Abstract
Chemical genetics in yeast has shown great potential in clarifying the pharmacology of various drugs. Investigating these results from a systems perspective has uncovered many facets of natural chemical tolerance, but many cellular interactions of chemicals still remain poorly understood. To uncover previously overlooked players in resistance to chemical stress we integrated several independent chemical genetics datasets with protein-protein interactions and a comprehensive collection of yeast protein complexes. As consequence we were able to identify the potential targets and mode of action of certain poorly understood compounds. However, most complexes recovered in our analysis appear to perform indirect roles in countering deleterious effects of chemicals by constituting an underlying intricate buffering system that has been so far underappreciated. This buffering role appears to be largely contributed by complexes pertaining to chromatin and vesicular dynamics. The former set of complexes seems to act by setting up or maintaining gene expression states necessary to protect the cell against chemical effects. Among the latter complexes we found an important role for specific vesicle tethering complexes in tolerating particular sets of compounds indicating that different chemicals might be routed via different points in the intracellular trafficking system. We also suggest a general operational similarity between these complexes and molecular capacitors (e.g. the chaperone Hsp90). Both have a key role in increasing the system’s robustness, although at different levels, through buffering stress and mutation, respectively. Therefore, it is conceivable that some of these complexes identified here might have roles in molding the evolution of chemical resistance and response.
INTRODUCTION
In the past decade chemical genetics of the budding yeast Saccharomyces cerevisiae has greatly advanced due to remarkable technological developments in robotic automation, imaging and high-throughput molecular biology1-4. These studies have tackled the genetics of cell-chemical interactions of an enormous catalog of chemicals on a genomic scale. An important objective of these studies is the identification of the genetic basis for the natural resistance or survival capability of yeast to a test substance present in its growth medium. Such investigations work under the assumption that deletion or haploinsufficiency (i.e. reduced copy number) of a gene which is required for natural resistance to the test chemical would diminish the fitness of yeast in the presence of that chemical. This reduction in fitness is typically tested by measuring the growth (a proxy for fitness) of homozygous or heterozygous gene deletion strains in the presence of the test chemical relative to that in the absence of the chemical5, 6. Genes uncovered in such screens have been used to identify pharmacological targets, mode of action, off-target effects and stress responses pertaining to various chemicals1-4. Although direct targets of chemicals and multi-drug resistance genes (e.g. ABC transporters) have been the primary concerns of such studies, they have consistently uncovered a large array of genes whose direct relationship to the chemical to which they offer resistance is rather unclear 7, 8. In some cases these genes have been investigated for off-target effects or have been ignored as pleiotropic manifestations of chemical stress 1, 4, 7. It is also possible that within this apparently confounding mass of genes that score as positives in the resistance to a given chemical there are genuine components of a deeper cellular infrastructure required for survival. These could represent underlying survival adaptations that act over and beyond the direct targets of drugs or the multi-drug resistance factors. We hoped to use the chemical genetics data that has accumulated over the years to tease out different underappreciated mechanisms of survival that are active in yeast when presented with chemical challenges.
The data generated by the numerous chemical genetics studies is scattered across the literature and is often presented in different formats (Additional file 1). Hence, we had to survey the chemical genetics literature systematically to collect the available data, integrate the results from different studies and reorganize them into a consistent format that we could use for further investigations. We then successively integrated this chemical-gene interaction network with protein-protein interaction data and with a database of manually curated protein complexes (CYC2008) 9. Thus, we obtained an integrated network that linked chemicals to different protein complexes. We used this chemical-protein complex network in conjunction with what is understood of the pharmacology of the chemicals to disentangle the components of the cellular infrastructure used to handle diverse chemical stresses. As a consequence, we obtained evidence for a multi-level network connecting distinct cellular functions such as vacuolar pH, mitochondrial protein synthesis, vesicular transport and chromatin remodeling-dependent gene regulation, which act as potent buffers against chemical insults. We also present evidence for functional differentiation among different chromatin remodeling and vesicular transport complexes in responding to different types of chemical stress.
RESULTS AND DISCUSSION
Construction of chemical-protein complex network and its biological significance
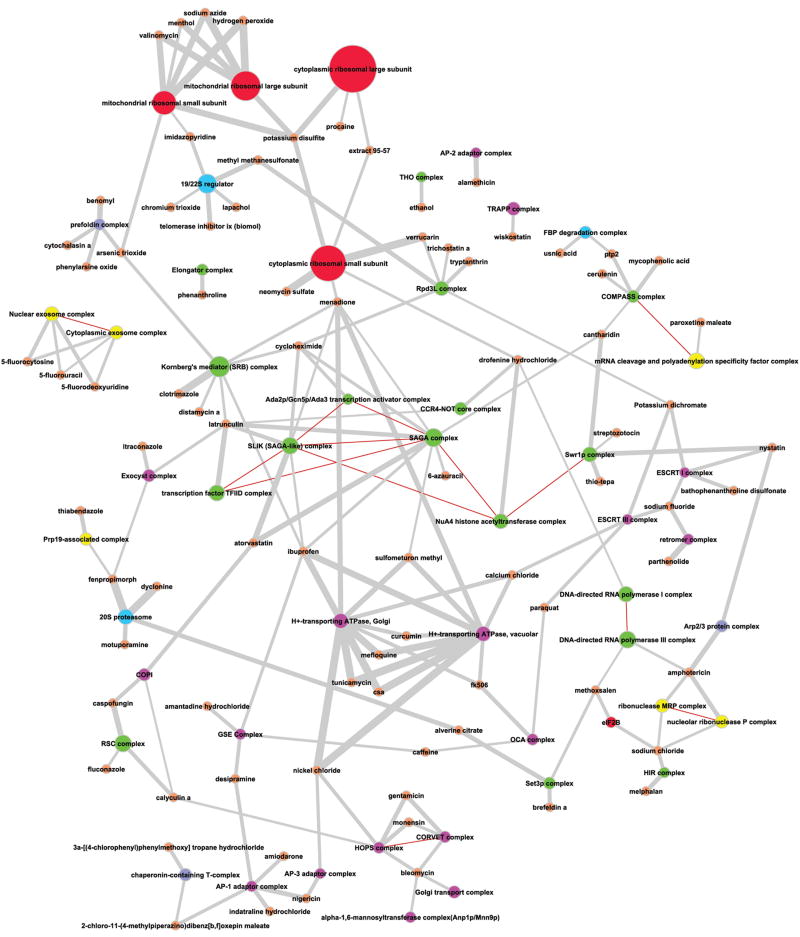
We integrated the data from 34 publications reporting distinct chemical genetics studies (Additional file 1) to assemble a graph termed the chemical phenotype (CP) network (T. M. Venancio, S. Balaji, S. Geetha, L. Aravind; manuscript in preparation), representing the entire set of non-redundant chemical-gene interactions recovered in these studies. Thus, in the CP network the nodes are either genes or chemicals; susceptibility of yeast to a chemical when a given gene is deleted or haploinsufficient results in an edge between the gene and the chemical in the CP network. The CP network was further integrated with the protein-protein interaction data (BioGrid database, version 2.0.47) 10 to construct a hybrid network. The hybrid network contains the significant interactions between chemicals and gene products, which were identified through simulations using degree-preserving random CP networks (see methods for details). This network contained 1,754 proteins and 331 chemical compounds (nodes) and 3,970 edges (Additional file 2). To reduce the dimensionality of the data, we mapped these protein-chemical interactions on to protein complexes from the CYC2008 curated protein complex database 9. We then identified and retained only the highly significant interactions (p ≤ 10-4) between protein complexes and chemicals by assessing the enrichment of chemical-gene product linkages in a given complex (see methods for details). This chemical-protein complex network (CPCnet) is a directed, bimodal network that contains 134 nodes (83 chemical and 51 protein complexes) and 182 edges representing the interactions between them (Fig. 1). The CPCnet contains only 12.5% of the total number of complexes characterized in yeast, suggesting that a relatively small fraction of the known complexes are have significant roles in resistance to specific chemicals. By rendering the network using the edge-weighted spring embedded (Kamada-Kawai) algorithm11, we were able to visualize several dense sub-graphs, which grouped together functionally related complexes such as those in involved in mitochondrial protein synthesis, vesicular transport, the exosome and chromatin remodeling (Fig. 1).
Figure 1. Network of chemical compounds and protein complexes.
High-Confidence interactions (Fisher Exact Test; p ≤ 10-4) between proteins and chemical compounds were obtained through the integration of chemical-genetic, protein-protein interaction and protein complex data. Although the CPCnet is a directed graph, the arrowheads are not shown, since the directionality is not considered in this analysis. Edges connecting chemicals and protein complexes (gray) were weighted according to the negative log10(p) of each interaction. Red edges connect complexes which compositions have at least one protein in common and are all unweighted. The sizes of nodes representing protein complexes are proportional to the number of unique proteins in the complex, whereas all chemical nodes have fixed and equal sizes. Color codes: Orange: chemical compounds; Green: chromatin-related complexes; Pink: Golgi/vacuolar/vesicular complexes; Yellow: RNA processing apparatus; Red: Translation machinery; Blue: proteasome; Purple: cytoskeleton-related complexes. The subgraph mentioned in the text interlinking several vesicular dynamics complexes is at the bottom of the figure.
In constructing this network we have used data from both homozygous as well as heterozygous deletions. Previous studies have proposed that chemical profiling via haploinsufficiency tends to recover direct interactions between chemicals and their targets 1, 3. In contrast, profiling of homozygous deletion mutants is believed to be less capable of recovering direct targets, because in this case the product of the deleted gene is absent. Instead, homozygous deletions are believed to recover interactions that might have an indirect buffering role against the deleterious effects of a compound, although there have been no objective tests differentiating direct from indirect effects in either type of deletion screen. When considering a protein complex, homozygous deletion of one of its components could “weaken” it and potentially expose a direct protein target of a chemical in the complex. Thus, in principle, homozygous deletions do have the potential to be informative regarding direct interactions between a chemical and a protein complex. Hence, we used data from both homozygous and heterozygous deletions in constructing the CPCnet. At the same time, we kept in the mind the caveat that interpreting this network requires a case-by-case analysis of its structure. We then inspected the CPCnet for recovery of the known effects of different chemicals and found that indeed several previously known chemical effects are represented in it (see below for further discussion of examples). This increased our confidence in the CPCnet as a predictive tool in deciphering poorly understood chemical effects. However, examination of the results showed that, as expected from the above discussion, at least two distinct effects contributed to the edges in this network: 1) Direct interactions between chemicals and particular protein complexes and 2) indirect buffering effects wherein a complex appears to confer natural resistance to a compound by virtue of its action which might be proximal or distal to the actual effect of the chemical. By further studying these latter effects we uncovered several previously poorly characterized buffering systems that might act at different levels in limiting the deleterious effects of substances.
Potential examples of direct interactions between chemicals and protein complexes
The most obvious interpretation of edges in this network is that reflective of direct interactions between chemicals and protein components of a particular complex. This applies only to a subset of the edges in the network. One example is the connection between 5-fluorouracil and the exosome complexes (Fig. 1). This is consistent with previous reports showing the inhibition of the exosome by this compound in different eukaryotes, including yeast 2, 3. This helped us to better understand other interactions of the exosome in the CPCnet, namely connections to 5-fluorocytosine and 5-fluorodeoxyuridine (Fig. 1). Being related to the base analog 5-fluorouracil, both these compounds could directly interact with the exosome via incorporation into the RNA substrates of this complex. Alternatively, they could be converted to 5-fluorouracil via a base metabolism pathway that has been observed in fungi 12.
Similarly, in other cases comparison of the interactions of less-understood compounds with previously known direct interactions recovered in the CPCnet helped in elucidating the mode of action. We found that two structurally distinct compounds, neomycin, an aminoglycoside and verrucarin A, a trichothecene are strongly connected to the cytoplasmic ribosomal small subunit (Fig. 1). Neomycin is known to bind ribosomal A-site (which recognizes the aminoacyl-tRNA) and affect the fidelity of translation13. Early studies have shown that verrucarin A binds eukaryotic ribosomes and has a negative effect on translation elongation14. Interestingly, the similar mode-of-action of verrucarin A and neomycin was also previously suggested by the integration of genetic interactions with the chemical genetics data 8. By inspecting the ribosomal proteins connected to both drugs, we found that neomycin and verrucarin A are respectively linked to 16 and 15 proteins, of which 12 are shared by both compounds. This is a significant overlap, especially given that the cytoplasmic ribosomal small subunit has 57 proteins. Mutations in some of these ribosomal proteins (e.g. Rps4) are known to affect accuracy of translation15, an effect similar to that of neomycin. Hence, we suggest that is possible that despite their structural distinctness, both neomycin sulfate and verrucarin A have a similar mode of action. This mode of action of verrucarin A, which is seen in the CPCnet, cannot be easily recovered from the unprocessed CP network because the relevant interactions are drowned in a mass of lower significance interactions. This example demonstrates the utility of the CPCnet in generating a specific hypothesis on drug action which can be tested via direct pharmacological experiments.
Indirect interactions: buffers act at several levels to protect cells against deleterious substances
The role of protein complexes as buffers which protect against chemical insults was highlighted by the recovery of mitochondrion-drug interactions in the CPCnet. Sodium azide and hydrogen peroxide were found to be strongly linked to the small and large subunits of the mitochondrial ribosome (Fig. 1). However, it is known that neither of these substances directly affects those complexes. Instead, sodium azide binds irreversibly to heme to inhibit cytochrome oxidase and disrupt the electron transport chain in the mitochondrial membrane16. H2O2 causes oxidative stress due to release of nascent oxygen which affects mitochondrial function (e.g. down-regulation of mitochondrial ribosomal RNAs) 17. Thus, the connection of the mitochondrial ribosomal subunits to these chemicals is an indirect one: it represents the role of these subunits as potential buffers against the above compounds by sustaining the production of mitochondrial proteins, which are either directly targeted or repressed by the action of these chemicals.
Further examination of the CPCnet revealed that it is dominated by certain functional groups of complexes (Fig 1, note color coding). The most prominent of these functional groups are the complexes involved in chromatin structure and dynamics (17 distinct complexes) and those involved in Golgi, vesicular and vacuolar dynamics (17 distinct complexes), which together constitute ~66.5% of the complexes represented in the network. In most cases, the pharmacological targets of a particular chemical (when these are known) are not part of the protein complexes linked to that chemical in the CPCnet (Fig. 1). Although off-target effects are possible, it should be noted that there is no shared group of proteins or likely homologs of the known targets among the complexes linked to these chemicals. Furthermore, even though some of these complexes share several protein subunits (e.g. TFIID and SAGA), this is not correlated with the number or the type of the chemicals to which they are connected. Taken together these observations suggest that complexes involved in chromatin and Golgi/vesicular and vacuolar dynamics actually function as two important classes of buffers, which form basis for the natural resistance to a diversity of chemical insults. Previous studies had noted a generic role for the vesicular and vacuolar systems as a possible basis for generic multi-drug resistance phenotypes7. However, to our knowledge these prior studies have not attempted explore the individual role of the various complexes involved in vesicular dynamics in natural resistance to specific groups of mechanistically diverse chemicals as seen in the CPCnet (Fig. 1). Hence, we examined in detail the roles of the complexes related to chromatin and vesicular/vacuolar dynamics to understand how they might help in protecting against chemical effects.
Vesicular/vacuolar dynamics and natural resistance to drugs
We observed that a subgraph of the CPCnet contains several distinct chemicals and multiple complexes such as the related HOPS and CORVET, the Golgi transport, the AP-3 and AP-1 adaptors and the mannosyltransferase, all of which are related to vesicular dynamics and Golgi function (Fig. 1). The majority of compounds in this subgraph are only connected to these functional systems and none else. This suggests that the actions of protein complexes at different points within the functional guild of vesicular dynamics are central to buffering the deleterious effects of this diverse set of chemicals. For example, we noticed that three mechanistically different antibiotics, namely monensin (a membrane cation-channel former), gentamicin (an aminoglycoside believed to inhibit protein synthesis) and bleomycin (a DNA-damaging agent) were connected to the two homologous complexes HOPS and CORVET18 (Fig. 1). This is consistent with prior experimental evidence that the intracellular toxicity of gentamicin in eukaryotic cells is related to its transit via the vesicular system19-21. In the case of monensin too its presence in the vesicular system is supposed to be a major aspect of its toxicity, presumably because of its membrane-pore forming capability19-21. Interestingly, though bleomycin’s main action is on DNA, an early study of its effect in mammalian fibroblasts and lymphoblasts indicated alterations in Golgi vesicle morphology 22. Though these studies were conducted in an animal system, in general terms it is consistent with our above observations. In addition to HOPS and CORVET we observed that the CPCnet contains the conserved Golgi transport (COG), TRAPP and exocyst complexes, all of which are tethers that link vesicles and target membranes prior to fusion 23. This observation suggests that the natural resistance to a variety of chemicals depends on an intact vesicle fusion machinery that could potentially play a vital role in transmitting the drugs to the lysosome/vacuole. In support of such a vacuolar targeting mechanism in eliminating deleterious chemicals, we find that the two proton-pump ATP complexes namely the Golgi and vacuolar H+ pumps are major hubs connected to a wide range of chemicals differing in structure and mode of action (Fig. 1). It is possible that the pH change brought about by the action of these pumps has a major detoxification function. However, given that the different tethering complexes are connected to different sets of chemicals we propose that each set of chemicals is intercepted and channelized via a distinct point in the vesicular trafficking system.
The OCA (Oxidant-induced Cell-cycle Arrest) complex was previously identified on the basis of proteomic and genetic studies as a complex required for survival in face of various redox stresses 24, 25. It is comprised of 5 paralogous protein phosphatases of the PTPase superfamily (Oca1, Oca2, Siw14, Oca4 and Oca6; two of which appear to lack the catalytic cysteine) and one protein with a TBC (Tre-2, Bub2 and Cdc16) superfamily domain (Oca5). While it is suspected to have protein phosphatase activity, its precise role remains unclear. Proteins with the TBC domain interact with RAB GTPases26, which are key proteins in mediating the interactions between the vesicular tether and the fusion complexes. Although the TBC domains are known to act as GTPase activating proteins (GAPs), the version in Oca5 lacks the key arginine and glutamine fingers required for this activity suggesting that it might merely bind RAB GTPases as a regulator. Disruption of one of the phosphatase subunits of this complex, Siw14p, has been implicated in a potential endocytosis defect under nutrient deprived conditions27. More speculatively, it remains to be seen if these phosphatases can also act on phosphatidyl inositol derivatives – a comparable phosphatase complex, the myotubularins in animals have been proposed to regulate vesicular transport by acting as phosphatases for phosphatidyl inositol derivatives 28. Thus, based on sequence analysis the proteins of the OCA complex and the preponderance of vesicle-related complexes in the CPCnet we suggest that the complex might regulate vesicular dynamics in distinct ways via action of both the TBC and phosphatase domains.
Role of chromatin complexes in innate resistance to diverse chemicals
Detailed examination of the 17 distinct complexes involved in chromatin structure and dynamics also indicated that their links to chemicals are primarily due to indirect interactions. Some striking examples of such connections of chromatin complexes are those to: 1) the poriferan toxin latrunculin, which disrupts actin polymerization, 2) sulfometuron methyl, an inhibitor of the enzyme acetohydroxyacid synthase, involved in amino acid biosynthesis, 3) NaCl, a general osmolarity determinant, 4) ethanol, which perhaps affects signaling GTPases and 5) clotrimazole, an inhibitor of the cytochrome p450 oxidase. Rather than their primary target complexes they show statistically significant linkage to different chromatin complexes such as SAGA, THO and HIR in the CPCnet (Fig. 1). In contrast, connections between chemicals and chromatin complexes emerging by virtue of the direct targets in those complexes are far fewer. Two examples are the link between a histone deacetylase inhibitor trichostatin A and the Rpd3L complex and that between the DNA-bending agent distamycin A and the mediator (SRB) complex. As noted above, there is a stark difference in the degree distribution of TFIID and SAGA in the CPCnet, despite the fact that they are partially redundant both in function and composition29, 30 (Fig. 1). Though TFIID and SAGA together contribute to the expression of most of the RNA Pol II transcribed genes in yeast 29, it is known that the former is largely related to house-keeping functions whereas the latter tends to be involved in environmental stress responses 29, 30. Further, all the interactions of the “SAGA-like complex” (SLIK)31 with chemicals are shared with SAGA suggesting that that the two complexes are functionally redundant with respect to surviving chemical insults. SAGA, a hub in the CPCnet, is implicated in resistance to 10 chemicals, which are structurally are mechanistically distinct (Fig. 1). However, we also found that multi-drug resistance genes show no statistically significant enrichment for SAGA binding in their promoter regions (T. M. Venancio, S. Balaji, S. Geetha, L. Aravind; manuscript in preparation). Hence, the role of the SAGA complex in resistance to chemicals appears to be due to its activation of numerous individual genes required to specifically counter different chemicals rather than in activating some multi-drug resistance pathways.
Like SAGA, the RNA Pol II associated mediator complex is another highly connected chromatin complex in the CPCnet (Fig. 1). While it shares several links to chemicals with the SAGA complex, it has unique connections of its own in the network. Interestingly, previous studies have implicated the mediator complex in response to non-chemical stresses, both by itself and in cooperation with the SAGA complex 30, 32. Thus, our results suggest a major role for both the mediator and SAGA in specifically enhancing the transcription of batteries of genes that are not only required for responding to heat, osmotic and nutrient deprivation stress but also for surviving deleterious chemical effects. All other chromatin complexes, for instance the SWR1 involved in deposition of the variant histone H2AZ, the COMPASS histone methylase complex, the NuA4 histone acetylase complex and the Rpd3L histone deacetylase complex are connected to far fewer chemicals. Most of these chemicals are generally not shared with the SAGA complex, even though in some cases the complexes share subunits with the SAGA complex (Fig. 1). This suggests that certain chromatin complexes might have specialized functions in regulating gene expression in context of countering the deleterious effects of smaller sets of chemicals. It is conceivable that these complexes act by generating or removing epigenetic marks in the vicinity of genes whose products are related to countering the effects of chemicals and thereby maintain them in a particular transcriptional state that is suitable for survival. Interestingly, Rpd3L complex was recently shown to be involved in the environmental stress response initiation 33. Our results suggest that this role might more generally even be required for responding to certain types of chemical stress. Thus, these chromatin complexes might be viewed as distally acting buffers which stabilize and allow a persistent response to chemicals at the level of transcription.
The cytoskeleton and the chemical response
Several chemicals in the CPCnet such as benomyl, cytochalasin A, phenylarsine oxide and arsenic trioxide are known to disrupt key cytoskeletal components of either or both the microtubule and microfilament systems 34-36. Interestingly, with exception of the Arp2/3 complex none of the target complexes affected by these substances were represented in the network. Instead the network contains multiple connections between such drugs and the two key chaperone complexes, prefoldin and CCT, implicated in the assembly of both microtubules and microfilaments36, 37 (Fig. 1). Inclusion of these chaperone complexes in the CPCnet suggests that they are likely to be the critical for resisting chemical attacks on the cytoskeleton by facilitating their repeated re-assembly upon disruption. This again indicates that as in the above cases the buffering effect of key cytoskeletal chaperone complexes, rather than direct interaction with chemicals, is the main process captured by the CPCnet.
Interestingly, the potent actin-depolymerizing agent latrunculin was not connected to any of the cytoskeletal chaperones. Rather it was connected primarily to the chromatin complexes TFIID, SAGA, SLIK, mediator and CCR4-NOT. This suggests that even in the case of the cytoskeleton the distal chromatin hubs, SAGA/SLIK and the mediator, rather than the proximal chaperone complexes could have a major protective function. It would be straightforward to assume that this is due to the above-proposed role of these complexes in maintenance of particular transcriptional states, perhaps in this case of the cytoskeletal components themselves or their chaperones. However, the fact that latrunculin is unique in being specifically linked to multiple chromatin complexes including TFIID, which is apparently limited to basal transcription does raise the possibility of a more direct effect. Given the roles of filamentous actin in the constitution and networking of chromatin complexes38-40, we believe that latrunculin induced depolymerization might impinge directly affect these complexes. Hence, it is possible that these chromatin-actin interactions are even more critical in natural resistance to latrunculin than the buffering of cytoplasmic cytoskeleton against actin depolymerization.
GENERAL CONCLUSIONS
We report herein a novel network representation of the chemical genetics data to investigate the roles of different protein complexes play in the natural chemical resistance of yeast. We find that the majority of complexes recovered in our analysis function indirectly to counter deleterious effects of chemicals, though a few of them appear to be the primary targets of the connected chemicals. By investigating the likely direct connections we were able to throw light on the potential targets of some compounds whose pharmacology was previously unclear. On the other hand, the indirect connections appear to involve a diverse set of complexes, dominated by those involved in chromatin or vesicular dynamics in buffering the cells against chemical attacks. The role of chromatin complexes in chemical survival may be compared to that of the trithorax and polycomb complexes in stabilizing and maintaining initially established patterns of gene expression during metazoan and plant development 41. This observation might also help in explaining the presence of a large number of functionally comparable and partially overlapping complexes involved in chromatin dynamics throughout the eukaryotic superkingdom – they are likely to be buffers that help in maintaining gene expression patterns to provide resistance against various extrinsic stresses which are not uncommon in fluctuating environments. Recovery of cytoskeletal chaperones in the CPCnet as protectors of the cytoskeleton against chemical disruption is reminiscent of the role of chaperones as “capacitors” against mutational disruption cellular networks 42. Hence, more generally, the buffering effect that we argue for the majority of the complexes recovered in the CPCnet could be compared to the concept of capacitors in the context of protection against mutations. Like capacitors, these complexes might be seen as innate shields that allow the organism to survive in the presence of the diverse chemicals, while allowing the more gradual evolution of specific responses. Hence, as in the case of capacitors, studying evolutionary changes in the CPCnet complexes might help in understanding the differing levels of noise (i.e. natural variability) to withstand chemical stress in a population or between different organismic lineages.
MATERIALS AND METHODS
We created an extensive collection of chemical genetics datasets, comprising homozygous and heterozygous profiling experiments (Additional file 1). The data was processed to assemble a basic network, the chemical phenotype (CP) network, which is a non-redundant compilation of all the datasets. This structure was then integrated with protein-protein interactions obtained from the BioGrid database (BioGrid database, version 2.0.47) 10 to assemble a hybrid network (Additional file 2). We found the term hybrid appropriate here, since protein-protein and chemical genetics interactions were integrated to create the network. The procedure involved the comparison of the set of interaction partners of a given protein with the genes required for growth in the presence of a particular chemical compound. Some attempts have been successfully tried before, such as the method devised by Parsons et al. to analyze chemical-genetic with genetic interactions and pathways 8. We reasoned that if the proteins required for growth in the presence of a chemical compound A significantly overlap with the interaction partners of a particular protein B, then A and B are likely to present some functional relationship that may help in understanding the mode of action of the compound A given the molecular functions the protein B. We used 1,000 randomized CP networks to assess the significance of each interaction and retained only the statistically significant interactions (p-value ≤ 10-3) with direct support in the CP network. Protein-protein interaction studies typically aim to depict the landscape of protein complexes in the cell. Thus, we further explored the hybrid network at a higher-level, by integrating a high-quality collection of manually curated protein complexes 9. We calculated the enrichment of the protein partners of each chemical in the hybrid network with the annotated components of each complex. To extract a high confidence network for the detailed study presented here we retained only interactions with a high statistical significance (Fisher Exact Test; p-value ≤ 10-4), thereby creating the CPCnet (Fig. 1). The protein complexes were than manually annotated with higher-order functional categories depicted in different colors. The graph was rendered using the Cytoscape software 43.
Supplementary Material
Acknowledgments
We acknowledge the Intramural Research Program of the National Institutes of Health, USA for funding our research. Due to space constraints we apologize for not citing all the manuscripts from which we extracted data but provide them in the supplementary material.
List of abbreviations
- CP network
Chemical phenotype network
- CPCnet
Chemical-protein complex network
Footnotes
Supplementary material is available at:
ftp://ftp.ncbi.nih.gov/pub/aravind/CPCNET/CPC_additional_file1.pdf
ftp://ftp.ncbi.nih.gov/pub/aravind/CPCNET/CPC_additional_file2.txt
ftp://ftp.ncbi.nih.gov/pub/aravind/CPCNET/CPC_additional_file3.pdf
Competing interests None declared.
References
- 1.Ericson E, Gebbia M, Heisler LE, Wildenhain J, Tyers M, Giaever G, Nislow C. PLoS Genet. 2008;4:e1000151. doi: 10.1371/journal.pgen.1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giaever G, Flaherty P, Kumm J, Proctor M, Nislow C, Jaramillo DF, Chu AM, Jordan MI, Arkin AP, Davis RW. Proc Natl Acad Sci U S A. 2004;101:793–798. doi: 10.1073/pnas.0307490100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lum PY, Armour CD, Stepaniants SB, Cavet G, Wolf MK, Butler JS, Hinshaw JC, Garnier P, Prestwich GD, Leonardson A, Garrett-Engele P, Rush CM, Bard M, Schimmack G, Phillips JW, et al. Cell. 2004;116:121–137. doi: 10.1016/s0092-8674(03)01035-3. [DOI] [PubMed] [Google Scholar]
- 4.Parsons AB, Lopez A, Givoni IE, Williams DE, Gray CA, Porter J, Chua G, Sopko R, Brost RL, Ho CH, Wang J, Ketela T, Brenner C, Brill JA, Fernandez GE, et al. Cell. 2006;126:611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 5.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, et al. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 6.Hoon S, St Onge RP, Giaever G, Nislow C. Trends Pharmacol Sci. 2008;29:499–504. doi: 10.1016/j.tips.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, Altman RB, Davis RW, Nislow C, Giaever G. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parsons AB, Brost RL, Ding H, Li Z, Zhang C, Sheikh B, Brown GW, Kane PM, Hughes TR, Boone C. Nat Biotechnol. 2004;22:62–69. doi: 10.1038/nbt919. [DOI] [PubMed] [Google Scholar]
- 9.Pu S, Wong J, Turner B, Cho E, Wodak SJ. Nucleic Acids Res. 2009;37:825–831. doi: 10.1093/nar/gkn1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bahler J, Wood V, Dolinski K, Tyers M. Nucleic Acids Res. 2008;36:D637–640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamada T, Kawai S. Information Processing Letters. 1989;31:7–15. [Google Scholar]
- 12.Hayes JD, Wolf CR. Harwood Academic; Amsterdam, the Netherlands: 1997. pp. 52–53. [Google Scholar]
- 13.Tor Y. Biochimie. 2006;88:1045–1051. doi: 10.1016/j.biochi.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Ehrlich KC, Daigle KW. Biochim Biophys Acta. 1987;923:206–213. doi: 10.1016/0304-4165(87)90005-5. [DOI] [PubMed] [Google Scholar]
- 15.Synetos D, Frantziou CP, Alksne LE. Biochim Biophys Acta. 1996;1309:156–166. doi: 10.1016/s0167-4781(96)00128-5. [DOI] [PubMed] [Google Scholar]
- 16.Keilin D, Keilin J. Prepared for publication by Joan Keilin. University Press; Cambridge[Eng.]: 1966. The history of cell respiration and cytochrome. [Google Scholar]
- 17.Crawford DR, Wang Y, Schools GP, Kochheiser J, Davies KJ. Free Radic Biol Med. 1997;22:551–559. doi: 10.1016/s0891-5849(96)00380-2. [DOI] [PubMed] [Google Scholar]
- 18.Peplowska K, Markgraf DF, Ostrowicz CW, Bange G, Ungermann C. Dev Cell. 2007;12:739–750. doi: 10.1016/j.devcel.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 19.Blackburn AS, Avery SV. Antimicrob Agents Chemother. 2003;47:676–681. doi: 10.1128/AAC.47.2.676-681.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dinter A, Berger EG. Histochem Cell Biol. 1998;109:571–590. doi: 10.1007/s004180050256. [DOI] [PubMed] [Google Scholar]
- 21.Sundin DP, Sandoval R, Molitoris BA. J Am Soc Nephrol. 2001;12:114–123. doi: 10.1681/ASN.V121114. [DOI] [PubMed] [Google Scholar]
- 22.Krishan A. Cancer Res. 1973;33:777–785. [PubMed] [Google Scholar]
- 23.Kummel D, Heinemann U. Curr Protein Pept Sci. 2008;9:197–209. doi: 10.2174/138920308783955252. [DOI] [PubMed] [Google Scholar]
- 24.Addinall SG, Downey M, Yu M, Zubko MK, Dewar J, Leake A, Hallinan J, Shaw O, James K, Wilkinson DJ, Wipat A, Durocher D, Lydall D. Genetics. 2008;180:2251–2266. doi: 10.1534/genetics.108.092577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pu S, Vlasblom J, Emili A, Greenblatt J, Wodak SJ. Proteomics. 2007;7:944–960. doi: 10.1002/pmic.200600636. [DOI] [PubMed] [Google Scholar]
- 26.Neuwald AF. Trends Biochem Sci. 1997;22:243–244. doi: 10.1016/s0968-0004(97)01073-6. [DOI] [PubMed] [Google Scholar]
- 27.Care A, Vousden KA, Binley KM, Radcliffe P, Trevethick J, Mannazzu I, Sudbery PE. Genetics. 2004;166:707–719. doi: 10.1534/genetics.166.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lecompte O, Poch O, Laporte J. Trends Biochem Sci. 2008;33:453–460. doi: 10.1016/j.tibs.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 29.Huisinga KL, Pugh BF. Mol Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- 30.Zapater M, Sohrmann M, Peter M, Posas F, de Nadal E. Mol Cell Biol. 2007;27:3900–3910. doi: 10.1128/MCB.00089-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pray-Grant MG, Schieltz D, McMahon SJ, Wood JM, Kennedy EL, Cook RG, Workman JL, Yates JR, 3rd, Grant PA. Mol Cell Biol. 2002;22:8774–8786. doi: 10.1128/MCB.22.24.8774-8786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan X, Struhl K. PLoS ONE. 2009;4:e5029. doi: 10.1371/journal.pone.0005029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alejandro-Osorio AL, Huebert DJ, Porcaro DT, Sonntag ME, Nillasithanukroh S, Will JL, Gasch AP. Genome Biol. 2009;10:R57. doi: 10.1186/gb-2009-10-5-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ling YH, Jiang JD, Holland JF, Perez-Soler R. Mol Pharmacol. 2002;62:529–538. doi: 10.1124/mol.62.3.529. [DOI] [PubMed] [Google Scholar]
- 35.Singh P, Rathinasamy K, Mohan R, Panda D. IUBMB Life. 2008;60:368–375. doi: 10.1002/iub.42. [DOI] [PubMed] [Google Scholar]
- 36.Vainberg IE, Lewis SA, Rommelaere H, Ampe C, Vandekerckhove J, Klein HL, Cowan NJ. Cell. 1998;93:863–873. doi: 10.1016/s0092-8674(00)81446-4. [DOI] [PubMed] [Google Scholar]
- 37.Geissler S, Siegers K, Schiebel E. Embo J. 1998;17:952–966. doi: 10.1093/emboj/17.4.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gieni RS, Hendzel MJ. Biochem Cell Biol. 2009;87:283–306. doi: 10.1139/O08-133. [DOI] [PubMed] [Google Scholar]
- 39.Andrin C, Hendzel MJ. J Biol Chem. 2004;279:25017–25023. doi: 10.1074/jbc.M401805200. [DOI] [PubMed] [Google Scholar]
- 40.Hu Q, Kwon YS, Nunez E, Cardamone MD, Hutt KR, Ohgi KA, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG, Fu XD. Proc Natl Acad Sci U S A. 2008;105:19199–19204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Breiling A, Sessa L, Orlando V. Int Rev Cytol. 2007;258:83–136. doi: 10.1016/S0074-7696(07)58002-2. [DOI] [PubMed] [Google Scholar]
- 42.Bergman A, Siegal ML. Nature. 2003;424:549–552. doi: 10.1038/nature01765. [DOI] [PubMed] [Google Scholar]
- 43.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.