Abstract
The independent evolution of similar morphologies has long been a subject of considerable interest to biologists. Does phenotypic convergence reflect the primacy of natural selection, or does development set the course of evolution by channelling variation in certain directions? Here, we examine the ontogenetic origins of relative limb length variation among Anolis lizard habitat specialists to address whether convergent phenotypes have arisen through convergent developmental trajectories. Despite the numerous developmental processes that could potentially contribute to variation in adult limb length, our analyses reveal that, in Anolis lizards, such variation is repeatedly the result of changes occurring very early in development, prior to formation of the cartilaginous long bone anlagen.
Keywords: evo-devo, parallel evolution, convergent evolution, allometry
1. Introduction
From some of the earliest evolutionary speculation to the present day, developmental drivers of evolution have been viewed as an alternative to adaptationist explanations for evolutionary patterns (e.g. [1–3]; reviewed in [4,5]). The basic argument has revolved around the issue of which is more important in determining evolutionary patterns: developmental factors that constrain the variation that is produced, or selective factors that determine how genetic variation is selected across generations (e.g. [6–9])?
A particular case of contention is that of convergent evolution. The independent evolution of similar phenotypes (morphologies) in similar environments has long been taken as evidence of the primacy of natural selection (reviewed in [8]). However, in recent years, workers have argued that convergence—and, in particular, what is sometimes called ‘parallel evolution’, in which convergent patterns are produced through similar developmental changes—is evident for the biasing role of development in shaping evolutionary patterns [7,10–15]. But these views are not necessarily mutually exclusive. On one hand, convergence per se does not necessarily imply the similar action of natural selection; only when convergence occurs in similar selective environments is adaptive evolution implied [8]. Even in such cases, however, the channelling effects of shared developmental programmes may still be important by limiting the universe of possible variation upon which selection can operate. In this view, a key question is whether convergent phenotypes are produced by convergent developmental changes.
For most of the 20th century, the fields of evolutionary biology and developmental biology progressed in parallel, acquiring independent research objectives and experimental techniques [5,16]. But renewed interest in the developmental mechanisms of morphological evolution in the late 1970s and 1980s—stimulated, in part, by the works of Gould [17] and the discovery of pan-metazoan Hox gene conservation [18]—galvanized the field of evolutionary developmental biology. Recently, much attention in evolutionary developmental biology has been given to uncovering the molecular bases of morphological evolution and whether changes in the action of the same genes underlie phenotypic convergence [19–22]. But phenotypic convergence can result from non-convergent genotypic evolution (reviewed in [23–25]). To fully understand the developmental bases of convergent phenotypes, it is therefore critical that detailed analyses of developmental mechanisms be placed within the appropriate phenotypic context.
Evolutionary developmental biology has predominantly focused on studying dramatic morphological changes where the scoring of discrete phenotypes rarely requires detailed measurement [26,27]. By contrast, much of the study of natural selection and evolutionary diversification focuses on traits that vary quantitatively rather than qualitatively, such as body size, limb length and head shape [28–30]. While many of these qualitative shifts in morphology have been linked to changes in early development [19–21,31], variation in quantitative traits often has complex developmental origins: variation can arise through changes in morphogenesis, differential growth rates or both [32–37]. The most critical question to address first when studying the developmental bases of convergent quantitative phenotypes is whether the morphologies arise through convergent developmental trajectories representing the full developmental history of a structure [38].
Caribbean Anolis lizards provide an excellent and extensively studied group for investigating the repeated, independent evolution of similar morphologies (reviewed in [30]). Anoles have radiated, for the most part, independently on each island in the Greater Antilles (Cuba, Hispaniola, Jamaica and Puerto Rico), producing essentially the same set of habitat specialists—termed ‘ecomorphs’—on each island. Of particular importance in this repeated ecomorphological specialization is variation in relative limb length (i.e. limb length controlled for body size), which is known to play a significant role in adaptation to different parts of the structural habitat (e.g. tree trunks, twigs and grass blades). Despite many years of ecological and evolutionary research on this genus, the developmental bases of limb length variation have not yet been examined. Just how many different ways has variation in relative limb length evolved in this genus? Discovering that species converge morphologically via similar developmental trajectories might suggest that anoles have inherited common patterns of variation from their ancestor, potentially biasing the response to selection. But finding that different lineages converge morphologically using different developmental mechanisms would indicate that natural selection was the primary factor determining evolutionary trajectories. The goal of this work was to test the hypothesis that species possessing similar limb morphologies have converged on these morphologies through similar alterations to their growth trajectories.
2. Methods
(a). Overview
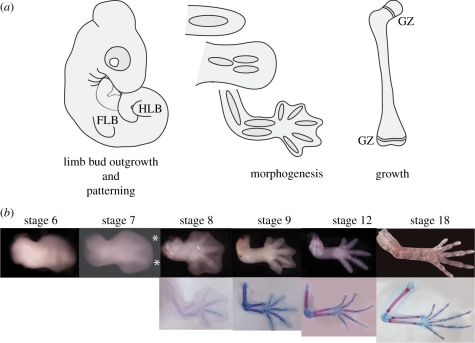
Predicting potential mechanisms of divergence first requires a general understanding of limb development (figure 1). During embryonic development, limb buds irrupt from the flank of the animal and are filled with an undifferentiated mesenchyme. During patterning, mesenchymal cells aggregate and differentiate to form a single cartilaginous anlagen, the precursor to the limb long bones. This cartilaginous rod then grows and bifurcates to form the limb skeleton. Ossification begins as a bony collar around each anlage, which expands towards the ends of the bone, establishing a growth zone. From this time on, proximodistal extension occurs solely at the growth zone, although the precise mechanism is unclear in reptiles (T. J. Sanger & C. E. Farnum 2004, personal observation). Variation in adult long bone length can therefore either result from variation generated during limb bud patterning, influencing the initial dimensions of the anlagen, or through differential rates of long bone elongation via the action of growth plates at either end of the bone.
Figure 1.
(a) Summary of limb and long bone development. Limb development proceeds through several stages: limb bud outgrowth and patterning, when the limb bud is only filled with undifferentiated mesenchyme; morphogenesis, where the cartilaginous anlagen of the limb bones form; and growth, mediated from epiphyseal growth zones. FLB, forelimb bub; HLB, hindlimb bud; and GZ, growth zone. (b) Limb development for Anolis sagrei. Asterisks denote visible condensations in the digital plate of stage 7 embryos. The earliest samples in our study were collected during stage 9, the earliest time when cartilage stains with Alcian blue.
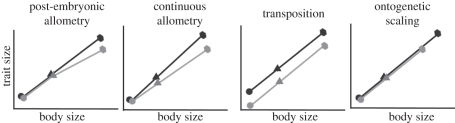
Interpreting evolutionary and developmental changes in relative size is best done in the context of allometry. Ontogenetic allometry, in particular, examines the relationship between local versus global rates of growth [39]. Several alternative allometric hypotheses could account for the patterns of limb length variation observed in adult Anolis lizards (figure 2). Variation in limb length may be due to localized changes in the rate of limb growth, either from the initial stages of morphogenesis (continuous allometry) or through stage-specific changes that occur later in ontogeny (post-embryonic allometry). These are both represented by a change in the slope of the allometric regression. Variation in adult morphology may also be the result of changes that occur during morphogenesis, represented by a transposition, or a shift in the intercept of the allometric regressions. Variation in adult morphology can also arise by scaling (extending or compressing) an allometric trajectory. However, in this case, we compare species that are roughly the same adult body size, indicating that this explanation cannot account for differences in relative limb length in these species. Long bone development is a complex process and, as a result, limb length variation can arise through many processes, whether by changing the patterning of the early limb bud or through time-specific changes in growth rate (e.g. [34,35,37,40–44]).
Figure 2.
Suite of alternative allometric hypotheses contrasting hypothetical species pairs. During post-embryonic allometry, species share similar embryonic trajectories, but diverge in relative size after hatching. Continuous allometry refers to a pattern in which species appear similar following morphogenesis, but subsequently diverge. A transposition of the allometric regressions reflects that a change has occurred prior to the earliest developmental stages measured. During scaling, the absolute size of the trait changes, but in proportion to body size. Circles, morphogenesis; triangles, hatching; hexagons, adulthood.
Detailed description of our methodology is included in the electronic supplementary material, appendix S1. Here, we briefly describe our study species, specimen preparation and statistical analyses.
(b). Study species and specimen preparation
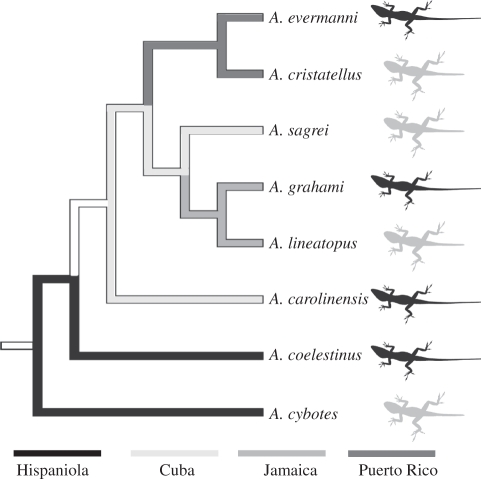
We collected growth series for eight Caribbean Anolis species representing one trunk-ground and one trunk-crown ecomorph from each island in the Greater Antilles (figure 3). Trunk-ground species have relatively longer limbs than trunk-crown species [30]. Limb patterning occurs early in development, and establishes the initial size and shape of the skeletal anlagen (figure 1). To extend juvenile patterns of allometric long bone growth closer to the period of morphogenesis, we also collected embryological series for four species—the trunk-crown and trunk-ground specialists from Cuba and Jamaica. These species were chosen because of their relative ease of care in captivity and the rate at which they lay eggs. Detailed methods of embryo collection have been described elsewhere [45,46].
Figure 3.
Phylogeny of eight anole species used in this study depicting independence of comparisons between island habitat specialists. Cartoons in the margin depict habitat specialist classification of each species (grey, trunk-ground; black, trunk-crown). Note that habitat specialists have evolved independently on each island.
As is common in herpetological studies in general, and previous studies of anoles in particular (e.g. [47,48]), we used snout-to-vent length (SVL) as a standard measure of body size. For embryological comparisons, we collected one to three individuals per stage for each of the four species examined, from the early stages of digital webbing reduction (stage 9/10) through hatching (stage 19 [45]). Combined with the post-hatching series, this represents nearly the entire period of long bone skeletal development and growth (figure 1).
We prepared post-hatching skeletons with standard clearing and staining protocols [49–51]. Scaled digital photographs of the limb elements were obtained using a Nikon DN100 camera mounted on a Nikon SMZ1000 dissecting microscope. We obtained individual long bone measurements with ImageJ using a single linear measurement [52,53].
(c). Statistical analysis
Log-transformed limb data for each species were best fit using a linear regression [54]. For each island pair, we tested for differences in slope and intercept between the two species with analysis of covariance (ANCOVA). To test for deviations from isometry (slope of the log–log regression = 1), we used a t-test for differences in slope for each species [55]. Embryonic and post-hatching data were treated independently.
To verify the results of this analysis using a second method, we also performed an ANCOVA using all post-hatching data, testing whether species across islands differed in slope. Because no significant differences in slope were found (see below), we then performed a phylogenetic ANOVA on the mean of the residuals for each species obtained from a single linear regression through all data. Residuals indicate the extent to which each species's limb elements, relative to body size, are larger or smaller than expected based on the data from all the species in the study; these residuals can thus be used to determine whether the ecomorphs differ in relative limb length. A phylogenetic ANOVA [56] controls for the relatedness of the taxa [57,58]. To perform the phylogenetic ANOVA, we first obtained an unbiased estimate of the evolutionary variance–covariance matrix [59]. Using multivariate Brownian motion simulations on a phylogeny of the eight species (figure 3) [60], we then generated 999 simulated datasets. The probability value for this analysis is the proportion of the simulations in which the simulated F-statistic is greater than the empirical F-statistic. All statistical procedures were run using SYSTAT v. 10.2 (Systat Software Inc., San Jose, CA, USA) or C code written expressly for this study and available from one of the authors (L.J.R.) upon request.
3. Results
(a). Relative growth of post-hatching limbs
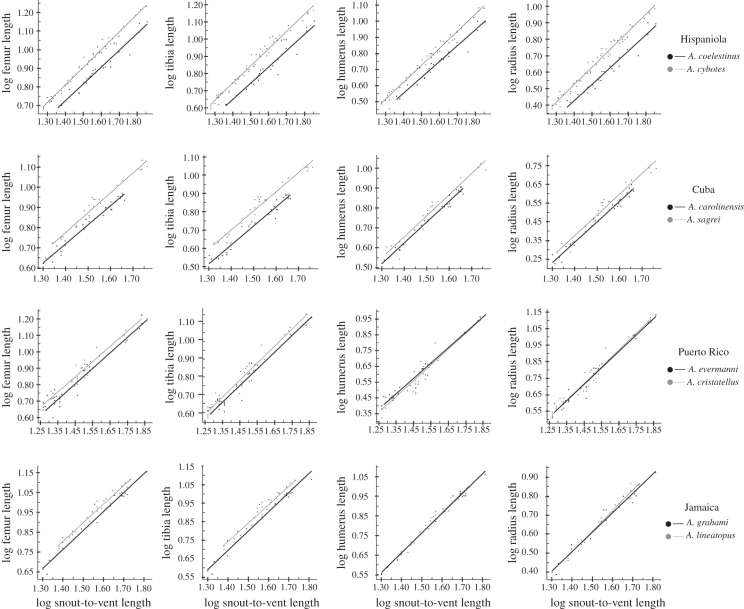
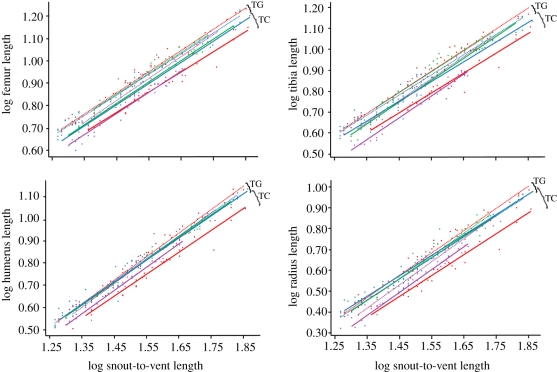
Variation in relative hindlimb length in adults is associated solely with changes that occur before hatching. This is clearly illustrated by significant differences in intercept, represented by a transposition in the post-hatching allometric regression (figure 4; table 1; electronic supplementary material, appendix S2). Both hindlimb elements examined (femur and tibia) exhibit the same pattern of post-hatching isometric growth. In no instance did we find significant differences in slope between species from the same island. Therefore, differences in relative hindlimb length cannot be attributed to differences in post-hatching allometric growth.
Figure 4.
Allometric plots of post-hatching growth series summarized by island and limb element. Note the parallel lines in all cases except for Jamaican and Puerto Rican forelimb elements.
Table 1.
Comparisons of relative limb growth between species from the same island. We found no significant differences in slope for any species pairs. Notice that post-hatching allometric trajectories for hindlimb elements show significant differences in intercept on all islands, while only Cuban and Hispaniolan forelimb elements show significant differences. Similar patterns were recovered in the embryonic analysis. Significant comparisons are highlighted in bold.
| slopes |
intercepts |
|||||
|---|---|---|---|---|---|---|
| d.f. | F | p | d.f. | F | p | |
| post-hatching allometry | ||||||
| Cuba | ||||||
| A. sagrei versus A. carolinensis | ||||||
| femur | 1,42 | 1.257 | 0.269 | 1,43 | 72.278 | p < 0.0001 |
| tibia | 1,42 | 0.34 | 0.563 | 1,43 | 94.614 | p < 0.001 |
| humerus | 1,42 | 0.361 | 0.551 | 1,43 | 17.427 | p < 0.0001 |
| radius | 1,42 | 0.417 | 0.522 | 1,43 | 27.108 | p < 0.0001 |
| Jamaica | ||||||
| A. lineatopus versus A. grahami | ||||||
| femur | 1,38 | 0.029 | 0.866 | 1,39 | 26.346 | p < 0.001 |
| tibia | 1,38 | 0.145 | 0.705 | 1,39 | 19.807 | p < 0.001 |
| humerus | 1,38 | 0.371 | 0.546 | 1,39 | 0.177 | 0.677 |
| radius | 1,38 | 1.685 | 0.202 | 1,39 | 0.449 | 0.507 |
| Hispaniola | ||||||
| A. cybotes versus A. coelestinus | ||||||
| femur | 1,70 | 1.098 | 0.298 | 1,71 | 248.036 | p < 0.0001 |
| tibia | 1,70 | 1.213 | 0.275 | 1,71 | 203.093 | p < 0.0001 |
| humerus | 1,70 | 2.54 | 0.115 | 1,71 | 158.09 | p < 0.0001 |
| radius | 1,70 | 1.638 | 0.205 | 1,71 | 231.776 | p < 0.0001 |
| Puerto Rico | ||||||
| A. cristatellus versus A. evermanni | ||||||
| femur | 1,57 | 0.178 | 0.675 | 1,58 | 21.87 | p < 0.001 |
| tibia | 1,57 | 0.147 | 0.703 | 1,58 | 11.226 | p < 0.001 |
| humerus | 1,57 | 0.233 | 0.631 | 1,58 | 0.292 | 0.591 |
| radius | 1,57 | 0.206 | 0.651 | 1,58 | 2.028 | 0.16 |
| embryonic limb allometry | ||||||
| Cuba | ||||||
| A. sagrei versus A. carolinensis | ||||||
| femur | 1,34 | 0.087 | 0.77 | 1,35 | 31.137 | p < 0.001 |
| tibia | 1,34 | 1.937 | 0.173 | 1,35 | 39.659 | p < 0.001 |
| humerus | 1,34 | 0.006 | 0.937 | 1,35 | 16.99 | p < 0.001 |
| radius | 1,34 | 0.782 | 0.383 | 1,35 | 8.884 | p < 0.001 |
| Jamaica | ||||||
| A. lineatopus versus A. grahami | ||||||
| femur | 1,31 | 0.394 | 0.535 | 1,32 | 5.515 | 0.025 |
| tibia | 1,31 | 0.003 | 0.956 | 1,32 | 5.694 | 0.023 |
| humerus | 1,31 | 0.591 | 0.448 | 1,32 | 0.993 | 0.326 |
| radius | 1,31 | 0.123 | 0.728 | 1,32 | 0.007 | 0.934 |
Forelimb elements exhibit different patterns of divergence on different islands. Significant differences in slope were never found between species from the same island. Only the forelimb elements from Cuban and Hispaniolan species have statistically distinguishable intercepts (figure 4; table 1; electronic supplementary material, appendix S2). Jamaican and Puerto Rican species vary neither in slope nor intercept, and the allometric trajectories vary only by ontogenetic scaling. Similar to the patterns shown by the hindlimbs, divergence of forelimb elements for Cuban and Hispaniolan species cannot be attributed to post-hatching allometric growth, but rather to a process that occurs prior to hatching.
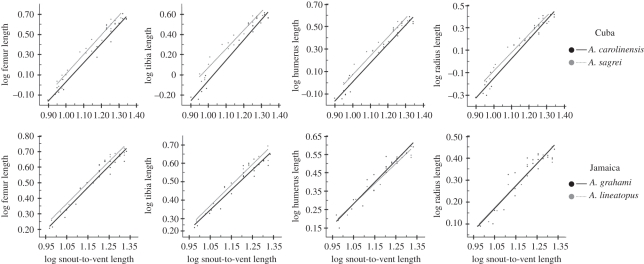
Phylogenetic ANOVAs using species residuals support these conclusions (figure 5; electronic supplementary material, appendix S3). Trunk-ground anoles have relatively longer hindlimb elements than trunk-crown anoles, but the relative size of the forelimb elements does not differ.
Figure 5.
Summary of allometric plots of post-hatching growth series organized by island and habitat. Despite variation in the degree of divergence in limb proportions on each island, trunk-ground anoles consistently have relatively longer hindlimbs compared with the trunk-crown anole from the same island. Ecomorph: solid line, trunk-crown; dotted line, trunk-ground. Islands: red lines, Hispaniola; purple lines, Cuba; blue lines, Puerto Rico; green line, Jamaica.
(b). Relative growth of embryonic limbs
By stage 10, ‘early digital web reduction’ [45] (figure 1), the relative size of the long bones is well established. Significant differences in intercept occur between the Cuban species' forelimbs and hindlimbs, and among the hindlimbs of Jamaican species (figure 6; table 1). Forelimbs are indistinguishable between the two Jamaican species, just as they are after hatching. Significant positive allometry exists for many of the embryonic long bones in these species (electronic supplementary material, appendix S4). But despite the decreased rate of long bone growth after hatching, these results indicate that the differences between ecomorphs have already been determined by the earliest stage at which repeatable measurements are possible.
Figure 6.
Plots of the relative growth of embryonic hindlimb elements highlighting the developmental origin of the ecomorphological variation early in development. Note the parallel allometric trajectories arising at the earliest stages of long bone development.
4. Discussion
Do properties of development affect the rate or direction of morphological evolution, or does natural selection simply remould variation to match current ecological conditions? Phenotypic convergence provides us with the opportunity to test this long-standing question by comparing the effects of natural selection and developmental biases during morphological evolution. If development were, in fact, channelling evolution along particular paths, one would expect that lineages converging phenotypically would use the same developmental trajectories.
Our analyses of post-hatching growth trajectories for Anolis trunk-ground and trunk-crown habitat specialists show that adult limb length variation is consistently the result of changes that occur prior to hatching (figures 4 and 5): differences in limb length are apparent at hatching, and limb long bones elongate in parallel relative to body size in different species. Examination of pre-hatching embryonic growth shows that species-specific morphologies are the result of changes that occur very early in limb development, prior to formation of the cartilaginous anlagen (figure 6). These results therefore suggest roles for both natural selection and the generative mechanisms regulating limb length variation during the diversification of this genus.
Several evolutionary scenarios can explain the repeated recruitment of early limb morphogenesis programmes for the generation of interspecific variation in adult limb length. Vavilov [61] and Haldane [62] observed that closely related species tend to vary in similar directions, suggesting commonalities in their developmental-genetic architecture. Such common genetic architectures might predispose evolutionary changes to occur along similar lines, following what has been termed ‘genetic lines of least resistance’ [63]. Under this scenario, while different developmental trajectories are possible, development serves as a channelling force by making some outcomes more easily attained by selection than others. Within a theoretical context, developmental constraints must align with genetic constraints to have a lasting evolutionary impact [64]. Although certainly possible in theory, Anolis is a very old genus (estimated age is at least 40 million years [30]); finding conserved genetic architectures over such great periods of time might be considered surprising ([65–68] and references therein; but see [69] for evidence of conserved phenotypic integration over long evolutionary timescales in anoles).
Alternatively, it may well be that, owing to the developmental architecture of the limb, there are only a limited number of ways that variation in adult morphology can be accomplished developmentally; in other words, the only way to modify an anole's limb morphology might be through changes in the size of the cartilaginous anlagen [44]. This would represent a form of absolute or global constraint, whereby some possibilities cannot be attained owing to the constraints of limb development. However, limbs are complex structures, and processes active during later development and growth are known to contribute to limb length variation, both within species (e.g. [34,35,37]) and among species (e.g. [41]; reviewed in [69]). In addition, forelimb and hindlimb elements appear to respond to selection independently on Jamaica (figures 3 and 4), and other lizard species exhibit non-isometric growth trajectories [70,71], which further indicates that no global constraints preclude allometric changes in limb dimensions.
A third possibility is that selection favouring differences in limb length between species in different habitats operates throughout their life histories. This would create strong functional and genetic correlations between early and late developmental stages [72]. If selection favours differences in limb length among juveniles, then it is possible that such differences might most easily be accomplished evolutionarily through changes in pre-hatching development, explaining why similar patterns recur repeatedly among species. Consistent selection throughout life would explain why there is no positive limb allometry in juvenile lizards; but because selection presumably does not act directly on limb length in developing embryos, this cannot explain why differences are repeatedly accomplished through early changes in limb morphogenesis time and time again.
At this time, it is not possible to distinguish between these competing evolutionary hypotheses. As mentioned above, deeply conserved developmental constraints may have channelled variation along certain dimensions, but the age and history of anoles make conserved genetic constraints seem unlikely. In addition, limb morphogenesis is highly variable in other vertebrate species, indicating that the overall developmental architecture of the limb is unlikely to channel the response to selection to particular developmental processes. Is it possible that the radiation of Anolis lizards used similar ‘genetic lines of least resistance’ to rapidly diversify? Could the genetic architecture of early limb patterning actually channel the response to selection? addressing these questions will lead to a more thorough understanding of the relative roles of selection and constraint in shaping the radiation of Anolis lizards, but will require further data on the underlying genetics of limb development among anole species, a greater understanding of the molecular underpinnings of limb variation within species, and further data on the habitat preferences of hatchlings and juvenile lizards.
Have anoles diversified in limb length through convergence or in parallel? While the distinction between these terms is contentious [8,23], a key factor often used to distinguish parallelism from convergence is whether the same developmental mechanisms underlie convergent phenotypes. In this light, parallelisms are a special case of convergence: parallelisms represent the interaction between selection and shared developmental constraints, while convergence is a more general phenomenon that can arise through multiple, unrelated evolutionary mechanisms (reviewed in [8]). Based on our results, we cannot rule out the possibility that anoles have evolved in parallel, by selection acting on a common set of developmental parameters controlling the size of the long bone anlagen. However, it is also possible that anoles have converged both morphologically and developmentally for reasons other than developmental constraint, as discussed above. The ultimate test of the parallel evolution hypothesis will be finding similarities in the genomic signature of natural selection to determine whether changes to the same loci have been independently selected on each island. Exploitation of the soon-to-be-published Anolis carolinensis genome, along with more detailed studies of natural history and development, should provide greater clarity regarding the relative importance of natural selection versus developmental constraint in guiding the evolutionary trajectory of Anolis.
Biologists are only just starting to bridge the divide between studies of natural selection and molecular genetics, providing a more detailed mechanistic understanding of the adaptive process [73]. But many of the best-studied examples of developmental evolution—where the specific genetic loci involved have been identified—have focused on discrete traits such as the presence or absence of specific characters (e.g. pelvic spines [19,74], wing spots [21] or vertebrate limbs [31]). Most studies of adaptation and divergence, however, focus on subtler, quantitative changes [75,76]. Thus, to connect studies of developmental evolution with the vast majority of studies of selection in nature, we need to shift our focus to quantitatively varying traits, the genetic basis of which is probably considerably more complicated than that in many of the model systems currently studied in evolutionary developmental biology.
Acknowledgements
Many people have helped with the collection and preparation of specimens examined in this article, most notably Michele Johnson, Paul Hime, Matt Edger, Alex Gamble and Kristin Sanger. Care and maintenance of the captive bred colonies would not have been nearly as successful without the assistance of Paul Hime, Jack Diani, Chip Deggendorf and Dan Piatchek, and other members of the Washington University in St Louis Animal Care and Use Committee. Jim Cheverud also aided in statistical analyses and interpretation. Hopi Hoekstra provided valuable comments on a previous version of this manuscript. We would also like to thank the governments of Jamaica, the Dominican Republic and Puerto Rico for permission to collect and export these specimens. This work was supported by a generous grant from the David and Lucile Packard Foundation, the National Science Foundation (award no. 0508696), Sigma xi, and the Department of Biology, Washington University in St Louis.
References
- 1.Bateson W. 1909. Mendel's principles of heredity. Cambridge, UK: Cambridge University Press [Google Scholar]
- 2.Goldschmidt R. 1940. The material basis of evolution. New Haven, CT: Yale University Press [Google Scholar]
- 3.Huxley J. 1942. Evolution: the modern synthesis. Cambridge, MA: MIT Press [Google Scholar]
- 4.Arthur W. 2004. Biased embryos and evolution. Cambridge, UK: Cambridge University Press [Google Scholar]
- 5.Amundson R. 2005. The changing role of the embryo in evolutionary thought. Cambridge, UK: Cambridge University Press [Google Scholar]
- 6.Simpson G. G. 1944. Tempo and mode in evolution. New York, NY: Columbia University Press [Google Scholar]
- 7.Maynard-Smith J. R., Burian R., Kaufffman S., Alberch P., Campbell J., Goodwin B., Lande R., Raup D., Wolpert L. 1985. Developmental constraints and evolution. Q. Rev. Biol. 60, 265–287 10.1086/414425 (doi:10.1086/414425) [DOI] [Google Scholar]
- 8.Losos J. B. 2011. Convergence, adaptation, and constraint. Evolution 65, 1827–1840 10.1111/j.1558-5646.2011.01289.x (doi:10.1111/j.1558-5646.2011.01289.x) [DOI] [PubMed] [Google Scholar]
- 9.Wake D. B., Wake M. H., Specht C. D. 2011. Homoplasy: from detecting pattern to determining process and mechanism of evolution. Science 331, 1032–1035 10.1126/science.1188545 (doi:10.1126/science.1188545) [DOI] [PubMed] [Google Scholar]
- 10.Wake D. B. 1991. Homoplasy: the result of natural selection, or evidence of design limitations? Am. Nat. 138, 543–567 10.1086/285234 (doi:10.1086/285234) [DOI] [Google Scholar]
- 11.Gould S. J. 2002. The structure of evolutionary theory. Cambridge, MA: Harvard University Press [Google Scholar]
- 12.Schluter D., Clifford E. A., Nemethy M., McKinnon J. S. 2004. Parallel evolution and inheritance of quantitative traits. Am. Nat. 163, 809–822 10.1086/383621 (doi:10.1086/383621) [DOI] [PubMed] [Google Scholar]
- 13.Whittall J. B., Voelckel C., Hodges S. A. 2006. Convergence, constraint and the role of gene expression during adaptive radiation: floral anthocyanins in Aquilegia. Mol. Ecol. 15, 4645–4657 10.1111/j.1365-294X.2006.03114.x (doi:10.1111/j.1365-294X.2006.03114.x) [DOI] [PubMed] [Google Scholar]
- 14.Atallah J., Liu N. H., Dennis P., Hon A., Larsen E. W. 2009. Developmental constraints and convergent evolution in Drosophila sex comb formation. Evol. Dev. 11, 205–218 10.1111/j.1525-142X.2009.00320.x (doi:10.1111/j.1525-142X.2009.00320.x) [DOI] [PubMed] [Google Scholar]
- 15.Des Marais D. L., Rausher M. D. 2010. Parallel evolution at multiple levels in the origin of hummingbird pollinated flowers in Ipomoea. Evolution 64, 2044–2054 [DOI] [PubMed] [Google Scholar]
- 16.Hamburger V. 1980. Embryology and the modern synthesis in evolutionary theory. In The evolutionary synthesis (eds Mayr E., Provine W. B.), pp. 97–112 Cambridge, MA: Harvard University Press [Google Scholar]
- 17.Gould S. J. 1977. Ontogeny and phylogeny Cambridge, MA: Harvard University Press [Google Scholar]
- 18.Duboule D., Dolle P. 1989. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J. 8, 1497–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shapiro M. D., Marks M. E., Peichel C. L., Blackman B. K., Nereng K. S., Jonsson B., Schluter D., Kingsley D. M. 2004. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature 428, 717–723 10.1038/nature02415 (doi:10.1038/nature02415) [DOI] [PubMed] [Google Scholar]
- 20.Colosimo P. F., et al. 2005. Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science 307, 1928–1933 10.1126/science.1107239 (doi:10.1126/science.1107239) [DOI] [PubMed] [Google Scholar]
- 21.Prud'homme B., Gompel N., Rokas A., Kassner V. A., Williams T. M., Yeh S., True J. R., Carroll S. B. 2006. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature 440, 1050–1053 10.1038/nature04597 (doi:10.1038/nature04597) [DOI] [PubMed] [Google Scholar]
- 22.Derome N., Duchesne P., Bernatchez L. 2006. Parallelism in gene transcription among sympatric lake whitefish (Coregonus clupeaformis Mitchill) ecotypes. Mol. Ecol. 15, 1239–1249 10.1111/j.1365-294X.2005.02968.x (doi:10.1111/j.1365-294X.2005.02968.x) [DOI] [PubMed] [Google Scholar]
- 23.Arendt J., Reznick D. 2008. Convergence and parallelism reconsidered: what have we learned about the genetics of adaptation? Trends Ecol. Evol. 23, 26–32 10.1016/j.tree.2007.09.011 (doi:10.1016/j.tree.2007.09.011) [DOI] [PubMed] [Google Scholar]
- 24.Manceau M., Domingues V. S., Linnen C. R., Rosenblum E. B., Hoekstra H. E. 2010. Convergence in pigmentation at multiple levels: mutations, genes and function. Phil. Trans. R. Soc. B 1552, 2439–2450 10.1098/rstb.2010.0104 (doi:10.1098/rstb.2010.0104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elmer K. R., Meyer A. 2011. Adaptation in the age of ecological genomics: insights from parallelism and convergence. Trends Ecol. Evol. 26, 298–306 10.1016/j.tree.2011.02.008 (doi:10.1016/j.tree.2011.02.008) [DOI] [PubMed] [Google Scholar]
- 26.Cooper W. J., Albertson R. C. 2008. Quantification and variation in experimental studies of morphogenesis. Dev. Biol. 321, 295–302 10.1016/j.ydbio.2008.06.025 (doi:10.1016/j.ydbio.2008.06.025) [DOI] [PubMed] [Google Scholar]
- 27.Klingenberg C. P. 2010. Evolution and development of shape: integrating quantitative approaches. Nat. Rev. Genet. 11, 623–635 [DOI] [PubMed] [Google Scholar]
- 28.Grant P. R. 1999. Ecology and evolution of Darwin's finches. Princeton, NJ: Princeton University Press [Google Scholar]
- 29.Schluter D. 2000. The ecology of adaptive radiation. Oxford, UK: Oxford University Press [Google Scholar]
- 30.Losos J. B. 2009. Lizards in an evolutionary tree: ecology and adaptive radiation of anoles. Berkeley, CA: University of California Press [Google Scholar]
- 31.Cohn M. J., Tickle C. 1999. Developmental basis of limblessness and axial patterning in snakes. Nature 399, 474–479 10.1038/20944 (doi:10.1038/20944) [DOI] [PubMed] [Google Scholar]
- 32.Abzhanov A., Kuo W. P., Hartmann C., Grant B. R., Grant P. R., Tabin C. J. 2006. The calmodulin pathway and evolution of elongated beak morphology in Darwin's finches. Nature 442, 563–567 10.1038/nature04843 (doi:10.1038/nature04843) [DOI] [PubMed] [Google Scholar]
- 33.Abzhanov A., Protas M., Grant B. R., Grant P. R., Tabin C. J. 2004. Bmp4 and morphological variation of beaks in Darwin's finches. Science 305, 1462–1465 10.1126/science.1098095 (doi:10.1126/science.1098095) [DOI] [PubMed] [Google Scholar]
- 34.Farnum C. E., Tinsley M., Hermanson J. W. 2008. Postnatal bone elongation of the manus versus pes: analysis of the chondrocytic differentiation cascade in Mus musculus and Eptesicus fuscus. Cells Tissues Organs 187, 48–58 10.1159/000109963 (doi:10.1159/000109963) [DOI] [PubMed] [Google Scholar]
- 35.Farnum C. E., Tinsley M., Hermanson W. 2008. Forelimb versus hindlimb skeletal development in the big brown bat, Eptesicus fuscus: functional divergence is reflected in chondrocytic performance in autopodial growth plates. Cells Tissues Organs 187, 35–47 10.1159/000109962 (doi:10.1159/000109962) [DOI] [PubMed] [Google Scholar]
- 36.Adams D. C., Nistri A. 2010. Ontogenetic convergence and evolution of foot morphology in European cave salamanders (Family: Plethodontidae). BMC Evol. Biol. 10, 216. 10.1186/1471-2148-10-216 (doi:10.1186/1471-2148-10-216) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanger T. J., Norgard E. Z., Pletscher L. S., Bevilacqua M., Brooks V. R., Sandell L. B., Cheverud J. M. 2010. Developmental and genetic origins of murine long bone length variation. J. Exp. Biol. 314B, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alberch P., Gould S. J., Oster G. F., Wake D. B. 1979. Size and shape in ontogeny and phylogeny. Paleobiology 5, 296–317 [Google Scholar]
- 39.Klingenberg C. P. 1998. Heterochrony and allometry: the analysis of evolutionary change in ontogeny. Biol. Rev. 73, 79–123 10.1017/S000632319800512X (doi:10.1017/S000632319800512X) [DOI] [PubMed] [Google Scholar]
- 40.Sears K. E., Behringer R. R., Rasweiler J. J., Niswander L. A. 2006. The development of bat flight: morphologic and molecular evolution of bat forelimb digits. Proc. Natl Acad. Sci. USA 103, 6581–6586 10.1073/pnas.0509716103 (doi:10.1073/pnas.0509716103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rolian C. 2008. Developmental basis of limb length in rodents: evidence for multiple divisions of labor in mechanisms of endochondral bone growth. Evol. Dev. 10, 15–28 10.1111/j.1525-142X.2008.00211.x (doi:10.1111/j.1525-142X.2008.00211.x) [DOI] [PubMed] [Google Scholar]
- 42.Ray R., Capecchi M. R. 2008. An examination of the chiropteran HoxD locus from an evolutionary perspective. Evol. Dev. 10, 657–670 10.1111/j.1525-142X.2008.00279.x (doi:10.1111/j.1525-142X.2008.00279.x) [DOI] [PubMed] [Google Scholar]
- 43.Hockman D., Mason M. K., Jacobs D. S., Illing N. 2009. The role of early development in mammalian limb diversification: a descriptive comparison of early limb development between the natal long-fingered bat (Miniopterus natalensis) and the mouse (Mus musculus). Dev. Dynam. 238, 965–979 10.1002/dvdy.21896 (doi:10.1002/dvdy.21896) [DOI] [PubMed] [Google Scholar]
- 44.Hall B. K. 2003. Unlocking the black box between genotype and phenotype: cell condensations as morphogenetic (modular) units. Biol. Philos. 18, 219–247 10.1023/A:1023984018531 (doi:10.1023/A:1023984018531) [DOI] [Google Scholar]
- 45.Sanger T. J., Losos J. B., Gibson-Brown J. J. 2008. A developmental staging series for the lizard genus Anolis: a new system for the integration of evolution, development, and ecology. J. Morph. 269, 129–137 10.1002/jmor.10563 (doi:10.1002/jmor.10563) [DOI] [PubMed] [Google Scholar]
- 46.Sanger T. J., Hime P. M., Johnson M. A., Diani J. 2008. Protocols for husbandry and embryo collection of Anolis lizards. Herpetol. Rev. 39, 58–63 [Google Scholar]
- 47.Beuttell K., Losos J. B. 1999. Ecological morphology of Caribbean anoles. Herpetol. Monogr. 13, 1–28 10.2307/1467059 (doi:10.2307/1467059) [DOI] [Google Scholar]
- 48.Calsbeek R., Smith T. B. 2007. Probing the adaptive landscape using experimental islands: density-dependent natural selection on lizard body size. Evolution 61–65, 1052–1061 10.1111/j.1558-5646.2007.00093.x (doi:10.1111/j.1558-5646.2007.00093.x) [DOI] [PubMed] [Google Scholar]
- 49.Hanken J., Wassersug R. 1981. The visible skeleton: a new double-stain technique reveals the nature of the ‘hard’ tissues. Funct. Photogr. 16, 22–26 [Google Scholar]
- 50.Taylor W. R., Dyke G. C. V. 1985. Revised procedures for staining and clearing small fishes and other vertebrates for bone and cartilage study. Cybium 9, 107–119 [Google Scholar]
- 51.Walker M. B., Kimmel C. B. 2007. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotechnic. Histochem. 82, 23–28 10.1080/10520290701333558 (doi:10.1080/10520290701333558) [DOI] [PubMed] [Google Scholar]
- 52.Rasband W. S. 1997. ImageJ. Bethesda, MD: US National Institutes of Health [Google Scholar]
- 53.Abramoff M. D., Magelhaes P. J., Ram S. J. 2004. Image processing with ImageJ. Biophotonics Int. 11, 36–42 [Google Scholar]
- 54.Huxley J. S. 1932. Problems of relative growth. London, UK: Methuen [Google Scholar]
- 55.Sokal R. R., Rohlf F. J. 1995. Biometry, 3rd edn. New York, NY: Freeman and Company [Google Scholar]
- 56.Garland T., Jr, Dickerman A. W., Janis C. M., Jones J. A. 1993. Phylogenetic analysis of covariance by computer simulation. Syst. Biol. 42, 265–292 [Google Scholar]
- 57.Felsenstein J. 1985. Phylogenies and the comparative method. Am. Nat. 125, 1–15 10.1086/284325 (doi:10.1086/284325) [DOI] [Google Scholar]
- 58.Harvey P. H., Pagel M. D. 1991. The comparative method in evolutionary biology. Oxford, UK: Oxford University Press [Google Scholar]
- 59.Revell L. J., Harmon L. J. 2008. Testing quantitative genetic hypotheses about the evolutionary rate matrix for continuous characters. Evol. Ecol. Res. 10, 311–321 [Google Scholar]
- 60.Nicholson K. E., Glor R. E., Kolbe J. J., Larson A., Hedges S. B., Losos J. B. 2005. Mainland colonization by island lizards. J. Biogeogr. 32, 929–938 10.1111/j.1365-2699.2004.01222.x (doi:10.1111/j.1365-2699.2004.01222.x) [DOI] [Google Scholar]
- 61.Vavilov N. 1922. The law of homologous series in variation. J. Genet. 12, 67–87 10.1007/BF02983073 (doi:10.1007/BF02983073) [DOI] [Google Scholar]
- 62.Haldane J. B. S. 1932. The causes of evolution. London, UK: Longman [Google Scholar]
- 63.Schluter D. 1996. Adaptive radiation along genetic lines of least resistance. Evolution 50, 1766–1796 10.2307/2410734 (doi:10.2307/2410734) [DOI] [PubMed] [Google Scholar]
- 64.Cheverud J. M. 1984. Quantitative genetics and developmental constraints on evolution by selection. J. Theor. Biol. 110, 155–171 10.1016/S0022-5193(84)80050-8 (doi:10.1016/S0022-5193(84)80050-8) [DOI] [PubMed] [Google Scholar]
- 65.Turelli M. 1998. Phenotypic evolution, constant covariances, and the maintenance of additive evolution. Evolution 42, 1342–1347 10.2307/2409017 (doi:10.2307/2409017) [DOI] [PubMed] [Google Scholar]
- 66.Agrawal A. F., Brodie E. D., Reiseberg L. H. 2001. Possible consequences of genes of major effect: transient changes in the G-matrix. Genetica 112–113, 33–43 10.1023/A:1013370423638 (doi:10.1023/A:1013370423638) [DOI] [PubMed] [Google Scholar]
- 67.Steppan S. J., Phillips P. C., Houle D. 2002. Comparative quantitative genetics: evolution of the G matrix. Trends. Ecol. Evol. 17, 320–327 10.1016/S0169-5347(02)02505-3 (doi:10.1016/S0169-5347(02)02505-3) [DOI] [Google Scholar]
- 68.Arnold S. J., Burger R., Hohenlohe P. A., Ajie B. C., Jones A. G. 2008. Understanding the evolution and stability of the G-matrix. Evolution 62, 2451–2461 10.1111/j.1558-5646.2008.00472.x (doi:10.1111/j.1558-5646.2008.00472.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kolbe J. J., Revell L. J., Szekely B., Brodie E. D., Losos J. B., III In press Convergent evolution of phenotypic integration and its alignment with morphological diversification in Caribbean Anolis ecomorphs. Evolution. (doi:10.1111/j.1558-5646.2011.01416.x) [DOI] [PubMed] [Google Scholar]
- 70.Pounds J. A., Jackson J. F., Shively S. H. 1982. Allometric growth of the hind limbs of some terrestrial Iguanid lizards. Am. Midl. Nat. 110, 201–207 10.2307/2425226 (doi:10.2307/2425226) [DOI] [Google Scholar]
- 71.Powell G. L., Russell A. P. 1992. Locomotor correlates of ecomorph designation in Anolis: an examination of three sympatric species from Jamaica. Can. J. Zool. 70, 725–739 10.1139/z92-107 (doi:10.1139/z92-107) [DOI] [Google Scholar]
- 72.Riedl R. 1978. Order in living organisms: a systems analysis of evolution. New York, NY: Wiley [Google Scholar]
- 73.Linnen C. R., Hoekstra H. E. 2009. Measuring natural selection on genotypes and phenotypes. Cold Spring Harbor Symp. Quant. Biol. 74, 155–168 10.1101/sqb.2009.74.045 (doi:10.1101/sqb.2009.74.045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shapiro M. D., Bell M. A., Kingsley D. M. 2006. Parallel genetic origins of pelvic reduction in vertebrates. Proc. Natl Acad. Sci. USA 103, 13 753–13 758 10.1073/pnas.0604706103 (doi:10.1073/pnas.0604706103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kingsolver J. G., Hoekstra H. E., Hoekstra J. M., Berrigan D., Vignieri S. N., Hill C. E., Hoang A., Gibert P., Beerli P. 2001. The strength of phenotypic selection in natural populations. Am. Nat. 157, 245–261 10.1086/319193 (doi:10.1086/319193) [DOI] [PubMed] [Google Scholar]
- 76.Kingsolver J. G., Pfennig D. W. 2007. Patterns and power of phenotypic selection in nature. Bioscience 57, 561–572 10.1641/B570706 (doi:10.1641/B570706) [DOI] [Google Scholar]