Abstract
Priority effects, in which the outcome of species interactions depends on the order of their arrival, are a key component of many models of community assembly. Yet, much remains unknown about how priority effects vary in strength among species in a community and what factors explain this variation. We experimented with a model natural community in laboratory microcosms that allowed us to quantify the strength of priority effects for most of the yeast species found in the floral nectar of a hummingbird-pollinated shrub at a biological preserve in northern California. We found that priority effects were widespread, with late-arriving species experiencing strong negative effects from early-arriving species. However, the magnitude of priority effects varied across species pairs. This variation was phylogenetically non-random, with priority effects stronger between closer relatives. Analysis of carbon and amino acid consumption profiles indicated that competition between closer relatives was more intense owing to higher ecological similarity, consistent with Darwin's naturalization hypothesis. These results suggest that phylogenetic relatedness between potential colonists may explain the strength of priority effects and, as a consequence, the degree to which community assembly is historically contingent.
Keywords: priority effect, historical contingency, Darwin's naturalization hypothesis
1. Introduction
The importance of history in determining the membership of species within and across communities has long been an area of active research in ecology. At large spatial and temporal scales there is now abundant evidence that historical events influence patterns of species richness and community composition [1,2]. However, at scales smaller than continents and inter-glacial cycles there is less consensus on whether historical or deterministic processes dominate community assembly [3–6]. Santayana [7] reasoned that ‘those who cannot remember the past are condemned to repeat it’, suggesting that human societies often repeat historical states, but the tendency for natural communities to repeat themselves or assume alternative states remains unclear [8–10].
One approach to resolving this apparent dichotomy is to identify the conditions that favour historically contingent assembly. A variety of conditions, including low dispersal rates, small ecosystem size and large species pools, have been thought to favour historically contingent assembly [4,5]. However, a common assumption underlying many of these conditions is the ability of early arriving species to competitively suppress late arriving ones, the phenomenon known as priority effects [11,12]. Therefore, a detailed understanding of the interspecific interactions that lead to strong or weak priority effects should shed light on why communities differ in the degree of historical contingency.
To gain such understanding, one potentially useful concept is Darwin's ‘naturalization hypothesis’ [13], i.e. that more closely related species are ecologically more similar and thus should compete more intensely [14–16]. If this hypothesis is correct, and if the intensity of competition is related to the strength of priority effects, phylogenetic relatedness between species may predict the degree of historical contingency in community assembly. Despite the increasing number of empirical tests of Darwin's hypothesis [17,18] and other phylogenetic approaches to understanding community assembly [19–21], little effort has been made to apply the hypothesis to explaining priority effects. Such application is presently difficult because most studies that quantified the strength of priority effects between species pairs [11,12,22,23] assessed only a limited portion of a natural community (generally two to three species). Although these studies provided valuable insights, the small number of species assayed makes it difficult to determine the range of variation in the strength of priority effects and its causes.
In this paper, we evaluate the use of Darwin's hypothesis in explaining priority effects, using nectar-inhabiting yeasts as a model system. Nectar yeast communities are low in species richness, vary in the phylogenetic relatedness between community members and occupy easily delineated, ephemeral habitats that require frequent dispersal [24–26]. For these reasons, this system provides an ideal opportunity for assessing priority effects across a whole community in light of Darwin's naturalization hypothesis. Furthermore, nectar yeasts are of broad ecological interest because they may affect plant–pollinator interactions, potentially decreasing floral attractiveness by consuming nectar resources [27] or attracting pollinators by producing volatiles or fermentation by-products [28].
Our approach involved experimental manipulation of the arrival order of nectar yeast species isolated from flowers of a hummingbird-pollinated shrub, Mimulus aurantiacus (sticky monkey flower). We used a fully factorial experimental design to investigate priority effects for six species (using a total of 30 pairwise combinations). These species together constitute the majority of endogenous yeast species identified during field surveys of M. aurantiacus flowers at a biological preserve in northern California and accounted for more than 90 per cent of all yeast observations in a recent study [29]. The experiment was carried out in laboratory microcosms using sterile nectar collected from greenhouse plants and molecular fingerprinting to assess yeast abundance and the strength of priority effects. Laboratory conditions ensured rigorous experimental control over species arrival order. We then used a phylogenetic approach to determine if relatedness could predict the strength of priority effects. In addition, we conducted growth experiments and used mass spectrometry to measure each species' nectar resource use. These data were used to estimate ecological similarity between species.
2. Methods
(a). Study system
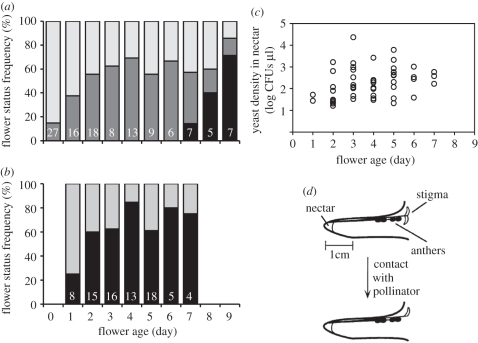
Our species occur in the chaparral ecosystem at Stanford University's Jasper Ridge Biological Preserve (JRBP). JRBP is located in the Santa Cruz Mountains in San Mateo County, California, USA (37°24′ N, 122°13′30″ W). Elevation ranges from 66 to 207 m. The climate is Mediterranean and receives an annual average of 662 mm of precipitation, falling almost exclusively during the winter months. Vegetation ranges from broadleaf evergreen to chaparral [30]. Mimulus aurantiacus is an evergreen shrub that occurs broadly at JRBP and along the coast of California and Oregon. At JRBP, M. aurantiacus plants flower from early April to early July [31], with individual flowers persisting between 7 and 10 days (figure 1).
Figure 1.
(a) Field observations on the effect of Mimulus aurantiacus flower age on stigma closure (see (d)) and flower wilting, (b) yeast presence in the nectar and (c) yeast abundance in the nectar from which yeast was detected at the Jasper Ridge Biological Preserve in May 2010. Numbers in (a,b) indicate sample sizes (number of flowers analysed). Data points in (c) represent individual flowers from which nectar was extracted and diluted for plating. (d) As indicated in the cross-section diagram of a M. aurantiacus flower with its stigma in the open and closed positions, stigmatic lobes respond to touch by pollinators (modified from Fetscher & Kohn [32]). (a) Light-grey filled bars: flower open, stigma open; grey filled bars: flower open, stigma closed; black filled bars, flower wilted. (b) Grey filled bars, yeast not detected; black filled bars, yeast detected.
(b). Yeast strains
The strains used in this study were isolated in 2010 during the course of a field study on nectar yeast community structure at JRBP. In brief, nectar from M. aurantiacus flowers was removed and streaked onto yeast media agar (YMA; Difco, Sparks, MD, USA). Individual colonies were plated and identified by sequencing a portion of the large subunit of the nuclear ribosomal RNA genes using the primer combination NL1–NL4 [33]. Species were identified by searching against GenBank using the Basic Local Alignment Search Tool (BLAST) and placement in phylogenetic trees with reference sequences. We identified seven species of ‘endogenous’ nectar yeasts, i.e. those capable of growth in nectar according to Brysch-Herzberg [24], all of which belong to the order Saccharomycetales. Epiphytic species of fungi (Aureobasidium, Cryptococcus and Rhodotorula spp.) were occasionally isolated from Mimulus nectar, but appear unable to grow in nectar owing to osmotic stress [24,25]. Preliminary tests supported this conclusion and these species were excluded from our experiments. Because one of our cultures of nectar-growing yeast species (Metschnikowia cibodasensis) became contaminated, our experiment used six species of nectivorous yeasts instead of seven (table 1). However, M. cibodasensis was rare in the field, and its exclusion is unlikely to affect our results.
Table 1.
Taxonomic assignments of nectar yeasts used for this study.
| species identity | top BLAST match (accession no.) | % match | accession number |
|---|---|---|---|
| Metschnikowia reukaufii | M. reukaufii (AJ716113) | 99.8 | JF809868 |
| M. koreensis | M. koreensis (HM627077) | 97.9 | JF809867 |
| M. kunwiensis | M. kunwiensis (FR819703) | 100 | JF809869 |
| Candida rancensis | C. rancensis (EU523604) | 99.5 | JF809871 |
| C. floricola | C. cf. floricola (AF313349) | 99.4 | JF809870 |
| Starmerella bombicola | S. bombicola (HQ111047) | 99.4 | JF809866 |
(c). Microcosm establishment and inoculation
To quantify the strength of priority effects, we simulated sequential yeast dispersal events to M. aurantiacus plants using mock flowers, i.e. plastic microtubes loaded with sterile M. aurantiacus nectar. This approach allowed us to maintain the chemical complexity of the nectar environment under conditions amenable to experimental manipulation. To accomplish this, we extracted nectar from individual flowers of multiple M. aurantiacus plants grown in the greenhouse using a sterile pipette tip. The plants derived from seeds collected at JRBP and were propagated in a greenhouse. Nectar from individual flowers was pooled (each flower generally contained between 1 and 10 µl of nectar), passed through a 0.5 µm filter and stored at −80°C until used in the experiment. Preliminary tests showed that this method of filtering and storing nectar successfully removed occasional mould contamination, did not adversely affect yeast growth and did not markedly change nectar sucrose concentration over time (electronic supplementary material, figure S1). At the start of the experiment, 9 µl aliquots of nectar were transferred into 200 µl GeneMate PCR tubes (BioExpress, UT, USA). One species of yeast was inoculated into the nectar on day 0 and another species on day 2, and they were allowed to grow at 25°C until the end of the experiment on day 5. Field observations suggest that flowers often go days between pollinator visits, and thus these time periods were chosen because they represented a reasonable approximation of flower longevity and pollinator visitation rates for M. aurantiacus at JRBP (figure 1).
Fresh yeast cultures were streaked 2 days in advance of each inoculation (except for C. floricola and S. bombicola which required 72 h for colonies to become visible). On the day of each inoculation, single colonies for each species were diluted into distilled H2O to a concentration of 400 cells µl−1 using a haemocytometer. Nectar was inoculated with 200 cells (in 0.5 µl H2O). Viability of each inoculant was confirmed by plating two replicates of 200 cells onto YMA plates [34]. Prior to the experiment, identity of each culture was confirmed using polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) (see §2e).
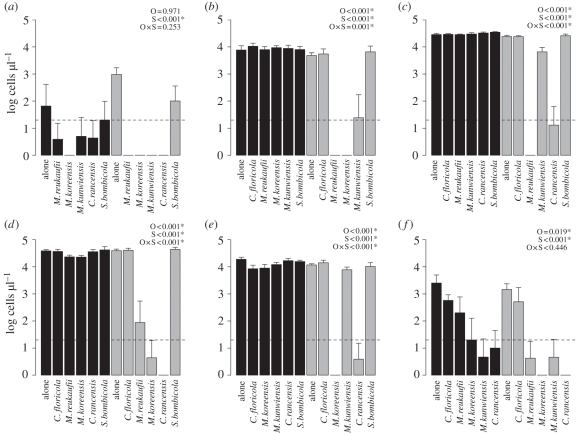
Most yeast species grew to match maximum densities observed in field studies (approx. 104 cells µl−1; figure 1) under the mock flower conditions in this experiment. The two exceptions to this, S. bombicola and C. floricola, both increased in abundance but achieved relatively low densities (figure 2).
Figure 2.
Final cell density of the focal species, including (a) C. floricola; (b) C. rancensis; (c) M. koreensis; (d) M. kunwiensis; (e) M. reukaufii; and (f) S. bombicola when introduced with another species (indicated on x-axis). Bars show the logarithmic mean of five replicates ±1 s.e., measured on day 5. Dashed line shows the initial inoculation density. Black bars show final cell densities when the focal species was introduced on day 0, and grey bars when introduced on day 2. Results of two-way ANOVA are shown in the upper right. Introduction O, order; S, species identity; O × S, interaction between order and species main effects. Asterisks indicate statistical significance after sequential Bonferonni correction for multiple tests.
(d). Experimental design
We used a fully factorial experimental design involving seven inoculations (six yeast species + one H2O treatment as negative control) and two orders of arrival (day 0 + day 2) for each of the seven inoculations. The H2O treatments allowed us to compare single species growth (for day 0 or day 2) against growth in competition. Each experimental replicate involved 42 treatment combinations (21 two-species combinations × 2 orders of introduction = 42). The experiment was replicated five times for a total of 5 × 42 = 210 replicates plus 10 negative controls (H2O only).
(e). Harvest and molecular identification of species
After 5 days, 1.5 µl of nectar from each treatment was diluted 1 : 10 and stored at −20°C. Cell density per microlitre of nectar was estimated at a later time using a haemocytometer and 200× magnification on a compound microscope. Another 1 µl of fresh nectar was serially diluted and plated on YMA at final concentrations containing 0.1 or 0.01 µl of nectar in order to obtain 50–200 colonies. Plates were cultured in an incubator at 25°C for 4 days, after which the number of colonies was counted. We estimated the number of colony forming units (CFUs) per treatment based on this and the dilution factor. Across all single species treatments, there was an approximately 1 : 1 relationship between CFUs and cell densities (r2 = 0.76, slope = 1.02; electronic supplementary material, figure S2), indicating good concordance between our estimates of yeast abundance. The remaining nectar from each experiment was stored at −80°C for later use in mass spectrometry (see below).
To determine the proportion of yeast species present, we extracted DNA from yeast colonies (12 for two species treatments and two for single species treatments) and used RFLP fingerprinting to determine species identity (for details see electronic supplementary material). In total, PCR–RFLP typing was carried out on approximately 1891 colonies (some two species treatments had less than 12 colonies).
(f). Measuring functional characteristics of yeast species
To characterize yeast functional traits, we measured two aspects of yeast physiology. First, to determine whether yeast species varied in resource use, we used liquid chromatography coupled with mass spectrometry (LC-MS/MS) to assess the chemical composition of nectar for all single-species and half of the negative-control treatments at the end of our experiment (n = 65). Specifically, LC-MS/MS was used to determine the concentration of sucrose, glucose, fructose and amino acids including 18 standard and six non-protein amino acids (for details see the electronic supplementary material).
Second, to assess their growth capabilities in different environments, we measured the population growth rates of each yeast species in four different types of media: 20 per cent sugar solutions (sucrose, glucose or fructose) amended with 0.2 per cent Difco Yeast Nitrogen Base (Becton Dickinson, Sparks, MD, USA) and a liquid version of the standard yeast media used for culturing. Growth of each treatment was measured by optical density (OD600) every 15 min for 23 h using an automated microplate reader (for culture details see electronic supplementary material). We used the R package grofit [35] to estimate two parameters used to compare population growth rates: the length of the lag phase (λ) and the cell doubling time [36].
(g). Data analysis
The effects of arrival order on the growth of each species was assessed individually using two-way analysis of variance (ANOVA) with abundance of the focal species as the response variable and the predictor variables as arrival order (order), competitor species identity (species) and their interaction (order × species). Cell density and CFU density were log-transformed to improve normality and reduce heteroscedasticity. To control for inflation of type I error rates from conducting multiple ANOVAs, we used sequential Bonferonni correction [37] to set the familywise error rate. Sequential Bonferonni correction does not assume independence between tests, which may be an important consideration if species responses are phylogenetically non-independent.
To test whether functional characteristics varied between species, we conducted a series of ANOVAs. For population growth parameters (lag time and doubling time), we used a two-way ANOVA with species identity (species), substrate type (substrate) and their interaction (species × substrate) as the predictor variables. For nectar chemistry, we used a two-way ANOVA with species identity (species), inoculation time (day 0 or day 2; time) and their interaction (species × order) as the predictor variables. Results were considered significant after sequential Bonferonni correction was used to adjust the significance threshold based on the number of ANOVAs per experiment.
To explain variation in the magnitude of priority effects, we looked for correlations between phylogenetic relatedness, ecological similarity and the strength of the priority effect for all species pairs. To determine phylogenetic relatedness between species, we built a phylogeny using our sequence data along with environmental sequences [24] and a recent phylogeny of the Saccharomycetales [38]. We used the Geneious interface to align sequences using the program MAFFT [39], estimate phylogeny using maximum likelihood with the program PhyML [40] and extract patristic distances from the resulting most likely phylogeny. Our reconstruction of the yeast phylogeny matched well with a previously published multi-gene phylogeny [38] and recovered all 12 major yeast clades identified in that study (electronic supplementary material, figure S4).
To measure ecological similarity, we used principal component analysis (PCA) to simplify species growth characteristics and resource use into a manageable number of resource axes and calculated the total pairwise distance between species along these axes. Because growth rate and resource use were measured in separate experiments, we conducted PCAs separately for each experiment. For each PCA, we retained only the axes that cumulatively explained more than 90 per cent of variation. Average PCA scores for each species were combined into a single dataset from which we calculated the total Euclidean distance between species pairs. To see whether equal weighting of PCA axes affected our results, we re-ran all statistical analyses with PCA scores standardized to unit variance. This did not change our results and is not reported further.
To measure the strength of priority effects, we estimated a species' relative ability to invade a competitive environment (invasion score) as a standard effect size comparison with its density when inoculated in monoculture on day 2 of the experiment, i.e. (μx− yi)/σp. For each species, we calculated mean (μx) and standard deviation (σx) of final density when inoculated in monoculture on day 2. Density in each invasion treatment (yi) was then subtracted from the monoculture mean (μx) and divided by the pooled standard deviation (σp). Thus, an invasion score of 0 represents no difference with growth in monoculture and negative and positive values represent deceases or increases relative to monoculture in units of standard deviation.
We used a combination of regression and Mantel tests to examine the correlations between invasion score, phylogenetic relatedness and ecological similarity. We used linear regression to test whether ecological distance or phylogenetic distance could predict invasion score. Because multiple comparisons using the same species pairing are not independent replicates of a given phylogenetic distance, we used the mean invasion score for each species pairing in these regressions. To make sure that averaging did not affect our results, we also ran the same analyses with a linear mixed effects model using replicates within a treatment as nested random effects. To test whether there was a relationship between phylogenetic distance and ecological distance, we used the Mantel test as implemented by the Vegan package in R [41]. To see if we could increase power over the Mantel test, we analysed the same data by decomposing distance matrices into principal coordinates and testing for correlation using the canonical correlation approach outlined in Peres-Neto & Jackson [42] and Vegan [41]. Because the results were similar they are not reported further. We also repeated all analyses excluding two species (C. floricola and S. bombicola) to see how this affected our results.
3. Results
(a). Interspecific effects on growth
All species tested experienced negative growth effects from the introduction of another species (figure 2 and electronic supplementary material, figure S4). Negative effects were characterized by strong dependence on arrival order, a hallmark of priority effects. Specifically, we found the absence of negative effects on the focal species when the other species were introduced late (figure 2; day 0 alone versus day 0 competitors) and a dramatic decrease in growth in the converse situation (figure 2; day 2 alone versus day 2 competitors). This was supported statistically by highly significant order × species effects for four out of six species (p < 0.001). However, the magnitude of negative effects was not equal across all species. This was particularly obvious in the case of S. bombicola and C. floricola, where priority effects were absent (order × species n.s.) and interspecific interactions were defined primarily by species identity (species effect; p < 0.001) rather than the order of arrival.
(b). Functional profiles
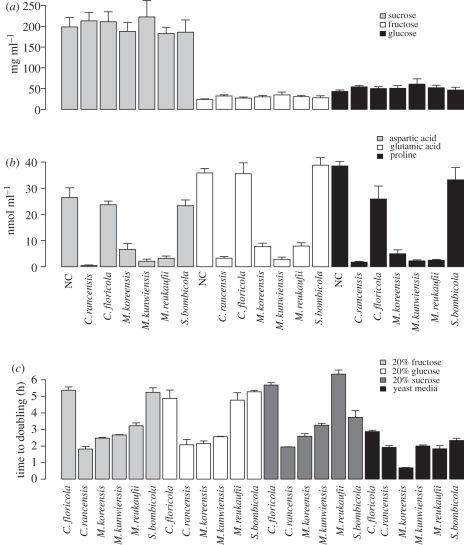
Chemical characterization showed high sugar concentrations and the presence of 13 amino acids (including two non-protein amino acids, citrulline and γ-aminobutyric acid (GABA)) in Mimulus nectar. Yeast colonization did not appear to have any effect on sugar concentrations (table 2 and figure 3a), but did significantly reduce amino acid concentrations with differences between species in the degree of consumption (figure 3b). While there was no effect of inoculation on day 0 versus day 2, species effects were significant for seven amino acids (ARG, ALA, ASP, GLU, PRO, TYR, VAL) after sequential Bonferonni correction (table 2). Despite the lack of yeast effects on sugar concentrations in nectar, there were significant differences between species in the doubling time and the length of the lag phase on different carbon substrates (ANOVA for doubling time: species × substrate interaction F15,48 = 15.9, p < 0.0001; ANOVA for lag time: species × substrate interaction F15,48 = 3.0, p = 0.002; figure 3c).
Table 2.
Significance of experimental treatments on amino acid and sugar concentrations in nectar. (Columns show p-values for two-way ANOVA testing for the effects of yeast species and the day of inoculation (day 0 versus day 2) on 13 amino acids and three sugars found in the nectar of M. aurantiacus. Bold text with an asterisk indicates significance after sequential Bonferonni correction for multiple tests.)
| response | species | day | species × day |
|---|---|---|---|
| arginine (ARG) | 0.0042* | 0.6450 | 0.7840 |
| citrulline (CIT) | 0.0312 | 0.1925 | 0.0751 |
| serine (SER) | 0.0060 | 0.3023 | 0.3609 |
| threonine (THR) | 0.0096 | 0.4859 | 0.1001 |
| alanine (ALA) | 0.0004* | 0.5125 | 0.5737 |
| γ-aminobutyric acid (GABA) | 0.2361 | 0.5916 | 0.7898 |
| proline (PRO) | < 0.001* | 0.2248 | 0.0227 |
| aspartic acid (ASP) | < 0.001* | 0.3919 | 0.6476 |
| histidine (HIS) | 0.0123 | 0.3502 | 0.2461 |
| valine (VAL) | < 0.001* | 0.6664 | 0.3119 |
| glutamic acid (GLU) | < 0.001* | 0.4865 | 0.5402 |
| leucine (LEU) | 0.0079 | 0.4543 | 0.9183 |
| tyrosine (TYR) | 0.0004* | 0.3998 | 0.9580 |
| sucrose | 0.3878 | 0.2704 | 0.9518 |
| glucose | 0.4167 | 0.2926 | 0.1392 |
| fructose | 0.4111 | 0.3622 | 0.3918 |
Figure 3.
Characteristics of nectar chemistry and yeast growth under different experimental conditions. (a) Concentrations of sucrose, glucose and fructose in Mimulus nectar with and without (NC) the presence of nectar yeasts. (b) Concentrations of three protein amino acids in Mimulus nectar that showed significant changes in abundance after yeast colonization. (c) Effects of different media on the doubling time of yeast species.
(c). Phylogenetic relatedness, ecological similarity and priority effects
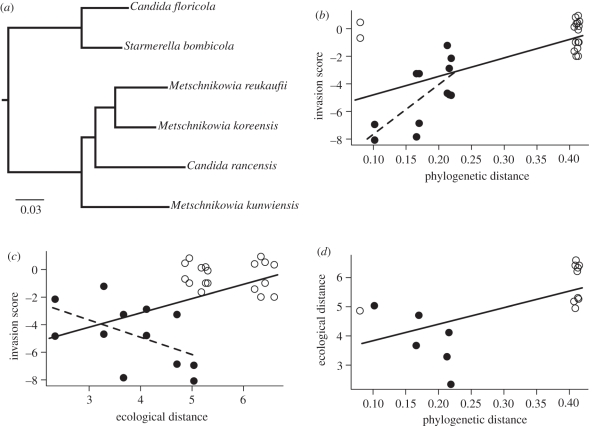
We found a significant relationship between phylogenetic distance and invasion score (r2 = 0.42, p < 0.0001; solid line in figure 4b). When S. bombicola and C. floricola were removed from the analysis, the relationship remained positive and significant (r2 = 0.47, p = 0.01; dashed line in figure 4b). The positive slope of this correlation indicates that flowers occupied by distant phylogenetic relatives are more easily invaded than those occupied by close relatives. There was also a significant relationship between overall ecological similarity and invasion score (r2 = 0.22, p = 0.01; solid line in figure 4c), indicating that competitive exclusion was stronger among ecologically similar species. However, this relationship appeared to shift orientation and was only marginally significant when S. bombicola and C. floricola were removed (r2 = 0.27, p = 0.086; dashed line in figure 4c). Linear mixed-effects models for the analyses presented in figure 4b,c gave identical results (electronic supplementary material, table S1). The relationship between total ecological similarity and phylogenetic relatedness was slightly weaker and depended somewhat on the scale of analysis. In general, close relatives were more similar ecologically as indicated by the positive correlation between ecological and phylogenetic distance (Mantel r = 0.62, p = 0.05; solid line in figure 4d). The direction of this relationship also appeared to reverse when S. bombicola and C. floricola were excluded, so that close relatives appeared to have more divergent ecological characteristics, but the relationship was not statistically significant (Mantel r = −0.75, p = 0.164; dashed line in figure 4d).
Figure 4.
Relationship between phylogeny, ecological similarity and the strength of priority effects (see text for details). (a) Molecular phylogeny illustrating the relationship between yeasts used in the experiment. Actual phylogenetic distances used in the analyses were calculated using the full tree shown in the electronic supplementary material, figure S4. (b) The strength of the priority effect decreases with phylogenetic distance. Each point represents the mean of five replicates for a given species invasion pairing. The invasion score represents species growth when invading an occupied habitat relative to growth in a monoculture over the same time period measured in units of standard deviation. The solid line represents correlation across the entire dataset (i.e. both open and filled circles), whereas filled circles and the dashed line indicate a subset of species that grew well in nectar. (c) The strength of the priority effect also decreases with increasing ecological distance, e.g. decreasing similarity in nectar resource use and growth rates. (d) Ecological distance also increases with phylogenetic distance, although the result depends on the phylogenetic scale.
4. Discussion
Our results show that priority effects are likely to be strong and widespread in the yeast community inhabiting flowers of M. aurantiacus. In general, early arriving species experienced little negative effects from later arriving species, whereas late-arriving species attained final densities far below their monoculture growth potential (figure 2). However, we observed considerable variation in the strength of priority effects across the nectar yeast community. Two species, S. bombicola and C. floricola, were easily excluded but unable to exclude other species even with priority. Although these species grew relatively poorly, there was also variation between species pairs that did grow well in nectar. For example, M. kunwiensis reached final densities of more than 20 000 cells µl−1 but was invaded by C. rancensis, whereas M. koreensis and M. reukaufii completely excluded each other when they were introduced earlier than the other (figure 2).
We were able to explain some of this variation based on phylogenetic distance and ecological similarity among species (figure 4). Thus, our results are consistent with Darwin's naturalization hypothesis, with close relatives being more ecologically similar and competing more intensely, ultimately resulting in stronger priority effects. Some caution should be used when evaluating these results as they were at least partially driven by two species (S. bombicola and C. floricola) that grew relatively poorly in the competition experiment. While both species did increase in abundance, it is possible that Mimulus nectar is not their primary habitat—S. bombicola, for example, was originally isolated from honey [43]. This type of habitat filtering has previously been shown to result in phylogenetic clustering of yeasts in nectar [44]. For this reason, the pattern we observe in this study probably results from both phylogenetic signal in adaptation to particular environmental conditions (e.g. habitat filtering) and phylogenetic signal related to resource competition (e.g. limiting similarity). Greater knowledge of the natural history of nectar yeasts, such as natural succession patterns and habitat specialization, will help to contextualize these results in the future.
(a). Implications for yeast community assembly
Taken along with previous work, our results provide a coherent framework of yeast community assembly in floral nectar. New flowers are initially sterile and yeasts are thought to colonize mainly via pollinators [24]. While pollinators vector a wide array of fungi, nectar presents a strong habitat filter and only a small subset of these species are able to colonize [44]. This filter probably acts in two ways. First, species must have high osmotolerance to grow in approximately 20–50% sugar concentrations found in nectar [44]. In addition, nectar often contains strong anti-microbial chemicals that may require specific adaptations to tolerate [45,46]. Our study suggests this is the case as S. bombicola and C. floricola grew poorly in nectar but achieved densities greater than 10 000 cells µl−1 under identical laboratory conditions in 20 per cent sugar solutions (data not shown). For those species that successfully disperse and are capable of growth in nectar, the outcome of arrival depends on biotic context. In an unoccupied flower, species grow rapidly to their carrying capacity and exhaust the available resources. If another species is already present, the extent to which later arrivals can colonize depends on the niche overlap between themselves and previous arrivals, with more intense competition between close relatives with similar resource requirements.
This picture of community assembly contains both stochastic and deterministic elements. Because nectar represents a relatively strong habitat filter there is a predictable (and phylogenetically non-random) subset of fungal species that are commonly found in floral nectar. In other words, given a pool of fungal propagules, it would be possible to predict which species could colonize a given nectar patch. However, within this subset of potential colonists, the strong priority effects we found demonstrate a high potential for historically contingent assembly based on the arrival order. The strong priority effects we describe here are probably responsible for the relatively low per-flower species richness found in field surveys of nectar yeast [24,25]. Strong priority effects may lead to a positive feedback where abundant species become more abundant, and such a feedback might explain the extreme dominance of M. reukaufii in our system, which accounts for about 75 per cent of yeast occurrences at JRBP [29]. Determining how the flower-scale dynamics we describe here influence community assembly at larger spatial scales will be an important challenge [47].
Our results have implications for understanding the ecosystem-level consequences of yeast colonization of floral nectar. While previous studies have focused on sugar consumption [27], our results suggest that amino acids may be a limiting resource for yeast growth. This seems reasonable when considering that the abundance of sugar in nectar may be a source of osmotic stress rather than a limiting resource. Although we did observe differences in growth rates across carbon substrates, the total consumption by yeasts was so minimal relative to starting concentrations in nectar that we were unable to detect these preferences in colonized nectar. The presence of amino acids in floral nectar and their role in pollinator visitation have been known for some time [48]. The differences between yeast species in amino acid consumption may thus have important consequences for plant pollinator visitation rates and determining the balance between mutualism and parasitism in the yeast-floral nectar symbiosis. For example, the non-protein amino acid GABA has been previously detected in nectar and is known to play a role in mammalian neural physiology, but was not affected at all by nectar yeast. On the other hand, glutamic acid, aspartic acid and proline were almost completely removed by some species (figure 3). An important further step in this research will be measuring sugar and amino acid composition from new and old flowers in the field to look for similar patterns of depletion.
(b). General implications
Our finding that priority effects are strongest between close relatives may have general implications beyond the specific yeast communities we studied. For example, phylogenetic perspectives on priority effects may increase our general ability to gauge whether communities are likely to experience historical contingency and to predict community assembly even under conditions favourable to historical contingency [5]. If priority effects are weak among distant relatives, communities where major players are from distant lineages may be unlikely to experience historically contingent assembly. Conversely, communities with many closely related species may be more susceptible to historical contingency. Similarly, if phylogenetic knowledge can be used to predict which species will have strong priority effects, given an initial colonist we may be able to predict a priori which other species are likely to co-invade. Thus, we may be able to predict common pathways of species assembly even if the order of arrival is difficult to predict. One important caveat is that this study was restricted to pairwise interactions owing to the large number of treatments involved. Generalizing these results will require consideration of possible indirect higher order effects from multispecies interactions.
Our results also indicate that caution should be used given that the degree of ecological similarity depends on the phylogenetic scale of investigation and the traits being examined [49]. While phylogeny was a significant predictor of competitive outcomes at all phylogenetic scales, the correlation between overall ecological similarity and phylogenetic relatedness broke down (possibly even changing sign) at smaller phylogenetic scales (figure 4). This finding may partly reflect the small number of comparisons in this particular comparison (particularly in our data subset) and the low power of Mantel tests. While phylogeny has been found to be a better predictor of ecological outcome than functional traits in some other studies [50], our result indicates that a better mechanistic understanding of which traits are responsible for competition outcomes is required. Greater information about the natural history and habitats of nectar-associated yeasts also will be important in choosing relevant physiological tests to measure ecological similarity among species and will help in designing future experiments.
5. Conclusion
The use of nectar yeast communities as a model system has enabled us to provide the first experimental evidence, to our knowledge, for the use of Darwin's naturalization hypothesis in explaining variation in the strength of priority effects. Overall, our results suggest that phylogenetic relatedness can be a powerful framework not just for understanding competitive interactions, but also for determining which communities are susceptible to historical contingency and which assembly pathways are common within those communities.
Acknowledgements
The authors would like to acknowledge Nona Chiariello and JRBP for supporting the field portions of this work; students of the 2010 Biology 44Y pilot class at Stanford University for helping to collect the data presented in figure 1; Nathan Kim, Katrina Luna, Christine Kyauk, Safiyyah Abdul-Khabir and Grace Goldberg for laboratory assistance; Karolina Krasinska, Yuen Tam and the Vincent Coats Foundation Mass Spectrometry Laboratory at Stanford University for chemical characterization of nectar; Daria Hekmat-Scafe and Patricia Chandler Seawell for helping to set up growth experiments; Annette Golonka and Carlos Herrera for helpful comments; and Stanford University for financial support. T.F. was supported by Stanford University's Terman Fellowship. M.B. was supported by the US National Science Foundation's Graduate Research Fellowship.
References
- 1.Davis M. B. 1986. Climatic instability, time lags, and community disequilibrium. In Community ecology (eds Diamond J. M., Case T. J.), pp. 269–284 New York, NY: Harper & Roy [Google Scholar]
- 2.Latham T. E., Ricklefs R. E. 1993. Continental comparisons of temperate-zone tree species diversity. In Species diversity in ecological communities: historical and geographical perspectives (eds Ricklefs R. E., Schluter D.). Chicago, IL: University of Chicago Press [Google Scholar]
- 3.Clements F. E. 1936. Nature and structure of the climax. J. Ecol. 24, 252–284 10.2307/2256278 (doi:10.2307/2256278) [DOI] [Google Scholar]
- 4.Fukami T. 2010. Community assembly dynamics in space. In Community ecology: processes, models and applications (eds Verhoef H. A., Morin P. J.), pp. 45–54 Oxford, UK: Oxford University Press [Google Scholar]
- 5.Chase J. M. 2003. Community assembly: when should history matter? Oecologia 136, 489–498 10.1007/s00442-003-1311-7 (doi:10.1007/s00442-003-1311-7) [DOI] [PubMed] [Google Scholar]
- 6.Gleason H. A. 1926. The individualistic concept of the plant association. Bull. Torrey Botanic. Club 53, 7–26 10.2307/2479933 (doi:10.2307/2479933) [DOI] [Google Scholar]
- 7.Santayana G. 1905. Life of reason: reason in common sense. New York, NY: Charles Scribner's Sons [Google Scholar]
- 8.Connell J. H., Sousa W. P. 1983. On the evidence needed to judge ecological stability or persistence. Am. Nat. 121, 789–824 10.1086/284105 (doi:10.1086/284105) [DOI] [Google Scholar]
- 9.Samuels C. L., Drake J. A. 1997. Divergent perspectives on community convergence. Trends Ecol. Evol. 12, 427–432 10.1016/S0169-5347(97)01182-8 (doi:10.1016/S0169-5347(97)01182-8) [DOI] [PubMed] [Google Scholar]
- 10.Schröder A., Persson L., De Roos A. M. 2005. Direct experimental evidence for alternative stable states: a review. Oikos 110, 3–19 10.1111/j.0030-1299.2005.13962.x (doi:10.1111/j.0030-1299.2005.13962.x) [DOI] [Google Scholar]
- 11.Alford R. A., Wilbur H. M. 1985. Priority effects in experimental pond communities: competition between Bufo and Rana. Ecology 66, 1097–1105 10.2307/1939161 (doi:10.2307/1939161) [DOI] [Google Scholar]
- 12.Wilbur H. M., Alford R. A. 1985. Priority effects in experimental pond communities: responses of Hyla to Bufo and Rana. Ecology 66, 1106–1114 10.2307/1939162 (doi:10.2307/1939162) [DOI] [Google Scholar]
- 13.Darwin C. 1859. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. London, UK: John Murray; [PMC free article] [PubMed] [Google Scholar]
- 14.Park T. 1954. Experimental studies of interspecies competition. II. Temperature, humidity, and competition in two species of Tribolium. Phys. Zool. 27, 177–238 [Google Scholar]
- 15.Tilman D. 1976. Ecological competition between algae: experimental confirmation of resource-based competition theory. Science 192, 463–465 10.1126/science.192.4238.463 (doi:10.1126/science.192.4238.463) [DOI] [PubMed] [Google Scholar]
- 16.Hutchinson G. E. 1961. The paradox of the plankton. Am. Nat. 95, 137–145 10.1086/282171 (doi:10.1086/282171) [DOI] [Google Scholar]
- 17.Burns J. H., Strauss S. Y. 2011. More closely related species are more ecologically similar in an experimental test. Proc. Natl Acad. Sci. USA 108, 5302–5307 10.1073/pnas.1013003108 (doi:10.1073/pnas.1013003108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang L., Tan J., Pu Z. 2010. An experimental test of Darwin's naturalization hypothesis. Am. Nat. 175, 415–423 10.1086/650720 (doi:10.1086/650720) [DOI] [PubMed] [Google Scholar]
- 19.Webb C. O., Ackerly D. D., McPeek M. A., Donoghue M. J. 2002. Phylogenies and community ecology. Ann. Rev. Ecol. Syst. 33, 475–505 10.1146/annurev.ecolsys.33.010802.150448 (doi:10.1146/annurev.ecolsys.33.010802.150448) [DOI] [Google Scholar]
- 20.Anderson T. M., Lachance M. A., Starmer W. T. 2004. The relationship of phylogeny to community structure: the cactus yeast community. Am. Nat. 164, 709–721 10.1086/425372 (doi:10.1086/425372) [DOI] [PubMed] [Google Scholar]
- 21.Cavender-Bares J., Kozak K. H., Fine P. V. A., Kembel S. W. 2009. The merging of community ecology and phylogenetic biology. Ecol. Lett. 12, 693–715 10.1111/j.1461-0248.2009.01314.x (doi:10.1111/j.1461-0248.2009.01314.x) [DOI] [PubMed] [Google Scholar]
- 22.Kennedy P. G., Peay K. G., Bruns T. D. 2009. Root tip competition among ectomycorrhizal fungi: are priority effects a rule or an exception? Ecology 90, 2098–2107 10.1890/08-1291.1 (doi:10.1890/08-1291.1) [DOI] [PubMed] [Google Scholar]
- 23.Louette G., De Meester L. 2007. Predation and priority effects in experimental zooplankton communities. Oikos 116, 419–426 10.1111/j.2006.0030-1299.15381.x (doi:10.1111/j.2006.0030-1299.15381.x) [DOI] [Google Scholar]
- 24.Brysch-Herzberg M. 2004. Ecology of yeasts in plant-bumblebee mutualism in central Europe. FEMS Microbiol. Ecol. 50, 87–100 10.1016/j.femsec.2004.06.003 (doi:10.1016/j.femsec.2004.06.003) [DOI] [PubMed] [Google Scholar]
- 25.Pozo M. I., Herrera C. M., Bazaga P. 2010. Species richness of yeast communities in floral nectar of southern Spanish plants. Microbial Ecol. 10, 82–91 [DOI] [PubMed] [Google Scholar]
- 26.Lachance M. A., Starmer W. T., Rosa C. A., Bowles J. M., Barker J. S. F., Janzen D. H. 2001. Biogeography of the yeasts of ephemeral flowers and their insects. FEMS Yeast Res. 1, 1–8 [DOI] [PubMed] [Google Scholar]
- 27.Herrera C. M., Garcia I. M., Perez R. 2008. Invisible floral larcenies: microbial communities degrade floral nectar of bumblebee-pollinated plants. Ecology 89, 2369–2376 10.1890/08-0241.1 (doi:10.1890/08-0241.1) [DOI] [PubMed] [Google Scholar]
- 28.Pozo M. I., de Vega C., Canto A., Herrera C. M. 2009. Presence of yeasts in floral nectar is consistent with the hypothesis of microbial-mediated signaling in plant–pollinator interactions. Plant Signal. Behav. 4, 1102–1104 10.4161/psb.4.11.9874 (doi:10.4161/psb.4.11.9874) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beslisle M., Peay K. G., Fukami T. Submitted Flowers as micro-islands: distribution of nectar-living microfungi in a California landscape. Microb. Ecol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cornwell W. K., Ackerly D. D. 2009. Community assembly and shifts in plant trait distributions across an environmental gradient in coastal California. Ecol. Monogr. 79, 109–126 10.1890/07-1134.1 (doi:10.1890/07-1134.1) [DOI] [Google Scholar]
- 31.Mooney H. A., Ehrlich P. R., Lincoln D. E., Williams K. S. 1980. Environmental controls on the seasonality of a drought deciduous shrub, Diplacus aurantiacus and its predator, the checkerspot butterfly, Euphydryas chalcedona. Oecologia 45, 143–146 10.1007/BF00346452 (doi:10.1007/BF00346452) [DOI] [PubMed] [Google Scholar]
- 32.Fetscher A. E., Kohn J. R. 1999. Stigma behavior in Mimulus aurantiacus (Scrophulariaceae). Am. J. Bot. 86, 1130–1135 10.2307/2656976 (doi:10.2307/2656976) [DOI] [PubMed] [Google Scholar]
- 33.O'Donnell K. 1993. Fusarium and it's near relatives. In The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics (eds Reynolds D. R., Taylor J. W.), pp. 225–233 Wallingford, UK: CAB International [Google Scholar]
- 34.Yarrow D. 1998. Methods for the isolation, maintenance and identification of yeasts. In The yeasts, a taxonomic study (eds Kurtzman C. P., Fell J. W.), pp. 77–100 Amsterdam, The Netherlands: Elsevier [Google Scholar]
- 35.Kahm M., Hasenbrink G., Lichtenberg-Frate H., Ludwig J., Kshischo M. 2010. grofit: fitting biological growth curves with R. J. Statist. Softw. 33, 1–21 [Google Scholar]
- 36.Toussaint M., Conconi A. 2006. High-throughput sensitive assay to measure yeast cell growth: a bench protocol for testing genotoxic agents. Nat. Protocols 1, 1922–1928 10.1038/nprot.2006.304 (doi:10.1038/nprot.2006.304) [DOI] [PubMed] [Google Scholar]
- 37.Rice W. R. 1989. Analyzing tables of statistical tests. Evolution 43, 223–225 10.2307/2409177 (doi:10.2307/2409177) [DOI] [PubMed] [Google Scholar]
- 38.Suh S. O., Blackwell M., Kurtzman C. P., Lachance M. A. 2006. Phylogenetics of Saccharomycetales, the ascomycete yeasts. Mycologia 98, 1006–1017 10.3852/mycologia.98.6.1006 (doi:10.3852/mycologia.98.6.1006) [DOI] [PubMed] [Google Scholar]
- 39.Katoh K., Misawa K., Kuma K., Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 10.1093/nar/gkf436 (doi:10.1093/nar/gkf436) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guindon S., Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 10.1080/10635150390235520 (doi:10.1080/10635150390235520) [DOI] [PubMed] [Google Scholar]
- 41.Oksanen J., et al. 2008. vegan: community ecology package. R package version 1.15-1. See http://cc.oulu.fi/∼jarioksa/opetus/metodi/vegantutor.pdf
- 42.Peres-Neto P. R., Jackson D. A. 2001. How well do multivariate data sets match? The robustness and flexibility of a Procrustean superimposition approach over the Mantel test. Oecologia 129, 169–178 10.1007/s004420100720 (doi:10.1007/s004420100720) [DOI] [PubMed] [Google Scholar]
- 43.Rosa C. A., Lachance M. A. 1998. The yeast genus Starmerella gen. nov. and Starmerella bombicola sp. nov., the teleomorph of Candida bombicola (Spencer, Gorin & Tullock) Meyer & Yarrow. Int. J. Syst. Bacteriol. 48, 1413–1417 10.1099/00207713-48-4-1413 (doi:10.1099/00207713-48-4-1413) [DOI] [PubMed] [Google Scholar]
- 44.Herrera C. M., Canto A., Pozo M. I., Bazaga P. 2009. Inhospitable sweetness: nectar filtering of pollinator-borne inocula leads to impoverished, phylogenetically clustered yeast communities. Proc. R. Soc. B 277, 747–754 10.1098/rspb.2009.1485 (doi:10.1098/rspb.2009.1485) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carter C., Thornburg R. W. 2004. Is the nectar redox cycle a floral defense against microbial attack? Trends Plant Sci. 9, 320–324 10.1016/j.tplants.2004.05.008 (doi:10.1016/j.tplants.2004.05.008) [DOI] [PubMed] [Google Scholar]
- 46.Lokvam J., Braddock J. F. 1999. Anti-bacterial function in the sexually dimorphic pollinator rewards of Clusia grandiflora (Clusiaceae). Oecologia 119, 534–540 10.1007/s004420050816 (doi:10.1007/s004420050816) [DOI] [PubMed] [Google Scholar]
- 47.Levin S. A. 1992. The problem of pattern and scale in ecology. Ecology 73, 1943–1967 10.2307/1941447 (doi:10.2307/1941447) [DOI] [Google Scholar]
- 48.Baker H. G., Baker I. 1986. The occurrence and significance of amino-acids in floral nectar. Plant Syst. Evol. 151, 175–186 10.1007/BF02430273 (doi:10.1007/BF02430273) [DOI] [Google Scholar]
- 49.Cavender-Bares J., Kitajima K., Bazzaz F. A. 2004. Multiple trait associations in relation to habitat differentiation among 17 Floridian oak species. Ecol. Monogr. 74, 635–662 10.1890/03-4007 (doi:10.1890/03-4007) [DOI] [Google Scholar]
- 50.Cadotte M. W., Cardinale B. J., Oakley T. H. 2008. Evolutionary history and the effect of biodiversity on plant productivity. Proc. Natl Acad. Sci. USA 105, 17 012–17 017 10.1073/pnas.0805962105 (doi:10.1073/pnas.0805962105) [DOI] [PMC free article] [PubMed] [Google Scholar]