Abstract
Within large sequence repositories such as GenBank there is a wealth of metadata providing contextual information that may enhance search and retrieval of relevant sequences for a range of subsequent analyses. One challenge is the use of free-text in these metadata fields where approaches are needed to extract, structure, and encode essential information. The goal of the present study was to explore the feasibility of using a combination of existing resources for annotating unstructured GenBank metadata, initially focusing on the “host” and “isolation_source” fields. This paper summarizes early results for 10 host organisms that include a characterization of associated isolation sources with respect to biomedical ontologies and semantic types. The findings from this preliminary study provide insights to the rich amount of information captured within these unstructured metadata, guidance for addressing the challenges and issues encountered, and highlight the potential value for enriching comparative biological studies towards improving human health.
INTRODUCTION
The availability of molecular sequence data for a broad range of organisms in centralized resources such as GenBank presents great opportunities for advancing biological discoveries1. Given the exponential growth of such repositories, there is an increasing need to organize information within metadata fields in order to facilitate the identification and retrieval of relevant sequences for biological and biomedical studies.
Each entry in GenBank is associated with a detailed set of information about a sequence including a description, scientific name of the source organism, bibliographic references, and a table of features2. This “Feature Table” provides contextual information through a series of biological annotations for each sequence. Collectively, these metadata fields represent both structured and unstructured data. For example, “organism” contains the formal scientific name for the source organism and can be considered a structured field since it is organized according to the NCBI Taxonomy3. There are also numerous unstructured (free-text) fields such as “host” and “isolation_source” in the Feature Table, which are respectively defined as “natural (as opposed to laboratory) host to the organism from which sequenced molecule was obtained” and “describes the physical, environmental and/or local geographical source of the biological sample from which the sequence was derived”4.
There have been some efforts for identifying and standardizing key terms in such free-text fields. Towards the creation of Habitat-Lite for use in relevant specifications for habitat information, the isolation_source field in GenBank was examined5,6. The approaches used revealed a variety of information in this field with a majority of values falling into the broad “organism-associated” category where further work is needed to extract more specific information such as organism and anatomy. Another recent study explored the use of existing biomedical ontologies and annotation services available through the National Center for Biomedical Ontology (NCBO) for identifying anatomical sources in the GenBank isolation_source and note fields for ten domesticated mammalian species towards enabling comparative microbiome hypotheses7. Other studies and resources further highlight the value of capturing these contextual data in a structured format8,9.
METHODS & RESULTS
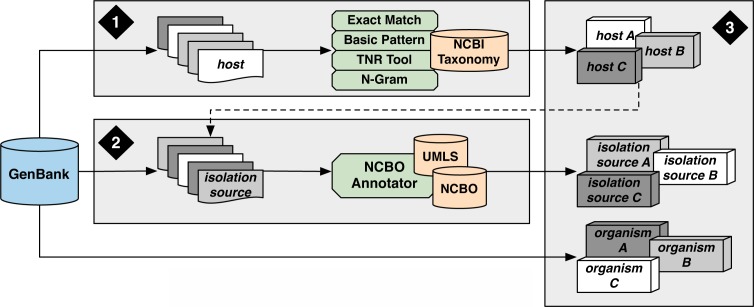
Building upon the aforementioned previous work, the goal of this feasibility study was to explore and develop approaches for annotating information within the unstructured “host” and “isolation_source” metadata in GenBank. Using a local GenBank database (Release 175), the following approach was followed (Figure 1): (1) identify and map host organisms to the NCBI Taxonomy, (2) annotate and characterize information in the isolation_source field using the NCBO BioPortal and UMLS Metathesaurus, and (3) describe how the structured host, isolation_source, and organism fields might be combined to enable host-oriented or cross-species studies.
Figure 1:
Overview of Methods
1. Identifying and Merging Organism Names in Host Metadata
All host values were extracted from the local GenBank database (n=1,350,040) and a list of unique values along with frequency counts was generated (n=28,907). In addition to including organism names (scientific, common, and synonyms) as anticipated, a manual review of this list revealed other types of information and varying formats. Given this, a combination of approaches was initially explored for identifying and mapping organism information to the NCBI Taxonomy (downloaded on June 23, 2010) that would facilitate the merging of values through the Taxonomy ID:
- Exact Match – find a completely exact match for the host value in the NCBI Taxonomy database. For example, the following map to ID 9796:horse Equus caballusequine domestic horse
- Basic Pattern Match – use basic rules to find organism names relative to specific delimiters (e.g., ‘;’, ‘,’, and ‘(’). For example, the following map to ID 9606:Homo sapiens (Human)Homo sapiens; gender M; age 55Homo sapiens, juvenile bloodhuman, South East Asia
- Taxonomic Name Recognition (TNR) Tool10 – this named entity recognition approach identifies taxonomic names in text and maps to universal identifiers if possible, which may link to Taxonomy IDs. Examples of those with no links:Poa rigidifoliaSerianthes calycina
- N-Gram – each host value is viewed as a sequence of n words and an attempt is made to find a match for n-grams from size 1 to n. The following examples map to ID 9913:bovine fetusHolstein dairy cowAustralian feedlot cattlecattle with eperythrozoonosis
These four approaches were applied sequentially where each was meant to build upon the results of the previous one (while recognizing that the subsequent approaches may introduce noise or inaccuracies). For the 28,907 unique host values, organism names were identified for 40.5% of the values with exact matching, 60.5% with basic pattern matching, 87.9% with the TNR tool, and 94.97% with the n-gram approach. Given that a portion of the organism names identified by the TNR tool could not be mapped to the NCBI Taxonomy, a final total of 75% of the values could be mapped to Taxonomy IDs. These values were subsequently merged according to these identifiers in order to identify a more comprehensive set of sequences for a given host organism. For example, the single value Homo sapiens is associated with 504,967 sequences; through the mapping process, there were found to be over 600 different host values that mapped to Homo sapiens resulting in a total of 545,470 sequences when merged.
For the purposes of this study, the top 10 host organisms ranked at the species-level were considered for further examination (thus excluding those ranked as genus, family, or subspecies as defined in the NCBI Taxonomy). Table 1A lists each organism along with the total number of host values (roughly equivalent to the number of GenBank entries) and number of unique host values (after manual review and removal of false positives).
Table 1:
Top 10 Host Organisms with Frequencies for Host (A), Isolation Source (B), and Organism (C).
| A. HOST | B. ISOLATION SOURCE | C. ORGANISM | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Taxonomy ID | Scientific Name | GenBank Common Name | Total Values | Unique Values | Total Values | Unique Value | Ontologies | Semantic Types | Total Values | Unique Values |
| 9606 | Homo sapiens | human | 545470 | 609 | 337437 | 3628 | 123 | 83 | 545357 | 19645 |
| 10116 | Rattus norvegicus | Norway rat | 156894 | 19 | 77399 | 30 | 71 | 20 | 80888 | 184 |
| 10118 | Rattus sp. | 76008 | 7 | 75933 | 4 | 20 | 5 | 75963 | 13 | |
| 9805 | Diceros bicornis | black rhinoceros | 49500 | 3 | 49494 | 3 | 12 | 2 | 49499 | 7 |
| 9796 | Equus caballus | horse | 27582 | 38 | 4338 | 59 | 71 | 32 | 27575 | 323 |
| 9792 | Equus grevyi | Grevy’s zebra | 23280 | 3 | 23270 | 4 | 14 | 4 | 23276 | 4 |
| 10090 | Mus musculus | house mouse | 21088 | 33 | 14710 | 44 | 80 | 26 | 21071 | 172 |
| 9913 | Bos taurus | cattle | 19540 | 78 | 10454 | 191 | 96 | 47 | 19462 | 884 |
| 9891 | Antilocapra americana | pronghorn | 12951 | 1 | 12950 | 1 | 12 | 2 | 12951 | 2 |
| 9844 | Lama glama | llama | 11582 | 2 | 11579 | 3 | 31 | 5 | 11582 | 7 |
2. Analyzing, Characterizing, and Merging Information in Isolation Source Metadata
A preliminary analysis of all isolation_source values in GenBank (n=1,837,706) consisting of 35,980 unique values revealed more complex semantics and syntax than the host field. Given this, a different approach was used that involved focusing on host-specific sets of values. The rationale for this was that these subsets may be used to develop a generalizable approach that could then be applied for all values.
For the 10 host organisms identified in the first step of this study, isolation_source values were extracted and each set of unique host-specific values was annotated using the NCBO Annotator Web service11. The default settings for this service were used for most parameters with the exception of “longestValue” (set to true), “mappingTypes” (set to inter-cui), and “format” (set to text). Each annotation includes a score, source ontology ID (e.g., 42789 = SNOMED Clinical Terms), concept ID, preferred name, synonym(s), and semantic type(s).
As an initial pass, annotations with a score of less than 10 were removed and the remaining annotations underwent further semantic analysis that involved summarizing the source ontologies (from NCBO BioPortal12 or UMLS Metathesaurus13) and semantic types (from the UMLS Semantic Network). Since a given value may map to multiple concepts and semantic types in one or multiple ontologies, a unique list of ontologies and semantic types was identified for each value and the total counts were calculated by summarizing across all values. For each host organism, Table 1B presents the total number of isolation_source values, number of unique isolation_source values, number of source ontologies, and number of semantic types. As these results demonstrate, there is variation across host organisms, which highlights the potential differences in the content and format of isolation_source values.
When combining results across the host organisms, the top 10 ontologies (out of a total of 124) were found to be: NCI Thesaurus, SNOMED CT, LOINC, Galen, BRENDA Tissue/Enzyme Source, MeSH, Uber Anatomy Ontology, Foundational Model of Anatomy, Mouse Adult Gross Anatomy, and RadLex. Other top host-specific ontologies included: HL7, ICNP, and Environment Ontology.
Across the 10 host organisms, the top 5 UMLS semantic types (out of a total of 88 and excluding “NCBO BioPortal Concept”) were: Qualitative Concept, Body Substance, Disease or Syndrome, Patient or Disabled Group, and Body Part. The following two examples depict multiple semantic types within a given isolation_source value:
lymph node of patient with sarcoidosis
Body Part = “lymph node”
Patient or Disabled Group = “patient”
Qualitative Concept = “with”
Disease or Syndrome = “sarcoidosis”
milk from cow suffering from mastitis
Body Substance = “milk”
Mammal = “cow”
Qualitative Concept = “from”
Disease or Syndrome = “mastitis”
Semantic types were used to further categorize the host-specific isolation_source values. For three of the top semantic types (Body Part, Body Substance, and Disease or Syndrome), the preferred names associated with each annotation were extracted (regardless of source ontology) and used to generate a preliminary ranked list of values in each category (recognizing that future efforts should involve use of the concept IDs and linkages between ontologies to generate such lists). With this strategy, the following are example isolation_source values that map to the single preferred name of “plasma” (semantic type = Body Substance) for Homo sapiens:
human serum or plasma
plasma from a 42-year old male
host plasma
plasma from Hodgkin lymphoma patient
plasma from bone marrow recipient
Table 2 (shaded rows) highlights the total number of isolation_source values for the three semantic types (along with the proportion of all isolation_source values) and the number of unique preferred names for five of the host organisms.
Table 2:
Top 5 Body Parts, Body Substances, Diseases or Syndromes, and Organisms for Selected Host Organisms.
| Homo sapiens | Rattus norvegicus | Equus caballus | Mus musculus | Bos taurus | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| ISOLATION SOURCE | Body Part | Total: 34950 (0.104) | Total: 132 (0.002) | Total: 71 (0.016) | Total: 3303 (0.225) | Total: 1056 (0.101) | |||||
| Unique: 94 | Unique: 15 | Unique: 8 | Unique: 13 | Unique: 20 | |||||||
|
| |||||||||||
| esophagus | 0.212 | lung | 0.432 | brain | 0.775 | cecum | 0.540 | rumen | 0.729 | ||
| external auditory canal | 0.143 | rat colon | 0.326 | vagina | 0.113 | ileum | 0.449 | teat | 0.050 | ||
| hoof | 0.028 | spleen | 0.002 | omasum | 0.028 | ||||||
| umbilicus | 0.140 | ileum | 0.068 | gastric mucosa | 0.028 | lung | 0.002 | brain | 0.031 | ||
| manubrium | 0.128 | caecum | 0.030 | intestinal | 0.002 | nasal | 0.026 | ||||
| glabella | 0.123 | kidney | 0.023 | uterus | 0.014 | ||||||
|
| |||||||||||
| Body Substance | Total: 44991 (0.133) | Total: 32209 (0.416) | Total: 3958 (0.912) | Total: 444 (0.030) | Total: 6084 (0.582) | ||||||
| Unique: 59 | Unique: 3 | Unique: 10 | Unique: 3 | Unique: 11 | |||||||
|
| |||||||||||
| saliva | 0.317 | feces | >99.999 | feces | 0.959 | feces | 0.980 | feces | 0.947 | ||
| feces | 0.259 | blood | <0.001 | semen | 0.022 | blood | 0.011 | blood | 0.021 | ||
| plasma | 0.166 | isolate | <0.001 | blood | 0.014 | lysate | 0.009 | milk | 0.014 | ||
| serum | 0.142 | peripheral blood | 0.003 | serum | 0.006 | ||||||
| blood | 0.027 | serum | <0.001 | exudate | 0.004 | ||||||
|
| |||||||||||
| Disease or Syndrome | Total: 3363 (0.010) | Total: 0 | Total: 14 (0.003) | Total: 983 (0.067) | Total: 445 (0.430) | ||||||
| Unique: 137 | Unique: 0 | Unique: 4 | Unique: 1 | Unique: 9 | |||||||
|
| |||||||||||
| subgingival plaque | 0.161 | sarcoid | 0.714 | Salmonella | 1.000 | interdigital necrobacillosis | 0.892 | ||||
| encephalitis | 0.143 | ||||||||||
| chronic hepatitis b | 0.140 | valvular endocarditis | 0.071 | mastitis | 0.070 | ||||||
| dermatitis | 0.020 | ||||||||||
| pneumococcal infection | 0.121 | endometritis | 0.071 | septicemia | 0.004 | ||||||
| warts | 0.004 | ||||||||||
| liver abscess | 0.050 | ||||||||||
| acute hepatitis b | 0.049 | ||||||||||
|
| |||||||||||
| ORGANISM | uncultured bacterium (0.589) | uncultured bacterium (0.986) | uncultured Neocallimastigales (0.897) | uncultured bacterium (0.957) | uncultured Neocallimastigales (0.280) | ||||||
| Human immunodeficiency virus 1 (0.112) | uncultured Escherichia sp. (0.002) | Equine infectious anemia virus (0.022) | Lactobacillus Reuteri (0.005) | uncultured bacterium (0.277) | |||||||
| Hepatitis C virus (0.027) | Seoul virus (0.002) | Burkholderia mallei PRL-2 (0.010) | uncultured Clostridiales Bacterium (0.005) | Rabies virus (0.055) | |||||||
| uncultured organism (0.020) | Lactobacillus reuteri (0.001) | Burkholderia mallei GB8 horse 4 (0.007) | Lymphocytic choriomeningitis virus (0.005) | uncultured rumen archaeon (0.036) | |||||||
| Hepatitis B virus (0.018) | uncultured Bacillus sp. (0.001) | Equine arteritis virus (0.005) | Hepatitis C virus (0.004) | uncultured rumen bacterium (0.035) | |||||||
3. Enabling Comparative Biology Inquiries
The ability to extract, structure, and encode contextual information captured within the host and isolation_source fields in GenBank may be valuable for a range of subsequent uses. As suggested in a previous study7, the organization of data within GenBank could potentially facilitate initiatives like the Human Microbiome Project (study variation in the human microbiome and its impact on disease) or comparative microbiome studies (compare microbiomes in similar environments across species). An essential component of such studies is the identification of relevant sequences for a given host organism and a better understanding of the context or environment in which they were collected.
As shown earlier, the identification of organism names within the host field and their subsequent mapping to Taxonomy IDs can enhance the number of relevant sequences for a given host (e.g., there was almost a 10% increase for Homo sapiens). Based on the enhanced sets of host-specific sequences, Table 2 depicts the top 5 body parts, body substances, diseases or syndromes, and organisms associated with five of the hosts based on the isolation_source field (along with the proportion of total values for the host-specific semantic type). With respect to microbiome studies, a potential use of this contextual information is enabling comparisons between organism sequences obtained from different body parts of the same host organism (e.g., “cecum” versus “ileum” for Mus musculus).
In addition to the aforementioned host-specific implications, the organization of unstructured GenBank fields may ultimately be used to enrich and facilitate cross-species studies by enabling context-specific questions such as: (1) For organism X, what are possible host organisms to study; (2) For body substance Y, what host organisms have been sources; or, (3) across a specified set of host organisms, how do the isolation sources and organisms compare? For example, as shown in Table 2, “feces” and “blood” are both among the top 5 body substances across the five host organisms.
DISCUSSION
Through this feasibility study, we have gained valuable insights to the richness and variation of information captured within two unstructured metadata fields in GenBank (host and isolation_source). The methods and results presented in this paper represent early attempts to structure this information towards enriching subsequent analyses. Next steps include performing extensive evaluations, addressing the various challenges and issues encountered, refining the techniques accordingly towards a more generalized approach, and demonstrating the potential impact on biological and biomedical studies.
The analysis of GenBank host metadata involved using four consecutive approaches for identifying organism names and mapping those names to NCBI Taxonomy IDs. While organism names were identified in 97% of the values (and 75% could be mapped to the NCBI Taxonomy), host organisms could not be identified or mapped for the remaining values for several reasons including: organism is not in the NCBI Taxonomy (e.g., Pachnoda ephippiata and Thamnomys rutilans), common name or synonym for an organism is not in the NCBI Taxonomy (e.g., snail, white-fronted wallaby, and avian), and typographical errors (e.g., Licopersicon esculentum instead of Lycopersicon esculentum and Biompahalaria pfeifferi instead of Biomphalaria pfeifferi). Further evaluation of the results from each approach is needed to quantify and further examine both the false negatives and false positives in order to improve the techniques. In addition, techniques will be needed to extract other contextual information that is captured in the host field aside from organism names such as organism attributes (e.g., “adult two-spotted spider mite” and “female Ixodes persulcatus”), diseases (e.g., diabetes-prone (BB-DP) rat), and relationships (e.g., Scolytus ratzeburgi on Betula pendula).
For isolation_source metadata in GenBank, a key goal was to gain a better understanding of the types of information found within this field. The NCBO Annotator Web service was used to annotate host-specific values where no restrictions to ontologies or semantic types were applied. The initial semantic analysis provided insights to the coverage of concepts for guiding next steps for both host-specific and host-independent analysis. Future work includes evaluating the annotations produced by NCBO Annotator to determine if and how parameters should be adjusted. For example, limiting to specific ontologies (e.g., guided by NCBO Recommender14) and focusing on particular semantic types.
CONCLUSION
This study involved examining the free-text host and isolation_source metadata fields in GenBank towards organizing key contextual information using a combination of existing biomedical ontology and annotation resources. Preliminary results for ten host organisms demonstrate how the structuring of these fields may contribute to comparative studies.
Acknowledgments
The authors thank Dr. Rama Kocherlakota for assistance with the local GenBank database. This work was supported in part by NIH grant R01-LM009725.
References
- 1.Baxevanis AD. The importance of biological databases in biological discovery. Curr Protoc Bioinformatics. 2009. Sep, Chapter 1:Unit 1. [DOI] [PubMed]
- 2.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008 Jan;36(Database issue):D25–30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. Jan;38(Database issue):D5–16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. http://www.ncbi.nlm.nih.gov/projects/collab/FT/.
- 5.Hirschman L, Clark C, Cohen KB, Mardis S, Luciano J, Kottmann R, et al. Habitat-Lite: a GSC case study based on free text terms for environmental metadata. OMICS. 2008 Jun;12(2):129–36. doi: 10.1089/omi.2008.0016. [DOI] [PubMed] [Google Scholar]
- 6.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008 May;26(5):541–7. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarkar IN. Leveraging biomedical ontologies and annotation services to organize microbiome data from mammalian hosts. AMIA Annu Symp Proc. 2010:717–21. [PMC free article] [PubMed] [Google Scholar]
- 8.Hankeln W, Buttigieg PL, Fink D, Kottmann R, Yilmaz P, Glockner FO. MetaBar - a tool for consistent contextual data acquisition and standards compliant submission. BMC Bioinformatics. 11:358. doi: 10.1186/1471-2105-11-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schriml LM, Arze C, Nadendla S, Ganapathy A, Felix V, Mahurkar A, et al. GeMInA, Genomic Metadata for Infectious Agents, a geospatial surveillance pathogen database. Nucleic Acids Res. Jan;38(Database issue):D754–64. doi: 10.1093/nar/gkp832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarkar IN. Biodiversity informatics: organizing and linking information across the spectrum of life. Brief Bioinform. 2007 Sep;8(5):347–57. doi: 10.1093/bib/bbm037. [DOI] [PubMed] [Google Scholar]
- 11.Shah NH, Bhatia N, Jonquet C, Rubin D, Chiang AP, Musen MA. Comparison of concept recognizers for building the Open Biomedical Annotator. BMC Bioinformatics. 2009;10(Suppl 9):S14. doi: 10.1186/1471-2105-10-S9-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noy NF, Shah NH, Whetzel PL, Dai B, Dorf M, Griffith N, et al. BioPortal: ontologies and integrated data resources at the click of a mouse. Nucleic Acids Res. 2009 Jul 1;37(Web Server issue):W170–3. doi: 10.1093/nar/gkp440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004 Jan 1;32(Database issue):D267–70. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jonquet C, Musen MA, Shah NH. Building a biomedical ontology recommender web service. J Biomed Semantics. 1(Suppl 1):S1. doi: 10.1186/2041-1480-1-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]