS, Jeffery G, McGonnell IM. The retinal pigment epithelium of the eye regulates the development of scleral cartilage. Dev Biol. 2010;347(1):40–52. doi: 10.1016/j.ydbio.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nusslein-Volhard C, Dahm R, editors. Zebrafish. First ed. Oxford University Press; New York: 2002. Practical Approach; No. 261. [Google Scholar]
- 30.Voronina VA, Kozhemyakina EA, O'Kernick CM, et al. Mutations in the human RAX homeobox gene in a patient with anophthalmia and sclerocornea. Hum Mol Genet. 2004;13(3):315–322. doi: 10.1093/hmg/ddh025. [DOI] [PubMed] [Google Scholar]
- 31.Langenberg T, Kahana A, Wszalek JA, Halloran MC. The eye organizes neural crest cell migration. Dev Dyn. 2008;237(6):1645–1652. doi: 10.1002/dvdy.21577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wada N, Javidan Y, Nelson S, Carney TJ, Kelsh RN, Schilling TF. Hedgehog signaling is required for cranial neural crest morphogenesis and chondrogenesis at the midline in the zebrafish skull. Development. 2005;132(17):3977–3988. doi: 10.1242/dev.01943. [DOI] [PubMed] [Google Scholar]
- 33.Lin C-Y, Chen W-T, Lee H-C, Yang P-H, Yang H-J, Tsai HJ. The transcription factor Six1a plays an essential role in the craniofacial myogenesis of zebrafish. Dev Biol. 2009;331:152–166. doi: 10.1016/j.ydbio.2009.04.029. [DOI] [PubMed] [Google Scholar]
- 34.Lin CY, Yung RF, Lee HC, Chen WT, Chen YH, Tsai HJ. Myogenic regulatory factors Myf5 and Myod function distinctly during craniofacial myogenesis of zebrafish. Dev Biol. 2006;299(2):594–608. doi: 10.1016/j.ydbio.2006.08.042. [DOI] [PubMed] [Google Scholar]
- 35.Luo R, An M, Arduini BL, Henion PD. Specific pan-neural crest expression of zebrafish crestin throughout embryonic development. Dev Dyn. 2001;220:169–174. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1097>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]