Cryo–electron tomography of Chlamydomonas flagella reveals previously uncharacterized features of the radial spokes, including structural heterogeneity and direct interactions with dyneins and between the spoke heads. A “radial spoke 3 stand-in” occupies what would be the site of a third spoke in organisms with spoke triplets.
Abstract
Radial spokes (RSs) play an essential role in the regulation of axonemal dynein activity and thus of ciliary and flagellar motility. However, few details are known about the complexes involved. Using cryo–electron tomography and subtomogram averaging, we visualized the three-dimensional structure of the radial spokes in Chlamydomonas flagella in unprecedented detail. Unlike many other species, Chlamydomonas has only two spokes per axonemal repeat, RS1 and RS2. Our data revealed previously uncharacterized features, including two-pronged spoke bases that facilitate docking to the doublet microtubules, and that inner dyneins connect directly to the spokes. Structures of wild type and the headless spoke mutant pf17 were compared to define the morphology and boundaries of the head, including a direct RS1-to-RS2 interaction. Although the overall structures of the spokes are very similar, we also observed some differences, corroborating recent findings about heterogeneity in the docking of RS1 and RS2. In place of a third radial spoke we found an uncharacterized, shorter electron density named “radial spoke 3 stand-in,” which structurally bears no resemblance to RS1 and RS2 and is unaltered in the pf17 mutant. These findings demonstrate that radial spokes are heterogeneous in structure and may play functionally distinct roles in axoneme regulation.
INTRODUCTION
Motile cilia and flagella are highly ordered organelles that are used by a great variety of species and cell types to generate movement. Defects in the assembly or function of these organelles are linked to several human diseases called ciliopathies, such as primary ciliary dyskinesia (reviewed in Fliegauf et al., 2007). The remarkably conserved architecture of these machines is built on a microtubule-based 9 + 2 scaffold called the axoneme (Figure 1A). Dynein ATPases, large minus end–directed microtubule motors, generate forces that produce relative sliding between subsets of the nine doublet microtubules (DMTs), which drives propagated bending of cilia and flagella (Satir, 1968; Sale and Satir, 1977; Summers and Gibbons, 1971; Gibbons, 1981). These axonemal dyneins are anchored to the A tubule of each doublet in two rows—the outer dynein arms (ODAs) and inner dynein arms (IDAs)—which bind in an ATP-sensitive manner reversibly to the B tubule of the neighboring doublet (reviewed in Kamiya, 2002). The generation of traveling waves and beating patterns typical for cilia and flagella requires precise regulation and coordination of the activity of the tens of thousands of dyneins in a single axoneme; if all dyneins on the nine DMTs were active simultaneously, the resulting forces would balance, and the axoneme would enter a rigor-like state.
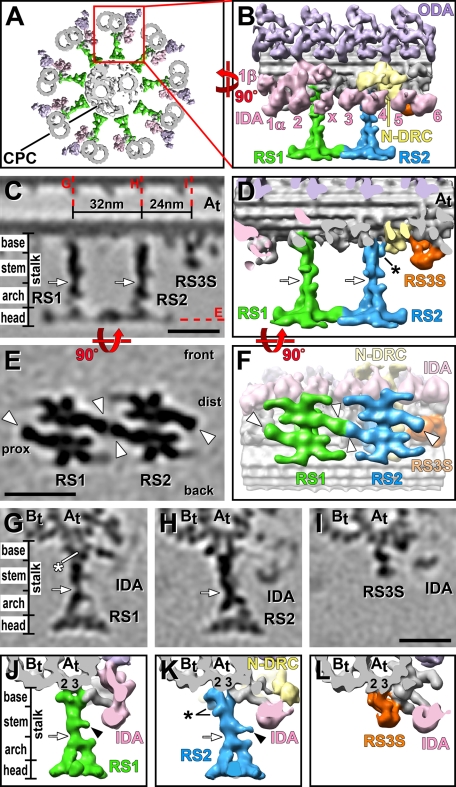
FIGURE 1:
3D structure of RSs in pWT. (A) Composite isosurface rendering of an axoneme cross section viewed from proximal (flagellar base) toward distal (flagellar tip). The CPC is surrounded by nine DMTs, one of which is boxed in red and shown in longitudinal orientation in B. (B–L) Isosurface renderings (B, D, F, J–L) and tomographic slices (C, E, G–I) show the averaged axonemal repeat in longitudinal views (B–F) from the front (B–D) and bottom (E–F; i.e., looking from the central pair toward the DMT), as well as in cross-sectional views (G–L). The structure of both RS1 (green) and RS2 (blue) can be separated into four regions: the two-pronged base closest to the DMT, the stem, the arch with two pillars, and the head domain closest to the CPC (C, D, G, H, J, K). Base, stem, and arch together represent the portion of the RS called the “stalk” in previous publications. White arrows in C, D, G, H, J and K indicate the waist, a narrow region that separates stem and arch; note the small protrusion from the stem (black arrowheads in J and K). (E, F) Bottom views of the two RS heads, revealing that each head consists of two rotationally symmetric halves; white arrowheads point to a region found in each of these four halves named the “ear.” The front, back, proximal (prox), and distal (dist) sides of the average are indicated in E for easier orientation in relation to the front view shown in C. Red dotted lines in C indicate the locations of the tomographic slices shown in E and G–I. Although both RSs and RS3S (orange) are attached to the same protofilaments A2 and A3 of the A tubule (At, 2, 3; G–L), the RS3S structure does not resemble any regions of RS1 or RS2. In contrast, the structures of RS1 and RS2 are very similar overall but also show several small differences: the base of RS2 is slightly larger than the RS1 base. The additional densities in RS2 are labeled with a black asterisk in D and K. The white asterisk in G points to a density present only in RS1. Other labels: B tubule (Bt), inner dynein arm heads (IDA 1α, 1β, 2–6, and x; rose), nexin–dynein regulatory complex (N-DRC, yellow), outer dynein arm (ODA, purple). Color coding of structures is preserved in all of the figures. Inner arm dyneins are numbered as in Nicastro et al. (2006). Scale bars, 25 nm; scale bar in I is applicable to G–I.
Radial spokes (RSs), which project from the DMTs toward the central pair complex (CPC; Figure 1A), are key players in this regulatory system that coordinates dynein activity (Smith and Sale, 1992; Howard et al., 1994). In both the unicellular alga Chlamydomonas reinhardtii and in humans, failure to assemble RSs results in flagellar and ciliary paralysis (Witman et al., 1978; Sturgess et al., 1979). Ultrastructural studies reported several cases of human respiratory disease or immotile cilia syndrome in which RSs are shortened or not assembled at all (Sturgess et al., 1979; Afzelius, 2004), highlighting their role in ciliary motility and human disease. Genetic evidence revealed that dynein regulation in axonemes is mediated in part by transient mechanical interactions between the CPC and RSs, which ultimately result in altering the phosphorylation state of the dynein arms through a biochemical signal transduction pathway involving kinases and phosphatases associated with the DMTs (Warner, 1970; Warner and Satir, 1974; Goodenough and Heuser, 1985; Porter and Sale, 2000; Yang et al., 2000; Yang and Sale, 2000; Smith and Yang, 2004; Elam et al., 2011).
Depending on the species, sets of either two (e.g., Chlamydomonas) or three (e.g., Tetrahymena, Trypanosoma, sea urchins, vertebrates) RSs repeat every 96 nm along the axoneme length (Dentler and Cunningham, 1977; Olson and Linck, 1977; Goodenough and Heuser, 1985; Nicastro et al., 2005, 2006; Koyfman et al., 2011). The location of the RSs is highly conserved and in precise register with the dyneins and other axonemal structures. RS1 is proximally located adjacent to the two-headed I1 inner dynein complex; RS2 is located 32 nm further distal, adjacent to the nexin–dynein regulatory complex (N-DRC); and in organisms with RS triplets, RS3 is located 24 nm distal of RS2 (Goodenough and Heuser, 1985; Nicastro et al., 2005,; Heuser et al., 2009). Both I1 dynein and the N-DRC are key targets of the RS/CPC signal transduction pathways that control microtubule sliding (Huang et al., 1982; Porter et al., 1992; Smith and Sale, 1992; Habermacher and Sale, 1997). RSs are large, multisubunit complexes with at least 23 radial spoke proteins (RSP1–23). RSP1–17 were identified through two-dimensional gel analysis of the Chlamydomonas mutant pf14, which contains a mutation in RSP3 and lacks any identifiable RS structure in electron microscopy (EM) images, whereas RSP18–23 were identified from purified RS 20S fractions (Huang et al., 1981; Piperno et al., 1981; Curry and Rosenbaum, 1993; Yang et al., 2001, 2006). Many of the spoke proteins contain domains associated with signal transduction, either directly, for example as transducers of calcium- or cAMP-mediated signals, or indirectly, as scaffolds binding transducer molecules (Yang et al., 2001; Patel-King et al., 2004; Dymek and Smith, 2007). Although we know some of the RS regulatory elements, the molecular interactions and mechanisms for controlling ciliary and flagellar motility are unknown.
Previous studies showed that the RSs are T-shaped structures, with the stalk projecting from the DMTs toward the CPC and the “T bar”–shaped spoke heads connecting to the CPC (Warner, 1970; Warner and Satir, 1974; Goodenough and Heuser, 1985; Nicastro et al., 2006). So far, structural studies of RSs have been limited by the specimen preparation and the ability to resolve structural details by classic EM of chemically fixed and stained specimens. However, structural analysis at greater resolution is essential for a better understanding of both the three-dimensional (3D) structure and subunit organization of radial spokes and of how their components connect to allow regulatory signal transduction between the CPC and the RS head, as well as between the RS bases and the remainder of the axoneme. Recent cryo–electron tomography (cryo-ET) studies analyzed the 3D structures of axonemal complexes in greater detail (Nicastro et al., 2006; Bui et al., 2008; Heuser et al., 2009). Here we focus on the ultrastructure of the RSs and how this informs our understanding of their function. Our 3D reconstructions of the RSs in Chlamydomonas axonemes—wild type (WT) and mutants—have a resolution as good as 3.3 nm, allowing the visualization of several previously uncharacterized features.
RESULTS
Cryo-ET reveals the overall structure of the two RSs in Chlamydomonas axonemes
To determine the structure of RS1 and RS2, we used cryo-ET to obtain 3D reconstructions of axonemes from Chlamydomonas WT and a mutant rescue with WT phenotype named pseudo WT (pWT) (for more details, see Table 1). The subtomographic averages of the axonemal repeats revealed the same RS structures in both strains, but greater detail is visible in the averages of the pWT data that reached the highest resolution of 3.3 nm (Supplemental Figure S1 and Table 1). To correlate the region that was previously named the “RS head” with our 3D structure of the RSs, we compared the WT RS structures with headless RSs of the Chlamydomonas mutant pf17 (see later discussion).
TABLE 1:
Strains used in this study.
| Name | Strain | Missing RSPs | Phenotype | Reference | Number of tomograms | Averaged repeats | Resolution (nm) |
|---|---|---|---|---|---|---|---|
| WT | CC-125, 137c mt+ | None | Normal | Harris (2009) | 5 | 750 | 3.9 |
| pWT | pf2-4::PF2-GFP | None | Normal | R. Bowera | 5 | 750 | 3.3 |
| pf17 | CC-262, mt- | 1, 4, 6, 9, 10 | RS head missing, paralyzed | Huang et al. (1981) | 5 | 800 | 4.0 |
aThe pWT strain was obtained by rescue of the N-DRC defective pf2 mutant strain (Rupp and Porter, 2003). It was generated by transformation of the pf2 mutant with the green fluorescent protein–tagged PF2 gene from WT and restores motility and biochemical and structural defects to levels indistinguishable from WT (R. Bower and M. Porter, unpublished results).
Both RSs are 42 nm long as measured from the A tubule to the tips of the CPC projections (Figure 1A). The overall shapes of RS1 and RS2 are very similar, but an in-depth analysis of the tomographic reconstructions and averages revealed several small differences in morphology and size (see later discussion). The width of each RS varies from 3 to 7 nm along the stalk to a maximum of 32 nm across each RS head when viewed in longitudinal orientation (Figure 1, B–F). Based on the electron densities in the axonemal averages, RS1 and RS2 have an estimated mass of ∼3.1 and ∼3.4 MDa, respectively, making RS2 slightly larger than RS1.
Three-dimensional structure of the RS stalk and heterogeneity between RS1 and RS2
Previous studies of RSs described them as T-shaped structures consisting of two parts—the stalk and, perpendicularly oriented to it, the spoke head (Warner, 1970; Warner and Satir, 1974; Goodenough and Heuser, 1985). Although we keep this terminology, our data provide sufficient details and unique morphological features to distinguish three major subregions within the stalk as follows: a two-pronged spoke base that facilitates RS attachment to the DMT, and a stem in the middle, which is followed by an arch that connects to the spoke head (Figure 1, C, D, G, H, J, and K, and Supplemental Video S1). The distance between the bases of RS1 and RS2 is 32 nm (Figure 1C), as previously reported (Goodenough and Heuser, 1985).
Most of the observed mass difference between RS1 and RS2 is due to structural heterogeneity between the RS bases, which are the ∼8-nm-long, two-pronged regions of the stalk that attach the RSs to the A tubule (Figure 1, C, D, G, H, J, and K, and Supplemental Video S1). The bases of RS1 and RS2 have approximate mass of 500 and 700 kDa, respectively. Each RS makes two connections to two protofilaments of the A tubule (Figures 1, G, H, J, and K, and 2, H and I; also see Supplemental Video S2). Cross-sectional views of the DMT show that the front prong of each RS base attaches to protofilament A2 (protofilaments are numbered according to the most widely accepted numbering system; reviewed in Linck and Stephens, 2007), and the back prong to A3 (Figures 1, G, H, J, and K, and 2, A–D). In addition to the DMT connection, we found that the tail domain of an IDA connects to the front prong of each RS base, that is, inner arm 2 (IA2) (dynein a or d connects to RS1, and IA3 (dynein c) to RS2 (Figures 1, J and K, and 2, A–D; also see Supplemental Video S3; inner arm dyneins are numbered according to structural location described in Nicastro et al., 2006).
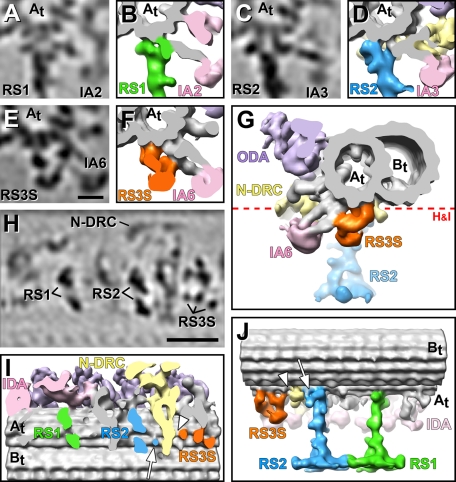
FIGURE 2:
Connections between RSs and neighboring structures. Tomographic slices (A, C, E, H) and isosurface renderings (B, D, F, G, I, J) of the averaged axonemal repeat from pWT show the connections of the bases of RS1, RS2, and RS3S. (A–G) Cross-sectional views from proximal (A–F) and distal (G) reveal direct connections between the tails of several inner dynein arms and the RSs or RS3S: inner arm dynein IA2 attaches to the front of the RS1 base (A, B) and dynein IA3 to the front of the RS2 base (C, D). The RS3S forms a connection to the tail of inner arm dynein IA6 (E–G); this dynein tail also attaches to the A-tubule. (H, I) Longitudinal bottom views of the axonemal repeat cut through the base of the RSs, as indicated by a red dotted line in G, show that both RS1 and RS2 have two attachment sites to the A tubule (At). These attachment points lie within a plane that is roughly perpendicular to the DMT axis. In contrast, RS3S has three attachment sites to the A tubule, which are aligned roughly parallel to the DMT axis, that is, in a plane rotated ∼90° compared with the docking sites of RS1 and RS2. (J) A longitudinal view from the back side reveals the three docking points of the RS3S and connections to the N-DRC from both the base of RS2 (white arrows in I and J) and of RS3S (white arrowhead in I and J). Other labels: B tubule (Bt), inner dynein arm heads (IDA 2, 3, 6; rose), nexin–dynein regulatory complex (N-DRC, yellow), outer dynein arms (ODA, purple). Scale bars, 10 nm in E, applicable for A–E, and 25 nm in H.
Although the structure of both RS bases and their connections with the DMT and dyneins are remarkably similar, we found two differences between the basal regions of RS1 and RS2. First, the space between the two prongs and the beginning of the stem density is slightly smaller for RS1 than for RS2, suggesting that there might be additional density present in RS1 that is missing in RS2 (Figure 1, compare G to H; Supplemental Video S1). Second, RS2 has additional densities attached to the back prong of the base and the neighboring stem region (asterisk in Figure 1, D and K; Supplemental Video S1). One of these additional densities is part of a unique connection between the back prong of RS2 and the N-DRC (white arrow in Figure 2, I and J; Supplemental Video S2), which was previously described (connection 8 in Heuser et al., 2009).
The stem region of the RS stalk is ∼15 nm long and has an approximate mass of 400 and 500 kDa in RS1 and RS2, respectively (Figure 1, C, D, G, H, J, and K). The slightly larger mass of the RS2 stem is mostly due to the aforementioned additional density at the basal back prong/stem transition (Figures 1K and 2J; Supplemental Video S1). A small protrusion from the mid region of the stem appears slightly shorter in RS1 than RS2 of pWT (Figure 1, compare J to K), but this difference is ambiguous because it was not consistently visible in other studied strains (Supplemental Figures S1 and S2), and the size of the difference is at the resolution limit. Aside from these small differences, both RS stems share the same general shape, including the constriction at the stem/arch intersection, named here the RS waist (white arrows in Figure 1, C, D, G, H, J, and K).
At our resolution, the arch structures of RS1 and RS2 appear identical (Figure 1, D, J, and K, and Supplemental Video S1). Each arch is ∼12 nm long, has an estimated mass of ∼550 kDa, and contains two pillars linking the stem to the RS head at two distinct connection sites (Figure 1, J and K, and Supplemental Video S1). The two pillars are arranged in a plane almost perpendicular to the DMT axis. Therefore the widths of the arch and the spacing between the pillars is invisible in longitudinal views of the axoneme, giving the RSs—including the spoke heads—their typical T-shaped appearance (Figure 1, C and D). In contrast, in DMT cross sections the arches are clearly visible, giving the RSs a stirrup shape in side views (Figure 1, G, H, J, and K, and Supplemental Videos S1 and S3).
The pf17 mutant defines the location of the RS head
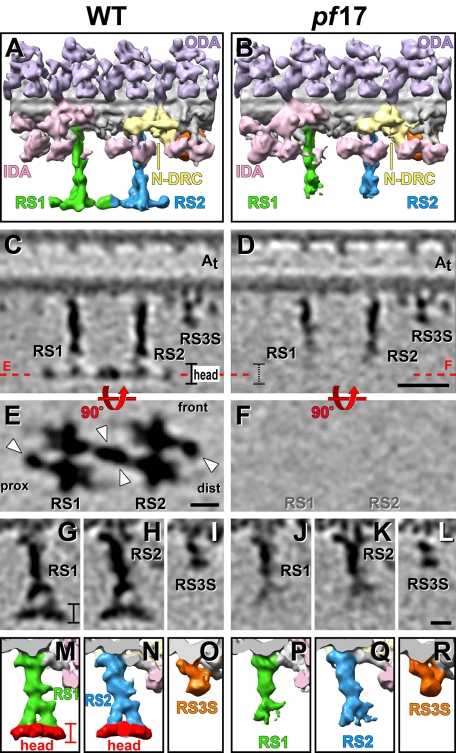
The RS head is the portion of the RSs closest to the CPC. Historically, the spoke head has been defined as the missing T bar in classic EM images of RSs in the Chlamydomonas mutant pf1, which is a mutation of RSP4 but assembles RSs that lack biochemically a subset of 5 RS proteins (RSP1, 4, 6, 9, 10; Luck et al., 1977; Piperno et al., 1981). The same subset of 5 RS proteins is also missing in the Chlamydomonas mutant pf17, with the affected gene product RSP9 (Huang et al., 1981). Comparing our WT RS structures with published classic EM images of WT and RS head-deficient mutants was not sufficient to unambiguously determine the extent of the RS head. Therefore we reconstructed tomograms and used subtomogram averaging of the 96-nm repeat of pf17 axonemes to determine the 3D structure of the RSs in this mutant at improved resolution.
In pf17, both RSs are missing very similar amounts of electron densities at the distal end of the RS structure (Figure 3, B and D, and Supplemental Video 4). A WT-pf17 mutant comparison shows that the entire plate-like density, which usually extends perpendicular to the RS stalk, is missing in RS1 and RS2 of pf17 (Figure 3, compare A, C, and E to B, D, and F; Supplemental Video S4). This observation agrees with the previously reported missing T bar structure (Huang et al., 1981). In addition to this completely missing density, the arch pillars of both RSs appear also shorter in isosurface rendering representations (Figure 3, B, P, and Q, and Supplemental Video S4). However, in tomographic slices of the pf17 average the pillars are still visible, albeit with reduced density compared with the WT structure (Figure 3, D, J, and K). Overall the RS structures of pf17 and WT/pWT are identical for the first ∼25 nm of the stalk, followed by an ∼10-nm-long arch region that appears weaker in density, and after ∼35 nm no electron density can be recognized in pf17. Owing to the nature of subtomogram averaging, it is difficult to draw a precise border where the RS heads begin, but previous studies reported the five RS head proteins completely missing in pf17 rather than just reduced (Huang et al., 1981). Therefore we interpret the last ∼7 nm of the 42-nm-long RSs, which completely lack electron density in our pf17 averages, as the RS heads (colored red in Figure 3, M and N). It is likely that most if not all parts of the arch structures are present in the RSs of pf17, despite appearing reduced in the averages; this would occur because with the arch pillars no longer stably connected to the RS head complex in pf17, the position of the pillars could become more flexible, causing their electron density to get blurred out and appear reduced in the averages, especially toward the axoneme center.
FIGURE 3:
Comparison of the RS structure in WT and the RS-headless mutant pf17. Isosurface renderings (A, B, M–R) and tomographic slices (C–L) from the averaged axonemal repeats of WT and pf17 viewed from the front (A–D), the bottom (E, F), and the proximal side (G–R) show a region defined as the RS head (colored red in M and N) missing in pf17. White arrowheads in E point to the ears of the RS head. The front, proximal (prox), and distal (dist) sides of the average are indicated in E for easier orientation in relation to the front view shown in C and D. Red dotted lines in C and D indicate the locations of the tomographic slices shown in E and F. Although both RS heads are missing in pf17, the RS3S (L, R) and other axonemal structures such as ODA, IDA, and N-DRC are unaffected. Other labels: inner dynein arm heads (IDA, rose), nexin–dynein regulatory complex (N-DRC, yellow), outer dynein arms (ODA, purple). Scale bars, 25 nm in D, 10 nm in E and L. Scale bars in D, E, and L are applicable for C, F, and G–K, respectively. Supplemental Figure S2 demonstrates that the defect observed in pf17 is due to lack of electron density in the region of the RS heads and is not caused by differences in the averaging quality and resolution.
The quality of averaged RS structures, especially of the RS heads, is usually a good measure for data quality and use of uncompressed axonemes for tomographic reconstructions and averaging (Nicastro, 2009). The RSs are long extensions from the DMTs, and in compressed axonemes they become distorted, leading to cone-shaped averages with blurred-out RS heads and decreasing resolution the further away from the DMT one measures. Several lines of evidence demonstrate that the observed RS defects in pf17 axonemes are not due to low-resolution or distortion artifacts. First, the subtomographic averages include only repeats from noncompressed or mildly compressed axonemes. Second, a comparison of axonemal averages from WT/pWT and a number of non-RS mutants with varying qualities and resolutions places the averaged repeat of pf17 at medium quality, similar to the WT average (Supplemental Figure S2). Third, the base and stem region of RS1 and RS2 in pf17 show well-defined details, including the constricted waist, whereas the quality drops abruptly below the waist (Figure 3, D, J, and K). This indicates that the described defects of the pf17 RSs are structural defects caused by the mutation rather than averaging artifacts.
At our resolution, the head structures of RS1 and RS2 appear identical (Figures 1, C–H, J, and K, 2J, and 3, A, C, E, G, H, M, and N; Supplemental Video S2). Each RS head has an estimated mass of ∼1.7 MDa and measures ∼7 nm along the RS axis and up to 32 nm along the DMT axis (Figure 1, C–F). In cross-sectional views of the DMT, both RS heads are up to 25 nm wide and show a curved shape with the concave surface facing the projections of the CPC (Figure 1, G, H, J, and K). Each RS head is composed of two halves that are related by twofold rotational symmetry around the RS stalk and are connected in the spoke head center (Figures 1, E and F, and 3E; Supplemental Video S2). All halves appear identical in shape and mass, suggesting a dimeric nature of each RS head. Each monomer (half) of the RS head contains three small extensions and one major lobe, the “RS ear,” which projects either distal from each front half or proximal from each back half, giving a total of four RS ears (two of RS1 and two of RS2) per 96-nm repeat (white arrowheads in Figures 1, E and F, and 3E). The front ear of RS1 and the back ear of RS2 form a unique connection between RS1 and RS2 (Figures 1, E and F, and 3E), which confirms previous observations made in plastic section EM of a thin connection between the two RSs (Witman et al., 1978). Although the heads of RS1 and RS2 appear structurally identical, on the level of connectivity this connection between the front of RS1 and the back of RS2 introduces heterogeneity between the two RS heads.
In Chlamydomonas a “radial spoke 3 stand-in” occupies the site of RS3
Although Chlamydomonas has only a RS pair per axonemal repeat, many species have RS triplets, with RS3 usually located 24 nm distal of RS2 (Goodenough and Heuser, 1985). An electron density considerably shorter than that for RS1 and RS2 was previously observed at this site in Chlamydomonas (Bui et al., 2008). In our averages we see that this structure, here called radial spoke 3 stand-in (RS3S), bears no structural resemblance to RS1 and RS2 in regard to either shape or attachment to the A tubule (Figures 1 and 2 and Supplemental Video S1). Although RS3S is attached to the same protofilaments A2 and A3 of the DMT as RS1 and RS2, RS3S forms three connections with the A tubule, which are not arranged front to back but along a near-longitudinal axis (Figure 2, H–J, and Supplemental Video S2). The most proximal of these three attachment sites seems to also connect to the N-DRC (white arrowheads in Figure 2, I and J; Supplemental Video S2). RS3S has an estimated mass of ∼950 kDa over a length of ∼18 nm from the DMT to its tip. Three major domains of the RS3S can be distinguished as follows: a small, longitudinally oriented bow between the two proximal DMT docking sites (Figures 1C and 2J), followed by a bulbous knob with an estimated mass of ∼300 kDa (Figures 1, C, D, I, and L, and 2J). This core is embraced by a large arm that starts at the most distal docking side of the RS3S and projects forward to the DMT docking site of the tail domain of the IDA IA6 (dynein a or d; Figure 2, G and J). The arm is also connected to the front side of the RS3S knob (Figure 2, E–G). It is unclear whether the RS3S densities contain any of the previously identified RS proteins, but the structure is markedly distinct from that of both RS1 and RS2. Given the short length of RS3S, it is probably not surprising that the RS3S structure is unaltered in the RS head-deficient mutant pf17 compared with its structure in WT/pWT (Figure 3, compare D, L, and R to C, I, and Q; Supplemental Video S4).
DISCUSSION
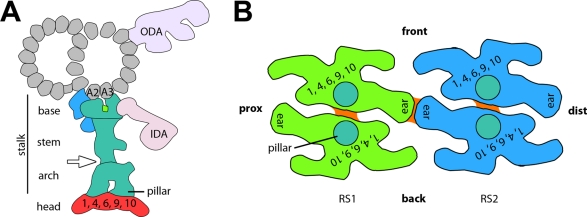
Our results revealed that RS1 and RS2 share overall the same shape but also exhibit small structural heterogeneities. Several features in the base and stem region are different between RS1 and RS2, including a connection between RS2 and the N-DRC. The two RS heads seem structurally identical but are heterogeneous in their connection between the front part of RS1 and the back part of RS2. RS3S shows no structural resemblance with the RS pair. Figure 4 summarizes our findings regarding the RS structure and heterogeneity between RS1 and RS2 in a simplified model.
FIGURE 4:
Schematic model of the RS structure in Chlamydomonas flagella. (A) Cross-sectional view from proximal showing, in turquoise, the consensus structure of RSs, that is, the part of the RS that seems consistent between RS1 and RS2; colored in green is a density that is present in RS1 and absent in RS2, and shown in blue are densities that are present in RS2 and absent from RS1. The region depicted in red is the head that is missing in the mutant pf17. Combined with the results of previous comparative biochemical studies, our data suggest that RS proteins RSP1, 4, 6, 9, and 10 are in the red-colored head region. (B) Bottom view of the heads of RS1 (green) and RS2 (blue) in a similar orientation as shown in Figures 1, E and F, and 3E. The front, proximal (prox), and distal (dist) sides of the model are indicated for easier orientation. The head of each RS consists of two rotationally symmetric units, each of which likely contains at least one copy of each RS head protein. Turquoise circles indicate the positions where the arch's pillars connect to the RS heads. Connections between the symmetric halves are shown in orange. The heads of RS1 and RS2 connect through the front and back ears, respectively.
RS1 and RS2 are structurally and possibly functionally distinct
Our cryo-ET data provide the 3D structure of the two RSs in Chlamydomonas at unprecedented resolution, allowing us to define subregions of the RSs and structural heterogeneity between RS1 and RS2 (summarized in Figure 4A), which may underlie thus-far-uncharacterized functional differences between the two RSs. The RSs had been believed to be homogeneous, as defects observed in classic EM studies of Chlamydomonas RS mutants seemed to affect both RSs similarly (Huang et al., 1981; Goodenough and Heuser, 1985; Curry and Rosenbaum, 1993). This view was challenged recently when Dymek et al. (2011) demonstrated that RNA interference of components of the calmodulin and spoke associated complex (CSC) resulted in the absence of RS2 from many axonemal repeats. These results suggested both that the CSC is located at (or near) the base of RS2 and that it is required for stable DMT attachment of RS2.
The RS2-to–N-DRC connection (Figure 2, I and J; Heuser et al., 2009) is one density unique to RS2. Candidate proteins that might be part of this connection include subunits of the CSC (discussed earlier) and FAP206. FAP206 was recently identified as an N-DRC–associated component but is also significant reduced in the RS mutant pf14 (Lin et al., 2011), which is missing all RS structure visible by classic EM (Piperno et al., 1977). We demonstrated in previous studies that the N-DRC and the I1 inner dynein complex share many structural characteristics and are both major nodes of the dynein regulatory network (Nicastro et al., 2006; Heuser et al., 2009; T. Heuser, C. F. Barber, J. Lin, J. Krell, M. Rebesco, M. E. Porter, and D. Nicastro, unpublished results). It is therefore also interesting that RS1 does not seem to form a direct connection to the I1 complex (Figures 1D and 2J; T. Heuser et al., unpublished results). This heterogeneity in structure and connectivity between RS1 and RS2 indicates that subsets of RSs might have different functions in regulating ciliary and flagellar motility.
This study also produced the unexpected finding that the tails of the dyneins IA2 and IA3 attach to the bases of RS1 and RS2 in a way suggesting that these attachments are essential for docking of IA2 and IA3 to the axoneme. Because neither of these dyneins has been reported missing in the pf14 mutant, it is possible that a portion of the RS base remains in pf14 and stabilizes IA2 and IA3 docking. This is supported by the fact that in situ binding of WT RSs to pf14 axonemes is significantly more efficient after NaCl extraction normally used to remove dyneins (Yang et al., 2001). On the basis of biochemical evidence, Yang et al. (2001) postulated that RSP22 (light chain 8 [LC8]) is present at the base of RSs. Given that LC8 defects affect both RS and dynein arm assembly (Pazour et al., 1998), LC8 could be involved in the docking of the dynein tails to the front prong of the RS bases.
Previous classic EM studies described RSs in many different organisms as mushroom or T shaped (Chasey, 1972, 1974; Goodenough and Heuser, 1985; Curry and Rosenbaum, 1993). We observe the overall architecture of the RS head as two rotationally symmetric halves (Figure 4B), which corresponds well with the description of dimeric RS heads in Tetrahymena (Goodenough and Heuser, 1985). A previous study reported a thin connection between the heads of RS1 and RS2 (Witman et al., 1978), which we resolved in our structures as the ear-to-ear connection between the spoke heads of RS1 and RS2 (Figures 1E, 3E, and 4B). This connection explains previous observations of RS tilting patterns in mussel gill cilia, in which RS1 and RS2 were found to always tilt to the same extent, whereas RS3 did not (Warner and Satir, 1974). Although the RS1-to-RS2 connection made through the ears might merely provide structural stability, it could also be important for RS function by being part of signal transduction or feedback mechanisms. RS tilting is itself expected to be part of the regulatory signaling controlling dynein activity. Our axonemes were isolated in the absence of ATP, and nearly all the observed RSs are untitled. From the occasional tilted RSs observed in our preparations it appears that the entire length of the RS tilts proximal/distal, suggesting that the “hinge” must be at or near the base of the RS. Indeed, the orientation of the two-pronged base might allow flexibility in the proximal/distal direction that would be prohibited by a tripod base such as seen with the RS3S.
Putative location and stoichiometry of some RSPs
Yang et al. (2006) presented a model of RSP localization based on published biochemical data, including dimerization. Summing up the masses of all RSPs placed in this model, plus the recently added subunits RSP18 and 19, results in a total mass of ∼2 MDa, compared with ∼3.1 and ∼3.4 MDa of our RS structures. This suggests either that unidentified RS components exist that are not included in the model or that some of the proteins are present in higher-than-expected copy numbers. The latter case seems to be true for the five RS head proteins RSP1, 4, 6, 9, and 10. Their total mass of 316 kDa is significantly smaller than the ∼1.7-MDa estimate for the RS heads in our 3D structures, meaning that there is room for five to six copies of each RS head protein. Given the twofold rotational symmetry of each RS head (Figures 1, E and F, and 4B), it seems likely that all (or most) head proteins exist in a minimum of two copies per RS. A recent interaction study of RS head proteins demonstrated a central role for RSP4 in the complex binding network in which RSP4 binds RSP1, 9, and 10 and RSP6 binds RSP9 and 10. This suggests that all RS head proteins are likely present in each RS head half. We specifically expect RSP4 and 9 to be present in both halves, as mutants in either one abolish the entire head (mutants pf1 and pf17, respectively) despite the fact that each head half has an independent pillar connection to the stalk. RSP10 is the only protein in the interaction study that showed a propensity to dimerize (Kohno et al., 2011), suggesting that it could be located at a site where two RS head halves interact (orange in Figure 4B). To interpret the putative location of additional RSPs, we attempted to correlate the negative stain images of a RS “12S complex,” a RS assembly intermediate purified from the Chlamydomonas cytoplasm (Diener et al., 2011), with our 3D structures. Diener and coauthors pointed out a paradox between their biochemical and structural data, in that their 12S fraction contained both RS head proteins (RSP 1,4, 6, 9, 10) and RSP 3, which has been identified as part of the RS base by both genetic and biochemical evidence (Diener et al., 1993), whereas the visualized L-shaped protein complexes were considerably shorter (28 nm) than full-length (42 nm) RSs (Diener et al., 2011). The L-shaped 12S protein complex fits into different regions of our cryo-ET density, and without additional positional information an unambiguous correlation was not possible.
The RS3S occupies the site of RS3 but bears no resemblance to RS1 and RS2
Although a previous study (Bui et al., 2008) observed a density in Chlamydomonas axonemes 24 nm distal of RS2 at the location where a third RS would assemble in organisms with RS triplets, the similarity of this complex in structure and protein composition to that of RS1 and RS2 was unclear. Our results clearly demonstrate that RS3S is structurally distinct from RS1 and RS2, not only in length, but also in its morphology and anchoring to the DMT. This suggests either that the RS3S structure is not related to RS3 in other organisms or, if RS3S is an evolutionary remnant of RS3, then the RS3 stalk differs significantly from the structures of RS1 and RS2. The fact that RS3S is connected to inner dynein IA6 and the N-DRC (Figure 2, E–I) suggests that, like RS1 and RS2, RS3S too could be involved in the dynein regulatory network.
In Tetrahymena, no differences have been reported between the structure of RS3 and that of RS1 and RS2. However, if all three RS heads were the same (as is the case for RS1 and RS2 in Chlamydomonas), how would the smaller spacing of 24 nm between RS2 and RS3 accommodate a third full-size RS head? Either RS3 would have to be substantially tilted to fit the three heads next to each other or all three Tetrahymena RS heads would have to be substantially smaller than those of Chlamydomonas, which would break the connection between RS1 and RS2. Alternatively, if RS3 structure and/or connectivity differ significantly from that of RS1 and RS2, this could allow the three RS heads to fit. Future analysis of RS triplets is required to answer these questions. It would also be interesting to see whether the RS3S structure is affected in the Chlamydomonas pf14 mutants or whether the protein composition of RS3S is distinct from that of RS1 and RS2.
The RS heterogeneity found in our structural studies raises a number of interesting questions for further studies. Chief among them is determining the proteomic differences between RS1 and RS2 through identification and analysis of mutants/RNA interference knockdowns that affect one RS and not the other. Such information could provide new insights into what individual roles the two RSs play in regulating dynein activity. Analysis of mutants affecting subsets of radial spokes could also reveal the function of the RS1-to-RS2 head connection in regulatory pathways. In the immediate future, structural and proteomic analysis of mutants such as pf14 and CSC knockdowns will allow us to determine which proteins are involved in RS docking and transmission of RS regulatory signals to the IDAs. We also highly anticipate structural work on organisms with RS triplets, such as Tetrahymena or mammals, which should inform us on both the relationship between RS3S and RS3 and how structurally distinct RS3 is from RS1 and RS2, leading to a greater understanding of how cilia motility is regulated in organisms with RS pairs and triplets.
MATERIALS AND METHODS
Axoneme isolation and cryosample preparation
All C. reinhardtii strains used in this study are summarized in Table 1. Cells were grown in liquid Tris-acetate-phosphate medium (Gorman and Levine, 1965) under a 16-h-light/8-h-dark cycle and harvested by 5-min centrifugation at 1100 × g. After two washes, cells were resuspended in a minimal medium containing 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.4, 1 mM SrCl2, 4% sucrose, and 1 mM dithiothreitol. The pH-shock method (Witman et al., 1972) was used to detach flagella from cell bodies. After deflagellation, 5 mM MgSO4, 1 mM ethylene glycol tetraacetic acid (EGTA), 0.1 mM EDTA, and 100 μg/ml aprotinin, pepstatin, and leupeptin were added to the medium, and cell bodies were pelleted by a 10-min spin at 1800 × g at 4°C. The flagella present in the supernatant were purified in two additional centrifugation steps at 2400 × g and 4°C for 10 min using 5 ml of a 20% sucrose solution as a cushion. The flagellar membrane was removed by detergent treatment for 30 min with 0.1% Nonidet P-40 (Sigma-Aldrich, St. Louis, MO) or 1% IGEPAL CA-630 (Sigma-Aldrich) for 30 min at 4°C. The axonemes were collected by centrifugation at 35,000 × g for 1 h at 4°C. The pelleted axonemes were resuspended in 10 mM HEPES, pH 7.4, 25 mM NaCl, 4 mM MgSO4, 1 mM EGTA, and 0.1 mM EDTA and processed within 24 h.
Quantifoil holey carbon grids (Quantifoil Micro Tools, Jena, Germany) were glow discharged for 30 s at −40 mA before a drop of a 10-nm colloidal gold solution (Sigma-Aldrich) was applied. After drying, grids were washed by briefly dipping them into Milli-Q purified water (Millipore, Billerica, MA), and excess water was removed with a filter paper (Whatman, Piscataway, NJ). After grids dried completely, they were loaded in a home-made plunge-freezing device (Trachtenberg, 1993), and 3 μl of axoneme sample and 1 μl of a 10-fold-concentrated, 10-nm colloidal gold solution were added to the grid. The mixture was blotted with a filter paper (Whatman #1) from the front side for ∼2 s and immediately plunge frozen in liquid ethane to achieve sample vitrification. Grids were stored in liquid nitrogen until examined by EM.
Cryo-ET
Details of tilt series acquisition and tomogram reconstruction were published earlier (Heuser et al., 2009; Nicastro, 2009). In brief, cryosamples were transferred into a transmission electron microscope (Tecnai F30; FEI, Hillsboro, OR), which was operated at 300 keV, using a cryoholder (Gatan, Pleasanton, CA). Data acquisition was controlled by the microscope software SerialEM (Mastronarde, 2005). Tilt series were recorded under low-dose conditions by stepwise rotating the sample from −65 to +65° with 1.5–2.5° increments. The cumulative electron dose was limited to ∼100 e/Å2. All images were digitally recorded on a 2k × 2k charge-coupled device camera (Gatan) at a magnification of 13,500, resulting in a pixel size of ∼1 nm, at −6 or −8 μm defocus and using an energy filter (Gatan) in zero-loss mode with 20-eV slit width. The IMOD software package (Kremer et al., 1996) was used for fiducial alignment and cryotomogram reconstruction by weighted backprojection. Only tomograms of noncompressed or mildly compressed and intact axonemes were used for further image processing and data analysis.
Image processing
For subtomogram averaging of the 96-nm-long axonemal repeat along the axonemal DMTs we used the program PEET (Particle Estimation for Electron Tomography; Nicastro et al., 2006). The repeats were computationally cut from the raw tomograms, aligned in three dimensions, and averaged with missing wedge compensation, resulting in an average with increased signal-to-noise ratio and improved resolution (Nicastro et al., 2006; Nicastro, 2009). Two different alignment strategies were tested: in the “global alignment” approach the entire 96-nm repeat unit (volume size, 100 × 100 × 80 nm) was used for 3D alignment, whereas the “local alignment” method optimized the alignment for smaller regions (volume sizes were either 90 × 50 × 32 nm centering on RS1, RS2, and RS3S or 66 × 22 × 32 nm centering on the RS head, respectively). For pf17 the global alignment resulted in the best average and is therefore depicted in all figures of pf17 axonemes. In contrast, the local alignment averages resolved the radial spokes at slightly better detail for WT and pWT and are therefore depicted in all figures of WT/pWT except for Figure 3A and Supplemental Figure S1A. Some of the tomograms were previously used, focusing on other axonemal complexes and using different image processing methods (Heuser et al., 2009; T. Heuser et al., unpublished results). The resolution of all analyzed strains was estimated at the stem part of RS2 using the Fourier shell correlation method (Harauz and Van Heel, 1986) and the 0.5 criterion (Table 1). The UCSF Chimera software package (Pettersen et al., 2004) was used to measure volume sizes and to visualize the axonemal averages by isosurface rendering. Masses of RS parts were estimated by assuming an average protein density of 1.43 g/cm3 (Quillin and Matthews, 2000) and normalizing the isosurface rendering threshold to the mass of DMT (Heuser et al., 2009). The placement of boundaries between RS1/RS2/RS3S and the IDA tails is based on comparison of our images with single-particle structures of dynein (Roberts et al., 2009).
Supplementary Material
Acknowledgments
We are grateful to Mary Porter and Raqual Bower (University of Minnesota, Minneapolis, MN) for providing Chlamydomonas strains and discussion, to Chen Xu for outstanding management of the electron microscopy facility at Brandeis University (Waltham, MA), and Elizabeth Smith (Dartmouth College, Hanover, NH) for critically reading the manuscript. This work was supported by funding from the National Institutes of Health (5R01GM083122), the W. M. Keck Foundation, and the National Science Foundation (DMR-MRSEC-0820492 partially supporting T.H.), by a Pew Scholars Award to D.N., and by a Career Development Program Award from the Leukemia and Lymphoma Society to C.B.
Abbreviations used:
- CPC
central pair complex
- CSC
calmodulin and spoke associated complex
- DMT
doublet microtubule
- EM
electron microscopy
- ET
electron tomography
- IA
inner arm
- IDA
inner dynein arm
- N-DRC
nexin–dynein regulatory complex
- ODA
outer dynein arm
- pWT
pseudo wild type
- RS
radial spoke
- RS3S
radial spoke 3 stand-in
- RSP
radial spoke protein
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-08-0692) on November 9, 2011.
REFERENCES
- Afzelius BA. Cilia-related diseases. J Pathol. 2004;204:470–477. doi: 10.1002/path.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui KH, Sakakibara H, Movassagh T, Oiwa K, Ishikawa T. Molecular architecture of inner dynein arms in situ in Chlamydomonas reinhardtii flagella. J Cell Biol. 2008;183:923–932. doi: 10.1083/jcb.200808050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasey D. Further observations on the ultrastructure of cilia from Tetrahymena pyriformis. Exp Cell Res. 1972;74:471–479. doi: 10.1016/0014-4827(72)90403-x. [DOI] [PubMed] [Google Scholar]
- Chasey D. The three-dimensional arrangement of radial spokes in the flagella of Chlamydomonas reinhardtii. Exp Cell Res. 1974;84:374–380. doi: 10.1016/0014-4827(74)90418-2. [DOI] [PubMed] [Google Scholar]
- Curry AM, Rosenbaum JL. Flagellar radial spoke: a model molecular genetic system for studying organelle assembly. Cell Motil Cytoskeleton. 1993;24:224–232. doi: 10.1002/cm.970240403. [DOI] [PubMed] [Google Scholar]
- Dentler WL, Cunningham WP. Structure and organization of radial spokes in cilia of Tetrahymena pyriformis. J Morphol. 1977;153:143–151. doi: 10.1002/jmor.1051530110. [DOI] [PubMed] [Google Scholar]
- Diener DR, Ang LH, Rosenbaum JL. Assembly of flagellar radial spoke proteins in Chlamydomonas: Identification of the axoneme binding domain of radial spoke protein 3. J Cell Biol. 1993;123:183–190. doi: 10.1083/jcb.123.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener DR, Yang P, Geimer S, Cole DG, Sale WS, Rosenbaum JL. Sequential assembly of flagellar radial spokes. Cytoskeleton (Hoboken) 2011;68:389–400. doi: 10.1002/cm.20520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymek EE, Heuser T, Nicastro D, Smith EF. The CSC is required for complete radial spoke assembly and wild-type ciliary motility. Mol Biol Cell. 2011;22:2520–2531. doi: 10.1091/mbc.E11-03-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymek EE, Smith EF. A conserved CaM- and radial spoke associated complex mediates regulation of flagellar dynein activity. J Cell Biol. 2007;179:515–526. doi: 10.1083/jcb.200703107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elam CA, Wirschell M, Yamamoto R, Fox LA, York K, Kamiya R, Dutcher SK, Sale WS. An axonemal PP2A B-Subunit is required for PP2A localization and flagellar motility. Cytoskeleton. 2011;68:363–372. doi: 10.1002/cm.20519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- Gibbons IR. Transient flagellar waveforms during intermittent swimming in sea urchin sperm. II. Analysis of tubule sliding. J Muscle Res Cell Motil. 1981;2:83–130. doi: 10.1007/BF00712063. [DOI] [PubMed] [Google Scholar]
- Goodenough UW, Heuser JE. Substructure of inner dynein arms, radial spokes, and the central pair/projection complex of cilia and flagella. J Cell Biol. 1985;100:2008–2018. doi: 10.1083/jcb.100.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman DS, Levine RP. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1965;54:1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermacher G, Sale WS. Regulation of flagellar dynein by phosphorylation of a 138-kD inner arm dynein intermediate chain. J Cell Biol. 1997;136:167–176. doi: 10.1083/jcb.136.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harauz G, Van Heel M. Exact filters for general geometry three dimensional reconstruction. Optik. 1986;73:146–156. [Google Scholar]
- Harris EH. The genus Chlamydomonas. In: Harris EH, editor. The Chlamydomonas Sourcebook: Introduction to Chlamydomonas and Its Laboratory Use. Vol. 2. Oxford: Academic Press; 2009. pp. 1–24. [Google Scholar]
- Heuser T, Raytchev M, Krell J, Porter ME, Nicastro D. The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella. J Cell Biol. 2009;187:921–933. doi: 10.1083/jcb.200908067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard DR, Habermacher G, Glass DB, Smith EF, Sale WS. Regulation of Chlamydomonas flagellar dynein by an axonemal protein kinase. J Cell Biol. 1994;127:1683–1692. doi: 10.1083/jcb.127.6.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Piperno G, Ramanis Z, Luck DJ. Radial spokes of Chlamydomonas flagella: genetic analysis of assembly and function. J Cell Biol. 1981;88:80–88. doi: 10.1083/jcb.88.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Ramanis Z, Luck DJ. Suppressor mutations in Chlamydomonas reveal a regulatory mechanism for flagellar function. Cell. 1982;28:115–124. doi: 10.1016/0092-8674(82)90381-6. [DOI] [PubMed] [Google Scholar]
- Kamiya R. Functional diversity of axonemal dyneins as studied in Chlamydomonas mutants. Int Rev Cytol. 2002;219:115–155. doi: 10.1016/s0074-7696(02)19012-7. [DOI] [PubMed] [Google Scholar]
- Kohno T, Wakabayahsi K, Diener DR, Rosenbaum JL, Kamiya R. Subunit interactions within the Chlamydomonas flagellar spokehead. Cytoskeleton (Hoboken) 2011;68:237–246. doi: 10.1002/cm.20507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyfman AY, Schmid MF, Gheiratmand L, Fu CJ, Khant HA, Huang D, He CY, Chiu W. Structure of Trypanosoma brucei flagellum accounts for its bihelical motion. Proc Natl Acad Sci USA. 2011;108:11105–11108. doi: 10.1073/pnas.1103634108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Lin J, Tritschler D, Song K, Barber CF, Cobb JS, Porter ME, Nicastro D. Building blocks of the nexin-dynein regulatory complex in Chlamydomonas flagella. J Biol Chem. 2011;286:29175–29191. doi: 10.1074/jbc.M111.241760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linck RW, Stephens RE. Functional protofilament numbering of ciliary, flagellar, and centriolar microtubules. Cell Motil Cytoskeleton. 2007;64:489–495. doi: 10.1002/cm.20202. [DOI] [PubMed] [Google Scholar]
- Luck D, Piperno G, Ramanis Z, Huang B. Flagellar mutants of Chlamydomonas: studies of radial spoke-defective strains by dikaryon and revertant analysis. Proc Natl Acad Sci USA. 1977;74:3456–3460. doi: 10.1073/pnas.74.8.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Nicastro D. Cryo-electron microscope tomography to study axonemal organization. Methods Cell Biol. 2009;91:1–39. doi: 10.1016/S0091-679X(08)91001-3. [DOI] [PubMed] [Google Scholar]
- Nicastro D, McIntosh JR, Baumeister W. 3D structure of eukaryotic flagella in a quiescent state revealed by cryo-electron tomography. Proc Natl Acad Sci USA. 2005;102:15889–15894. doi: 10.1073/pnas.0508274102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- Olson GE, Linck RW. Observations of the structural components of flagellar axonemes and central pair microtubules from rat sperm. J Ultrastruct Res. 1977;61:21–43. doi: 10.1016/s0022-5320(77)90004-1. [DOI] [PubMed] [Google Scholar]
- Patel-King RS, Gorbatyuk O, Takebe S, King SM. Flagellar radial spokes contain a Ca2+-stimulated nucleoside diphosphate kinase. Mol Biol Cell. 2004;15:3891–3902. doi: 10.1091/mbc.E04-04-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Wilkerson CG, Witman GB. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT) J Cell Biol. 1998;141:979–992. doi: 10.1083/jcb.141.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Piperno G, Huang B, Luck DJ. Two-dimensional analysis of flagellar proteins from wild-type and paralyzed mutants of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1977;74:1600–1604. doi: 10.1073/pnas.74.4.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Huang B, Ramanis Z, Luck DJ. Radial spokes of Chlamydomonas flagella: polypeptide composition and phosphorylation of stalk components. J Cell Biol. 1981;88:73–79. doi: 10.1083/jcb.88.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME, Power J, Dutcher SK. Extragenic suppressors of paralyzed flagellar mutations in Chlamydomonas reinhardtii identify loci that alter the inner dynein arms. J Cell Biol. 1992;118:1163–1176. doi: 10.1083/jcb.118.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME, Sale WS. The 9 + 2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J Cell Biol. 2000;151:F37–F42. doi: 10.1083/jcb.151.5.f37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillin ML, Matthews BW. Accurate calculation of the density of proteins. Acta Crystallogr D Biol Crystallogr. 2000;56:791–794. doi: 10.1107/s090744490000679x. [DOI] [PubMed] [Google Scholar]
- Roberts AJ, et al. AAA+ Ring and linker swing mechanism in the dynein motor. Cell. 2009;136:485–495. doi: 10.1016/j.cell.2008.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp G, Porter ME. A subunit of the dynein regulatory complex in Chlamydomonas is a homologue of a growth arrest-specific gene product. J Cell Biol. 2003;162:47–57. doi: 10.1083/jcb.200303019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale WS, Satir P. Direction of active sliding of microtubules in Tetrahymena cilia. Proc Natl Acad Sci USA. 1977;74:2045–2049. doi: 10.1073/pnas.74.5.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir P. Studies on cilia. 3. Further studies on the cilium tip and a “sliding filament” model of ciliary motility. J Cell Biol. 1968;39:77–94. doi: 10.1083/jcb.39.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EF, Sale WS. Regulation of dynein-driven microtubule sliding by the radial spokes in flagella. Science. 1992;257:1557–1559. doi: 10.1126/science.1387971. [DOI] [PubMed] [Google Scholar]
- Smith EF, Yang P. The radial spokes and central apparatus: mechano-chemical transducers that regulate flagellar motility. Cell Motil Cytoskeleton. 2004;57:8–17. doi: 10.1002/cm.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgess JM, Chao J, Wong J, Aspin N, Turner JA. Cilia with defective radial spokes: a cause of human respiratory disease. N Engl J Med. 1979;300:53–56. doi: 10.1056/NEJM197901113000201. [DOI] [PubMed] [Google Scholar]
- Summers KE, Gibbons IR. Adenosine triphosphate-induced sliding of tubules in trypsin-treated flagella of sea-urchin sperm. Proc Natl Acad Sci USA. 1971;68:3092–3096. doi: 10.1073/pnas.68.12.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachtenberg S. Fast-freezing devices for cryo-electron-microscopy. Micron. 1993;24:1–12. [Google Scholar]
- Warner FD. New observations on flagellar fine structure. The relationship between matrix structure and the microtubule component of the axoneme. J Cell Biol. 1970;47:159–182. doi: 10.1083/jcb.47.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner FD, Satir P. The structural basis of ciliary bend formation. Radial spoke positional changes accompanying microtubule sliding. J Cell Biol. 1974;63:35–63. doi: 10.1083/jcb.63.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman GB, Carlson K, Berliner J, Rosenbaum JL. Chlamydomonas flagella. I. Isolation and electrophoretic analysis of microtubules, matrix, membranes, and mastigonemes. J Cell Biol. 1972;54:507–539. doi: 10.1083/jcb.54.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman GB, Plummer J, Sander G. Chlamydomonas flagellar mutants lacking radial spokes and central tubulesStructure, composition, and function of specific axonemal components. J Cell Biol. 1978;76:729–747. doi: 10.1083/jcb.76.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Diener DR, Rosenbaum JL, Sale WS. Localization of calmodulin and dynein light chain LC8 in flagellar radial spokes. J Cell Biol. 2001;153:1315–1326. doi: 10.1083/jcb.153.6.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, et al. Radial spoke proteins of Chlamydomonas flagella. J Cell Sci. 2006;119:1165–1174. doi: 10.1242/jcs.02811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Fox L, Colbran RJ, Sale WS. Protein phosphatases PP1 and PP2A are located in distinct positions in the Chlamydomonas flagellar axoneme. J Cell Sci. 2000;113((Pt 1),):91–102. doi: 10.1242/jcs.113.1.91. [DOI] [PubMed] [Google Scholar]
- Yang P, Sale WS. Casein kinase I is anchored on axonemal doublet microtubules and regulates flagellar dynein phosphorylation and activity. J Biol Chem. 2000;275:18905–18912. doi: 10.1074/jbc.M002134200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.