Abstract
Cytochrome P450 3A4 (CYP3A4) metabolizes ~50% of all clinically used drugs. Although CYP3A4 expression varies widely between individuals, the contribution of genetic factors remains uncertain. In this study, we measured allelic CYP3A4 heteronuclear RNA (hnRNA) and mRNA expression in 76 human liver samples heterozygous for at least one of eight marker SNPs and found marked allelic expression imbalance (1.6–6.3-fold) in 10/76 liver samples (13%). This was fully accounted for by an intron 6 SNP (rs35599367, C>T), which also affected mRNA expression in cell culture on minigene transfections. CYP3A4 mRNA level and enzyme activity in livers with CC genotype were 1.7- and 2.5-fold, respectively, greater than in CT and TT carriers. In 235 patients taking stable doses of atorvastatin, simvastatin, or lovastatin for lipid control, carriers of the T allele required significantly lower statin doses (0.2–0.6-fold, P=0.019) than non-T carriers for optimal lipid control. These results indicate that intron 6 SNP rs35599367 markedly affects expression of CYP3A4 and could serve as a biomarker for predicting response to CYP3A4-metabolized drugs.
Keywords: polymorphism, gene expression, CYP3A4, statin, allelic expression imbalance, cytochrome P450
Introduction
Cytochrome P450 (CYP) enzymes metabolize endogenous and xenobiotic compounds. Belonging to the CYP3A subfamily, CYP3A4 is the most abundant CYP enzyme, involved in metabolizing 45–60% of all currently used drugs,1 including several statins—cholesterol-lowering HMG-CoA reductase inhibitors. However, CYP3A4 activity or protein content shows 10–100-fold inter-individual variations,2–5 influencing drug response and toxicity. Although the expression of CYP3A4 can be affected by non-genetic factors (like diet, inducer or depressor, age, sex), genetic factors acting in cis- and/or trans- are thought to be the main contributors to inter-individual differences in CYP3A4 activity.6 Genetic factors acting in trans-, for example, polymorphisms in transcription factors,7 different splice variants in transcription factors8,9 and differences in microRNA regulation10,11 have been reported to account for a portion of inter-individual variability in CYP3A4 expression/activity. However, whether and how cis-acting polymorphisms in CYP3A4 contribute to inter-person variability in CYP3A4 expression remains unresolved.
Currently known genetic variants in CYP3A4 that change the amino-acid sequence are rare (<1%) (http://www.cypalleles.ki.se/cyp3a4.htm), and therefore, can only account for a small portion of the observed variability. A more common variant, CYP3A4*1B, in the 5′-flanking region, has been associated with drug response and diseases,4,12 but results are inconsistent,13–15 and its function remains controversial.12,16–18 Moreover, CYP3A4*1B is in linkage disequilibrium (LD) with the CYP3A5-expressing allele CYP3A5*1 in African Americans,19,20 raising the possibility that the expression of CYP3A5 could have accounted for any linked clinical phenotype.21 Further suspected CYP3A4 polymorphisms include a TGT insertion (rs34401238),22 an enhancer region SNP (rs2737418),23 and an intron 7 SNP (rs4646437).24 Although reporter gene assays suggested an effect for the TGT insertion and for rs2737418, the in vivo significance of TGT remains unresolved,22 and results on CYP3A4 mRNA and enzyme activity were contradictory for rs2737418.23 The intron 7 SNP rs4646437 was found to be associated with CYP3A4 protein/enzyme activity, but only in livers from males.24 Therefore, the role of functional polymorphisms in CYP3A4 remains uncertain.
The purpose of this study was to search for common cis-acting functional polymorphism(s) in CYP3A4 and evaluate their effects on CYP3A4-metabolized drugs in vivo. To search for common functional polymorphisms, we measured allelic heteronuclear RNA (hnRNA)/mRNA expression in human autopsy livers. A detectable allelic RNA expression imbalance (AEI) is a direct measure of cis-acting regulatory factors in CYP3A4 that affect RNA expression, processing, or turnover. Although Hirota et al.25 had observed an AEI for CYP3A4 using two marker SNPs and a semi-quantitative method, their extensive search for the functional polymorphism(s) responsible for the observed AEI was inconclusive. In this study, we used eight marker SNPs with a quantitative AEI analysis of high precision, followed by use of the AEI status (AEI positive or AEI negative) as phenotypic trait to scan for regulatory polymorphisms. Using this systematic approach, we identified a single functional SNP located in intron 6 of CYP3A4, which lacks LD with any known frequent polymorphisms, probably accounting for difficulties encountered previously in the search for the functional variants.23,24 The intron 6 SNP fully accounted for the observed allelic mRNA expression pattern and correlated with CYP3A4 total mRNA level and enzyme activity in human livers, whereas previously suggested polymorphisms had no effect.
The in vivo effect of intron 6 SNP on CYP3A4 substrate drug metabolism was assessed in a cohort of patients with coronary artery diseases who were taking CYP3A4-metabolized statins for lipid control. Statins (3-hydroxy-3-methylglutaryl coenzyme A reductase inhibitors) are widely used for the treatment of hypercholesterolemia. Among several statins, simvastatin, lovastatin, and atorvastatin are metabolized by CYP3A4 and display similar pharmacokinetics parameters. Atorvastatin is more potent than simvastatin and lovastatin, owing to its status as a substrate for OATP1B1, which transports the drug into the hepatocyte.26 As there is a strong correlation between statin dose, blood drug concentration, and lipid response,26,27 genetic polymorphisms that alter CYP3A4 enzyme activity are expected to affect blood drug level and lipid response or side effects. Using several CYP3A4 polymorphisms, earlier studies have reported contradictory results,26,28–31 possibly because the mechanism that could have resulted in altered enzyme activities remain uncertain. On the other hand, functional polymorphism in CYP3A4 would be expected to affect the stable statin dose requirement to reach a cholesterol reduction target. On the basis of this assumption, we tested the association between intron 6 SNP genotype and stable statin dose requirement to reach an optimal lipid control goal. The results indicate that intron 6 SNP was significantly associated with statin dosage in patients undergoing standard therapy, suggesting that intron 6 SNP affects statin metabolism in vivo.
Materials and methods
Tissue samples
Three sets of human autopsy/biopsy tissue samples were obtained from The Cooperative Human Tissue Network Midwestern and Western Division. Liver cohort 1 (maintained at Eli Lilly) consisted of 43 livers and cohort 2 consisted of 93 livers, whereas cohort 3 consisted of 106 small intestine samples, taken mostly from the duodenum, with a few samples also from ileum and jejunum. All samples were obtained for this study under a protocol approved by the Ohio State University Institutional Review Board. Of cohort 1, collected over 10 years earlier, 23 had been measured for CYP3A4 enzyme activities when the tissues were fresh (data not reported earlier), and all tissues were screened to select heterozygotes for allelic mRNA ratios measurement. Livers in cohort 1 had several fold lower total mRNA levels of both CYP3A4 and the house keeping gene GADPH compared with cohort 2 (measured by quantitative reverse transcriptase polymerase chain reaction (RT-PCR); possibly because of longer storage time), and they were excluded from association analysis with total mRNA as phenotype. In contrast, allelic mRNA ratios have proven more robust even with partially decayed mRNA, assuming that there are no allelic differences during post-mortem decay; therefore, we considered cohort 1 eligible for AEI analysis. Cohort 2 was assayed for allelic and total mRNA levels, whereas cohort 3 was genotyped for the intron 6 SNP, and heterozygous tissues were analyzed for allelic mRNA expression.
Patients
Subjects were participants in the Ohio State University Coronary Artery Disease Study, who presented to the OSU Heart Center with symptomatic cardiovascular disease with at least 75% angiographic luminal stenosis (newly diagnosed or established) requiring percutaneous coronary intervention. Two hundred seventy-three patients documented to be taking stable doses (same dose for at least 6 months) of an HMG-CoA reductase inhibitor (statin) for lipid control were selected for this study. Statin doses were titrated for each patient to reach predetermined cholesterol control goals as described.32 Lipid levels were measured at the time of enrollment and after reaching stable dose of statins. However, the lipid levels at the time of enrollment did not represent the basal level in all patients (without medication), because the documentation of any prior medication was incomplete. After enrolling into the study, the patients did not use other lipid-lowering drugs. Enrollment and trial conditions had been approved by the Ohio State University Institutional Review Board, with written informed consent obtained from each patient. The study population reflects demographics of the Columbus area and surrounding rural counties of Ohio.
DNA and RNA preparation
Preparation of genomic DNA (gDNA), RNA, cDNA from tissues and blood samples, and plasmid DNA from cultured cells, was performed as described.33–35 To avoid gDNA contamination in tissue RNA, samples were treated with DNase I. To avoid plasmid DNA contamination in RNA extracted from transfected cells, samples were treated with DNase I and two restriction enzymes (DpnI and XbaI, to linearize plasmid DNA so it can be degraded by DNase I). All samples were tested by real-time PCR, showing no amplification after 40 cycles in cDNA preparation without RT.
Quantitative analysis of allelic ratios in gDNA and RNA using SNaPshot
The detailed method has been published.33,34 Briefly, a fragment of DNA or RNA (after conversion to cDNA) surrounding a marker SNP was PCR amplified, followed by a primer extension assay (SNaPshot) that targets the polymorphic site. Eight marker SNPs (including the functional intron 6 SNP; Supplementary Figure 1) located in either 3′UTR or intronic regions were used to measure allelic ratios of mature mRNA (3′UTR markers) or hnRNA (intronic markers) in 76 out of the 136 livers heterozygous for at least one marker SNP. gDNA allelic ratios, normalized to 1, served as internal control; none of the subjects displayed gDNA copy number variants, indicated by a significant deviation from unity, although genomic deletions would not have been detectable. Deviations of allelic RNA ratios from 1 (after normalization to DNA ratios), that is AEI, indicate the presence of cis-acting polymorphisms in CYP3A4 that affect mRNA expression levels. Three independent AEI measurements were performed in each sample for each marker SNP.
Genotyping
Thirteen SNPs in CYP3A4 (including the eight marker SNPs) (Table 1) were genotyped in gDNA from liver samples with a multiplex SNaPshot assay36 or allele-specific real-time PCR.37 Seven SNPs in CYP3A4/3A5 were genotyped in gDNA from 273 patients. PCR conditions and primer sequences are provided in Supplementary Table 1. Selection of genotyping methods was guided by the suitability of the assay for the specific polymorphism and cost; each method was quality controlled by common procedures, including replications, assessment of Hardy–Weinberg equilibrium, and validation with a different method including sequencing.
Table 1.
Polymorphisms tested in liver samples
| SNP # | SNP ID | Position | MAF |
|---|---|---|---|
| 1 | rs34401238TGT ins | −11231 | 0.023 |
| 2 | rs2737418G>T | −7310 | 0.030 |
| 3 | rs2740574A>G(*1B) | −392 | 0.076 |
| 4 | rs2687105A>T | Intron 2 | 0.080 |
| 5* | rs28988579T>G | Inton 4 | 0.019 |
| 6* | rs35599367C>T | Intron 6 | 0.042 |
| 7* | rs2246709C>T | Intron 7 | 0.269 |
| 8 | rs4646437C>T | Intron 7 | 0.159 |
| 9* | rs2242480G>A | Intron 10 | 0.133 |
| 10* | rs3735451A>G | Intron 12 | 0.133 |
| 11* | rs28988604C>T | 3′UTR | 0.030 |
| 12* | rs28969391delT | 3′UTR | 0.148 |
| 13* | rs28371763A>T | 3′UTR | 0.023 |
Abbreviations: AEI, allelic expression imbalance; MAF, minor allele frequency; SNP, single nucleotide polymorphism.
MAF was derived from all liver samples (cohorts 1 and 2) (reference sequence AF280107).
indicates marker SNPs for AEI measurement.
Quantitative mRNA and hnRNA analysis
mRNA levels of CYP3A4, transcription factors PXR, RXRa, CAR, and HNF4a, and splicing protein SF2/ASF in human livers or small intestines were measured with real-time PCR33 using gene-specific primers38 (Supplementary Table 1) and SYBR Green (Applied Biosystems), with GAPDH mRNA as an internal control as described.33 hnRNA was also measured in minigene-transfected cells using similar real-time PCR method as applied to mRNA, with primers targeting the intronic regions (Supplementary Table 1).
Sequencing CYP3A4
The region from ~10 000 bp upstream of the transcription start site to the last exon (from 50013 to 89410 in AF280107, total length 39 397 bp) was sequenced in two liver samples (A43 and B76) that showed allele-specific RNA expression. PCR and sequencing primers were shown in Supplementary Table 1.
CYP3A4 enzymatic activity assay
CYP3A4 activities were quantified from liver microsomes with testosterone as a probe as described.39
Reporter gene assay to test enhancer activity of intron 6 SNP
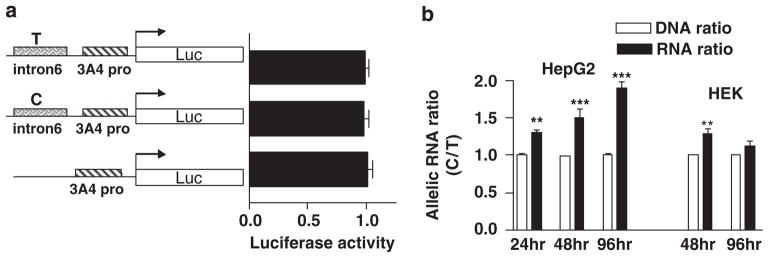
A DNA fragment of ~1700 bp in length (from 60256 to 61990, AF280107) was PCR amplified from the CYP3A4-promoter region and cloned into PGL3 reporter gene vector using XhoI and Hind III cloning sites. To test whether intron 6 contains enhancer/repressor elements that could be affected by intron 6 SNP, an intron 6 fragment (~1000 bp from 76686 to 77738) harboring either the C or T allele was PCR amplified and ligated upstream of promoter fragment40,41 using KpnI and XhoI sites (see Figure 7 for details). The promoter and intron 6 fragments were sequenced, showing the expected sequences. These plasmids were transfected into HepG2 cells. As transfection efficiency control, Renilla luciferase constructs driven by a TK promoter were co-transfected with the PGL3 fused constructs at a 1:3 ratio, and luciferase activities measured at 48 h post-transfection with Dual-Glo luciferase assays kit (Promega) on a fluorescence plate reader (PerkinElmer Life and Analytical Science, Waltham, MA, USA). For each construct, three clones were selected for DNA preparation.
Figure 7.
In vitro cell transfection assays. (a) Effect of intron 6 region on CYP3A4-promoter activity tested in a luciferase reporter gene assay. The intron 6 region was inserted upstream of the proximal CYP3A4-promoter region, and both C and T alleles were tested using bioluminescence output. (b) Effects of intron 6 SNP on CYP3A4 minigene RNA expression. Minigene constructs, consisting of exon 6, intron 6, and exon 7, harboring the C or T allele of intron 6 SNP, were co-transfected into HepG2 or HEK293 cells, and allelic DNA and RNA ratios measured at 24, 48, and 96 h post-transfection, using intron 6 SNP as the marker. The intron 6 SNP plasmid DNA ratio was normalized to 1 for each experiment. Compared with plasmid DNA ratio, **P<0.01, ***P<0.001 analysis of variance with Bonferroni post-test.
Minigene assays
CYP3A4 gene fragments from intron 4 to intron 7 (~2300 bp, from 75700 to 77994, AF280107), harboring either the C or T alleles of intron 6 SNP were PCR amplified from gDNA and cloned into pcDNA3 vector using KpnI and XhoI sites. The expected DNA sequences were confirmed by sequencing. We selected three clones of each construct for DNA preparation. Same amounts of minigenes harboring C or T allele were co-transfected into HepG2 or HEK293 cells and cells harvested at 24, 48, and 96 h post-transfection for plasmid DNA and RNA preparation. Allelic DNA and RNA ratios were measured as described above.
Cell culture and transfection
Cells were cultured at 37 °C in a humidified incubator at 5% CO2 in DMEM (HepG2) or DMEM/F12 (HEK) supplemented with 10% fetal bovine serum, 100Uml−1 penicillin and 100 μgml−1 streptomycin. The day before transfection, cells were plated into 12-well plates. Transfection was performed using lipofectamine 2000 according to the manufacture’s protocol (Invitrogen Life Technologies, Carlsbad, CA, USA).
Data analysis
Haplotype structure and LD plots were generated using Helix-Tree software (Golden Helix, Bosman MT) and Haploview. A multiple linear regression model was used for testing genotype effects on RNA expression, enzyme activity, and statin dose requirement using SPSS or Minitab software. We used forward and backward stepwise regression to select the best set of predictors in the multiple linear regression models with cutoff P-value ≤0.05. For mRNA data, sex and expression levels of PXR and RXR were included as covariates, whereas for enzyme activity data, sex, age, and exposure of inducer were included as covariates. For statin dose data, pretreatment total cholesterol level was a covariate. Genotype effects were adjusted for these covariates. The goodness-of-fit was judged by residual plots and normal quartile plot, testing the fulfillment of linear regression assumptions, that is, constant variance, normality, and independency. The association between three stable statin dose levels (≤10 mg, 20 mg, and ≥40 mg) and genotype was analyzed using proportional odds logistics regression model performed using SAS 9.1 software. The suitability of model fitting was judged by deviance goodness-of-fit statistics P-value and score test P-value, both of which should exceed 0.05. Mann–Whitney test was used to analyze the differences in median statin doses between the two groups where the data are not normally distributed, whereas Student’s t-test was used to compare the proportions or means between the two groups where the data are normally distributed.
Results
Scanning for cis-acting CYP3A4 polymorphisms that affect mRNA levels
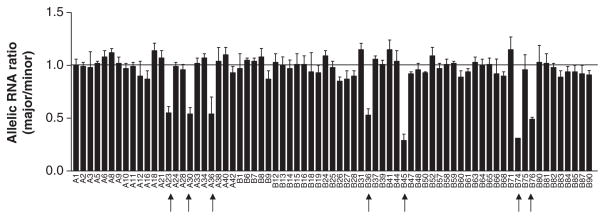
To search for cis-acting regulatory polymorphisms in CYP3A4 that affect mRNA levels, we measured allelic mRNA expression of CYP3A4 using three frequent marker SNPs located in the 3′UTR (#11, 12, 13, Table 1, SNP #12 is a T-deletion polymorphism, but can be measured in the same way as a SNP). Because of the high CYP3A4 expression in liver, we also used four common intronic SNPs (#5, 7, 9, 10, Table 1, intron 6 SNP #6 was not included in this initial screen; Supplementary Figure 1) as markers to measure allelic expression of CYP3A4 hnRNA as described.25 Among 136 liver samples screened (43 from cohort 1 and 93 from cohort 2), 73 were heterozygous for at least one of the seven marker SNPs and therefore suitable for allelic RNA expression measurement. Normalized allelic RNA ratios for 66 of the 73 samples were closed to 1 (range from 0.87 to 1.15; Figure 1; Supplementary Table 2), indicating RNA levels derived from each of the two alleles are similar, arguing against the presence of cis-acting regulatory polymorphisms in these samples. In contrast, seven tissues (A23, A30, A36, B36, B45, B74, and B76) showed allelic RNA ratios significantly deviating from 1 (ratio represent main allele/variant allele, range 0.29–0.55), demonstrating significant AEI. In each case, the variant allele of the marker SNPs is present at 2–3-fold higher levels than the main allele, indicating that one or more cis-acting regulatory polymorphic sites must be heterozygous in these samples (Figure 1; Supplementary Table 2). Twenty-eight samples including two samples showing AEI (AEI positive samples A30 and B45) were measured with both intronic marker SNPs and 3′UTR SNPs. Allelic RNA ratios obtained from intronic and 3′UTR marker SNPs did not differ significantly between each other in the same subject (small s.d., Supplementary Table 2), indicating that a cis-acting polymorphism(s) in CYP3A4 affects both mRNA and hnRNA levels equally.
Figure 1.
Allelic mRNA/hnRNA expression ratios of CYP3A4 in human livers measured with a primer extension assay (SNaPshot) using seven marker SNPs (SNP 5, 7, 9, 10, 11, 12, and 13 in Table 1). Allelic RNA ratios were normalized to genomic DNA ratios set at 1. Data represent the average of three measurements per marker using single or multiple marker SNPs (mean±s.d.). Arrow indicates samples with AEI ratios significant different from 1 (analysis of variance with Dunnett post-test, P<0.05).
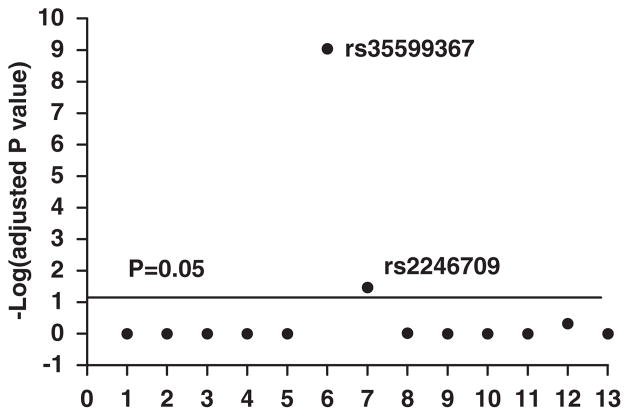
To search for responsible polymorphism(s), 13 CYP3A4 polymorphisms (Table 1) were genotyped and the association between AEI status (AEI positive or AEI negative) and genotype tested in 73 samples with AEI data available. A single SNP (#6, rs35599367, C>T; Table 1; Supplementary Figure 1) located in intron 6 showed highly significant association with AEI status (adjusted P-value 9.12×10−10) (Figure 2). Each sample showing AEI (that is AEI-positive sample) was heterozygous for intron 6 SNP, and each intron 6 SNP heterozygous tissue was AEI positive (Supplementary Table 2). None of the other tissues was AEI positive. This result implicates intron 6 SNP as the only causative factor. SNP rs2246709 (#7) also scored with moderate significance (P=0.034, Figure 2), likely because of partial LD with the intron 6 SNP (Supplementary Figure 2). Other SNPs including previously identified promoter SNP *1B rs2740574 (#3), rs34401238 (#1, TGT insertion), rs2737418 (#2), and rs4646437 (#8) did not show significant association (P>0.05, Figure 2). These results indicate that intron 6 SNP is functional per se or in high LD with a functional SNP.
Figure 2.
Association between genotypes and allelic RNA expression imbalance (AEI) status (AEI positive or AEI negative). Only intron 6 SNP rs35599367 scored with high significance, whereas SNP 7 (rs2246709 in intron 7) was marginally significant. The solid line indicates P=0.05 level for the association, without adjusting for multiple comparisons. 1. rs34401238 (TGT insertion); 2. rs2737418; 3. rs2740574(*1B); 4. rs2687105; 5. rs28988579; 6. rs35599367 (intron 6); 7. rs2246709; 8. rs4646437; 9. rs2242480; 10. rs3735451; 11. rs28988604; 12. rs28969391; 13. rs28371763.
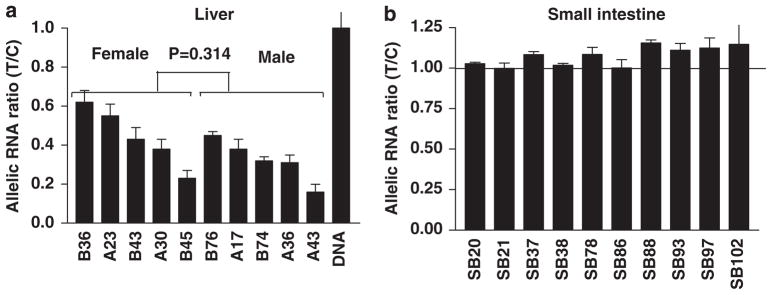
If intron 6 were the only cause of AEI, we would expect all intron 6 SNP heterozygous samples show AEI when using intron 6 SNP itself as a marker. To test this, we screened all 136 liver samples (cohorts 1 and 2) for intron 6 SNP genotype, and identified 10 heterozygous samples, including three that were heterozygous only for intron 6 SNP (A17, A43, and B43), but not any of the other 13 SNPs tested (Supplementary Table 2). AEI was measured in these 10 samples using intron 6 SNP as a marker. As expected, all 10 samples showed significant AEI with allelic ratios (minor T allele/major C allele) ranging from 0.16 to 0.62 (Figure 3a), with no differences between males and females (t-test, P=0.314). Therefore, the minor T allele of intron 6 SNP is linked to reduced mRNA/hnRNA levels (the inverse allelic mRNA ratio of major C/minor T ranges from 1.6 to 6.25). The minor T allele is exclusively linked to the main alleles of all other SNPs tested (haplotype 5), except for low LD with rs2246709 (Supplementary Table 3; LD plot Supplementary Figure 2). This finding accounts for the allelic mRNA ratios below unity observed with all other marker SNPs (major/minor allele; that is the main allele is expressed less) in tissues that are heterozygous for intron 6 SNP.
Figure 3.
Allelic hnRNA expression ratios of CYP3A4 in human livers (a) and intestines (b) measured with a primer extension assay (SNaPshot) using intron 6 SNP as marker. Allelic RNA ratios were normalized to genomic DNA ratios set at 1. Data represent the average of three independent measurements for each sample (mean±s.d.). All allelic RNA ratios in panel (a) were significantly different from 1 (analysis of variance with Dunnett post-test, P<0.05), whereas allelic RNA ratios in panel (b) were all close to 1.
To search for other possible polymorphisms that may account for the observed AEI, we sequenced the entire CYP3A4 locus, from ~10 kb upstream of the transcription start site to the last exon (from 50013 to 89410 in AF280107, total length 39 397 bp) in two AEI-positive samples (B43 and B76). Any possible functional polymorphisms causing AEI must be heterozygous in both samples. However, except for intron 6 SNP in both two samples and intron 7 SNP rs2246709 in B76 sample, there was not a single additional heterozygous polymorphic site present in the entire CYP3A4 locus in these two samples. This is consistent with our genotyping results (Supplementary Table 2), that is both samples were homozygous for major alleles of the 13 SNPs genotyped except for intron 6 and intron 7 SNP (rs2246709, in B76 only), as expected from the strong LD of intron 6 with the major alleles of all other SNPs (Supplementary Table 3). As the AEI ratio analysis was unambiguous, this result rules out other SNPs within the sequenced region and strongly indicates that intron 6 SNP is the only cause of AEI. We cannot exclude that a causative SNP could have resided outside the sequenced region, but any such variant would have to be in complete LD with the intron 6 SNP, an unlikely scenario (large LD block of over 40 kb) given the 40 kb region surveyed, and the haplotype structure of CYP3A4 shown in Supplementary Figure 2.
As the regulation of mRNA levels by a polymorphism is often tissue specific, as we have observed with VKORC1,42 we determined whether intron 6 SNP is also active in tissues other than liver by measuring AEI in human small intestine samples using intron 6 SNP as marker. Of 106 small intestines from cohort 3, 10 were heterozygous for intron 6 SNP (nine duodenums, one ileum SB86) and selected for AEI measurement. In contrast to liver samples, where all heterozygous tissues showed AEI when using intron 6 SNP as marker (Figure 3a), none of the 10 small intestine samples showed AEI (Figure 3b), demonstrating intron 6 SNP has no effect in small intestines. This result supports tissue-specific regulation affected by intron 6 SNP.
Intron 6 SNP associates with decreased CYP3A4 mRNA level and enzyme activity in human livers
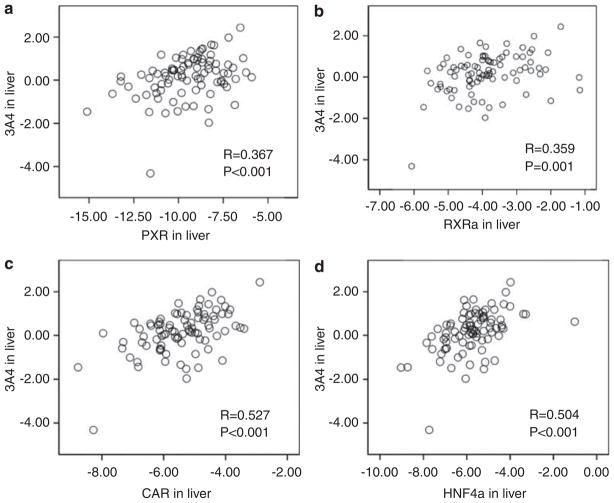
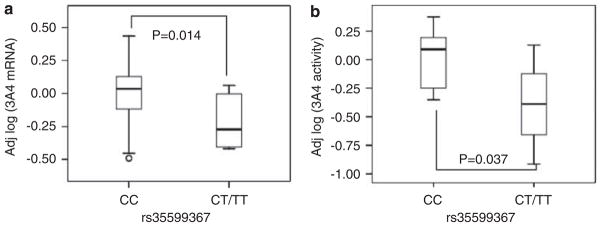
Total CYP3A4 mRNA levels were measured by quantitative RT-PCR in 93 liver samples from cohort 2. Although mRNA levels did not differ between Caucasians and African Americans, females had 1.3-fold higher levels than males (95% confidence interval (CI): 1.00–1.68, two-sided P=0.042) as reported.43 Livers with the main CC genotype of intron 6 SNP had 1.7-fold (95% CI: 1.1–2.8) higher levels than CT and TT carriers combined (t-test, two-sided P=0.028), with no interactions between genotypes and sex. To test the effect of CYP3A4 transcription factors,8,9,44,45 mRNA levels were also measured for pregnane X receptor (PXR, NR1I2), constitutive androstane receptor (CAR, NR1I3), retinoid receptor (RXRa), and hepatocyte nuclear factor (HNF4α1A). CYP3A4 mRNA expression positively correlated with all four transcription factors, as reported8,9,44,45 (Figure 4). After adjusting for sex and transcription factors, intron 6 SNP remained significantly associated with CYP3A4 expression (1.67-fold CC over CT and TT combined, 95%, CI: 1.11–2.46, P=0.014) (Figure 5a), showing that the genotype effect was not confounded by these transcription factors. Moreover, there is no interaction between intron 6 SNP genotype and transcription factors. Intron 6 SNP, sex, and the expression of transcription factors account for 32% of CYP3A4 mRNA variability, whereas intron 6 SNP alone explained 7% of the variability.
Figure 4.
Correlation between mRNA expression of four transcription factors PXR (a), RXRα (b), CAR (c), and HNF4α (d) and CYP3A4 in 93 human livers (cohort 2). CYP3A4 mRNA levels were positively correlated with mRNA levels of each of the four transcription factors, with correlation coefficients R ranging from 0.37 to 0.53 (P≤0.001). Data are in Log10 scale.
Figure 5.
Box plots of CYP3A4 mRNA levels (a) (cohort 2) and enzyme activity (b) (cohort 1) in human liver samples, grouped by intron 6 SNP genotype. The y axis shows adjusted CYP3A4 mRNA levels or enzyme activities. mRNA levels were adjusted for sex and mRNA levels of PXR and RXR transcription factors, whereas enzyme activities were adjusted for age, sex, and inducer exposure.
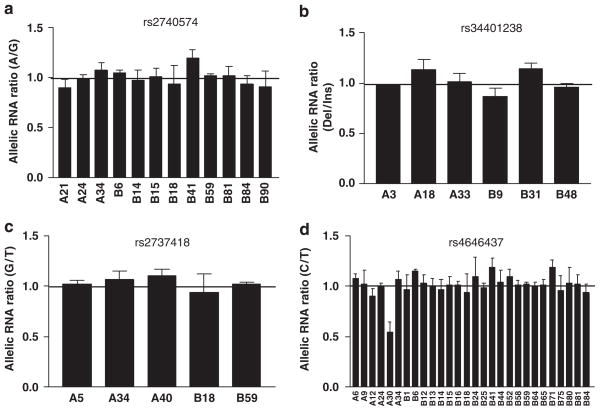
CYP3A4 enzyme activity was measured using a testosterone 6β-hydroxylation assay39 in 23 liver samples from cohort 1 (performed upon collection before storage). Of these 23 liver samples, 4 were intron 6 SNP heterozygous carriers, 3 had been exposed to CYP3A4 inducers (phenobarbital, carbamazepine, nifedipine, and dexamethasone), 10 were female, and 5 from children (age <15 years) (Supplementary Table 4). To control for these covariates, we fitted the data into a multiple linear regression model with age, sex, and inducer exposure as covariates. As shown in Figure 5b, CYP3A4 enzyme activity was 2.5-fold higher for intron 6 SNP CC than CT carriers, after adjusting for age (<15 years child, >15 years adult), sex, and inducers (two-sided P=0.037, 95% CI: 1.1–5.6). Consistent with allelic RNA expression, these results show that intron 6 SNP decreases both CYP3A4 mRNA levels and enzyme activities (protein levels) in vivo. Intron 6 SNP, age, sex, and exposure to inducer account for 45% of CYP3A4 protein variability, whereas intron 6 SNP along explained 12% of the variability. In contrast, CYP3A4*1B, TGT insertion, rs2737418, and rs4646437 had no effects on AEI (Figure 6), total CYP3A4 mRNA level, and enzyme activity (P>0.05).
Figure 6.
Allelic RNA ratios in liver samples heterozygous for SNPs rs2740574 (CYP3A4*1B) (a), rs34401238 (TGT insertion) (b), rs2737418 (c), and rs4646437 (d). None of the samples showed allelic RNA ratios deviating from 1, except A30 in panel (d), which was also heterozygous for intron 6 SNP.
Molecular genetic mechanisms underlying intron 6 SNP regulation
Intron 6 SNP is located 192 bp upstream of exon 7, within several serine/arginine-rich protein-binding motifs. Among these, a predicted SF2/ASF-binding site implicated in splicing scored highest (ESEfinder searching, http://rulai.cshl.edu/tools/ESE/), which is deleted by the T allele (CAGCGTA to CAGTGTA). However, RT-PCR amplification of transcripts from exon 5 to exon 7 of CYP3A4 did not reveal any splice variants, regardless of C or T alleles. Moreover, the allelic RNA ratios measured with marker SNPs located both in the 3′UTR (mostly mature RNA) and within intronic regions (premature hnRNA) in two samples co-heterozygous for intron 6 SNP and 3′UTR marker SNP (A30 and B45) consistently showed the same allele to be less well expressed, considering the LD pattern in CYP3A4. This result argues against altered splicing as primary mechanism, because if intron 6 were to affect pre-mRNA splicing and thereby causing AEI, we would expect to see different allelic RNA ratios obtained with intronic and 3′UTR marker SNPs in these two samples, and allelic mRNA ratios should be in opposite directions. Therefore, the defect appears to reside at an earlier step in hnRNA expression and processing.
The SF2/ASF protein has additional functions beyond splicing, such as regulating translation and stabilizing mRNA.46,47 To test whether SF2/ASF is involved in intron 6 SNP regulation, the expression of SF2/ASF mRNA in liver and small intestine (duodenum) was measured using real-time PCR, to account for tissue-specific effects of intron 6 SNP on CYP3A4 expression. However, similar SF2/ASF expression (cycle threshold were 26.3 and 26.8 for liver and small intestine, respectively) argues against a role for SF2/ASF, but differences in SF2/ASF splice variants in liver and small intestine cannot be ruled out, among other possible mechanisms.
Some intronic regions contain enhancer/attenuator elements that regulate transcription.40,41 To test whether intron 6 contains regulatory elements, a reporter gene assay was developed suitable for testing enhancer regions within the transcribed gene locus, as described.40,41 The CYP3A4-promoter region (~1700 bp, from 60256 to 61990, AF280107) was amplified and cloned into PGL3 reporter gene vector. An intron 6 fragment (~1000 bp, containing intron region only, from 76686 to 77738) harboring either the C or T allele was added upstream of the CYP3A4-promoter fragment (Figure 7a). These plasmids were transfected into HepG2 cells and luciferase activity measured 48 h post-transfection. The intron 6 region did not affect the activity of the CYP3A4 promoter, regardless of the presence of the C or T allele. This result suggests that intron 6 is unlikely to regulate CYP3A4-promoter activity, consistent with a lack of predicted transcription factor-binding sites in the intron 6 region, assessed with Promolign (http://polly.wustl.edu/promolign/main.html).
Intronic regions can further influence RNA levels by regulating transcriptional elongation rate or RNA processing/turnover.48–50 To test this possibility, a minigene was constructed that contained the DNA sequence from intron 4 to intron 7 (~2300 bp, from 75700 to 77994, AF280107), harboring either the C or T allele of intron 6 SNP. After transfection into HepG2 or HEK293 cells, both hnRNA and correctly spliced mature mRNA were detectable, with the level of hnRNA being ~5% of mature RNA. Again, we did not detect any splice variants after either C or T minigene transfection, consistent with liver results. Transcription levels of mature RNA and hnRNA resulting from the minigene carrying the C allele were higher and peaked earlier than the T allele, but inter-transfection variability did not permit accurate quantitation. To compare the transcript levels of minigene constructs with C and T alleles, the two constructs were co-transfected in equal amounts and allelic RNA and plasmid DNA ratios measured in HepG2 or HEK293 cells as described earlier,33,42 at 24, 48, or 96 h post-transfection. To avoid interference from endogenously expressed CYP3A4 RNA, a PCR primer was used matching transcribed pCDNA vector sequence 3′downstream of the minigene for cDNA synthesis. As shown in Figure 7b, after normalization to the plasmid DNA ratios of the transfected minigenes at each time point, allelic RNA ratios (C/T) consistently increased with post-transfection time in HepG2 cells. At 96 h post-transfection, allelic RNA ratios C/T were ~2 (normalized to the plasmid DNA ratios) (Figure 7b), reaching a similar level as allelic RNA ratios observed in human livers. In HEK293 cells, the allelic RNA ratio was only 1.2 C/T at 48 h post-transfection, likely due to the differential regulatory protein expression in HEK293 and HepG2 cells.
Intron 6 SNP (C>T) associates with statin dose requirement
The in vivo effect of intron 6 SNP was assessed by linking it to the titrated dosage of CYP3A4-metabolized statins required for reaching an optimal cholesterol control goal as described. 32 General clinical characteristics of the study population of 273 patients in this observational investigation are shown in Supplementary Table 5. Patients were either on a CYP3A4-statin (atorvastatin n=142, lovastatin n=9, simvastatin n=84) or non-CYP3A4-statin (fluvasatatin, pravastatin, rosuvastatin, total n=38). The majority of subjects were Caucasian (89%), male sex (67%), and on a CYP3A4-statin (86%). Intron 6 SNP was genotyped in all 273 patients. Additional known functional non-synonymous SNPs in CYP3A4 (*17 rs4987161 and *18 rs28371759), although with low frequency, were also genotyped. Moreover, we genotyped the common SNP CYP3A4*1B, previously suggested to be functional,4,12,15 and TGT insertion rs34401238, reported to be active in in vitro reporter gene assays, to evaluate any effects on statin metabolism. Two commonly known functional SNPs in CYP3A5 (*3 rs776746 and *5 rs41303343) were also genotyped (Table 2), because CYP3A5 often shares the same substrates with CYP3A4. Therefore, the variability in the metabolism of CYP3A substrates could result from genetic variability of CYP3A5. Three SNPs (rs2740574, rs776746, and rs1303343) deviated from Hardy–Weinberg equilibrium, because of different allele frequency in Caucasian and Africa American populations, whereas all SNPs followed Hardy–Weinberg equilibrium when analyzed separately for each group (Table 2). Therefore, genotype association was calculated separately for these three SNPs in Caucasians and African Americans, and results were reported only for Caucasians, because the African American group size was insufficient. The allele frequency of intron 6 SNP is ~5%, consistent with that reported in NCBI database (4–8%). Absence of CYP3A4 SNPs *17 and *18 is consistent with reported low-allele frequency.
Table 2.
CYP3A4 polymorphisms genotyped in 273 patients
| SNP | Alleles | Location | All Patients, n = 273
|
Caucasian, n = 243
|
Non-Caucasian, n = 30
|
|||
|---|---|---|---|---|---|---|---|---|
| MAF | HWE P | MAF | HWE P | MAF | HWE P | |||
| rs34401238 | TGT Del/Ins | CYP3A4 enhancer | 0.016 | 0.78 | 0.017 | 0.79 | 0.018 | 0.92 |
| rs2740574 | A>G, *1B | CYP3A4 promoter | 0.105 | 6.06E–07 | 0.054 | 0.71 | 0.350 | 0.77 |
| rs35599367 | C>T | CYP3A4 intron 6 | 0.047 | 0.061 | 0.052 | 0.08 | 0 | NA |
| rs4987161 | T>C, *17 | CYP3A4 exon 7 | 0 | NA | 0 | NA | 0 | NA |
| rs28371759 | T>C, *18 | CYP3A4 exon 10 | 0 | NA | 0 | NA | 0 | NA |
| rs776746 | G>A, *3 | CYP3A5 intron 3 | 0.138 | 2.81E–08 | 0.088 | 0.09 | 0.376 | 0.28 |
| rs1303343 | T Del/Ins, *7 | CYP3A5 exon 11 | 0.012 | 4.53E–06 | 0.008 | 0.95 | 0.055 | 0.76 |
Abbreviations: HWE, Hardy–Weinberg equilibrium; MAF, minor allele frequency; SNP, single nucleotide polymorphism.
A subset of 235 patients was on CYP3A4-metabolized statins (atorvastatin, lovastatin, and simvastatin) with daily doses ranging from 5 to 80 mg. Of these, 22 were intron 6 SNP carrier (20 CT heterozygotes and 2 TT homozygotes). Table 3 shows the clinical characteristics of intron 6 SNP carriers and non-carriers. The stable titrated statin doses were significantly lower in intron 6 SNP carriers than in non-carriers (Mann–Whitney test, two-sided P=0.039), whereas there were no differences in other characteristics including lipid levels before and after statin treatment between these two groups. To further test the strength of association between intron 6 SNP and statin dose requirement, we divided statin doses into three levels: ≤10 mg, 20 mg, and ≥40 mg, and tested the association between intron 6 genotype and dose level using a proportional odds logistic regression model, and adjusting for different statins used (atorvastatin and simvastatin/lovastatin combined) (Table 4), because atorvastatin has higher potency in lowering lipid levels than simvastatin and lovastatin.26,27 After controlling for different statins, carriers of the intron 6 minor T allele were less likely taking higher statin dose with odds ratio of 0.355 (95% CI: 0.16–0.81, two-sided P=0.014, Table 4), as expected from lower hepatic expression of CYP3A4. Considering that the test represents a single hypothesis supported independently by molecular genetic results, and therefore not requiring multiple comparison adjustment, the P-value of 0.0136 is consider significant at α=0.05 level or a false-positive rate <5%. For a further quantitative assessment of statin dose requirement for intron 6 SNP carriers and non-carriers, we used a multiple linear regression model with total cholesterol level before treatment as covariate. The result shows that the stable statin dose for intron 6 T-allele carriers was only a 0.27 fraction compared with non-T carriers (P=0.019) (Table 5). Similar results were obtained when analyzing patients on atorvastatin and simvastatin separately (Table 5). Intron 6 SNP explains 5% of inter-individual variability in stable dose requirement. Therefore, intron 6 SNP is significantly linked to reduced statin dose requirements. These results are consistent with the mRNA level and enzyme activity data shown above, with liver from T-allele carriers having <50% mRNA level or enzyme activity compared with non-T-allele carriers. In contrast, in Caucasian patients, CYP3A4*1B and CYP3A5*3 did not show any significant associations (Table 4), consistent with AEI data. The low frequency of rs34401238 (TGT insertion) and rs1303343 did not permit statistical analysis of these data (Table 4). Moreover, because of the low number of patients receiving the non-CYP3A4 substrate statins, any effect of intron 6 SNP cannot be evaluated in this group.
Table 3.
Characteristics of the study population grouped by intron 6 SNP genotype
| Intron 6 SNP genotypes
|
P-value | ||
|---|---|---|---|
| CC, n = 213 | CT+TT, n = 22 | ||
| Statin dose (mg per day) | 40 (20, 40) | 20 (10, 25) | 0.039* |
| Age (years) | 62±11 | 64±12 | 0.40 |
| Male (%) | 67.6 | 63.6 | 0.71 |
| Caucasian (%) | 87.3 | 100 | 0.09 |
| LDL (mg per 100 ml) | |||
| Before | 106±51 | 91±25 | 0.17 |
| After | 83±33 | 78±31 | 0.52 |
| HDL (mg per 100 ml) | |||
| Before | 33±10 | 35±10 | 0.38 |
| After | 35±10 | 35±9 | 0.93 |
| Triglyceride (mg per 100 ml) | |||
| Before | 263±453 | 202±165 | 0.22 |
| After | 156±109 | 149±73 | 0.71 |
| Cholesterol (mg per 100 ml) | |||
| Before | 187±82 | 166.8±47.5 | 0.11 |
| After | 149±42 | 143.2±33.3 | 0.50 |
| Hypertension (%) | 76 | 82 | 0.51 |
| Diabetes mellitus (%) | 49 | 32 | 0.10 |
| Family history of CAD (%) | 44 | 46 | 0.87 |
| Tobacco use (%) | 54 | 68 | 0.19 |
Abbreviations: CAD, coronary artery disease; HDL, high-density lipoprotein; LDL, low-density lipoprotein; SNP, single nucleotide polymorphism.
Data for statin dose represent median (first quartile, third quartile), and statistics P-value was obtained from Mann–Whitney test. For other continuous variables, data are mean±s.d. and P-values were obtained from t-test. For discrete variables, data represent proportion, and the differences between the two groups were tested by two-sample proportion t-test. Lipid ‘before’ and ‘after’ levels represent the lipid levels at the time of enrollment and after reaching stable statin dose.
P<0.05.
Table 4.
CYP3A4/3A5 genotype distribution and odds ratios for association between higher statin dose requirement and genotype, after adjusting for different statins used
| SNP | Genotype | Drug | No. of patients in each group
|
Odds ratio (95% confidence interval) | P-value | ||
|---|---|---|---|---|---|---|---|
| ≤10 mg | 20 mg | ≥40 mg | |||||
| rs35599367 (CYP3A4, intron 6) | CC | Ator | 32 | 38 | 59 | 1 (Reference) | 0.014* |
| Sim+Lov | 6 | 28 | 50 | ||||
| CT+TT | Ator | 5 | 5 | 2 | 0.36 (0.16–0.81) | ||
| Sim+Lov | 2 | 5 | 2 | ||||
| rs2740574 (CYP3A4, *1B) | CC | Ator | 32 | 31 | 55 | 1 (Reference) | 0.37 |
| Sim+Lov | 7 | 21 | 38 | ||||
| CT+TT | Ator | 3 | 7 | 4 | 0.71 (0.33–1.51) | ||
| Sim+Lov | 0 | 8 | 5 | ||||
| rs34401238 (CYP3A4, TGT ins) | DD | Ator | 37 | 43 | 58 | Not applicable | |
| Sim+Lov | 8 | 31 | 15 | ||||
| DI | Ator | 0 | 0 | 2 | |||
| Sim+Lov | 0 | 2 | 0 | ||||
| rs776746 (CYP3A5, *3) | GG | Ator | 32 | 32 | 48 | 1 (Reference) | 0.17 |
| Sim+Lov | 6 | 27 | 37 | ||||
| GA+AA | Ator | 3 | 6 | 11 | 1.72 (0.80–3.74) | ||
| Sim+Lov | 1 | 2 | 6 | ||||
| rs1303343 (CYP3A5, *7) | DD | Ator | 35 | 37 | 58 | Not applicable | |
| Sim+Lov | 7 | 29 | 43 | ||||
| DI | Ator | 0 | 1 | 2 | |||
| Sim+Lov | 0 | 0 | 0 | ||||
Abbreviations: Ator, atorvastatin; Lov, lovastatin; Sim, simvastatin; SNP, single nucleotide polymorphism.
Data for SNPs rs2740574, rs776746, and rs1303343 are from Caucasian patients only.
P<0.05.
Table 5.
CYP3A4 intron 6 SNP genotype and stable statin dose
| Statins | N | Dose ratio (T carrier/non-T carrier) | 95% confidence interval | P-value |
|---|---|---|---|---|
| CYP3A4 substrates | 235 | 0.27 | 0.19–0.66 | 0.019* |
| Atorvastatin | 142 | 0.22 | 0.14–0.54 | 0.024* |
| Simvastatin | 84 | 0.6 | 0.37–0.97 | 0.042* |
Abbreviations: CYP, cytochrome p450; hnRNA, heteronuclear RNA; SNP, single nucleotide polymorphism.
P<0.05.
Discussion
This study shows that intron 6 SNP rs35599367 is significantly linked to reduced CYP3A4 mRNA expression and enzyme activity in human livers, and importantly, it fully accounts for differences in allelic mRNA expression. As intron 6 SNP is not in substantial LD with any other SNPs, it had escaped detection by association studies using haplotype tag SNPs.23,24 None of the previously reported CYP3A4 SNPs, including promoter *1B,4,12 enhancer TGT insertion,22 enhancer rs2737418,23 and intron 7 SNP rs464643724 had detectable effects on allelic mRNA expression, mRNA level, and enzyme activity, arguing against a contribution of these SNPs to CYP3A4 variability in the liver.
The allele frequency of intron 6 SNP in the examined groups (95% of the samples were from Caucasian) was 5–7%, resulting in 10–13% heterozygosity in this study, consistent with previously reported allele frequencies51 of 0.043, 0.043, and 0.083, for African Americans, Chinese, and Caucasians, respectively.
Common molecular mechanisms for an intronic SNP to alter mRNA levels are to affect transcription, RNA elongation, splicing, or maturation.40,41,48,49 As the allelic ratios were similar for mRNA and hnRNA in livers co-heterozygous for both exonic and intronic marker SNPs, splicing and mRNA turnover are unlikely to account for the different expression. Moreover, CYP3A4 mRNA and hnRNA levels were shown to vary in parallel in human livers,52 arguing for a defect early in transcription and RNA processing. The reporter gene assay results argue against a role in transcriptional regulation as an enhancer element that could reside within the transcribed region. On the other hand, intron 6 SNP could affect the folding of single-stranded DNA or nascent RNA and hence RNA elongation. This mechanism was consistent with the minigene results, showing higher C than T allele expression levels in HepG2 cells (Figure 7b) when only the introns and exons surrounding the intron 6 region were expressed by transfection. In silico DNA or RNA folding analysis (Mfold program) shows that intron 6 SNP changes the folding of single-stranded DNA and RNA (not shown), potentially affecting the binding of regulatory proteins. However, this hypothesis requires further investigation.
The molecular genetics results reported here lead to predictions about dosage requirements for drugs primarily metabolized by CYP3A4. Although the main emphasis in this study is on the molecular genetics of CYP3A4, a first exploratory clinical study was completed by assessing intron 6 SNP effects on stable statin dosage requirements to attain a target lipid control level in CAD patients. Consistent with reduced expression of the minor allele, intron 6 SNP was significantly associated with reduced stable dose requirements of statin drugs that are predominantly metabolized by CYP3A4 (atorvastatin, lovastatin, and simvastatin). As statin doses are titrated to reach a desired LDL, the target can be achieved at lower doses in carriers of the intron 6 SNP T allele conveying reduced metabolism. In support of this notion, pharmacokinetic studies have shown that inhibition of CYP3A4 activity drastically increases plasma concentrations of simvastatin and lovastatin,53,54 suggesting that CYP3A4 activity is a major determinant of plasma concentration of CYP3A4-metabolized statins. Reports on the association between previously identified CYP3A4-promoter SNP (*1B, −392 A>G) with lipid-lowering efficacy and safety of simvastin or atorvastatin treatment has been contradictory.28,29 Our clinical association study of CYP3A4* 1B agrees with the results reported in Fiegenbaum et al.28 and is consistent with the molecular genetics results, showing that this promoter SNP has no effect on hepatic CYP3A4 mRNA expression. However, whether CYP3A4*1B affects CYP3A4 mRNA level in tissues other than liver, for example small intestines affecting statin oral bioavailability, requires further study. Similarly, results on the association between CYP3A5 null allele *3 and CYP3A4-statin substrates and lipid response are contradictory, with increased, decreased, or unchanged results reported,28,55,56 possibly due to small sample size and different experimental designs. Our current results showing no effect of CYP3A5 SNPs on statin dose is consistent with the reports that CYP3A5 does not have a major function in statin metabolism.57 This result also suggests that the association between intron 6 SNP with statin dosage is not confounded by CYP3A5*3 SNP. Therefore, intron 6 SNP is the only frequent CYP3A polymorphism shown to affect statin dosages in our study.
There are several limitations in our observational clinical association study. First, the basal untreated lipid levels were not uniformly available (the lipid levels were obtained at the time of enrollment before treatment was initiated at Ohio State University Medical Center, but treatment with lipid-lowering drugs before entry into the study was not ascertained); therefore, the association between intron 6 SNP and lipid response cannot be evaluated. Second, to estimate the contribution of intron 6 SNP to statin dose requirement, genetic variants in other genes that are also involved in pharmacokinetics processes of statins, for example, ABCB1 and OATP1B1, should be tested, requiring a larger cohort. Third, evaluation of CYP3A4*1B and CYP3A5*3 on statin dose requirement may have been limited by the small sample size, with statistical analysis restricted to Caucasians. Large differences in allele frequency between Caucasian and non-Caucasian for CYP3A4*1B and CYP3A5*3 would therefore require cohorts including more non-Caucasians (Table 2). Finally, as CYP3A4 activity is subject to induction or inhibition by many other drugs including statins themselves,58 the concomitant medications and duration of statin usage should be considered. To fully evaluate the predictive value of intron 6 SNP in statin dose or side effects, a prospective larger cohort study will be needed in the future to test the association between intron 6 SNP and lipid response, dose requirement and rare adverse effects, such as rhabdomyolysis.
CYP3A4 mRNA expression was also significantly affected by the expression of four transcription factors tested (PXR, RXRa, CAR, and HNF4a). The effect of intron 6 SNP was independent of the influence of these trans-acting factors, but it remains to be determined whether and to what extent genetic factors determine the activity of these transcription factors, using an approach similar to the one used for CYP3A4. Also, the contribution of intron 6 SNP to the overall variability of metabolic activity in vivo, and variability of metabolic clearance for each statin, or any other CYP3A4 drug substrate, must be addressed in future studies.
Even though CYP3A4 activity shows considerable inter-individual variability, new drugs are often targeted for metabolism by CYP3A4, to avoid problems arising from null mutations in other drug metabolizing CYP enzymes, such as CYP2D6. The results presented here show that a portion of the variability in CYP3A4 can be accounted for by the intron 6 SNP. The clinical relevance of this finding is shown by the impact of intron 6 SNP on the titrated dose of two statin drugs that depend on CYP3A4 for their elimination. As CYP3A4 is involved in the metabolism of approximately half of all clinically used drugs, the intron 6 SNP is likely to affect dosing requirements, response, and toxicity of numerous drugs, including anticancer agents with narrowly defined dosage regimens. Therefore, CYP3A4 intron 6 SNP has the potential to become a valuable biomarker in clinical practice, and in drug discovery and development.
Supplementary Material
Acknowledgments
This study was supported by a grant from Eli Lilly and a grant from National Institute of Allergy and Infectious Diseases NIH/NIAID (1R21AI074399 to DW). GEC was supported by National Heart, Lung, and Blood Institute NIH/NHLBI career development grant (K23 HL004483).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on The Pharmacogenomics Journal website (http://www.nature.com/tpj)
References
- 1.Danielson PB. The cytochrome P450 superfamily: biochemistry, evolution and drug metabolism in humans. Curr Drug Metab. 2002;3:561–597. doi: 10.2174/1389200023337054. [DOI] [PubMed] [Google Scholar]
- 2.Westlind-Johnsson A, Malmebo S, Johansson A, Otter C, Andersson TB, Johansson I, et al. Comparative analysis of CYP3A expression in human liver suggests only a minor role for CYP3A5 in drug metabolism. Drug Metab Dispos. 2003;31:755–761. doi: 10.1124/dmd.31.6.755. [DOI] [PubMed] [Google Scholar]
- 3.Shimada T, Yamazaki H, Mimura M, Inui Y, Guengerich FP. Inter-individual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharmacol Exp Ther. 1994;270:414–423. [PubMed] [Google Scholar]
- 4.Westlind A, Lofberg L, Tindberg N, Andersson TB, Ingelman-Sundberg M. Interindividual differences in hepatic expression of CYP3A4: relationship to genetic polymorphism in the 5′-upstream regulatory region. Biochem Biophys Res Commun. 1999;259:201–205. doi: 10.1006/bbrc.1999.0752. [DOI] [PubMed] [Google Scholar]
- 5.Lamba JK, Lin YS, Schuetz EG, Thummel KE. Genetic contribution to variable human CYP3A-mediated metabolism. Adv Drug Deliv Rev. 2002;54:1271–1294. doi: 10.1016/s0169-409x(02)00066-2. [DOI] [PubMed] [Google Scholar]
- 6.Ozdemir V, Kalow W, Tang BK, Paterson AD, Walker SE, Endrenyi L, et al. Evaluation of the genetic component of variability in CYP3A4 activity: a repeated drug administration method. Pharmacogenetics. 2000;10:373–388. doi: 10.1097/00008571-200007000-00001. [DOI] [PubMed] [Google Scholar]
- 7.Lamba J, Lamba V, Strom S, Venkataramanan R, Schuetz E. Novel single nucleotide polymorphisms in the promoter and intron 1 of human pregnane X receptor/NR1I2 and their association with CYP3A4 expression. Drug Metab Dispos. 2008;36:169–181. doi: 10.1124/dmd.107.016600. [DOI] [PubMed] [Google Scholar]
- 8.He P, Court MH, Greenblatt DJ, von Moltke LL. Human pregnane X receptor: genetic polymorphisms, alternative mRNA splice variants, and cytochrome P450 3A metabolic activity. J Clin Pharmacol. 2006;46:1356–1369. doi: 10.1177/0091270006292125. [DOI] [PubMed] [Google Scholar]
- 9.Lamba J, Lamba V, Schuetz E. Genetic variants of PXR (NR1I2) and CAR (NR1I3) and their implications in drug metabolism and pharmacogenetics. Curr Drug Metab. 2005;6:369–383. doi: 10.2174/1389200054633880. [DOI] [PubMed] [Google Scholar]
- 10.Takagi S, Nakajima M, Mohri T, Yokoi T. Post-transcriptional regulation of human pregnane X receptor by micro-RNA affects the expression of cytochrome P450 3A4. J Biol Chem. 2008;283:9674–9680. doi: 10.1074/jbc.M709382200. [DOI] [PubMed] [Google Scholar]
- 11.Pan YZ, Gao W, Yu AM. MicroRNAs regulate CYP3A4 expression via direct and indirect targeting. Drug Metab Dispos. 2009;37:2112–2117. doi: 10.1124/dmd.109.027680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rebbeck TR, Jaffe JM, Walker AH, Wein AJ, Malkowicz SB. Modification of clinical presentation of prostate tumors by a novel genetic variant in CYP3A4. J Natl Cancer Inst. 1998;90:1225–1229. doi: 10.1093/jnci/90.16.1225. [DOI] [PubMed] [Google Scholar]
- 13.Wojnowski L, Kamdem LK. Clinical implications of CYP3A polymorphisms. Expert Opin Drug Metab Toxicol. 2006;2:171–182. doi: 10.1517/17425255.2.2.171. [DOI] [PubMed] [Google Scholar]
- 14.Lamba JK, Lin YS, Thummel K, Daly A, Watkins PB, Strom S, et al. Common allelic variants of cytochrome P4503A4 and their prevalence in different populations. Pharmacogenetics. 2002;12:121–132. doi: 10.1097/00008571-200203000-00006. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Martin E, Martinez C, Pizarro RM, Garcia-Gamito FJ, Gullsten H, Raunio H, et al. CYP3A4 variant alleles in white individuals with low CYP3A4 enzyme activity. Clin Pharmacol Ther. 2002;71:196–204. doi: 10.1067/mcp.2002.121371. [DOI] [PubMed] [Google Scholar]
- 16.Amirimani B, Walker AH, Weber BL, Rebbeck TR. RESPONSE: remodification of clinical presentation of prostate tumors by a novel genetic variant in CYP3A4. J Natl Cancer Inst. 1999;91:1588–1590. doi: 10.1093/jnci/91.18.1588. [DOI] [PubMed] [Google Scholar]
- 17.Spurdle AB, Goodwin B, Hodgson E, Hopper JL, Chen X, Purdie DM, et al. The CYP3A4*1B polymorphism has no functional significance and is not associated with risk of breast or ovarian cancer. Pharmacogenetics. 2002;12:355–366. doi: 10.1097/00008571-200207000-00003. [DOI] [PubMed] [Google Scholar]
- 18.Ball SE, Scatina J, Kao J, Ferron GM, Fruncillo R, Mayer P, et al. Population distribution and effects on drug metabolism of a genetic variant in the 5′ promoter region of CYP3A4. Clin Pharmacol Ther. 1999;66:288–294. doi: 10.1016/S0009-9236(99)70037-8. [DOI] [PubMed] [Google Scholar]
- 19.Zeigler-Johnson C, Friebel T, Walker AH, Wang Y, Spangler E, Panossian S, et al. CYP3A4, CYP3A5, and CYP3A43 genotypes and haplotypes in the etiology and severity of prostate cancer. Cancer Res. 2004;64:8461–8467. doi: 10.1158/0008-5472.CAN-04-1651. [DOI] [PubMed] [Google Scholar]
- 20.Miao J, Jin Y, Marunde RL, Kim S, Quinney S, Radovich M, et al. Association of genotypes of the CYP3A cluster with midazolam disposition in vivo. Pharmacogenomics J. 2009;9:319–326. doi: 10.1038/tpj.2009.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27:383–391. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- 22.Matsumura K, Saito T, Takahashi Y, Ozeki T, Kiyotani K, Fujieda M, et al. Identification of a novel polymorphic enhancer of the human CYP3A4 gene. Mol Pharmacol. 2004;65:326–334. doi: 10.1124/mol.65.2.326. [DOI] [PubMed] [Google Scholar]
- 23.Perera MA, Thirumaran RK, Cox NJ, Hanauer S, Das S, Brimer-Cline C, et al. Prediction of CYP3A4 enzyme activity using haplotype tag SNPs in African Americans. Pharmacogenomics J. 2009;9:49–60. doi: 10.1038/tpj.2008.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schirmer M, Rosenberger A, Klein K, Kulle B, Toliat MR, Nurnberg P, et al. Sex-dependent genetic markers of CYP3A4 expression and activity in human liver microsomes. Pharmacogenomics. 2007;8:443–453. doi: 10.2217/14622416.8.5.443. [DOI] [PubMed] [Google Scholar]
- 25.Hirota T, Ieiri I, Takane H, Maegawa S, Hosokawa M, Kobayashi K, et al. Allelic expression imbalance of the human CYP3A4 gene and individual phenotypic status. Hum Mol Genet. 2004;13:2959–2969. doi: 10.1093/hmg/ddh313. [DOI] [PubMed] [Google Scholar]
- 26.Shitara Y, Sugiyama Y. Pharmacokinetic and pharmacodynamic alterations of 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase inhibitors: drug-drug interactions and interindividual differences in transporter and metabolic enzyme functions. Pharmacol Ther. 2006;112:71–105. doi: 10.1016/j.pharmthera.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 27.Jones PH, Davidson MH, Stein EA, Bays HE, McKenney JM, Miller E, et al. Comparison of the efficacy and safety of rosuvastatin versus atorvastatin, simvastatin, and pravastatin across doses (STELLAR* Trial) Am J Cardiol. 2003;92:152–160. doi: 10.1016/s0002-9149(03)00530-7. [DOI] [PubMed] [Google Scholar]
- 28.Fiegenbaum M, da Silveira FR, Van der Sand CR, Van der Sand LC, Ferreira ME, Pires RC, et al. The role of common variants of ABCB1, CYP3A4, and CYP3A5 genes in lipid-lowering efficacy and safety of simvastatin treatment. Clin Pharmacol Ther. 2005;78:551–558. doi: 10.1016/j.clpt.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 29.Kajinami K, Brousseau ME, Ordovas JM, Schaefer EJ. CYP3A4 genotypes and plasma lipoprotein levels before and after treatment with atorvastatin in primary hypercholesterolemia. Am J Cardiol. 2004;93:104–107. doi: 10.1016/j.amjcard.2003.08.078. [DOI] [PubMed] [Google Scholar]
- 30.Gao Y, Zhang LR, Fu Q. CYP3A4*1G polymorphism is associated with lipid-lowering efficacy of atorvastatin but not of simvastatin. Eur J Clin Pharmacol. 2008;64:877–882. doi: 10.1007/s00228-008-0502-x. [DOI] [PubMed] [Google Scholar]
- 31.Becker ML, Visser LE, van Schaik RH, Hofman A, Uitterlinden AG, Stricker BH. Influence of genetic variation in CYP3A4 and ABCB1 on dose decrease or switching during simvastatin and atorvastatin therapy. Pharmacoepidemiol Drug Saf. 2010;19:75–81. doi: 10.1002/pds.1866. [DOI] [PubMed] [Google Scholar]
- 32.Grundy SM, Cleeman JI, Merz CN, Brewer HB, Jr, Clark LT, Hunninghake DB, et al. Implications of recent clinical trials for the National Cholesterol Education Program Adult Treatment Panel III Guidelines. J Am Coll Cardiol. 2004;44:720–732. doi: 10.1016/j.jacc.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 33.Wang D, Johnson AD, Papp AC, Kroetz DL, Sadee W. Multidrug resistance polypeptide 1 (MDR1, ABCB1) variant 3435C>T affects mRNA stability. Pharmacogenet Genomics. 2005;15:693–704. [PubMed] [Google Scholar]
- 34.Pinsonneault J, Nielsen CU, Sadee W. Genetic variants of the human H+/dipeptide transporter PEPT2: analysis of haplotype functions. J Pharmacol Exp Ther. 2004;311:1088–1096. doi: 10.1124/jpet.104.073098. [DOI] [PubMed] [Google Scholar]
- 35.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dai Z, Papp AC, Wang D, Hampel H, Sadee W. Genotyping panel for assessing response to cancer chemotherapy. BMC Med Genomics. 2008;1:24. doi: 10.1186/1755-8794-1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papp AC, Pinsonneault JK, Cooke G, Sadee W. Single nucleotide polymorphism genotyping using allele-specific PCR and fluorescence melting curves. Biotechniques. 2003;34:1068–1072. doi: 10.2144/03345dd03. [DOI] [PubMed] [Google Scholar]
- 38.Leeder JS, Gaedigk R, Marcucci KA, Gaedigk A, Vyhlidal CA, Schindel BP, et al. Variability of CYP3A7 expression in human fetal liver. J Pharmacol Exp Ther. 2005;314:626–635. doi: 10.1124/jpet.105.086504. [DOI] [PubMed] [Google Scholar]
- 39.Kolwankar D, Vuppalanchi R, Ethell B, Jones DR, Wrighton SA, Hall SD, et al. Association between nonalcoholic hepatic steatosis and hepatic cytochrome P-450 3A activity. Clin Gastroenterol Hepatol. 2007;5:388–393. doi: 10.1016/j.cgh.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 40.Giacopelli F, Rosatto N, Divizia MT, Cusano R, Caridi G, Ravazzolo R. The first intron of the human osteopontin gene contains a C/EBP-beta-responsive enhancer. Gene Expr. 2003;11:95–104. doi: 10.3727/000000003108748991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hugo H, Cures A, Suraweera N, Drabsch Y, Purcell D, Mantamadiotis T, et al. Mutations in the MYB intron I regulatory sequence increase transcription in colon cancers. Genes Chromosomes Cancer. 2006;45:1143–1154. doi: 10.1002/gcc.20378. [DOI] [PubMed] [Google Scholar]
- 42.Wang D, Chen H, Momary KM, Cavallari LH, Johnson JA, Sadee W. Regulatory polymorphism in vitamin K epoxide reductase complex subunit 1 (VKORC1) affects gene expression and warfarin dose requirement. Blood. 2008;112:1013–1021. doi: 10.1182/blood-2008-03-144899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolbold R, Klein K, Burk O, Nussler AK, Neuhaus P, Eichelbaum M, et al. Sex is a major determinant of CYP3A4 expression in human liver. Hepatology. 2003;38:978–988. doi: 10.1053/jhep.2003.50393. [DOI] [PubMed] [Google Scholar]
- 44.Vyhlidal CA, Gaedigk R, Leeder JS. Nuclear receptor expression in fetal and pediatric liver: correlation with CYP3A expression. Drug Metab Dispos. 2006;34:131–137. doi: 10.1124/dmd.105.005967. [DOI] [PubMed] [Google Scholar]
- 45.Goodwin B, Redinbo MR, Kliewer SA. Regulation of cyp3a gene transcription by the pregnane × receptor. Annu Rev Pharmacol Toxicol. 2002;42:1–23. doi: 10.1146/annurev.pharmtox.42.111901.111051. [DOI] [PubMed] [Google Scholar]
- 46.Michlewski G, Sanford JR, Caceres JF. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell. 2008;30:179–189. doi: 10.1016/j.molcel.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 47.Lemaire R, Prasad J, Kashima T, Gustafson J, Manley JL, Lafyatis R. Stability of a PKCI-1-related mRNA is controlled by the splicing factor ASF/SF2: a novel function for SR proteins. Genes Dev. 2002;16:594–607. doi: 10.1101/gad.939502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baralle M, Pastor T, Bussani E, Pagani F. Influence of Friedreich ataxia GAA noncoding repeat expansions on pre-mRNA processing. Am J Hum Genet. 2008;83:77–88. doi: 10.1016/j.ajhg.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buratti E, Brindisi A, Pagani F, Baralle FE. Nuclear factor TDP-43 binds to the polymorphic TG repeats in CFTR intron 8 and causes skipping of exon 9: a functional link with disease penetrance. Am J Hum Genet. 2004;74:1322–1325. doi: 10.1086/420978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grabczyk E, Usdin K. The GAA*TTC triplet repeat expanded in Friedreich’s ataxia impedes transcription elongation by T7 RNA polymerase in a length and supercoil dependent manner. Nucleic Acids Res. 2000;28:2815–2822. doi: 10.1093/nar/28.14.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thompson EE, Kuttab-Boulos H, Witonsky D, Yang L, Roe BA, Di Rienzo A. CYP3A variation and the evolution of salt-sensitivity variants. Am J Hum Genet. 2004;75:1059–1069. doi: 10.1086/426406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodriguez-Antona C, Sayi JG, Gustafsson LL, Bertilsson L, Ingelman-Sundberg M. Phenotype-genotype variability in the human CYP3A locus as assessed by the probe drug quinine and analyses of variant CYP3A4 alleles. Biochem Biophys Res Commun. 2005;338:299–305. doi: 10.1016/j.bbrc.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 53.Neuvonen PJ, Kantola T, Kivisto KT. Simvastatin but not pravastatin is very susceptible to interaction with the CYP3A4 inhibitor itraconazole. Clin Pharmacol Ther. 1998;63:332–341. doi: 10.1016/S0009-9236(98)90165-5. [DOI] [PubMed] [Google Scholar]
- 54.Jalava KM, Olkkola KT, Neuvonen PJ. Itraconazole greatly increases plasma concentrations and effects of felodipine. Clin Pharmacol Ther. 1997;61:410–415. doi: 10.1016/S0009-9236(97)90191-0. [DOI] [PubMed] [Google Scholar]
- 55.Willrich MA, Hirata MH, Genvigir FD, Arazi SS, Rebecchi IM, Rodrigues AC, et al. CYP3A53A allele is associated with reduced lowering-lipid response to atorvastatin in individuals with hypercholesterolemia. Clin Chim Acta. 2008;398:15–20. doi: 10.1016/j.cca.2008.07.032. [DOI] [PubMed] [Google Scholar]
- 56.Kivisto KT, Niemi M, Schaeffeler E, Pitkala K, Tilvis R, Fromm MF, et al. Lipid-lowering response to statins is affected by CYP3A5 polymorphism. Pharmacogenetics. 2004;14:523–525. doi: 10.1097/01.fpc.0000114762.78957.a5. [DOI] [PubMed] [Google Scholar]
- 57.Park JE, Kim KB, Bae SK, Moon BS, Liu KH, Shin JG. Contribution of cytochrome P450 3A4 and 3A5 to the metabolism of atorvastatin. Xenobiotica. 2008;38:1240–1251. doi: 10.1080/00498250802334391. [DOI] [PubMed] [Google Scholar]
- 58.Willrich MA, Hirata MH, Hirata RD. Statin regulation of CYP3A4 and CYP3A5 expression. Pharmacogenomics. 2009;10:1017–1024. doi: 10.2217/pgs.09.42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.