Abstract
Translocations and mutations in the core binding factor genes, RUNX1 or CBFB, are found in acute myeloid and lymphocytic leukemia, therapy-related myeloid leukemia, myelodysplastic syndrome, chronic myelomonocytic leukemia, and in familial platelet disorder with predisposition to acute myeloid leukemia. Here we review the biochemical and biological properties of the normal Runx1 protein, discuss the nature of RUNX1 mutations in myeloid leukemia, their prognostic significance, and the mutations that cooperate or co-exist with them in these various diseases.
Keywords: Runx1, AML, MDS, Hematopoietic Stem Cells
I. INTRODUCTION
In 1973 Janet Rowley, using new chromosome banding techniques, identified a reciprocal translocation between chromosomes 8 and 21 in two female patients with acute myeloid leukemia (AML).1 That same month, Rowley published another paper demonstrating that the end of the long arm of chromosome 22 thought to be missing in the Philadelphia chromosome, a cytogenetic abnormality frequently associated with chronic myeloid leukemia, had in fact not been lost but was instead relocated to the end of chromosome 9.2 These contemporaneous papers established that consistent chromosomal translocations could be correlated with specific leukemia subtypes. Eighteen years later the Acute Myeloid Leukemia 1 (AML1) residing at the breakpoint in t(8;21)(q22;q22) was cloned,3 and later renamed RUNX1.4 Hence the discovery of RUNX1 mutations was an important milestone in the history of cancer genetics. Janet Rowley’s keen powers of observation and intuition that translocations were causative in leukemia, and not simply correlative, begat an era of intensive research in cancer genetics, which may have reached its zenith with the application of next generation sequencing technology.
Fast forward nearly 40 years, and we now know much about the RUNX1 gene and its encoded protein. Runx1 is sequence-specific DNA binding protein, and has an obligate non-DNA binding partner called core binding factor β (CBFβ), the gene for which is also targeted by translocations important in AML, the inv(16)(p13;q22) and t(16;16)(p13;q22).5 AML with any of these three translocations is often referred to as “core binding factor leukemia”. Runx1 has essential functions in normal hematopoiesis in the embryo and the adult. In addition to the t(8;21), translocations and mutations in RUNX1 have been found in de novo and therapy-related AML, myelodysplastic syndrome (MDS), chronic myelomonocytic leukemia (CMML), acute lymphocytic leukemia (ALL), and in the autosomal dominant pre-leukemia syndrome familial platelet disorder with predisposition to acute myeloid leukemia (FPD/AML).
II. THE RUNX1 PROTEIN - DOMAIN STRUCTURE AND FUNCTION, AND INTERACTING PROTEINS
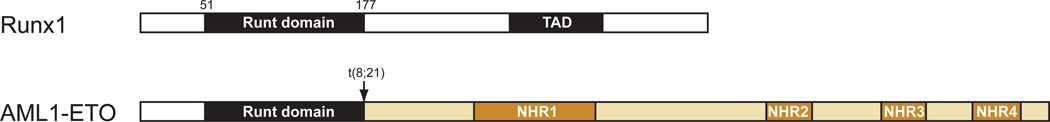
Runx1 is a sequence-specific DNA binding protein, and like most proteins of its ilk contains effecter domains linked by less structured sequences. By far the most well characterized domain of Runx1 is its DNA binding “Runt” domain, named after the first member of the family to be cloned, the Drosophila runt protein.6; 7 (Figure 1) Multiple structures of the Runt domain have been solved.8–11 The DNA and CBFβ interacting interfaces are on opposite sides of the Runt domain and do not overlap, and CBFβ does not touch the DNA. The primary role of CBFβ is to increase binding of Runx1 to DNA by stabilizing a particular conformation of the Runt domain.11; 12 As will be described later, many missense mutations in the Runt domain have been identified in AML, MDS, CMML, and FPD/AML, the vast majority of which involve residues at the DNA binding interface.
Figure 1.
Schematic diagram of Runx1 and AML1-ETO. White/black represent sequences from Runx1, and gold from ETO. TAD, transactivation domain ; NHR1-4, nervy homology domains 1–4.
The second most well-characterized domain in Runx1 is the transactivation domain, which is located midway between the Runt domain and the C-terminus, and is essential for Runx1’s in vivo functions.13–16 No structures of this domain have been solved, although computational analysis17 predicts that parts of the transactivation domain and an adjacent inhibitory domain are likely to be structured. Multiple proteins have been identified that interact with sequences C-terminal to the Runt domain that presumably mediate its activities.18–28 Mutations C-terminal to the Runt domain are also found in leukemia, and are primarily nonsense or frameshift mutations that result in the production of proteins lacking all or part of the transactivation domain. A few missense mutations have also been found, but their functional significance has not been established.29; 30
Less well-characterized sequences in the C-terminus of Runx1 affect Runx1’s DNA binding potential. Specifically, deletion of C-terminal sequences causes Runx1 to bind DNA with an affinity approximately 40 fold greater than that of the full-length protein.14; 31 Therefore Runx1 proteins lacking the inhibitory sequences can presumably out-compete binding of the functional full-length protein to DNA, and dominantly inhibit its activity. For simplicity’s sake we will use the term “Runx1 mutations” to refer to all mutations other than translocations, including loss of function (amorphic) mutations, hypomorphic mutations, and antimorphic mutations that create dominant negative RUNX1 alleles.
III. RUNX1 FUNCTION IN NORMAL HEMATOPOIESIS
Runx1’s earliest role in development is for the differentiation of hematopoietic progenitors and stem cells (HSCs) from a small population of endothelial cells in the conceptus.32–34 Because mutations in the germline caused mid-gestation lethality, conditional deletion strategies were necessary to ascertain its role in adult hematopoiesis. Deletion of Runx1 in adult HSCs caused multi-lineage blocks in B and T lymphoid development and megakaryocyte maturation, and thus the mice are lymphopenic and thrombocytopenic.35–37 Notably, Runx1 loss in HSCs does not cause AML on its own, but establishes a pre-leukemic state that predisposes to AML following the acquisition of secondary mutations.38; 39 The effects of Runx1 loss on HSCs and progenitors are not entirely understood. One outcome is an increase in a population of cells in the mouse bone marrow that lacks lineage markers and expresses the HSC markers Sca-1 and c-Kit (LSK cells).32; 35; 36; 38; 39 Runx1 loss also increases the number of granulocyte and megakaryocyte progenitors in the bone marrow and causes what has been alternatively described as a myeloproliferative disease or myelodysplasia,35; 37 and in one study a lower penetrance lymphoma was noted.37 Runx1 loss does not, however, cause a notable decline in functional long-term repopulating HSCs,39; 40 which is probably a critical property contributing to the pre-leukemic state, as a mutant HSC that is rapidly lost from the bone marrow cannot provide a target population for secondary mutations. Runx1 loss enhances the ability of mouse hematopoietic progenitors to undergo serial replating in culture,36 which is regarded as a measure of self-renewal activity and may also contribute to the maintenance of a dysfunctional progenitor population.
IV. RUNX1 MUTATIONS IN AML
A. t(8;21)
AML can be subdivided into several subtypes including AML with recurrent cytogenetic abnormalities, AML with multi-lineage dysplasia (this includes patients with an antecedent MDS or myeloproliferative disease), AML and MDS therapy related (following chemotherapy or radiation exposure), and AML not otherwise categorized.41. Translocations or loss of function RUNX1 mutations have been found in all of these subtypes.
Characteristic genetic abnormalities include the t(8;21) and inv(16) in RUNX1 and CBFB, respectively, each of which defines subgroups within the category of recurrent cytogenetic abnormalities, and confer a favorable prognosis. The t(8;21) breaks the RUNX1 gene in intron 5, and results in fusion of the N-terminal portion of Runx1 (including the Runt domain, but minus the transactivation domain) to a protein most commonly known as ETO (encoded by RUNX1T1) (Figure 1)..3; 42–45 ETO contains four domains conserved with its Drosophila homologue nervy, the structures for all of which, along with their interacting proteins, have been solved.46–51 Mutations that specifically disrupt the interaction between individual domains in ETO and their associated proteins revealed that one domain in particular, nervy homology region 2 (NHR2, also known as hydrophobic heptad repeat or HHR) is critical for AML1-ETO’s leukemogenic activity in retroviral transduction based assays.52; 53 NHR2 forms a four-helix bundle (a dimer of dimers), and mutations that reduced the tetramer to dimer abrogated AML1-ETO’s leukemogenic activity.53 That oligomerization of AML1-ETO per se was important was demonstrated by the ability of an oligomerization domain from the forkhead binding protein to substitute for NHR2 and enable AML1-ETO to confer serial replating activity to primary bone marrow cells.54
On the other hand, the most C-terminal domain, NHR4, also known as the myeloid-Nervy-DEAF-1 (MYND), appears to restrain AML1-ETO’s leukemogenic activity, as mutations that severely disrupt the NHR4 fold promote AML1-ETO’s activity.55 In fact, the full-length unaltered form of AML1-ETO is not by itself leukemogenic, and can only cause AML in mice when combined with another oncogene such as Fms-like tyrosine kinase 3 internal tandem duplication (FLT3-ITD), TEL-PDGFBR, or activated KIT.56–58 But either the deletion or mutation of NHR4 will allow AML1-ETO to induce leukemia in the absence of a co-transduced oncogene.55 NHR4 binds the silencing mediator of retinoid and thyroid hormone receptor (SMRT) and nuclear receptor co-repressor (N-CoR) complexes, as well as a protein called SON, and presumably one or more of these complexes is responsible for dampening AML1-ETO’s activity.55; 59–61 Mutations involving NHR4 have not been found in t(8;21) leukemia, thus this does not appear to represent a common mechanism for augmenting AML1-ETO’s activity.62
Deletions of the NHR1 domain have yielded conflicting results, with one group reporting an effect on AML1-ETO’s activity63 and others not.52; 54 The NHR1 deletion in the former study63 was larger than in the latter two studies,52; 54 and included sequences C-terminal to the conserved NHR1 domain that were previously shown to bind N-CoR and to contribute to leukemogenesis.52; 64 The larger deletion prevented association with p300 and acetylation of two lysine residues within the Runx1 portion of the AML1-ETO protein.63 Substituting one of those lysines with an arginine impaired AML1-ETO’s leukemogenic activity, indicating that the ability to recruit p300 and become acetylated is important for AML1-ETO function.63
The two other interactions mediated by AML1-ETO that are essential for its activity are DNA and CBFβ binding by the Runt domain, although there is some disagreement about the importance of the latter.52; 54; 65–67 Thus AML1-ETO has several surfaces amenable to targeting with small molecule or peptide inhibitors: the Runt domain:DNA interface, potentially the Runt domain:CBFβ interface, and NHR2 oligomerization. Inhibition of p300 or other histone acetylases may also provide a therapeutic option.63
Multiple lines of evidence indicate that the acquisition of AML1-ETO confers a different phenotype than Runx1 mutations. Genetic experiments in Drosophila showed that AML1-ETO behaves as a constitutive repressor, blunting the transcription of genes the Runx1 homologue lozenge activates, but additionally those that lozenge would normally repress.68 The phenotype of conditional knock-in mice in which AML1-ETO expression is activated in the adult bone marrow resembles a somewhat milder version of Runx1 loss, as the mice had no evidence of lymphopenia or thrombocytopenia.69 AML1-ETO conditional knock-in mice did, however, display some of the aberrations in progenitors seen upon Runx1 loss, including increased numbers of granulocyte-monocyte (GM) progenitors and enhanced serial replating activity. AML1-ETO in the conditional knock-in mice cooperated with the HIP1-PDGFBR oncogene to induce a very rapid myeloproliferative disease (MPD) that was not observed with either mutation alone.70 As hematopoietic differentiation was not impeded in this AML1-ETO knock-in mouse model, this suggests that defects in the stem/progenitor pool caused by AML1-ETO are fundamental to the leukemic process. The phenotypes in mice correlate with the leukemic phenotypes, as AML1-ETO is found in the French-American-British (FAB) M2 subtype, also known as acute myeloblastic leukemia with maturation, whereas biallelic RUNX1 mutations have been found in minimally differentiated acute myeloblastic leukemia, AML M0. Finally, as discussed in more detail below, RUNX1 mutations confer a considerably worse prognosis than the t(8;21).
The t(8;21) is the most common translocation in pediatric AML patients (10%–20%).71–74 One study documented a prenatal origin of the t(8;21) from the Guthrie cards of half of the pediatric patients analyzed.75 Two of the positive patients were between 10–12 years of age at the time of diagnosis, therefore harbored a pre-leukemic clone for more than a decade before developing AML. Patients in long-term remission can also harbor residual t(8;21)-containing cells in their bone marrow for many years.45; 76 Eighteen percent of healthy individuals have t(8;21) containing cells detectable by polymerase chain reaction, and AML1-ETO transcripts were detected in 40% of cord blood samples.77; 78 Thus the t(8;21) results in the acquisition of a long-lived pre-leukemic HSC that has no overt clinical manifestations.
The mutations that cause AML are often divided into two classes: class I mutations which activate signaling pathways, hence proliferation and survival, and class II mutations that generally involve transcription factors and cause impaired differentiation, decreased apoptosis, and growth arrest. Another class of frequently mutated genes encodes epigenetic regulators. The class I mutations most frequently found in t(8;21) AML include KIT, NRAS, and KRAS. About 20% of t(8;21) patients have activating mutations in KIT, and exon 17 mutations have been found to confer an unfavorable prognosis in multiple studies.79–81 FLT3 is the most commonly mutated gene in AML, but FLT3 mutations occur at a relatively low rate in t(8;21) leukemia.
B. AMORPHIC AND ANTIMORPHIC RUNX1 MUTATIONS IN AML
RUNX1 mutations were first described in AML M0 and FPD/AML,82; 83 followed shortly thereafter in MDS,84 and more recently in CMML.85; 86 More recent larger-scale sequencing efforts are providing a more comprehensive picture of the frequency and scope of RUNX1 mutations, their prognostic significance, and the co-existing mutations.
Two groups recently analyzed large numbers of AML patients for the presence of RUNX1 mutations.29; 30 A report from the German-Austrian AML study group, which evaluated 18 to 60 year old AML patients (primarily de novo AML, but including a smaller number of secondary and therapy-related AML patients) found RUNX1 mutations in 53 of 945 (5.6%) cases.29 An earlier study of an older 15- to 90-year old Taiwanese patient population with de novo AML reported a higher incidence of RUNX1 mutations (13.2%, 62 of 470 patients).30 It was suggested by authors of the former study that the higher frequency of RUNX1 mutations in the Taiwanese study could be caused by their inclusion of older patients, as the mutation frequency increases with age. In both studies most RUNX1 mutations were frameshift mutations, the remainder included missense, nonsense, in-frame, or silent mutations, and the vast majority were mono-allelic. The mutations were primarily located in the Runt domain and C-terminal to the Runt domain. Both groups reported RUNX1 mutations were mainly found in the cytogenetic intermediate-risk group, and closely associated with trisomy 8. No RUNX1 mutations were found in the favorable risk group with characteristic genetic abnormalities that include t(8;21) and inv(16). In univariate analyses RUNX1 mutations were found to be associated with refractory disease and inferior event free, relapse free, and overall survival. Allogeneic hematopoietic stem cell transplant improved the outcome of patients with RUNX1 mutations,29; 30 while patients who instead received repetitive cycles of high-dose cytarabine or autologous hematopoietic stem cell transplant relapsed or died.29 RUNX1 mutations were associated with the presence of MLL- PTD mutations in both studies and IDH1/IDH2 in one study,29 but inversely correlated with CEBPA and NPM1 mutations.29; 30 No significant correlation was found with FLT3, NRAS, KRAS, KIT, PTPN11, or WT1 mutations, despite the fact that several of these, and in particular KIT mutations are frequent in t(8;21) AML. Both groups found RUNX1 mutations highly associated with AML M0; one group also reported association with M130 and the other with M2 morphologies.29
An analysis of 111 pediatric AML cases identified five with RUNX1 mutations, in addition to twenty t(8;21) and sixteen inv(16) cases, bringing the total of core binding factor mutations in this pediatric AML cohort to 36.9%.87 If one combines the frequencies of RUNX1 mutations in adult AML (13% in the unselected Taiwanese study), with the t(8;21) (7% in a Cancer and Leukemia Group B (CALGB) study with a median age of 52; and inv(16) (8% in the same CALGB study),88 this results in an overall frequency of core binding factor mutations in adult AML of approximately 28%. RUNX1 mutations, t(8;21), and inversion 16 are mutually exclusive.
V. RUNX1 MUTATIONS IN MDS
MDS is a clonal stem cell disorder characterized by ineffective production of myeloid lineage cells with associated dysplasia that can involve one or more myeloid lineages. There are multiple subcategories of MDS, including refractory cytopenia with unilineage dysplasia, refractory anemia with ringed sideroblasts with associated thrombocytosis, refractory cytopenia with multilineage dysplasia, refractory anemia with excess blasts I and II, 5q- syndrome, myelodysplasia unclassifiable, and refractory cytopenia of childhood. Approximately one third of MDS patients will progress to AML over time. Fewer than half of MDS patients have chromosomal abnormalities, and balanced translocations are rare.
Nevertheless the first report of a RUNX1 mutation in MDS was a balanced translocation, the t(3;21)(q26.2;q22).89 However loss of function RUNX1 mutations are far more common in MDS, and numerous reports have documented them.84; 90; 91 At the time of writing the most recent report was a mutational screen in 439 MDS patients for a broad array of cancer-associated genes, in which mutations in RUNX1 along with 17 other genes were identified.92 RUNX1 mutations were the third most frequent (8.7%), surpassed only by mutations in the epigenetic regulators TET2 (20.5%) and ASXL1 (14.4%). A multivariate analysis that included risk stratification using the International Prognostic Scoring System93 showed that mutations in RUNX1, ASXL1, TP53, EZH2, and ETV6 were independent predictors of poor overall survival in all but the highest risk category. Mutations in RUNX1, TP53, and NRAS correlated with severe thrombocytopenia and elevated blast counts, but not with neutropenia or anemia. Loss of function Runx1 mutations in mice affect megakaryocyte but not granulocyte or erythroid differentiation, consistent with the MDS phenotype seen in human patients with RUNX1 mutations. A 13.8% frequency of RUNX1 mutations was reported in an earlier study of 188 MDS + CMML patients.94 Samples from MDS patients who progressed to secondary AML (s-AML) were analyzed for mutations at both stages.94; 95 In most cases RUNX1 mutations were present in both the MDS and s-AML samples, and in a smaller number of cases RUNX1 mutations were found in the s-AML but not in the antecedent MDS. Thus RUNX1 mutations are likely to be early events in many cases, but can also be later events in disease progression. Conversion from mono- to biallelic RUNX1 mutations was also observed in several s-AML samples, either through acquisition of an independent mutation or uniparental disomy.95
In the fourteen samples in the Bejar et al.92 study that had mutations in addition to RUNX1, they were most often in TET2 (12), ASXL1 (12), EZH2 (8), and NRAS (6), and there was no overlap with mutations in TP53, JAK2, ETV6, IDH1/2, NPM1, GNAS, BRAF, PTEN, or CDKN2A. Thus although the types of mutations in RUNX1 found in AML and MDS were similar, the cooperating mutations were distinct. In general, very few activated tyrosine kinases were identified in MDS, confirming previous hypotheses that MDS is generally associated with class II mutations and mutations in epigenetic regulators, and MPD with class I mutations.
An intriguing observation is that loss of function RUNX1 mutations in MDS are highly correlated with previous exposure to radiation, both therapeutic and accidental, the latter in atomic bomb survivors and individuals who lived in close proximity to the Semipalatinsk nuclear test site in what today is Kazakhstan.90; 96 The close association of RUNX1 mutations with radiation suggest either that the RUNX1 gene is particularly sensitive to DNA damage following radiation, or that preexisting RUNX1 mutations may predispose patients to MDS following DNA damage.
RUNX1 mutations were also recently described in Fanconi anemia (FA) patients, who have a 30%–40% probability of developing MDS and AML by age 40.97 A screen of 57 FA bone marrows for chromosome copy number changes and mutations in commonly MDS/AML genes (TET2, CBL, NRAS, TP53, RUNX1, CEBPA, NPM1, FLT3, and MLL) found that the somatic acquisition of only three abnormalities correlated with MDS/AML in FA patients: 3q+, 7/7q−, and RUNX1 translocations, deletions, and mutations.98
VI. RUNX1 MUTATIONS IN CMML
CMML has overlapping features of MDS and myeloproliferative neoplasms, including peripheral monocytosis > 1×109/L, <20% blood or bone marrow blasts, and bone marrow dysplasia in one or more myeloid lineage, and progresses to AML in 15–20% of patients. CMML is a relatively rare disease, thus sequencing studies of the scale described above for AML and MDS have not been performed. In a smaller scale analysis by the Munich Leukemia Group, mutations in TET, CBL, NRAS, KRAS, JAK2, RUNX1, and MPL were interrogated in 81 CMML samples.99 The majority (72.8%) of CMML samples had a mutation in TET, CBL, NRAS, KRAS, JAK2, or RUNX1, with RUNX1 mutations in 8.6% of patients. RUNX1 mutations in this study were not found to be of prognostic relevance. Another analysis was performed in a Taiwanese population, and RUNX1 mutations were found in 30/81 patients (37%). Both were unselected groups with a preponderance of elderly patients. In the Taiwanese cohort, there was a trend toward faster progression to AML in the RUNX1 mutated group, which was especially pronounced when RUNX1 mutations occurred in the C-terminus.86
VII. RUNX1 MUTATIONS IN FPD/AML
FPD/AML is an autosomal dominant disorder caused by mutations in RUNX1. Many but not all FPD/AML patients have low platelet counts or platelet activation defects.100; 101 The penetrance of MDS/AML in FPD/AML patients is >40%, with a median age of incidence of 33 years.102 Large intragenic deletions in RUNX1 in FPD/AML established haploinsufficiency is one mechanism for the disease,83 but mutations are also frequently found in the Runt domain. FPD/AML is clearly an intriguing syndrome and an improved understanding of the pathogenesis of MDS and AML in this disorder would seem to be key for unraveling the mechanisms underlying RUNX1 mutant AML and MDS in general. However the ability to gain more insight into the pathogenesis of MDS/AML in FPD/AML has been hampered by small patient numbers and the heterogeneity of the disease presentation.
VIII. BIOCHEMICAL AND FUNCTIONAL ANALYSES OF RUNX1 MUTATIONS
The majority of RUNX1 mutations can be categorized based on their potential impact on the protein (Table 1). These include : 1) large deletions; 2) mutations resulting in truncation within the Runt domain; 3) missense mutations in the Runt domain at the DNA interface that affect DNA but not CBFβ binding; 4) missense mutations in the Runt domain at the CBFβ interface that affect CBFβ but not DNA binding; 5) missense mutations in the Runt domain that affect both DNA and CBFβ binding through destabilizing the Runt domain fold; 6) mutations that truncate Runx1 C-terminal to the Runt domain and remove all or part of the transactivation domain; and 7) missense mutations C-terminal to the Runt domain (rare). It has been hypothesized that these various mutations would have different biological effects, with some behaving as loss of function (amorphic) mutations, others as hypomorphic mutations, and some as antimorphic mutations that could create dominant interfering Runx1 proteins.
Table 1.
RUNX1 mutations in AML, MDS, and CMML, and FPD/AML
| Mutation | Affects | Type | |||
|---|---|---|---|---|---|
| DNA binding |
CBFβ binding |
Runt domain fold |
Transactivation | ||
| Large deletion | yes | yes | yes | yes | Amorphic |
| Truncation before or within Runt domain | yes | yes | yes | yes | Amorphic |
| Missense mutation in Runt domain at DNA interface1 | yes | yes or no | yes or no | yes | Antimorphic or Amorphic |
| Missense mutation in Runt domain at CBFβ interface2 | no | yes | no | yes | Hypomorphic |
| Missense mutation in Runt domain, not at DNA or CBFβ interface2 | yes | yes | yes | yes | Hypomorphic |
| Truncation C-terminal to Runt domain | no | no | no | yes | Antimorphic |
| Missense mutation C-terminal to Runt domain2 | no | no | no | yes3 | Antimorphic3 |
Common in leukemia. Mutations that affect DNA but not CBFβ binding result in antimorphic alleles, and are more common than those that affect both DNA and CBFβ binding.
Rare in leukemia
Presumed, not tested
Matheny et al.103 compared different categories of missense mutations in the Runt domain using both biophysical and genetic approaches, and could confirm that mutations that perturbed CBFβ binding or the Runt domain fold resulted in hypomorphic Runx1 alleles in mice, while a mutation that severely impaired DNA but not CBFβ binding generated a weakly antimorphic allele. The mechanism for the antimorphic activity was not clear, but could involve sequestering a limiting protein with a Runx1:CBFβ heterodimer that cannot bind DNA. The majority of leukemia mutations in the Runt domain are at the DNA interface, indicating that severe disruption of Runx1 activity is more likely to be pathogenic.
Watanabe et al.104 compared mutations in the Runt domain that affected DNA binding to a truncation C-terminal to the Runt domain that removed the transactivation domain by overexpressing the mutant proteins in a bone marrow transplant model. Both induced MDS, but with different properties, in that the DNA binding mutant caused leukocytosis while the C-terminal truncation mutant caused leukopenia. Thus, different mutations will indeed contribute different biological properties to the Runx1 protein, and presumably to disease phenotype.
Most RUNX1 mutations are mono-allelic, and unfortunately a disease caused by mono-allelic mutations has been very difficult to model in the mouse. Although moderately (15%) decreased platelet counts were reported in mice haploinsufficient for RUNX1,105 this was not reproduced in another lab,103 potentially due to differences in genetic background of the mice. More pronounced thrombocytopenia was observed in mice homozygous for a hypomorphic RUNX1 allele,103 suggesting that reducing the effective dosage by more than 50% may provide a strategy for more faithfully modeling at least some aspects of RUNX1 haploinsufficiency in human disease. Hence one of the more interesting unresolved questions is why mono-allelic RUNX1 mutations would confer a more adverse phenotype than t(8;21)? One possible explanation is that mono-allelic RUNX1 mutations tend to occur in older AML patients, who may have accumulated more cooperating mutations than younger patients with t(8;21).
IX. CONCLUSION
Patients with t(8;21) and inv(16) for the most part do well with standard induction and high dose cytarabine consolidation alone without the need for allogeneic stem cell transplantation. However even for these so-called favorable prognosis core binding factor leukemias, long-term leukemia-free survival is only 50%.106 Prognosis for patients with RUNX1 mutations, which typically fall into the intermediate risk cytogenetic categories (normal and non-complex), is even worse. Our increased knowledge of core binding factor mutations and function has not yet resulted in novel therapeutic approaches. This may be due, in part, to RUNX1’s role as a class II mutation that may be involved in altering the expression of a multitude of target genes. This is illustrated by the recognition that t(8;21) AML is associated with its own unique gene methylation profile that is predictive of outcome.107 Similarly, the presence or absence of a RUNX1 mutation in AML M0 can be ascertained through the use of gene expression profiling, because RUNX1 mutations result in altered expression of key target genes in a reproducible manner.108 Given the multitude of potential Runx1 targets, one approach would be to focus on therapies that alter expression of many target genes simultaneously, as is the case for hypomethylating agents in MDS. A second approach would be to focus on combinatorial therapies, simultaneously targeting the class II effects of RUNX1 and the class I mutations in tyrosine kinases that frequently accompany RUNX1, such as the FLT3-ITD or activated KIT. One such phase I trial is currently underway at MD Anderson Cancer Center, in which the hypomethylating 5-azacitadine is being tested in combination with the FLT3 inhibitor sorafenib in relapsed and refractory AML. A third approach would be to incorporate knowledge about specific targets of Runx1 in AML: a so-called “target the target” approach. For example, a recent study showed that AKT3 and RARA are upregulated in AML M0,108 providing a rationale for further investigation into the efficacy of small molecules targeting these pathways. Given the difficulties in modeling leukemias with mono-allelic Runx1 deficiency in the mouse (described earlier), more robust Runx1 deficient leukemia models may need to be created by crossing Runx1 haploinsufficient mice with mice carrying tyrosine kinase mutations that are known to cooperate with Runx1. Such improved models may ultimately allow us to better “target the target.”
The incentive to engage in research with the goal of unlocking the secrets of the pathogenesis of AML with RUNX1 mutations would also be increased if we had an improved understanding of the prognostic relevance of RUNX1 mutations in specific therapeutic situations. How does RUNX1 mutation status affect response to hypomethylating agents? FLT3 inhibitors? HDAC inhibitors? Therapy with high dose anthracyclines or high dose cytarabine? In MDS? In AML? These questions remain unanswered, as the prognostic relevance of RUNX1 mutations have really only been examined on a global level. Our knowledge about the role of RUNX1 in AML has clearly made great strides since Dr. Rowley identified the reciprocal translocation between chromosomes 8 and 21 nearly 40 years ago, but much remains undiscovered.
Acknowledgements
We gratefully acknowledge support from the National Institutes of Health (R01 CA149976 and T32 CA009615) and the Abramson Family Cancer Research Institute.
Abbreviations
- ALL
acute lymphocytic leukemia
- AML
acute myeloid leukemia
- AML1
acute myeloid leukemia 1
- CALGB
Cancer and Leukemia Group B
- CBFB
Core Binding Factor Beta
- CMML
chronic myelomonocytic leukemia
- FAB
French-American-British
- FLT3-ITD
Fms-like tyrosine kinase 3 internal tandem duplication
- FPD-AML
familial platelet disorder with predisposition to acute myeloid leukemia
- GM
granulocyte-monocyte
- HHR
hydrophobic heptad repeat
- HSCs
hematopoietic stem cells
- MDS
myelodysplastic syndrome
- MYND
myeloid-nervy-DEAF-1
- N-CoR
nuclear receptor co-repressor
- NHR1
nervy homology region 1
- NHR2
nervy homology region 2
- NHR4
nervy homology region 4
- s-AML
secondary AML
- SMRT
silencing mediator of retinoid and thyroid hormone receptor
Contributor Information
James K. Mangan, Email: james.mangan@uphs.upenn.edu.
Nancy A. Speck, Email: nancyas@exchange.upenn.edu.
REFERENCES
- 1.Rowley JD. Identification of a translocation with quinacrine fluorescence in a patient with acute leukemia. Ann Genet. 1973;16:109–112. [PubMed] [Google Scholar]
- 2.Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 3.Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc Natl Acad Sci. USA. 1991;88:10431–10434. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Wijnen A, Stein G, Lian J, Stein J, Hiebert S, Ito Y, Speck N, Komori T, Neil J, Quarles D. Systematic nomenclature for the runt homology transcription factors, CBFA/PEBP2α/AML, involved in hematopoiesis and skeletal development. J Cell Biochem. 1999 [Google Scholar]
- 5.Liu P, Tarle SA, Hajra A, Claxton DF, Marlton P, Freedman M, Siciliano MJ, Collins FS. Fusion between transcription factor CBFβ/PEBP2β and a myosin heavy chain in acute myeloid leukemia. Science. 1993;261:1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- 6.Kania MA, Bonner AS, Duffy JB, Gergen JP. The Drosophila segmentation gene runt encodes a novel nuclear regulatory protein that is also expressed in the developing nervous system. Genes Dev. 1990;4:1701–1713. doi: 10.1101/gad.4.10.1701. [DOI] [PubMed] [Google Scholar]
- 7.Kagoshima H, Shigesada K, Satake M, Ito Y, Miyoshi H, Ohki M, Pepling M, Gergen JP. The Runt-domain identifies a new family of heteromeric DNA-binding transcriptional regulatory proteins. Trends Genet. 1993;9:338–341. doi: 10.1016/0168-9525(93)90026-e. [DOI] [PubMed] [Google Scholar]
- 8.Berardi M, Sun C, Zehr M, Abildgaard F, Peng J, Speck NA, Bushweller JH. The Ig fold of the core binding factor a Runt domain is a member of a family of structurally and functionally related Ig fold DNA binding domains. Structure Fold Des. 1999;7:1247–1256. doi: 10.1016/s0969-2126(00)80058-1. [DOI] [PubMed] [Google Scholar]
- 9.Bravo J, Li Z, Speck NA, Warren AJ. The leukaemia-associated AML1 (Runx1)-CBFβ complex functions as a DNA-induced molecular clamp. Nat Struct Biol. 2001;8:371–377. doi: 10.1038/86264. [DOI] [PubMed] [Google Scholar]
- 10.Nagata T, Gupta V, Sorce D, Kim W-Y, Sali A, Chait BT, Shigesada K, Ito Y, Werner MH. Immunoglobulin motif DNA-binding and heterodimerization for the PEBP2/CBF Runt-domain. Nat Struct Biol. 1999;6:615–619. doi: 10.1038/10658. [DOI] [PubMed] [Google Scholar]
- 11.Tahirov TH, Inoue-Bungo T, Morii H, Fujikawa A, Sasaki M, Kimura K, Shiina M, Sato K, Kumasaka T, Yamamoto M, Ishii S, Ogata K. Structural analyses of DNA recognition by the AML1/Runx-1 Runt domain and its allosteric control by CBFβ. Cell. 2001;104:755–767. doi: 10.1016/s0092-8674(01)00271-9. [DOI] [PubMed] [Google Scholar]
- 12.Yan J, Liu Y, Lukasik SM, Speck NA, Bushweller JH. CBFbeta allosterically regulates the Runx1 Runt domain via a dynamic conformational equilibrium. Nat Struct Mol Biol. 2004;11:901–906. doi: 10.1038/nsmb819. [DOI] [PubMed] [Google Scholar]
- 13.Bae S-C, Ogawa E, Maruyama M, Oka H, Satake M, Shigesada K, Jenkins NA, Gilbert DJ, Copeland NG, Ito Y. PEBP2αB/Mouse AML1 consists of multiple isoforms that possess differential transactivation potentials. Mol Cell Biol. 1994;14:3242–3252. doi: 10.1128/mcb.14.5.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanno T, Kanno Y, Chen L-F, Ogawa E, Kim W-Y, Ito Y. Intrinsic transcriptional activation-inhibition domains of the polyomavirus enhancer binding protein 2/core binding factor α subunit revealed in the presence of the b subunit. Mol Cell Biol. 1998;18:2444–2454. doi: 10.1128/mcb.18.5.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okuda T, Takeda K, Fujita Y, Nishimura M, Yagyu S, Yoshida M, Akira S, Downing JR, Abe T. Biological characteristics of the leukemia-associated transcriptional factor AML1 disclosed by hematopoietic rescue of AML1-deficient embryonic stem cells by using a knock-in strategy. Mol Cell Biol. 2000;20:319–328. doi: 10.1128/mcb.20.1.319-328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dowdy CR, Xie R, Frederick D, Hussain S, Zaidi SK, Vradii D, Javed A, Li X, Jones SN, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Definitive hematopoiesis requires Runx1 C-terminal-mediated subnuclear targeting and transactivation. Hum Mol Genet. 2010;19:1048–1057. doi: 10.1093/hmg/ddp568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Kitabayashi I, Yokoyama A, Shimizu K, Ohki M. Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation. EMBO J. 1998;17:2294–3004. doi: 10.1093/emboj/17.11.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitabayashi I, Aikawa Y, Nguyen LA, Yokoyama A, Ohki M. Activation of AML1-mediated transcription by MOZ and inhibition by the MOZ-CBP fusion protein. Embo J. 2001;20:7184–7196. doi: 10.1093/emboj/20.24.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aikawa Y, Nguyen LA, Isono K, Takakura N, Tagata Y, Schmitz ML, Koseki H, Kitabayashi I. Roles of HIPK1 and HIPK2 in AML1- and p300-dependent transcription, hematopoiesis and blood vessel formation. Embo J. 2006;25:3955–3965. doi: 10.1038/sj.emboj.7601273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruhn L, Munnerlyn A, Grosschedl R. ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRα enhancer function. Genes Dev. 1997;11:640–653. doi: 10.1101/gad.11.5.640. [DOI] [PubMed] [Google Scholar]
- 22.Yagi R, Chen L-F, Shigesda K, Murakami Y, Ito Y. A WW domain-containing Yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lutterbach B, Westendorf JJ, Linggi B, Issac S, Seto E, Hiebert SW. A mechanism of repression by Acute Myeloid Leukemia-1, the target of multiple chromosomal translocations in acute leukemia. J Biol Chem. 2000;275:651–656. doi: 10.1074/jbc.275.1.651. [DOI] [PubMed] [Google Scholar]
- 24.McLarren KW, Lo R, Grbavec D, Thirunavukkarasu K, Karsenty G, Stifani S. The mammalian basic helix loop helix protein HES-1 binds to and modulates the transactivating function of the runt-related factor Cbfa1. J Biol Chem. 2000;275:530–538. doi: 10.1074/jbc.275.1.530. [DOI] [PubMed] [Google Scholar]
- 25.Javed A, Guo B, Hiebert S, Choi JY, Green J, Zhao SC, Osborne MA, Stifani S, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Groucho/TLE/R-esp proteins associate with the nuclear matrix and repress RUNX (CBF(alpha)/AML/PEBP2(alpha)) dependent activation of tissue-specific gene transcription. J Cell Sci. 2000;113(Pt 12):2221–2231. doi: 10.1242/jcs.113.12.2221. [DOI] [PubMed] [Google Scholar]
- 26.Schroeder TM, Kahler RA, Li X, Westendorf JJ. Histone deacetylase 3 interacts with runx2 to repress the osteocalcin promoter and regulate osteoblast differentiation. J Biol Chem. 2004;279:41998–42007. doi: 10.1074/jbc.M403702200. [DOI] [PubMed] [Google Scholar]
- 27.Vega RB, Matsuda K, Oh J, Barbosa AC, Yang X, Meadows E, McAnally J, Pomajzl C, Shelton JM, Richardson JA, Karsenty G, Olson EN. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119:555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 28.Chakraborty S, Sinha KK, Senyuk V, Nucifora G. SUV39H1 interacts with AML1 and abrogates AML1 transactivity. AML1 is methylated in vivo. Oncogene. 2003;22:5229–5237. doi: 10.1038/sj.onc.1206600. [DOI] [PubMed] [Google Scholar]
- 29.Gaidzik VI, Bullinger L, Schlenk RF, Zimmermann AS, Rock J, Paschka P, Corbacioglu A, Krauter J, Schlegelberger B, Ganser A, Spath D, Kundgen A, Schmidt-Wolf IG, Gotze K, Nachbaur D, Pfreundschuh M, Horst HA, Dohner H, Dohner K. RUNX1 mutations in acute myeloid leukemia: results from a comprehensive genetic and clinical analysis from the AML study group. J Clin Oncol. 2011;29:1364–1372. doi: 10.1200/JCO.2010.30.7926. [DOI] [PubMed] [Google Scholar]
- 30.Tang JL, Hou HA, Chen CY, Liu CY, Chou WC, Tseng MH, Huang CF, Lee FY, Liu MC, Yao M, Huang SY, Ko BS, Hsu SC, Wu SJ, Tsay W, Chen YC, Lin LI, Tien HF. AML1/RUNX1 mutations in 470 adult patients with de novo acute myeloid leukemia: prognostic implication and interaction with other gene alterations. Blood. 2009;114:5352–5361. doi: 10.1182/blood-2009-05-223784. [DOI] [PubMed] [Google Scholar]
- 31.Gu T-L, Goetz TL, Graves BJ, Speck NA. Autoinhibition and partner proteins, CBFβ and Ets-1, modulate DNA binding by CBFα2(AML1) Mol Cell Biol. 2000;20:91–103. doi: 10.1128/mcb.20.1.91-103.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457:889–891. doi: 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.North TE, Gu T-L, Stacy T, Wang Q, Howard L, Binder M, Marín-Padilla M, Speck NA. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 1999;126:2563–2575. doi: 10.1242/dev.126.11.2563. [DOI] [PubMed] [Google Scholar]
- 34.Yokomizo T, Ogawa M, Osato M, Kanno T, Yoshida H, Fujimoto T, Fraser S, Nishikawa S, Okada H, Satake M, Noda T, Nishikawa SI, Ito Y. Requirement of Runx1/AML1/PEBP2aB for the generation of haematopoietic cells from endothelial cells. Genes Cells. 2001;6:13–23. doi: 10.1046/j.1365-2443.2001.00393.x. [DOI] [PubMed] [Google Scholar]
- 35.Growney JD, Shigematsu H, Li Z, Lee BH, Adelsperger J, Rowan R, Curley DP, Kutok JL, Akashi K, Williams IR, Speck NA, Gilliland DG. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106:494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ichikawa M, Asai T, Saito T, Yamamoto G, Seo S, Yamazaki I, Yamagata T, Mitani K, Chiba S, Ogawa S, Kurokawa M, Hirai H. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med. 2004;10:299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- 37.Putz G, Rosner A, Nuesslein I, Schmitz N, Buchholz F. AML1 deletion in adult mice causes splenomegaly and lymphomas. Oncogene. 2006;25:929–939. doi: 10.1038/sj.onc.1209136. [DOI] [PubMed] [Google Scholar]
- 38.Motoda L, Osato M, Yamashita N, Jacob B, Chen LQ, Yanagida M, Ida H, Wee HJ, Sun AX, Taniuchi I, Littman D, Ito Y. Runx1 protects hematopoietic stem/progenitor cells from oncogenic insult. Stem Cells. 2007;25:2976–2986. doi: 10.1634/stemcells.2007-0061. [DOI] [PubMed] [Google Scholar]
- 39.Jacob B, Osato M, Yamashita N, Wang CQ, Taniuchi I, Littman DR, Asou N, Ito Y. Stem cell exhaustion due to Runx1 deficiency is prevented by Evi5 activation in leukemogenesis. Blood. 2009:1610–1620. doi: 10.1182/blood-2009-07-232249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ichikawa M, Goyama S, Asai T, Kawazu M, Nakagawa M, Takeshita M, Chiba S, Ogawa S, Kurokawa M. AML1/Runx1 negatively regulates quiescent hematopoietic stem cells in adult hematopoiesis. J Immunol. 2008;180:4402–4408. doi: 10.4049/jimmunol.180.7.4402. [DOI] [PubMed] [Google Scholar]
- 41.Vardiman JW, Harris NL, Brunning RD. The World Health Organization (WHO) classification of the myeloid neoplasms. Blood. 2002;100:2292–2302. doi: 10.1182/blood-2002-04-1199. [DOI] [PubMed] [Google Scholar]
- 42.Erickson P, Gau J, Chang K-S, Look T, Whisenant E, Raimondi S, Lasher R, Trujillo J, Rowley J, Drabkin H. Identification of breakpoints in t(8;21) AML and isolation of a fusion transcript with similarity to Drosophila segmentation gene runt. Blood. 1992;80:1825–1831. [PubMed] [Google Scholar]
- 43.Miyoshi H, Kozu T, Shimizu K, Enomoto K, Maseki N, Kaneko Y, Kamada N, Ohki M. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript. EMBO J. 1993;12:2715–2721. doi: 10.1002/j.1460-2075.1993.tb05933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nucifora G, Birn DJ, Erickson P, Gao J, LeBeau MM, Drabkin HA, Rowley JD. Detection of DNA rearrangement in the AML1 and ETO loci and of an AML1/ETO fusion mRNA in patients with t(8;21) Blood. 1993;81:883. [PubMed] [Google Scholar]
- 45.Chang KS, Fan YH, Stass SA, Estey EH, Wang G, Trujillo JM, Erickson P, Drabkin H. Expression of AML1-ETO fusion transcripts and detection of minimal residual disease in t(8;21)-positive acute myeloid leukemia. Oncogene. 1993;8:983–988. [PubMed] [Google Scholar]
- 46.Liu Y, Chen W, Gaudet J, Cheney MD, Roudaia L, Cierpicki T, Klet RC, Hartman K, Laue TM, Speck NA, Bushweller JH. Structural basis for recognition of SMRT/N-CoR by the MYND domain and its contribution to AML1/ETO's activity. Cancer Cell. 2007;11:483–497. doi: 10.1016/j.ccr.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Y, Cheney MD, Gaudet JJ, Chruszcz M, Lukasik SM, Sugiyama D, Lary J, Cole J, Dauter Z, Minor W, Speck NA, Bushweller JH. The tetramer structure of the Nervy homology two domain, NHR2, is critical for AML1/ETO's activity. Cancer Cell. 2006;9:249–260. doi: 10.1016/j.ccr.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 48.Park S, Chen W, Cierpicki T, Tonelli M, Cai X, Speck NA, Bushweller JH. Structure of the AML1-ETO eTAFH domain-HEB peptide complex and its contribution to AML1-ETO activity. Blood. 2009;113:3558–3567. doi: 10.1182/blood-2008-06-161307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corpora T, Roudaia L, Oo ZM, Chen W, Manuylova E, Cai X, Chen MJ, Cierpicki T, Speck NA, Bushweller JH. Structure of the AML1-ETO NHR3-PKA(RIIalpha) complex and its contribution to AML1-ETO activity. J Mol Biol. 2010;402:560–577. doi: 10.1016/j.jmb.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Plevin MJ, Zhang J, Guo C, Roeder RG, Ikura M. The acute myeloid leukemia fusion protein AML1-ETO targets E proteins via a paired amphipathic helix-like TBP-associated factor homology domain. Proc Natl Acad Sci U S A. 2006;103:10242–10247. doi: 10.1073/pnas.0603463103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei Y, Liu S, Lausen J, Woodrell C, Cho S, Biris N, Kobayashi N, Wei Y, Yokoyama S, Werner MH. A TAF4-homology domain from the corepressor ETO is a docking platform for positive and negative regulators of transcription. Nat Struct Mol Biol. 2007;14:653–661. doi: 10.1038/nsmb1258. [DOI] [PubMed] [Google Scholar]
- 52.Yan M, Ahn EY, Hiebert SW, Zhang DE. RUNX1/AML1 DNA binding domain and ETO/MTG8 NHR2 dimerization domain are critical to AML1-ETO9a leukemogenesis. Blood. 2008:883–886. doi: 10.1182/blood-2008-04-153742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wichmann C, Becker Y, Chen-Wichmann L, Vogel V, Vojtkova A, Herglotz J, Moore S, Koch J, Lausen J, Mantele W, Gohlke H, Grez M. Dimer-tetramer transition controls RUNX1/ETO leukemogenic activity. Blood. 2010;116:603–613. doi: 10.1182/blood-2009-10-248047. [DOI] [PubMed] [Google Scholar]
- 54.Kwok C, Zeisig BB, Qiu J, Dong S, So CW. Transforming activity of AML1-ETO is independent of CBFbeta and ETO interaction but requires formation of homo-oligomeric complexes. Proc Natl Acad Sci U S A. 2009;106:2853–2858. doi: 10.1073/pnas.0810558106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ahn EY, Yan M, Malakhova OA, Lo MC, Boyapati A, Ommen HB, Hines R, Hokland P, Zhang DE. Disruption of the NHR4 domain structure in AML1-ETO abrogates SON binding and promotes leukemogenesis. Proc Natl Acad Sci U S A. 2008;105:17103–17108. doi: 10.1073/pnas.0802696105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grisolano JL, O'Neal J, Cain J, Tomasson MH. An activated receptor tyrosine kinase, TEL/PDGFβR, cooperates with AML1/ETO to induce acute myeloid leukemia in mice. Proc Natl Acad Sci USA. 2003;100:9506–9511. doi: 10.1073/pnas.1531730100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang YY, Zhao LJ, Wu CF, Liu P, Shi L, Liang Y, Xiong SM, Mi JQ, Chen Z, Ren R, Chen SJ. C-KIT mutation cooperates with full-length AML1-ETO to induce acute myeloid leukemia in mice. Proc Natl Acad Sci U S A. 2011;108:2450–2455. doi: 10.1073/pnas.1019625108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schessl C, Rawat VP, Cusan M, Deshpande A, Kohl TM, Rosten PM, Spiekermann K, Humphries RK, Schnittger S, Kern W, Hiddemann W, Quintanilla-Martinez L, Bohlander SK, Feuring-Buske M, Buske C. The AML1-ETO fusion gene and the FLT3 length mutation collaborate in inducing acute leukemia in mice. J Clin Invest. 2005;115:2159–2168. doi: 10.1172/JCI24225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gelmetti V, Zhang J, Fanelli M, Minucci S, Pelicci PG, Lazar MA. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol Cell Biol. 1998;18:7185–7191. doi: 10.1128/mcb.18.12.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lutterbach B, Westendorf JJ, Linggi B, Patten A, Moniwa M, Davie JR, KD H, Bardwell VJ, Lavinsky RM, Rosenfeld MG, Glass C, Seto E, Hiebert SW. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol Cell Biol. 1998;18:7176–7184. doi: 10.1128/mcb.18.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang J, Hoshino T, Redner RL, Kajigaya S, Liu JM. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC complex. Proc Natl Acad Sci USA. 1998;95:10860–10865. doi: 10.1073/pnas.95.18.10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hackanson B, Abdelkarim M, Jansen JH, Lubbert M. NHR4 domain mutations of ETO are probably very infrequent in AML1-ETO positive myeloid leukemia cells. Leukemia. 2010;24:860–861. doi: 10.1038/leu.2009.291. [DOI] [PubMed] [Google Scholar]
- 63.Wang L, Gural A, Sun XJ, Zhao X, Perna F, Huang G, Hatlen MA, Vu L, Liu F, Xu H, Asai T, Deblasio T, Menendez S, Voza F, Jiang Y, Cole PA, Zhang J, Melnick A, Roeder RG, Nimer SD. The Leukemogenicity of AML1-ETO Is Dependent on Site-Specific Lysine Acetylation. Science. 2011;333:765–769. doi: 10.1126/science.1201662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Amann JM, Nip J, Strom DK, Lutterbach B, Harada H, Lenny N, Downing JR, Meyers S, Hiebert SW. ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol Cell Biol. 2001;21:6470–6483. doi: 10.1128/MCB.21.19.6470-6483.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Park S, Speck NA, Bushweller JH. The role of CBFbeta in AML1-ETO's activity. Blood. 2009;114:2849–2850. doi: 10.1182/blood-2009-07-231233. [DOI] [PubMed] [Google Scholar]
- 66.Roudaia L, Cheney MD, Manuylova E, Chen W, Morrow M, Park S, Lee CT, Kaur P, Williams O, Bushweller JH, Speck NA. CBFbeta is critical for AML1-ETO and TEL-AML1 activity. Blood. 2009;113:3070–3079. doi: 10.1182/blood-2008-03-147207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kwok C, Zeisig BB, Dong S, So CW. The role of CBFbeta in AML1-ETO's activity. Blood. 2010;115:3176–3177. doi: 10.1182/blood-2009-12-260570. [DOI] [PubMed] [Google Scholar]
- 68.Wildonger J, Mann RS. The t(8;21) translocation converts AML1 into a constitutive transcriptional repressor. Development. 2005;132:2263–2272. doi: 10.1242/dev.01824. [DOI] [PubMed] [Google Scholar]
- 69.Higuchi M, O'Brien D, Kumaravelu P, Lenny N, Yeoh EH, Downing JR. Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer Cell. 2002;1:63–74. doi: 10.1016/s1535-6108(02)00016-8. [DOI] [PubMed] [Google Scholar]
- 70.Oravecz-Wilson KI, Philips ST, Yilmaz OH, Ames HM, Li L, Crawford BD, Gauvin AM, Lucas PC, Sitwala K, Downing JR, Morrison SJ, Ross TS. Persistence of leukemia-initiating cells in a conditional knockin model of an imatinib-responsive myeloproliferative disorder. Cancer Cell. 2009;16:137–148. doi: 10.1016/j.ccr.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rubnitz JE, Raimondi SC, Halbert AR, Tong X, Srivastava DK, Razzouk BI, Pui CH, Downing JR, Ribeiro RC, Behm FG. Characteristics and outcome of t(8;21)-positive childhood acute myeloid leukemia: a single institution's experience. Leukemia. 2002;16:2072–2077. doi: 10.1038/sj.leu.2402633. [DOI] [PubMed] [Google Scholar]
- 72.Raimondi SC, Chang MN, Ravindranath Y, Behm FG, Gresik MV, Steuber CP, Weinstein HJ, Carroll AJ. Chromosomal abnormalities in 478 children with acute myeloid leukemia: clinical characteristics and treatment outcome in a cooperative pediatric oncology group study-POG 8821. Blood. 1999;94:3707–3716. [PubMed] [Google Scholar]
- 73.Pession A, Rondelli R, Basso G, Rizzari C, Testi AM, Fagioli F, De Stefano P, Locatelli F. Treatment and long-term results in children with acute myeloid leukaemia treated according to the AIEOP AML protocols. Leukemia. 2005;19:2043–2053. doi: 10.1038/sj.leu.2403869. [DOI] [PubMed] [Google Scholar]
- 74.Perel Y, Auvrignon A, Leblanc T, Michel G, Reguerre Y, Vannier JP, Dalle JH, Gandemer V, Schmitt C, Mechinaud F, Lejars O, Piguet C, Couillaud G, Pautard B, Landman-Parker J, Thuret I, Aladjidi N, Baruchel A, Leverger G. Treatment of childhood acute myeloblastic leukemia: dose intensification improves outcome and maintenance therapy is of no benefit--multicenter studies of the French LAME (Leucemie Aigue Myeloblastique Enfant) Cooperative Group. Leukemia. 2005;19:2082–2089. doi: 10.1038/sj.leu.2403867. [DOI] [PubMed] [Google Scholar]
- 75.Wiemels JL, Xiao Z, Buffler PA, Maia AT, Ma X, Dicks BM, Smith MT, Zhang L, Feusner J, Wiencke J, Pritchard-Jones K, Kempski H, Greaves M. In utero origin of t(8;21) AML1-ETO translocations in childhood acute myeloid leukemia. Blood. 2002;99:3801–3805. doi: 10.1182/blood.v99.10.3801. [DOI] [PubMed] [Google Scholar]
- 76.Nucifora G, Larson RA, Rowley JD. Persistence of the 8;21 translocation in patients with acute myeloid leukemia type M2 in long-term remission. Blood. 1993;82:712–715. [PubMed] [Google Scholar]
- 77.Song J, Mercer D, Hu X, Liu H, Li MM. Common leukemia- and lymphoma-associated genetic aberrations in healthy individuals. J Mol Diagn. 2011;13:213–219. doi: 10.1016/j.jmoldx.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Basecke J, Cepek L, Mannhalter C, Krauter J, Hildenhagen S, Brittinger G, Trumper L, Griesinger F. Transcription of AML1/ETO in bone marrow and cord blood of individuals without acute myelogenous leukemia. Blood. 2002;100:2267–2268. doi: 10.1182/blood-2002-06-1673. [DOI] [PubMed] [Google Scholar]
- 79.Paschka P, Marcucci G, Ruppert AS, Mrozek K, Chen H, Kittles RA, Vukosavljevic T, Perrotti D, Vardiman JW, Carroll AJ, Kolitz JE, Larson RA, Bloomfield CD. Adverse prognostic significance of KIT mutations in adult acute myeloid leukemia with inv(16) and t(8;21): a Cancer and Leukemia Group B Study. J Clin Oncol. 2006;24:3904–3911. doi: 10.1200/JCO.2006.06.9500. [DOI] [PubMed] [Google Scholar]
- 80.Cairoli R, Beghini A, Grillo G, Nadali G, Elice F, Ripamonti CB, Colapietro P, Nichelatti M, Pezzetti L, Lunghi M, Cuneo A, Viola A, Ferrara F, Lazzarino M, Rodeghiero F, Pizzolo G, Larizza L, Morra E. Prognostic impact of c-KIT mutations in core binding factor leukemias: an Italian retrospective study. Blood. 2006;107:3463–3468. doi: 10.1182/blood-2005-09-3640. [DOI] [PubMed] [Google Scholar]
- 81.Schnittger S, Kohl TM, Haferlach T, Kern W, Hiddemann W, Spiekermann K, Schoch C. KIT-D816 mutations in AML1-ETO-positive AML are associated with impaired event-free and overall survival. Blood. 2006;107:1791–1799. doi: 10.1182/blood-2005-04-1466. [DOI] [PubMed] [Google Scholar]
- 82.Osato M, Asou N, Abdalla E, Hoshino K, Yamasaki H, Okubo T, Suzushima H, Takatsuki K, Kanno T, Shigesada K, Ito Y. Biallelic and heterozygous point mutations in the Runt domain of the AML1/PEBP2aB gene associated with myeloblastic leukemias. Blood. 1999;93:1817–1824. [PubMed] [Google Scholar]
- 83.Song W-J, Sullivan MG, Legare RD, Hutchings S, Tan X, Kufrin D, Ratajczak J, Resende IC, Haworth C, Hock R, Loh M, Felix C, Roy D-C, Busque L, Kurnit D, Willman C, Gewirtz AM, Speck NA, Bushweller JH, Li FP, Gardiner K, Poncz M, Maris JM, Gilliland DG. Haploinsufficiency of CBFA2 (AML1) causes familial thrombocytopenia with propensity to develop acute myelogenous leukamia (FPD/AML) Nature Genet. 1999;23:166–175. doi: 10.1038/13793. [DOI] [PubMed] [Google Scholar]
- 84.Imai Y, Kurokawa M, Izutsu K, Hangaishi A, Takeuchi K, Maki K, Ogawa S, Chiba S, Mitani K, Hirai H. Mutations of the AML1 gene in myelodysplastic syndrome and their functional implications in leukemogenesis. Blood. 2000;96:3154–3160. [PubMed] [Google Scholar]
- 85.Gelsi-Boyer V, Trouplin V, Adelaide J, Aceto N, Remy V, Pinson S, Houdayer C, Arnoulet C, Sainty D, Bentires-Alj M, Olschwang S, Vey N, Mozziconacci MJ, Birnbaum D, Chaffanet M. Genome profiling of chronic myelomonocytic leukemia: frequent alterations of RAS and RUNX1 genes. BMC Cancer. 2008;8:299. doi: 10.1186/1471-2407-8-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kuo MC, Liang DC, Huang CF, Shih YS, Wu JH, Lin TL, Shih LY. RUNX1 mutations are frequent in chronic myelomonocytic leukemia and mutations at the C-terminal region might predict acute myeloid leukemia transformation. Leukemia. 2009;23:1426–1431. doi: 10.1038/leu.2009.48. [DOI] [PubMed] [Google Scholar]
- 87.Radtke I, Mullighan CG, Ishii M, Su X, Cheng J, Ma J, Ganti R, Cai Z, Goorha S, Pounds SB, Cao X, Obert C, Armstrong J, Zhang J, Song G, Ribeiro RC, Rubnitz JE, Raimondi SC, Shurtleff SA, Downing JR. Genomic analysis reveals few genetic alterations in pediatric acute myeloid leukemia. Proc Natl Acad Sci U S A. 2009;106:12944–12949. doi: 10.1073/pnas.0903142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Byrd JC, Mrozek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, Pettenati MJ, Patil SR, Rao KW, Watson MS, Koduru PR, Moore JO, Stone RM, Mayer RJ, Feldman EJ, Davey FR, Schiffer CA, Larson RA, Bloomfield CD. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–4336. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 89.Nucifora G, Birn DJ, Espinosa R, III, Erickson P, LeBeau MM, Roulston D, McKeithan TW, Drabkin H, Rowley JD. Involvement of the AML1 gene in the t(3;21) in therapy-related leukemia and in chronic myeloid leukemia in blast crisis. Blood. 1993;81:2728–2734. [PubMed] [Google Scholar]
- 90.Harada H, Harada Y, Tanaka H, Kimura A, Inaba T. Implications of somatic mutations in the AML1 gene in radiation-associated and therapy-related myelodysplastic syndrome/acute myeloid leukemia. Blood. 2003;101:673–680. doi: 10.1182/blood-2002-04-1010. [DOI] [PubMed] [Google Scholar]
- 91.Steensma DP, Gibbons RJ, Mesa RA, Tefferi A, Higgs DR. Somatic point mutations in RUNX1/CBFA2/AML1 are common in high-risk myelodysplastic syndrome, but not in myelofibrosis with myeloid metaplasia. Eur J Haematol. 2005;74:47–53. doi: 10.1111/j.1600-0609.2004.00363.x. [DOI] [PubMed] [Google Scholar]
- 92.Bejar R, Stevenson K, Abdel-Wahab O, Galili N, Nilsson B, Garcia-Manero G, Kantarjian H, Raza A, Levine RL, Neuberg D, Ebert BL. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364:2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Greenberg P, Cox C, LeBeau MM, Fenaux P, Morel P, Sanz G, Sanz M, Vallespi T, Hamblin T, Oscier D, Ohyashiki K, Toyama K, Aul C, Mufti G, Bennett J. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89:2079–2088. [PubMed] [Google Scholar]
- 94.Dicker F, Haferlach C, Sundermann J, Wendland N, Weiss T, Kern W, Haferlach T, Schnittger S. Mutation analysis for RUNX1, MLL-PTD, FLT3-ITD, NPM1 and NRAS in 269 patients with MDS or secondary AML. Leukemia. 2010;24:1528–1532. doi: 10.1038/leu.2010.124. [DOI] [PubMed] [Google Scholar]
- 95.Flach J, Dicker F, Schnittger S, Schindela S, Kohlmann A, Haferlach T, Kern W, Haferlach C. An accumulation of cytogenetic and molecular genetic events characterizes the progression from MDS to secondary AML: an analysis of 38 paired samples analyzed by cytogenetics, molecular mutation analysis and SNP microarray profiling. Leukemia. 2011;25:713–718. doi: 10.1038/leu.2010.304. [DOI] [PubMed] [Google Scholar]
- 96.Zharlyganova D, Harada H, Harada Y, Shinkarev S, Zhumadilov Z, Zhunusova A, Tchaizhunusova NJ, Apsalikov KN, Kemaikin V, Zhumadilov K, Kawano N, Kimura A, Hoshi M. High frequency of AML1/RUNX1 point mutations in radiation-associated myelodysplastic syndrome around Semipalatinsk nuclear test site. J Radiat Res (Tokyo) 2008;49:549–555. doi: 10.1269/jrr.08040. [DOI] [PubMed] [Google Scholar]
- 97.Alter BP. Cancer in Fanconi anemia, 1927–2001. Cancer. 2003;97:425–440. doi: 10.1002/cncr.11046. [DOI] [PubMed] [Google Scholar]
- 98.Quentin S, Cuccuini W, Ceccaldi R, Nibourel O, Pondarre C, Pages MP, Vasquez N, Dubois d'Enghien C, Larghero J, Peffault de Latour R, Rocha V, Dalle JH, Schneider P, Michallet M, Michel G, Baruchel A, Sigaux F, Gluckman E, Leblanc T, Stoppa-Lyonnet D, Preudhomme C, Socie G, Soulier J. Myelodysplasia and leukemia of Fanconi anemia are associated with a specific pattern of genomic abnormalities that includes cryptic RUNX1/AML1 lesions. Blood. 2011;117:e161–e170. doi: 10.1182/blood-2010-09-308726. [DOI] [PubMed] [Google Scholar]
- 99.Kohlmann A, Grossmann V, Klein HU, Schindela S, Weiss T, Kazak B, Dicker F, Schnittger S, Dugas M, Kern W, Haferlach C, Haferlach T. Next-generation sequencing technology reveals a characteristic pattern of molecular mutations in 72.8% of chronic myelomonocytic leukemia by detecting frequent alterations in TET2, CBL, RAS, and RUNX1. J Clin Oncol. 2010;28:3858–3865. doi: 10.1200/JCO.2009.27.1361. [DOI] [PubMed] [Google Scholar]
- 100.Owen C, Barnett M, Fitzgibbon J. Familial myelodysplasia and acute myeloid leukaemia--a review. Br J Haematol. 2008;140:123–132. doi: 10.1111/j.1365-2141.2007.06909.x. [DOI] [PubMed] [Google Scholar]
- 101.Owen C. Insights into familial platelet disorder with propensity to myeloid malignancy (FPD/AML) Leuk Res. 2010;34:141–142. doi: 10.1016/j.leukres.2009.07.037. [DOI] [PubMed] [Google Scholar]
- 102.Liew E, Owen CJ. Familial myelodysplastic syndromes - a review of the literature. Haematologica. 2011 doi: 10.3324/haematol.2011.043422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Matheny CJ, Speck ME, Cushing PR, Zhou Y, Corpora T, Regan M, Newman M, Roudaia L, Speck CL, Gu TL, Griffey SM, Bushweller JH, Speck NA. Disease mutations in RUNX1 and RUNX2 create nonfunctional, dominant-negative, or hypomorphic alleles. Embo J. 2007;26:1163–1175. doi: 10.1038/sj.emboj.7601568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Watanabe-Okochi N, Kitaura J, Ono R, Harada H, Harada Y, Komeno Y, Nakajima H, Nosaka T, Inaba T, Kitamura T. AML1 mutations induced MDS and MDS/AML in a mouse BMT model. Blood. 2008;111:4297–4308. doi: 10.1182/blood-2007-01-068346. [DOI] [PubMed] [Google Scholar]
- 105.Sun W, Downing JR. Haploinsufficiency of AML1 results in a decrease in the number of LTR-HSCs while simultaneously inducing an increase in more mature progenitors. Blood. 2004;104:3565–3572. doi: 10.1182/blood-2003-12-4349. [DOI] [PubMed] [Google Scholar]
- 106.Marcucci G, Mrozek K, Ruppert AS, Maharry K, Kolitz JE, Moore JO, Mayer RJ, Pettenati MJ, Powell BL, Edwards CG, Sterling LJ, Vardiman JW, Schiffer CA, Carroll AJ, Larson RA, Bloomfield CD. Prognostic factors and outcome of core binding factor acute myeloid leukemia patients with t(8;21) differ from those of patients with inv(16): a Cancer and Leukemia Group B study. J Clin Oncol. 2005;23:5705–5717. doi: 10.1200/JCO.2005.15.610. [DOI] [PubMed] [Google Scholar]
- 107.Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ, Schifano E, Booth J, van Putten W, Skrabanek L, Campagne F, Mazumdar M, Greally JM, Valk PJ, Lowenberg B, Delwel R, Melnick A. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Silva FP, Swagemakers SM, Erpelinck-Verschueren C, Wouters BJ, Delwel R, Vrieling H, van der Spek P, Valk PJ, Giphart-Gassler M. Gene expression profiling of minimally differentiated acute myeloid leukemia: M0 is a distinct entity subdivided by RUNX1 mutation status. Blood. 2009:3001–3007. doi: 10.1182/blood-2009-03-211334. [DOI] [PubMed] [Google Scholar]