Abstract
In the CTSA era there is great interest in aggregating and comparing populations across institutions. These sites likely represent data differently in their clinical data warehouses and other databases. Clinical data warehouses frequently are structured in a generalized way that supports many constituencies. For research, there is a need to transform these heterogeneous data into a shared representation, and to perform categorization and interpretation to optimize the data representation for investigators. We are addressing this need by extending an existing temporal abstraction-based clinical database query system, PROTEMPA. The extended system allows specifying data types of interest in federated databases, extracting the data into a shared representation, transforming it through categorization and interpretation, and loading it into a registry database that can be refreshed. Such a registry’s access control, data representation and query tools can be tailored to the needs of research while keeping local databases as the source of truth.
Background
The CTSA program supports comparative effectiveness research in part through promoting development of consortia of clinical sites with heterogeneous populations [1]. The Atlanta Clinical and Translational Science Institute (ACTSI) has multiple such sites with separate IT departments that manage different electronic health record (EHR) systems. Patient-related research that spans multiple ACTSI sites currently takes place but data integration requires one-off solutions. Other CTSAs face similar challenges. This limits the extent to which CTSAs can support large-scale studies of the real-world impact of clinical interventions.
Data integration is a policy challenge, but the technical obstacles are still great. Most institutions make limited use of standard data representations. They lack repositories of data models and common data elements that document their databases’ contents in computable form. They lack tools for extracting data from unstructured records. Institutions with tools that enable federated data access and integration [2] leverage data more completely for research, quality improvement and operations and for providing evidence-based decision support and health information exchange [3–6]. While support by EHR and data warehouse vendors for these capabilities is an important enabler, seamless access to multiple such systems requires additional infrastructure to bridge them that is not readily available.
To support multi-site clinical research, ACTSI is building a virtual data warehouse that retrieves data from source systems and transforms it into a consistent, comparable and structured view. Its architecture leverages software from the cancer Biomedical Informatics Grid (caBIG®, [2]) and CardioVascular Research Grid (CVRG, http://www.cvrgrid.org) projects. It provides services for common data elements, data models, and terminologies for a broad range of source databases with schemas that may be externally controlled and thus substantially not modifiable. These include clinical data warehouses that may return large numbers of rows in typical queries and have highly generalized schemas that may be substantially different from how researchers conceive of the data.
To put these databases on the grid, we are building an Extract, Transform and Load (ETL) system for creating topic-specific subsets of source databases, or registries, that refresh periodically from sources. The system uses an existing temporal abstraction implementation, PROTEMPA, as the transform part of the ETL process. It adds to PROTEMPA an enhanced data model, support for extracting data from multiple heterogeneous databases and flexible loading of the temporal abstraction output. Below we describe our architecture, our implementation progress, and an application that leverages this system to characterize the impact of co-morbidities on hospital readmissions.
Methods
Temporal Abstraction
PROTEMPA is described in detail in [8, 9]. PROTEMPA uses a temporal abstraction ontology [10] to represent broad classes of data that are stored in clinical databases and mechanisms for deriving or abstracting information from data. Supported broad data classes include constants (atemporal data such as demographics), observations with a timestamp (e.g., laboratory test results), and events with a timestamp or that occur during an interval of time (e.g., medication administration, diagnoses, procedures). Supported abstraction mechanisms include defining categories by specifying an isA relation between a data type and a category. It allows defining trends and states (e.g., rising serum sodium) in sequential data by specifying an abstractedFrom relation between the data and a specification of the state or trend. It supports defining temporal patterns in sequential data by specifying an abstractedFrom relation and the temporal relationship(s) (e.g., before, after) of interest. Categories and patterns are translated into rules that process source data in a rules engine. Filters may be specified to constrain retrieved data (e.g., by year). In the Registry Project, we are using PROTEMPA’s temporal abstraction ontology to specify a shared representation of data and derived information across multiple databases.
Virtual Data Model
We call this temporal abstraction ontology-based shared representation a virtual data model (VDM). VDMs include the features of the temporal abstraction ontology described above. They additionally support specifying properties of data types, e.g., a laboratory test result data type would have normal range and critical flag properties. They support specifying hasA relationships between data types (e.g., a patient has one or more visits). They also allow associating data types and properties with one or more terms from a controlled terminology. The architecture supports deploying multiple general-purpose or topic-specific VDMs that may be used in one or more registries.
The data types defined in VDMs are mapped to source database schemas in an XML document that defines “chains” of joins for each data type and its properties and relationships. SQL SELECT statements are generated from these mappings. A plugin mechanism supports implementing automated SQL generation for alternative database management systems. Separate VDM and mapping specifications allow VDMs to remain schema-agnostic.
Registry Project ETL Process
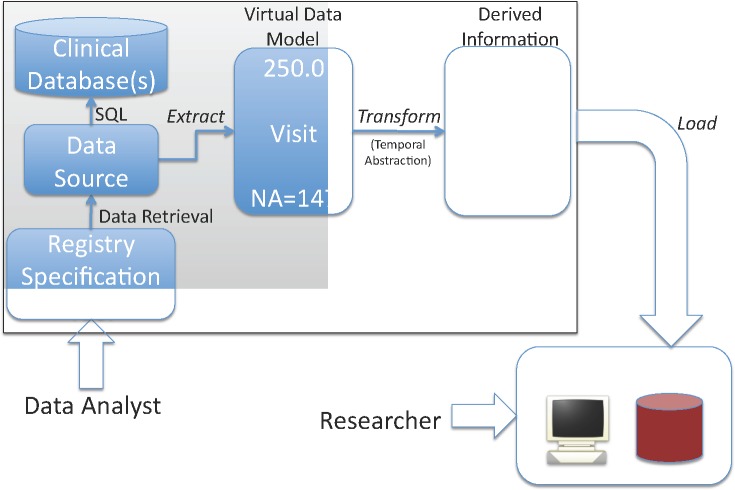
In our ETL process (Figure 1), a data analyst specifies a registry’s contents in terms of data types defined in a VDM. Data modelers create VDMs and define mappings from the VDMs’ data types to the source databases’ schemas as described above. The system generates SQL SELECT statements from the mappings that are defined for the selected data types, executes queries, and uses the mappings to extract the data into the form specified in the VDMs. Rules transform the data into derived information using temporal abstraction (see above). The system then loads the data and derived information into the open source i2b2 system (http://www.i2b2.org, [7]). For registry refreshes, the architecture provides interfaces that allow executing source system queries to retrieve only data added since a certain timestamp.
Figure 1.
Registry Project ETL Process
Loading into i2b2
We use version 1.5 of i2b2. It provides an easy-to-use web user interface for researchers to query clinical and research databases. It provides web service interfaces that we can leverage to load data and derived information from PROTEMPA. It supports defining taxonomies that populate its user interface’s pick lists for query. We have implemented automated loading of PROTEMPA output into i2b2. We also have implemented automated population of i2b2’s query pick lists using the data types that are selected in a registry’s specification. VDMs support this because the relationships between data types in VDMs (e.g., abstractedFrom) define a graph that allows representation of derived data types as “parents” of other derived data type and source data type “children.” Thus, i2b2 effectively serves as a data and metadata cache that investigators query, while source databases remain the source of truth.
Security and Access Control
Security and access control is provided at two levels. In the data extract and load phases (Figure 1), data is de-identified as required and encrypted for transfer over the network using data source-level access control. At the registry data access layer, we leverage i2b2’s user authentication and access control mechanisms. The architecture supports leveraging caGrid security infrastructure for federated authentication and authorization [2] and centralized policy management in the future.
Source Databases
Initial work has focused on two source systems. The Emory Healthcare Clinical Data Warehouse (CDW) is a home-grown Oracle (Oracle Corp., Redwood Shores, CA) 10g database that contains nearly 2 TB of clinical and administrative data from Emory’s hospitals and clinics. It contains over 90% of the data in Emory’s Cerner (Cerner Corp., Kansas City, MO) Millennium-based EHR. The Grady Health System Diabetes Patient Tracking System (DPTS) also is an Oracle database and contains both manually curated and electronically loaded data from Grady diabetes clinics. The DPTS includes diagnoses, procedures, laboratory test results, billing codes, risk factors and prognostic information.
Initial Registries
We are working with the Emory Healthcare Office of Quality and Emory and Grady researchers in diabetes and cardiovascular disease to develop an initial set of registries that can answer the following questions:
What diagnostic information in diagnosis codes, medication history, laboratory test results, procedure history, observations and free-text documents can contribute to predictive models of adverse outcomes such as unplanned re-hospitalizations within 30 days?
What center and/or ethic differences exist in cardiac catheterization patients with respect to diabetes and cardiovascular disease, e.g., frequency of cardiac interventions following catheterization after X length of time, changes in creatinine over time adjusted for co-morbidities, age, gender.
We are creating two registries to support these kinds of questions. The first, a co-morbidities registry, is extracted from the Emory CDW. It represents encounter, discharge diagnosis and medication, procedure and laboratory test result data. It represents 30-day readmissions (sequential hospital encounters within 30 days) as derived information. It includes multiple disease-specific classes of diagnoses (e.g., Cancer, Heart Failure, Diabetes) and medications (e.g., Diabetes medication). The co-morbidities registry has several goals. It aims to allow assessing levels of evidence for specific diagnoses as found in various data. It also aims to leverage the temporal sequence of events and observations to create models of disease severity and risk. This registry is partially implemented as described in Results.
The second diabetes/cardiovascular registry is extracted from the Emory CDW and the Grady DPTS. It is in planning and early implementation. It will contain encounter, co-morbidity, diabetes and cardiovascular disease medications, cardiac catheterization laboratory reports, and a subset of laboratory test results that are relevant to diabetes and cardiovascular disease. It will include derived medication and disease categories, and derived interpretations of laboratory test results.
Results
We have completed an initial round of development. The software is implemented in Java. VDMs are managed and deployed using the client-server version of the Protégé ontology editor (http://protege.stanford.edu) and its frames database format. The software is deployed on Linux virtual machines that are hosted in Emory Healthcare’s network environment. A simple command line Java program invokes the software to extract source data, transform and load it. We are loading data into Excel spreadsheets while the i2b2 integration is being implemented. These spreadsheets contain pivot table definitions that allow for data visualization and exploration including drill-down and filter. The spreadsheets contain the same data and derived information that we expect to load into i2b2. They have been useful for showing data to stakeholders and collecting requirements while we implement the full version of the software.
Co-morbidities Virtual Data Model
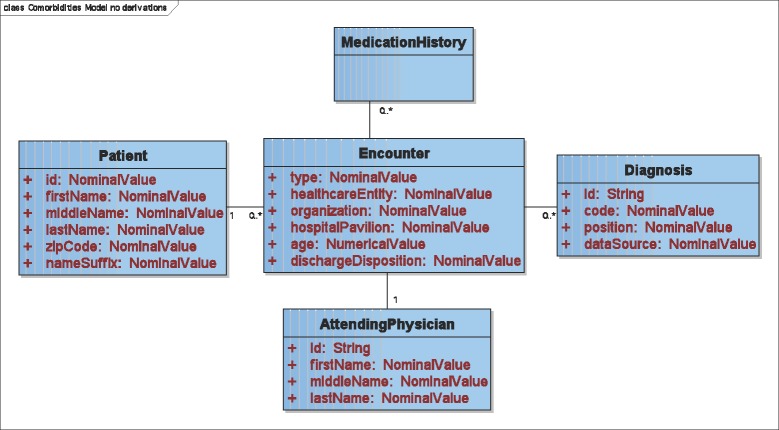
Our initial VDM for the co-morbidities registry (portions shown in Figure 2 and Figure 3) contains Patient, Encounter, AttendingPhysician, Diagnosis, Procedure, VitalSign and MedicationHistory data types, which are extracted from source data. Some of these VDM data types have properties. For example, Encounter has a dischargeDisposition property, and Diagnosis has a position property that encodes whether it is primary or secondary. Some data types have 1:N relationships to each other (e.g., Patients may have one or more Encounters). These data types, properties and relationships are mapped to the Emory CDW’s schema using the mechanism described above. An Oracle 10g SQL generation plugin that we have implemented generates queries. These queries are comparable to what our database personnel would have hand-coded were they retrieving the same data manually.
Figure 2.
UML diagram of the co-morbidities virtual data model’s data types (Procedure and VitalSign not shown).
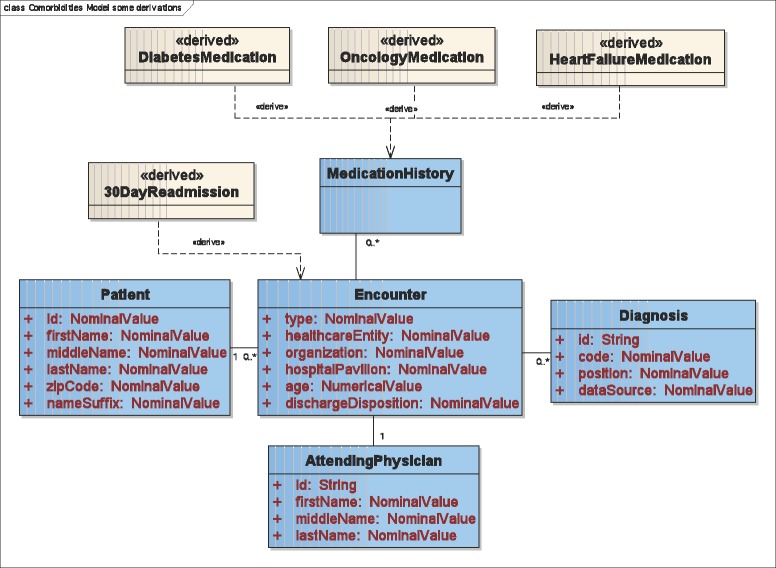
Figure 3.
UML diagram showing classes of medications and 30-day readmissions represented as derived data types.
The model also defines derived data types (straw-colored classes in Figure 3) that are computed. A 30DayReadmission data type (Figure 3) is derived from Encounter and is defined as two sequential encounters within 30 days of each other. Three categories of medications are specified (Figure 3) that define groupings of drugs in a medication history. Not shown are nine categories of ICD-9 discharge diagnosis codes such as DiabetesDiagnosis and HeartFailureDiagnosis.
Co-morbidities Data ETL
The software extracted all inpatient hospitalizations at Emory facilities in FY2009 and transformed the data using the temporal abstractions and categories that we defined in the co-morbidities VDM. It loaded the resulting categories, admission and discharge dates, patient demographics, and encounter location, discharge disposition, attending physician, procedure and vital sign information into Excel. Pivot tables allowed drill-down from categories to data and through Emory clinical sites from hospital to unit. A need to support temporal data exploration with drill down and filter while visualizing the sequence of hospital visits emerged from this analysis as a way to develop hypotheses about what variables might contribute to a predictive model of re-admissions.
Discussion
We expect that the Registry Project ETL process, when fully implemented, will enable creation of a broad range of databases for research across ACTSI that are associated with metadata describing their contents. For access to subsets of large data warehouses that contain EHR data, we expect that registry databases will be substantially easier for researchers to use and queries will be much more responsive than if we implemented direct federated query of source systems. Queries of such systems easily could return millions of rows. Executing federated queries during registry creation allows these potentially long queries to be executed in the background. This architecture also supports access to databases and data warehouses with broad constituencies, which necessitates more complex data access and query solutions. We expect that the smaller size of the registry databases will make federated query of their contents within the ACTSI virtual data warehouse feasible.
The registry project’s VDMs can represent data substantially differently from how it is represented in the source systems, including allowing representation of information that must be computed from the source data. This is substantially different from caBIG® tools, which provide support for mapping databases to object-oriented domain models but allow few data transformations. While such transformations could be implemented as stored procedures and views in the database layer, we expect that this solution would be comparatively difficult to maintain and less reusable as any implementation would be tied to the syntax of the source databases. We expect that the capability to perform such transformations in the software layer would be a valuable addition to caBIG® tooling for accessing clinical databases over the Grid that are similar to ours.
Leveraging i2b2 allows us to use its researcher-oriented user interface, data retrieval and security features. Though it is likely that i2b2 would require substantial modification to leverage our VDMs and data derivation mechanisms directly, we can precompute derived information prior to import into i2b2 and transform the VDMs into a taxonomy structure as described above. Some relationships between data types are lost upon import because i2b2 does not support them, e.g., a hospitalization’s primary diagnosis and discharge disposition are presented to users as independent data points. Supporting use cases that require specific relationships to be retained may necessitate the development of additional user interface options.
Achieving broad use of registries created in this fashion will require testing our VDM and mapping software with a variety of databases and implementing additional features to the SQL generation mechanism as needed. Also, we will develop tools for curating, browsing and querying VDMs including web sites and web services. Our future plans include building cancer registries for brain tumor and lymphoma research that will involve loading data into an existing caBIG® Silver-level compatible database at Emory. We expect to achieve compatibility with caBIG® by creating virtual data models that correspond to the target database’s caBIG® domain analysis model.
We are developing enhanced security and natural language processing mechanisms for future inclusion in the project. We are developing XACML (eXtensible Access Control Markup Language, http://www.oasis-open.org/committees/xacml)-based policy management that allows centrally managed policies while retaining local control over data access. We are implementing text de-identification based on HIDE [11], which combines rule-based methods and conditional random field-based entity recognition. We also are developing methods for concept extraction from text that extend cTAKES (Clinical Text Analysis and Knowledge Extraction System, http://ohnlp.sourceforge.net/cTAKES) with rule-based natural language processing and conditional random field-based machine learning. We will implement concept extraction as an additional kind of data derivation mechanism in PROTEMPA, thus allowing registry specifications to combine concepts extracted from text with retrieval of structured data from source databases.
Conclusion
The ACTSI Registry Project architecture allows subsets of large general-purpose databases to be extracted, transformed and loaded into a common schema that is substantially different from the source systems’ representations. We expect this to enable more intuitive query systems for direct use by researchers and to facilitate integration of data across systems with markedly different data representations.
Acknowledgments
This work was supported in part by PHS Grant UL1 RR025008, KL2 RR025009 and TL1 RR025010 from the CTSA program, NIH, NCRR; NHLBI grant R24 HL085343; and M01 RR-00039 from the GCRC program, NIH, NCRR.
References
- 1.Selker HP, Strom BL, Ford DE, Meltzer DO, Pauker SG, Pincus HA, et al. White paper on CTSA consortium role in facilitating comparative effectiveness research: September 23, 2009 CTSA consortium strategic goal committee on comparative effectiveness research. Clin Transl Sci. 2010 Feb;3(1):29–37. doi: 10.1111/j.1752-8062.2010.00177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oster S, Langella S, Hastings S, Ervin D, Madduri R, Phillips J, et al. caGrid 1.0: an enterprise Grid infrastructure for biomedical research. J Am Med Inform Assoc. 2008;15(2):138–49. doi: 10.1197/jamia.M2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chute CG, Beck SA, Fisk TB, Mohr DN. The Enterprise Data Trust at Mayo Clinic: a semantically integrated warehouse of biomedical data. J Am Med Inform Assoc. 2010 Mar-Apr;17(2):131–5. doi: 10.1136/jamia.2009.002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradshaw RL, Matney S, Livne OE, Bray BE, Mitchell JA, Narus SP. Architecture of a federated query engine for heterogeneous resources. AMIA Annu Symp Proc. 2009;2009:70–4. [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson PD, Tu SW, Musen MA, Purves I. A virtual medical record for guideline-based decision support. Proc AMIA Symp. 2001:294–8. [PMC free article] [PubMed] [Google Scholar]
- 6.German E, Leibowitz A, Shahar Y. An architecture for linking medical decision-support applications to clinical databases and its evaluation. J Biomed Inform. 2009 Apr;42(2):203–18. doi: 10.1016/j.jbi.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Murphy SN, Weber G, Mendis M, Gainer V, Chueh HC, Churchill S, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2) J Am Med Inform Assoc. 2010 Mar 1;17(2):124–30. doi: 10.1136/jamia.2009.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Post AR, Sovarel AN, Harrison JH. Abstraction-based temporal data retrieval for a Clinical Data Repository. AMIA Annu Symp Proc. 2007:603–7. [PMC free article] [PubMed] [Google Scholar]
- 9.Post AR, Harrison JH., Jr PROTEMPA: A Method for Specifying and Identifying Temporal Sequences in Retrospective Data for Patient Selection. J Am Med Inform Assoc. 2007 Jun 6;(14):674–83. doi: 10.1197/jamia.M2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shahar Y. A framework for knowledge-based temporal abstraction. Artif Intell. 1997;90:79–133. doi: 10.1016/0933-3657(95)00036-4. [DOI] [PubMed] [Google Scholar]
- 11.Gardner J, Xiong L. An integrated framework for de-identifying unstructured medical data. Data & Knowledge Engineering. 2009 Dec;68(12):1441–51. [Google Scholar]