Directional migration of MDCK cells is regulated by the ratio of the deposited basement membrane proteins laminin-511 and laminin-332. Knockdown of laminin-511 or its receptor integrin α3 inhibits directional migration and destabilizes cell–cell contacts, disturbing the polarization machinery.
Abstract
Sustained directional migration of epithelial cells is essential for regeneration of injured epithelia. Front–rear polarity of migrating cells is determined by local activation of a signaling network involving Cdc42 and other factors in response to spatial cues from the environment, the nature of which are obscure. We examined the roles of laminin (LM)-511 and LM-332, two structurally different laminin isoforms, in the migration of Madin–Darby canine kidney cells by suppressing expression of their α subunits using RNA interference. We determined that knockdown of LM-511 inhibits directional migration and destabilizes cell–cell contacts, in part by disturbing the localization and activity of the polarization machinery. Suppression of integrin α3, a laminin receptor subunit, in cells synthesizing normal amounts of both laminins has a similar effect as knockdown of LM-511. Surprisingly, simultaneous suppression of both laminin α5 and laminin α3 restores directional migration and cell–cell contact stability, suggesting that cells recognize a haptotactic gradient formed by a combination of laminins.
INTRODUCTION
Sustained directional migration of cells depends on establishment of stable front–rear polarity (Iden and Collard, 2008; Nelson, 2009; Petrie et al., 2009). Extracellular “spatial cues” (Drubin and Nelson, 1996) from the environment activate the small GTPase Cdc42, leading to a cascade of events that include assembly of polarity protein complexes and stable orientation of the cytoskeleton and membrane trafficking in the direction of migration (Iden and Collard, 2008; Petrie et al., 2009; Osmani et al., 2010). In epithelial cells, directional migration is of particular importance in wound healing, in which concerted migration of cells at the wound edge is required for efficient repair (Friedl and Gilmour, 2009). Dysregulation of directional migration, on the other hand, may contribute to metastasis in epithelial cancers (Marinkovich, 2007; Friedl and Gilmour, 2009).
The nature of spatial cues that initiate directional migration in epithelial cells is poorly understood but may involve proteins of the extracellular matrix such as isoforms of laminin (Zhang and Kramer, 1996; Plopper et al., 1998; Frank and Carter, 2004; Sehgal et al., 2006; Hartwig et al., 2007; Marinkovich, 2007). All laminins are heterotrimeric proteins composed of distinct α, β, and γ subunits (Miner and Yurchenco, 2004). Prototypical laminins (LMs) such as LM-511 (α5β1γ1) are cross shaped, with short arms contributed by the amino terminus of each subunit and a stem composed of a coiled coil of all three subunits (Miner and Yurchenco, 2004). Laminin receptors, including integrins α3β1 and α6β4, bind primarily to globular domains at the carboxy terminus of the laminin α subunit near the distal end of the coiled coil (Miner and Yurchenco, 2004). In prototypical laminins, LN or polymerization domains occur at the amino terminus of each subunit; these interact with LN domains of two other molecules to form a network that is the foundation of the basement membrane (Yurchenco, 2011). In contrast, a few laminins, including LM-332 (α3β3γ2), are composed of truncated subunits lacking one or more LN domains and are incapable of network formation and basement membrane assembly on their own (Cheng et al., 1997; Miner and Yurchenco, 2004; Marinkovich, 2007).
Although many cells synthesize more than one isoform of laminin, functional interactions between these have generally not been considered (Miner et al., 1997; Miner and Yurchenco, 2004; Sorokin et al., 1997; Aumailley and Rousselle, 1999; Calaluce et al., 2006; McMillan et al., 2006; Sugawara et al., 2007; Chiharu et al., 2010). The Madin–Darby canine kidney (MDCK) cell line, for example, expresses both LM-332 and LM-511 under certain conditions (Yu et al., 2005; Mak et al., 2006; Moyano et al., 2010). In this study we explore the role of LM-511 in migration of MDCK cells and unexpectedly discover that directional migration depends on the ratio of the LM-511 α5 subunit to that of the LM-332 α3 subunit.
RESULTS
MDCK cells constitutively express LM-511
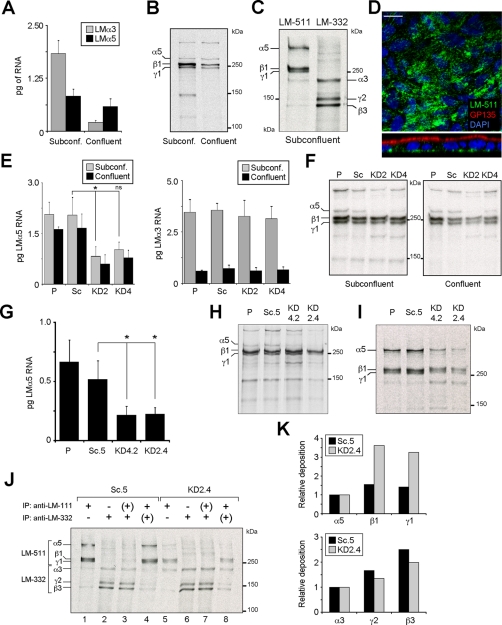
In MDCK cells, expression of LM-332 only occurs in subconfluent cells due to tight transcriptional regulation by transforming growth factor-β (Mak et al., 2006; Moyano et al., 2010). To determine the pattern of LM-511 expression, RNA was extracted from subconfluent and confluent MDCK cell cultures, and the amount of α3 (LM-332) and α5 (LM-511) mRNA was measured by quantitative reverse transcriptase PCR (qPCR). As shown in Figure 1A, levels of α3 mRNA were substantially reduced in confluent cultures relative to subconfluent cultures, as previously reported (Mak et al., 2006; Moyano et al., 2010). Laminin α5 mRNA, on the other hand, was present in significant and approximately equal amounts in both subconfluent and confluent cultures. These results were confirmed by examining synthesis and deposition of LM-511 by metabolic labeling and immunofluorescence (Figure 1, B–D). Thus, subconfluent MDCK cells simultaneously synthesize and deposit both LM-332 and LM-511, whereas confluent cells express only LM-511.
FIGURE 1:
Knockdown of LM-511 synthesis in MDCK cells. (A) Quantitative real-time PCR (qPCR) for LMα3 and LMα5 mRNA expression in either subconfluent (day 1) or confluent (day 4) MDCK cell cultures grown in growth medium on plastic. Bars represent the mean (±SD, n = 3). (B) Autoradiography of 35S-labeled and immunoprecipitated LM-511 (using an anti–LM-111 antibody) from subconfluent or confluent MDCK cell cultures. The experiment was repeated twice with similar results. (C) Representative autoradiography of two experiments of radiolabeled LM-511 and LM-332 extracted from ECM deposited by subconfluent MDCK cell cultures in serum-free, hormone-supplemented ExCell medium on collagen. (D) Confocal micrographs (XY, top; XZ, bottom) of confluent MDCK cell cultures stained for LM-511 using an anti–LM-111 antibody (green), GP135 (red), and nuclei (blue). No monospecific antibody against canine LMα5 is available. Scale bar, 10 μm. (E) qPCR for LMα5 (left) and LMα3 (right) expression (mean ± SD, n = 2) from parental MDCK cells (P), cells stably expressing scramble (Sc), or the two different LMα5 shRNA constructs (KD2 and KD4) cultured in growth medium on plastic. p = 0.0089 (multicomparison analysis); *p < 0.05; ns, not statistically significant. (F) Efficiency of LM-511 protein reduction, assessed by pulse labeling and immunoprecipitation using an anti–LM-111 antibody in both subconfluent and confluent KD2 and KD4 cell cultures compared with parental (P) and scramble (Sc). A representative autoradiography of two experiments is shown. (G) qPCR for LMα5 expression (mean ± SD, n = 2) of clones KD2.4, KD4.2, and Sc.5 and parental cells grown on collagen in ExCell. p = 0.0214 (multicomparison analysis); *p < 0.05. (H) Reduction of LM-511 protein biosynthesis and (I) deposition in the ECM of clones KD2.4 and KD4.2 compared with parental (P) and Sc.5 cell lines assessed by pulse-labeling and immunoprecipitation. (J) Reciprocal immunoprecipitation of LM-511 and LM-332 deposited into the ECM from radiolabeled subconfluent Sc.5 or KD2.4. Single immunoprecipitation of LM-511 (lanes 1 and 5) or LM-332 (lanes 2 and 6). From their respective unbound fractions (+) the reciprocal laminins were immunoprecipitated in a second round (lanes 3, 4, 7, and 8). (K) Deposition of β and γ laminin chains relative to α chains calculated from densitometry of lanes 1 and 2 (Sc.5) and lanes 5 and 6 (KD2.4) in J.
Suppression of LM-511 production with short hairpin RNA
To investigate the function of LM-511 in MDCK cells, synthesis of the α5 subunit was suppressed using retroviral-mediated expression of short hairpin RNA (shRNA) constructs. Initial experiments with pools of cells demonstrated that knockdown was effective using two different target sequences (KD2, KD4) at both the mRNA and protein level and was independent of culture confluency (Figure 1, E and F). Furthermore, knockdown of laminin α5 had no effect on either the synthesis or regulation of laminin α3 expression (Figure 1E). On the basis of these observations, individual stable clones were selected from the two pools and the scrambled sequence control, and clones KD2.4, KD4.2, and Sc.5 were chosen for further analysis. KD2.4 exhibited 57% suppression of laminin α5 mRNA and 75% suppression of α5 protein, with the values for KD4.2 intermediate between those of the Sc.5 control and KD2.4 for protein expression (59% mRNA; 24% protein; Figure 1, G and H). Note that for analysis of the LM-511 protein in this and subsequent experiments, a polyclonal anti–LM-111 antibody that reacts with the β1 and γ1 subunits was used because no specific anti-α5 antibody that reacts with the canine protein is available. Previous work indicated that MDCK cells do not express the α1 subunit (Yu et al., 2005; our unpublished data). The anti–LM-332 antibody used here is specific for LM-332, although reactivity is primarily against the β3 and γ2 subunits.
Levels of laminin α5 deposition into the matrix underlying the cells reflected the other measures of suppression, although unexpectedly high amounts of β1 and γ1 were deposited in the matrix by the suppressed lines (Figure 1, I–K). One potential explanation of this is that, in the absence of α5, the β1 and γ1 subunits associate with another laminin α subunit. Because LM-311 is known to be expressed in some cells, we first tested the possibility that LMα3 associated with β1 and γ1 in the KD2.4 suppressed line by sequential immunoprecipitations of matrix deposited by metabolically labeled cultures (Figure 1J). In control Sc.5 cells, only LM-511 was detected by immunoprecipitation with anti-LM-111, although a small amount of a band migrating at the position of LMα3 was also seen (lane 1; compare to lane 2). Immunoprecipitation with polyclonal anti–LM-332 detected primarily LM-332, although some faint higher–molecular weight bands were also visible (lane 2). Sequential precipitation of labeled matrix with first one antibody and then the other (with an intermediate protein-A precipitation step to eliminate residual amounts of the first antibody) gave essentially the same results as the single immunoprecipitations (lanes 3 and 4). When labeled matrix from KD2.4 was immunoprecipitated, the amount of LM-511 precipitated was much less, as expected, but there was also a clearly noticeable increase in the amount of LMα3 (lane 5; compare to lane 1). Both single and sequential immunoprecipitations of LM-332 resembled controls (lanes 6 and 7; compare to lanes 2 and 3). However, when first LM-332 and then LM-511 were immunoprecipitated, the anti–LM-111 antibody used to precipitate LM-511 detected not only β1 and γ1, but also α3 and some β3 and γ2 (lane 8). Thus, the sequential immunoprecipitation experiment showed that knockdown of LMα5 expression resulted in increased amounts of what was potentially LM-311 (lanes 5 and 8). The appearance of β3 and γ2 in lane 8 is, however, difficult to explain unless there is some physical association between different laminin molecules not disrupted by the conditions of immunoprecipitation. The conclusion that knockdown of LMα5 leads to the secretion of LM-311 is consistent with measurements of the relative deposition of subunits (Figure 1K). Deposition of β1 and γ1 relative to α5 in anti–LM-111 immunoprecipitates is higher in KD2.4 than in control Sc.5 (Figure 1K, top). In contrast, deposition of β3 and γ2 relative to α3 is lower in anti–LM-332 precipitates, possibly due to the association of a fraction of α3 with β1 and γ1 (Figure 1K, bottom).
Another possible explanation for the deposition of increased amounts of β1 and γ1 when expression of LMα5 is suppressed is that expression of a new laminin α subunit is induced by the knockdown. Of the other known laminin α chains, LMα4 was of particular interest because it is truncated like LMα3 and, most important, would likely migrate on reducing SDS gels with a mobility similar to that of β1 and γ1 and therefore would be difficult to detect. Furthermore, LM-411 is known to be expressed in the kidney under certain circumstances (Miner et al., 1997; Hansen and Abrass, 2003). To test this, RT-PCR was conducted on RNA prepared from control and suppressed lines using primers specific for the canine gene product, but no PCR product corresponding to LMα4 was detected (data not shown). In addition, deposited matrix from metabolically labeled control and KD2.4 cells was immunoprecipitated with anti–LM-111 and run on an SDS gel without reduction. Under these conditions, laminin trimers, which are stabilized by disulfide bonds, would migrate as intact molecules. However, no new trimeric species that migrated with mobility consistent with that of intact LM-411 were observed (data not shown). Despite these results, the possibility that some other variant of another laminin α chain is expressed upon knockdown of LMα5 cannot be ruled out.
Suppression of LM-511 reduces directional migration
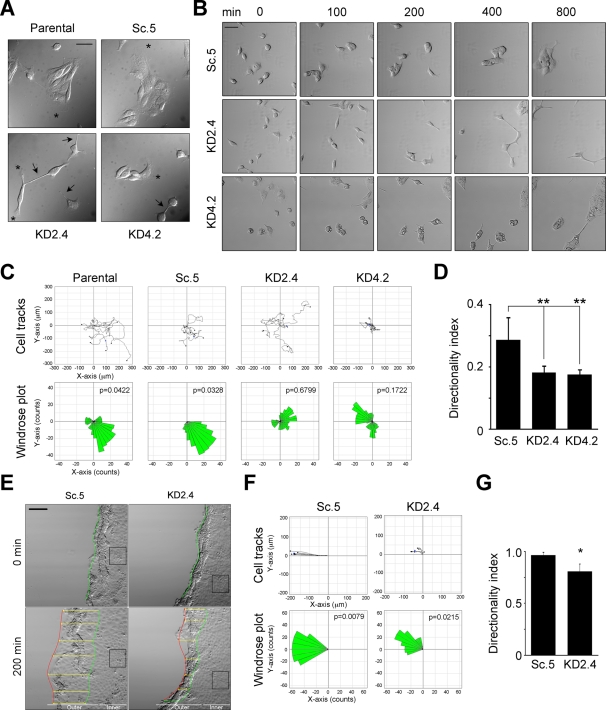
In cultures plated at subconfluent density on collagen type I–coated coverslips in serum-free, hormone-supplemented medium, KD2.4 and KD4.2 cells had distinctly different morphologies than control cells (Figure 2A). KD2.4 cells were more dispersed relative to parental and Sc.5 controls, which tended to form multicellular islands, with KD4.2 intermediate in appearance (Figure 2A). Furthermore, although all cell lines exhibited a variety of membrane extensions or processes, those in KD2.4 tended to be longer, thinner, and more numerous. Cell–cell contacts in control cells were extensive and continuous; in contrast, those in KD2.4 and to some extent KD4.2 were often limited to interactions mediated by fine processes (Figure 2A).
FIGURE 2:
Knockdown of LMα5 results in loss of directional migration. (A) Parental MDCK (P), Sc.5, KD2.4, and KD4.2 cells were grown on collagen-coated coverslips in hormone-supplemented, serum-free ExCell medium for 6 h and photographed using DIC microscopy. Arrows indicate cell protrusions and asterisks indicate lamellipodia. Scale bar, 20 μm. (B) Time-lapse videomicroscopy of Sc.5, KD2.4, and KD4.2 cells. Still DIC frames (in minutes, after 1 h of adhesion) of a representative video of at least four independent experiments are shown. Scale bar, 50 μm. (C) Migration of parental (P), Sc.5, KD2.4, or KD4.2 individual cells was tracked and plotted (top) and analyzed for persistence of migration (bottom, windrose plots). The p values for circular dispersion were calculated using the Rayleigh test for vectorial data. (D) Mean values for the directionality index (±SD, n = 5) are shown as a bar graph. p = 0.004 (multicomparison analysis); **p < 0.01. (E) Time-lapse videomicroscopy still images of confluent Sc.5 or KD2.4 cell cultures after wounding. The beginning (0 min) and the end locations of the wound edge (200 min) are indicated by the green and red lines, respectively, whereas the overall progression of the wound edge is indicated by the yellow arrows. The black boxes correspond to “inner” cells analyzed separately in Supplemental Figure S1. Scale bar, 100 μm. (F) Cell tracks (top) and windrose plots (bottom) corresponding to either Sc.5 or KD2.4 cells at the wound edge. (G) The mean directionality index (±SD, n = 3) of cells at the wound edge is represented as a bar graph. p = 0.0286 (multicomparison analysis); *p < 0.05.
Time-lapse studies were conducted to determine whether the observed morphological changes accompanying reduction in LM-511 expression affected cell migration. Control (parental and Sc.5) MDCK cells and the suppressed cell lines KD2.4 and KD4.2 were plated at subconfluent density in serum-free medium on collagen-coated chamber slides and their behavior recorded for 800 min (∼13 h). As shown in the selected still images in Figure 2B and in Supplemental Movie S1, Sc.5 cells gradually formed small, multicellular islands that continued to collectively migrate. By the end of the incubation period, Sc.5 cells and parental cells (not shown) had coalesced into a small number of islands (usually one to two per field). KD2.4 and to a lesser extent KD4.2 were usually slow to form islands of adherent cells and tended to remain more disperse, even after 800 min of incubation (Figure 2B). KD2.4 cells, in particular, often did not form broad lamellipodia but instead extended long, thin processes resembling filopodia in the direction of migration (Figure 2B). Cell–cell contacts, when present, were generally limited to contact between these thin, extended processes (Figure 2B; also see Figure 3).
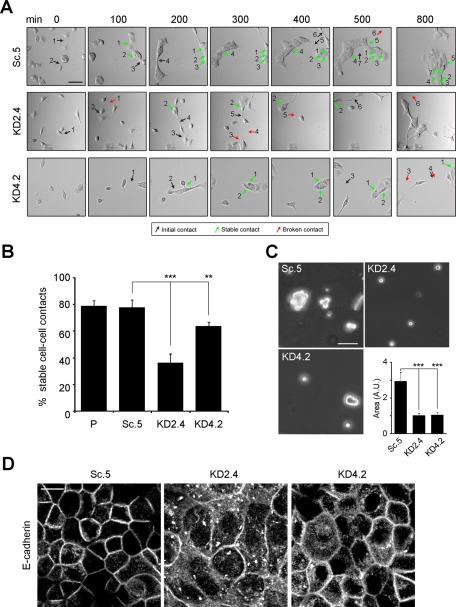
FIGURE 3:
Knockdown of LMα5 reduces cell–cell contact stability. (A) Sc.5, KD2.4, or KD4.2 cells were tracked by time-lapse videomicroscopy and scored for stability of cell–cell contacts over the length of the video (800 min). In the selected still DIC images, representative initial cell–cell contacts are indicated by black arrows, stable ones by green arrows, and failed ones (broken) by red arrows. Scale bar, 50 μm. (B) Cell–cell contacts were scored in four independent experiments as described in A, and the mean percentage (±SD) of stable cell–cell contacts is represented as a bar graph. p < 0.0001 (multicomparison analysis); **p < 0.01; ***p < 0.001. (C) Representative micrographs of Sc.5, KD2.4, and KD4.2 cells cultured in hanging-drop assays after trituration to dislodge cell–cell contacts. The area of cell aggregates was measured and the mean (±SD) of three independent experiments is shown as a bar graph. A.U., arbitrary units. Scale bar, 50 μm. p < 0.0001 (multicomparison analysis); ***p < 0.001. (D) Confocal fluorescence micrographs of confluent Sc.5, KD2.4, or KD4.2 cell cultures grown on permeable supports in ExCell and immunostained for E-cadherin. Scale bar, 10 μm.
Quantitative assessment of migratory behavior was obtained by tracking individual cells using image analysis software (Figure 2, B and C). Differences in migration between controls and suppressed cells were not immediately evident by examination of migratory tracks (Figure 2C, top). However, further analysis revealed that KD2.4 and KD4.2 had significantly reduced levels of directionality evident in windrose plots and calculated measures of directionality (Figure 2C, bottom, and D).
Loss of directional migration was even more apparent in wounded cultures. Confluent Sc.5 and KD2.4 cells were wounded with a pipette tip and advancement of the edge followed for 200 min by time-lapse microscopy. Under these conditions, the KD2.4 wound edge migrated only about one-third as far as the Sc.5 control (Figure 2E, yellow arrows). Cell migration tracks and windrose plots indicated that control cells at the wound edge exhibited highly directional migration toward the wound, whereas the directionality of KD2.4, and to a lesser extent KD4.2, was significantly reduced (Figure 2, F and G; and data not shown). Cells located a few cell layers interior from the wound edge also exhibited aberrant directional migration (Supplemental Figure S1).
Overall, analysis of cells expressing reduced amounts of LM-511 indicates that both individual and confluent cells responding to a wound lack the ability to efficiently migrate in a particular direction relative to controls. Migration itself is not, however, impaired; suppressed cells sometimes moved greater distances and at higher velocities than controls (Supplemental Information, Figure S1).
Suppression of LM-511 prevents formation of stable cell–cell contacts
As mentioned previously, adhesions between cells in KD2.4 often did not coalesce into contiguous zones of cell–cell contact but remained limited to puncta (Figure 2A). In time-lapse studies of subconfluent cultures, most initial cell–cell contacts between control cells (Sc.5 and parental) matured into permanent adhesive interactions as islands formed. In contrast, many cell–cell contacts in KD2.4 failed to mature and were lost as cells involved in the contact moved away from each other. This was apparent when individual initial contacts were tracked over the entire incubation period (Figure 3A, arrows). In Sc.5 and parental cells, nearly 80% of contacts remained stable during the time of observation. For KD2.4, this number was <40%, and with KD4.2 contact stability declined to ∼70% (Figure 3B).
These results indicated that either the contacts between suppressed cells were abnormal or contacts did not mature because cells quickly moved away from each other. To shed light on this point, the stability of cell–cell contacts was measured by hanging-drop assay (Ehrlich et al., 2002; Qin et al., 2005). When examined in this way, the area of KD2.4 aggregates was significantly less than that of Sc.5, consistent with formation of weaker cell–cell adhesions in the suppressed line (Figure 3C). The supposition that cell–cell contacts in laminin α5–suppressed cells were abnormal was also supported by immunofluorescence of E-cadherin in confluent cultures. In Sc.5, staining was confined mainly to the lateral edges of the cell (Figure 3D). In KD2.4 and to a lesser extent KD4.2, cell borders as demarcated by E-cadherin staining were diffuse, and extensive punctate staining was visible in the cytoplasm (Figure 3D).
In summary, observations in migrating cells, suspended aggregates, and confluent cultures suggest that knockdown of LM-511 affects the intrinsic integrity and stability of cell–cell adhesions.
Directional migration and cell–cell contact stability are rescued by LM-511–enriched matrices
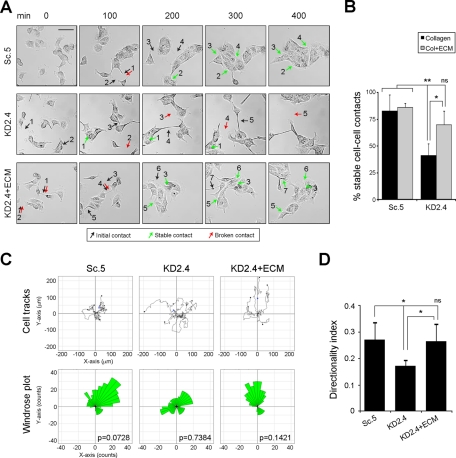
If reduction in LM-511 synthesis and deposition in suppressed cell lines is responsible for defects in directional migration and cell–cell contact stability, then replacement of LM-511 should restore the normal phenotype. To determine this, a rescue experiment was conducted using a preformed matrix. Normal MDCK cells were grown to confluence on collagen, and the cells were removed by treatment with ammonium hydroxide. Because LM-332 is not expressed in confluent cultures (Mak et al., 2006; Moyano et al., 2010), the residual matrix left after cell extraction was enriched in LM-511. When KD2.4 cells were plated on preformed matrix (KD2.4 plus extracellular matrix [ECM]), cell–cell contact stability, as well as contiguous island formation, was partially restored in comparison to the Sc.5 control and KD2.4 plated on collagen alone (Figure 4, A and B, and Supplemental Movie S2). Furthermore, directional migration determined by windrose plots and directionality index was nearly normal (Figure 4, C and D).
FIGURE 4:
LM-511–enriched preformed ECM rescues directional migration and cell–cell contact stability in LMα5-suppressed cells. (A) Time-lapse videomicroscopy still images of Sc.5 or KD2.4 cells plated on collagen (Sc.5, KD2.4) or KD2.4 cells plated on LM-511–enriched ECM (KD2.4+ECM). Examples of initial cell–cell contacts are indicated by black arrows, stable ones by green arrows, and failed ones (broken) by red arrows. Scale bar, 50 μm. (B) Contacts were scored in three independent experiments and the mean percentage (±SD) of stable cell–cell contacts represented. p = 0.0056 (multicomparison analysis); *p < 0.05; **p < 0.01; ns, not statistically significant. (C) Tracks of Sc.5, KD2.4, or KD2.4 + ECM cells were plotted (top), and the persistence of directional migration was analyzed (windrose plots, bottom). The p values for circular dispersion were calculated using the Rayleigh test for vectorial data. (D) The mean directionality index (±SD, n = 3) for Sc.5, KD2.4, or KD2.4 + ECM is shown. p = 0.0121 (multicomparison analysis); *p < 0.05; ns: not statistically significant.
Overall, these observations indicate that the two major phenotypic differences between control and suppressed cell lines in directional migration and cell–cell contact stability are rescued by a preformed LM-511 matrix, which is consistent with the idea that LM-511 is mechanistically involved in these cell behaviors.
Regulation of the polarization machinery is abnormal in cells expressing reduced amounts of LM-511
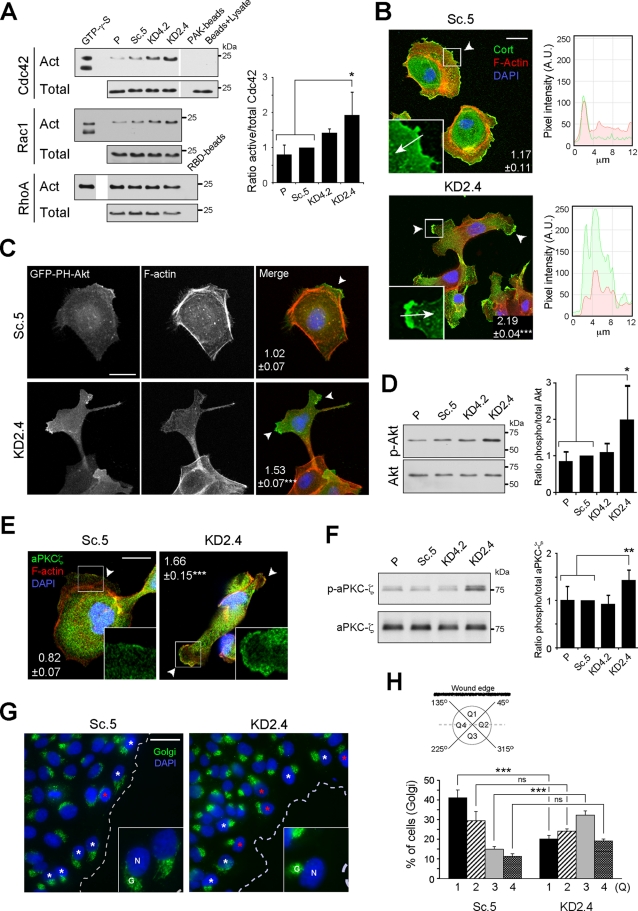
Sustained migratory polarity depends on localized activation of Cdc42, accompanied by oriented assembly of polarity complexes, the microtubule cytoskeleton and Golgi complex, and the branched actin network that forms the core of the lamellipodium (Etienne-Manneville and Hall, 2001, 2003; Etienne-Manneville et al., 2005; Nelson, 2009; Petrie et al., 2009; Osmani et al., 2010). To determine whether the polarity-signaling mechanism was disturbed by LM-511 suppression, the localization and activities of various components were examined. As shown in Figure 5A, the activity of Cdc42 was significantly increased in KD2.4 relative to controls, and also trended higher in KD4.2. The activity of Rac1 (but not of RhoA) was also increased, although it did not statistically differ from controls (Figure 5A).
FIGURE 5:
The polarization machinery is dysregulated in LMα5-suppressed cells. (A) Pull-down assays for active (Act) Cdc42, Rac1, and RhoA. A fraction of the lysates prior to pull-down was immunoblotted as total input. GTP-γ-S was a positive control for activation, whereas either PAK or RBD beads alone or only glutathione–agarose beads incubated with cell lysates (beads + lysate) were negative controls. The mean active/total ratio (±SD, n = 3) normalized to Sc.5 for Cdc42 is shown in the graph on the right. p = 0.0237 (multicomparison analysis); *p < 0.05. Differences in Rac1 and RhoA activities relative to controls were not significant, and quantitative data are not shown. (B) Confocal fluorescence micrographs of either Sc.5 or KD2.4 cells stained for cortactin (green), F-actin (red), and nuclei (blue). Arrowheads indicate lamellipodia/cell protrusions, and the mean number of positive protrusions (±SEM, n = 3) per cell is indicated. p < 0.0001 (multicomparison analysis); ***p < 0.001. Scale bar, 20 μm. Inset, detail of lamellipodium/cell protrusion showing only the green channel (cortactin); arrows indicate the direction of pixel intensity measurement for the histograms to the right, with red corresponding to actin staining. A.U., arbitrary units. (C) Confocal fluorescence micrographs of either Sc.5 or KD2.4 cells transiently transfected with GFP-PH-Akt (green) and stained for F-actin (red) and nuclei (blue). The mean number (±SEM, n = 2) of positive protrusions (arrowheads) per cell is indicated. p = 0.0055; **p < 0.01. Scale bar, 20 μm. (D) Immunoblot of phosphorylated and total Akt. The graph represents the mean ratio (±SD, n = 5) phospho/total Akt normalized to Sc.5. p = 0.0173 (multicomparison analysis); *p < 0.05. (E) Confocal fluorescence micrographs of either Sc.5 or KD2.4 cells stained for aPKCζ (green), F-actin (red), and nuclei (blue). The mean number (±SEM, n = 2) of positive protrusions (arrowheads) per cell is indicated. p < 0.0001; ***p < 0.001. Scale bar, 20 µm. Inset, detail of a lamellipodium/cell protrusion showing only the green channel (aPKCζ). (F) Immunoblot of phosphorylated and total aPKCζ. The graph represents the mean ratio (±SD, n = 6) phospho/total aPKCζ normalized to Sc.5. p = 0.0035 (multicomparison analysis); **p < 0.01. (G) Confocal fluorescence micrographs of either Sc.5 or KD2.4 cell monolayers wounded and stained 12 h later for Golgi (green) and nuclei (blue). White asterisks indicate examples of polarized Golgi complex orientation, and red asterisks indicate examples of Golgi oriented backward with respect the wound edge (dotted line). Scale bar, 20 μm. Inset, magnification of cells at the wound edge showing the nuclei (N) and Golgi (G). (H) Each cell at the wound edge was divided into four quadrants (Q1–Q4) centered on the nucleus (see scheme), and Golgi orientation was scored as percentage of cells with the Golgi complex in each quadrant (mean ± SEM, n = 3). p < 0.0001 (multicomparison analysis); ***p < 0.001; ns: not statistically significant.
Cortactin, a marker of the lamellipodium (Bryce et al., 2005), was usually localized to a single contiguous line along the plasma membrane in Sc.5 controls but appeared as a dense concentration of staining at more than one plasma membrane location per cell in KD2.4 (Figure 5B). Phosphatidylinositol 3,4,5-trisphosphate (PIP3), which is normally found at the single leading edge of migrating cells, was also enriched in more than one location in KD2.4, as determined by localization of transiently expressed green fluorescent protein (GFP)–PH-Akt (Figure 5C), and phosphorylation of Akt, which is activated by PIP3 binding, was increased in KD2.4 (although not in KD4.2) relative to controls (Figure 5D; Nelson, 2009; Petrie et al., 2009). aPKCζ, a component of the Par polarity complex (Shin et al., 2007; Nelson, 2009), was, like PIP3 and cortactin, localized to more than one location on the cell membrane in KD2.4 in a diffuse distribution relative to Sc.5 and exhibited abnormally high activity (Figure 5, E and F). Finally, in wounded monolayers the orientation of the Golgi complex toward the wound edge, which reflects both polarized assembly of microtubules and membrane trafficking, was inverted in KD2.4 in comparison to Sc.5 (Figure 5, G and H; Etienne-Manneville and Hall, 2001). In addition, wounded monolayers of KD2.4 cells, in a similar manner to subconfluent cultures, showed cortactin and aPKCζ in multiple plasma membrane locations per cell compared with Sc.5 (Supplemental Figure S2).
When these observations are considered together, it seems evident that suppression of LM-511 has effects not only on the migratory behavior of cells, but also on the intrinsic signaling machinery that determines the stable front–rear axis required for sustained directionality.
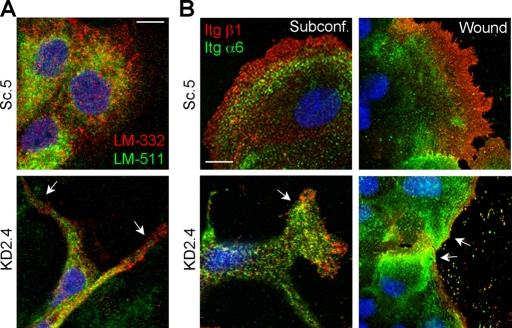
LM-511 and LM-332 and their integrin receptors α3β1 and α6β4 are spatially segregated in migrating cells
Because directional migration is determined by polarization of the intrinsic signaling machinery, we hypothesized that the distribution of laminins and their integrin receptors would be similarly polarized. To prove this, the relative localization of both LM-511 and LM-332 was assessed by confocal immunofluorescence in migrating cells. It is striking that in control cells (Sc.5), LM-332 was enriched in the leading edge of subconfluent cells, whereas LM-511 was mainly restricted to an area immediately behind LM-332 (Figure 6A). In KD2.4, the distribution of staining was much different, as expected, and did not show a similarly segregated pattern. Note that the putative staining of LM-511 in KD2.4 predominantly reflects secreted β1 and γ1 subunits, which the polyclonal antibody used in this experiment detects (Figure 1, J and K); no monospecific antibody against canine LMα5 is available.
FIGURE 6:
LM-511 and LM-332 and their receptors integrins α3β1 and α6β4 are spatially separated in control migrating cells but not in LMα5 knockdown cells. (A) Subconfluent cultures of either Sc.5 or KD2.4 cells were fixed and stained for LM-511 (using an anti–LM-111 antibody; green) and LM-332 (using a monoclonal antibody against the β3 subunit; red). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; blue). Arrows indicate locations on the cell periphery in KD2.4 where staining with the LM-111 antibody extends to the edge in contrast to Sc.5 controls. Scale bar, 10 μm. Note that the staining in KD2.4 cells for LM-511 (using the antibody anti–LM-111) is likely detecting only the β1 and γ1 subunits, which biochemical analysis suggests are secreted (Figure 1, J and K). No monospecific antibody against canine LMα5 is available. (B) Unpermeabilized cells from either subconfluent (left) or wounded cultures (right) were stained for β1 integrins (red) and for α6 integrin (green). Nuclei were stained with DAPI (blue). Arrows indicate locations on the cell periphery in KD2.4 where α6 integrin staining extends to the edge in contrast to Sc.5 controls. Scale bar, 10 μm.
LM-511 and LM-332 are ligands for both α3β1 and α6β4 integrins, which are expressed in MDCK cells (DiPersio et al., 2000; Nishiuchi et al., 2003, 2006; Stipp, 2010). As an initial assessment of integrin α3β1 and α6β4 roles in the migratory behavior of MDCK cells, integrin subunits α6 and β1 were localized in Sc.5 and KD2.4 cells by immunofluorescence. Integrin β1 served as a surrogate for α3 because no appropriate anti-canine α3 antibody was available. As shown in Figure 6B, β1 staining was enriched in the leading edge of subconfluent or wound-edge Sc.5 cells, with a concentration of α6 staining just behind that of β1 staining. In contrast, the staining pattern of the two integrins in KD2.4 in the cell periphery was much less distinct, with concentrated α6 staining sometimes reaching the cell edge (Figure 6B, arrows).
Taken together, these results are consistent with the idea that a polarized spatial distribution of LM-511 and LM-332 may form a haptotactic gradient recognized by α3β1 and α6β4 integrins that, in turn, promotes directional cell migration.
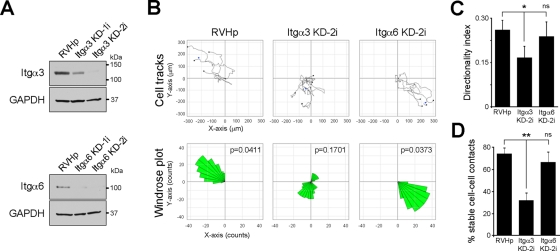
Integrin α3 plays a role in MDCK cell directional migration
To examine more directly individual functions of the two laminin-binding integrins α3 and α6, integrin expression was suppressed in parental cells by shRNA (Figure 7A) and the migratory behavior of the affected cell lines examined by time-lapse microscopy. It is surprising that cells in which integrin α3 expression was suppressed (Itgα3-KD) exhibited a migratory phenotype resembling that of cells with suppressed laminin α5 expression (Figure 7B). In contrast, suppression of integrin α6 (Itgα6-KD) had no apparent effect on migratory behavior. Both calculated directional migration (Figure 7, B and C) and the stability of cell–cell contacts (Figure 7D) were significantly reduced in Itgα3-KD relative to control cells and Itgα6-KD, suggesting that α3β1 plays an important role in these processes. As a whole, these results indicate that integrin α3β1 is essential for directional migration of MDCK cells, presumably through its interactions with one or more of the laminins synthesized by migrating cells.
FIGURE 7:
Suppression of integrin α3 expression affects directional migration and cell–cell contact stability. (A) Western blot to assess integrin α3 and α6 expression in parental MDCK cells stably transduced with either the empty vector as control (RVHp) or shRNA constructs against integrins α3 or α6 (Itgα3 KD-1i and 2i, or Itgα6 KD-1i and 2i, respectively). Glyceraldehyde-3-phosphate dehydrogenase is shown as loading control. (B) Migration of individual cells corresponding to RVHp, Itgα3 KD-2i, or Itgα6 KD-2i cell cultures was tracked by time-lapse videomicroscopy, plotted (top), and analyzed for persistence of migration (windrose plot, bottom). The p values for circular dispersion were calculated using the Rayleigh test for vectorial data. (C) Mean directionality index (±SD, n = 5). p = 0.0107 (multicomparison analysis); *p < 0.05; ns: not statistically significant. (D) Stability of cell–cell contacts was analyzed as described before, and the mean (±SD) of three independent experiments is shown. p = 0.0011 (multicomparison analysis); **p < 0.01; ns: not statistically significant.
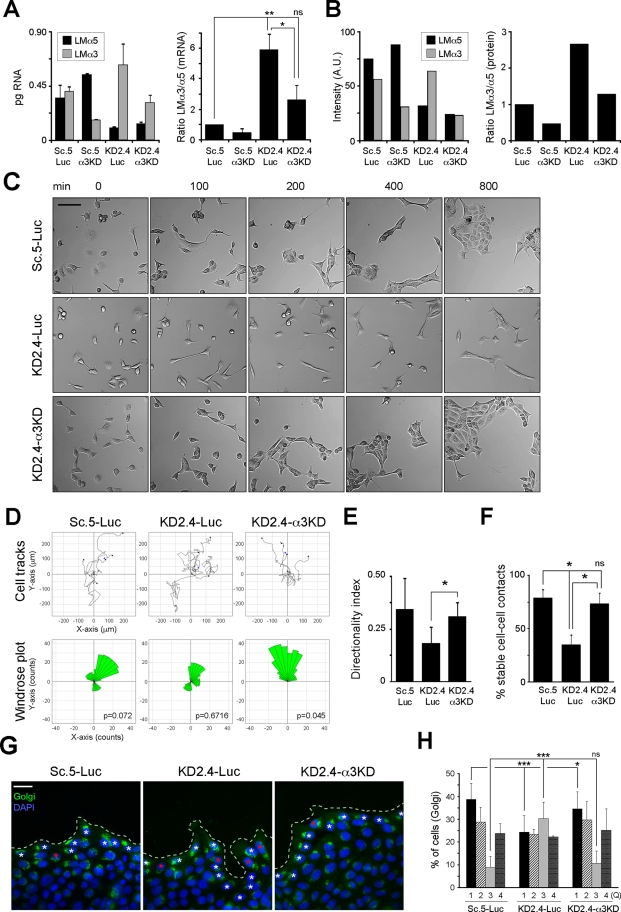
Reduction of laminin α3 expression in addition to that of laminin α5 restores a normal migratory phenotype and stable cell–cell adhesions
Because subconfluent MDCK cells express both LM-511 and LM-332, it seemed possible that the loss of directional migration and cell–cell contact stability in KD2.4 was a consequence of increased relative expression of LM-332 in the context of reduced LM-511 expression. To test this, KD2.4 cells were transiently transfected with small interfering RNA (siRNA) duplexes targeted to either luciferase (Luc) or laminin α3 (α3KD). As shown in Figure 8A, duplexes targeting laminin α3 effectively suppressed mRNA levels in both Sc.5 and KD2.4 compared with the luciferase control. Although KD2.4-Luc exhibited higher levels of laminin α3 expression than Sc.5-Luc, the degree of suppression by laminin α3 siRNA was similar in both cell lines. When mRNA expression ratios were calculated, the double knockdown (KD2.4-α3KD) had a ratio closer to the Sc.5-Luc control than the KD2.4-Luc (Figure 8A). Similarly, the ratio of deposited laminin α3 to α5 protein was closer to that of controls in the double knockdown (Figure 8B). Dramatically, when migratory behavior was examined, knockdown of laminin α3 in KD2.4 restored directionality (Figure 8, C–E, and Supplemental Movie S3), the stability of cell–cell contacts (Figure 8F), and Golgi orientation (Figure 8, G and H). Apparently, as this experiment demonstrated, expression of different laminins in a particular ratio is more important in determining directional migration than the absolute level of expression.
FIGURE 8:
Reduction of laminin α3 expression in KD2.4 rescues persistence of migration and stability of cell–cell contacts. (A) Sc.5 or KD2.4 cells were transiently transfected with either siRNA duplexes against LMα3 (α3KD) or luciferase (Luc) as negative control, and mRNA levels for both LMα5 and LMα3 were analyzed by qPCR. The mean expression levels (±SD) of one representative experiment of two are shown (left). The ratio LMα3/α5 for each group normalized to Sc.5-Luc is shown on the right. p = 0.0059 (multicomparison analysis); *p < 0.05; **p < 0.01; ns: not statistically significant. (B) Cells treated as in A were metabolically labeled, and LM-511 and LM-332 were immunoprecipated from the deposited ECM. After SDS–PAGE and autoradiography, the bands corresponding to LMα5 and LMα3 were scanned and intensities expressed in arbitrary units (A.U.; left). The ratio of LMα3/α5 normalized to Sc.5-Luc is also shown (right). The experiment was repeated twice with similar results. (C) Still images from representative time-lapse videomicroscopy (one of three experiments) of Sc.5-Luc, KD2.4-Luc, and KD2.4-α3KD. Scale bar, 40 μm. (D) Individual cells from Sc.5-Luc, KD2.4-Luc, or KD2.4-α3KD time-lapse videomicroscopy were tracked and plotted (top) and analyzed for persistence of migration (windrose plot, bottom). The p values for circular dispersion were calculated using the Rayleigh test. (E) Directionality index (mean ±SD, n ≥ 4) of Sc.5-Luc, KD2.4-Luc, or KD2.4-α3KD cells. p = 0.0138 (multicomparison analysis); *p < 0.05. (F) Cell–cell contacts were scored as indicated before and represented as the mean percentage (±SD) of stable cell–cell contacts of two independent experiments. p = 0.0315 (multicomparison analysis); *p < 0.05; ns, not statistically significant. (G) Confocal immunofluorescence of wounded cell monolayers of Sc.5-Luc, KD2.4-Luc, or KD2.4-α3KD stained for the Golgi complex (green) and nuclei (blue) 12 h after wounding. White asterisks indicate examples of polarized Golgi orientation, and red asterisks indicate examples of Golgi oriented backward with respect to the wound edge (dotted line). Scale bar, 20 μm. (H) Golgi orientation was assessed as previously shown, indicating the mean percentage (±SEM) of cells with a Golgi complex in each quadrant (Q) from three independent experiments. p < 0.0001 (multicomparison analysis); ***p < 0.001; ns: not statistically significant.
DISCUSSION
In this study we demonstrate a novel function of LM-511 in epithelial cell migration. When LM-511 production is suppressed, MDCK cells exhibit reduced directional migration and cell–cell adhesion stability. The former appears to be due at least in part to disruption of intrinsic cellular mechanisms that determine front–rear polarity. Most significantly, the aberrant phenotype exhibited by cells synthesizing reduced amounts of LM-511 is rescued not only by plating cells on preformed matrices enriched in LM-511, but also by suppressing laminin α3 in cells already expressing reduced quantities of laminin α5. To our knowledge, our results are the first description of a laminin function dependent on the cooperation of different endogenous isoforms.
Directional migration of cells depends on establishment of a front–rear axis (Iden and Collard, 2008; Nelson, 2009; Petrie et al., 2009). The polarization process is initiated by localized activation of Cdc42 on the plasma membrane in response to a spatial cue, followed by assembly of the Par3/Par6/aPKC polarity complex, generation of signals leading to the formation of a lamellipodium, and orientation of microtubules, Golgi, and the membrane trafficking apparatus in the direction of migration (Etienne-Manneville and Hall, 2001; Etienne-Manneville et al., 2005; Iden and Collard, 2008; Nelson, 2009; Petrie et al., 2009; Osmani et al., 2010). In MDCK cells synthesizing reduced amounts of LM-511, the activities of Cdc42, aPKCζ, and Akt are elevated relative to controls, and markers of the cell front including aPKCζ, cortactin, and PIP3 are abnormally localized to more than one area of the plasma membrane. Furthermore, these cells are unable to orient the Golgi complex in the direction of a wound (Figure 5, G and H). Taken together, these observations suggest that affected cells are unable to identify a distinct spatial cue and attempt to ectopically assemble multiple lamellipodia, resulting in increased activities of polarity factors. Ultimately, the cells are unable to establish a single front–rear axis and sustain migration in a particular direction.
In our experiments, the lack of directional migration was observed both in subconfluent cultures and wounded, confluent cultures. In both instances the precise nature of the spatial cues determining migratory direction is obscure. Experiments were conducted on collagen I–coated substrata in hormone-supplemented, serum-free medium to eliminate uncontrolled contributions from serum adhesion and growth factors. Hence directional input is likely generated endogenously in a kind of symmetry-breaking event (Li, 2009). On the basis of our observations, we hypothesize that this symmetry-breaking event may be the local deposition of LM-332 to the free edge of a wounded monolayer that, prior to wounding, was adherent to a matrix enriched in LM-511 but containing no LM-332 (Mak et al., 2006; Moyano et al., 2010). Deposition of LM-332 creates a haptotactic gradient that is then reflected in the distribution of integrins (Figure 6) and is then interpreted by the cell as a directional migratory signal (deHart et al., 2003; Sehgal et al., 2006; Kligys et al., 2007; Hamill et al., 2009). Disruption of this gradient by either suppression of LM-511 or knockdown of integrin α3 leads to loss of directional migration (Figure 7, B and C). These results are largely consistent with the observations of others in keratinocytes (Goldfinger et al., 1999; Hintermann et al., 2001; deHart et al., 2003; Choma et al., 2004; Sehgal et al., 2006; Margadant et al., 2009; Wen et al., 2010), although in those studies only LM-332 was examined. The most dramatic observation supporting our hypothesis is that reduction of both laminin α5 and laminin α3 expression rescues the phenotype exhibited by laminin α5 knockdown alone. If differential distribution and quantitative balance of laminin α5- and laminin α3-containing laminins is required to set up a haptotactic gradient, then it makes sense that suppression of laminin α3 in the context of laminin α5 reduction, which restores the ratio of expression to a more normal value (Figure 8, A and B), might provide the cell with sufficient spatial information to determine the front–rear axis.
In addition to its effect on directional migration, suppression of LM-511 also destabilizes cell–cell adhesion. In suppressed cells, nascent adhesions often do not coalesce into a continuous contact zone during migration and are unstable when measured in a hanging-drop assay (Figure 3, A–C). In confluent cultures, cell–cell adhesion in suppressed cells also appears abnormal, with significant amounts of intracellular E-cadherin in vesicles (Figure 3D). How directional migration and cell–cell adhesion are linked is unclear. One possible mechanism may involve the nectin family of adhesion molecules (Takai et al., 2008; Brakeman et al., 2009; Kitt and Nelson, 2011). In migrating cells, the nectin-like molecule Necl-5 is believed to participate in the development of front–rear polarity through cross-talk between growth factor receptors and integrins. When a migrating cell encounters another cell, Necl-5 facilitates formation of nectin-dependent cell–cell contacts, leading subsequently to more stable E-cadherin–based adhesions (Takai et al., 2008). If suppression of LM-511 causes dysregulation of Necl-5, then both directional migration and formation of stable cell–cell adhesions might be affected. It may also be significant in the context of our studies that disruption of afadin, a nectin- and actin-binding protein, has been reported to cause abnormal deposition of laminin and mislocalization of laminin receptors (Komura et al., 2008).
The two laminins discussed here, LM-511 and LM-332, are structurally distinct (Miner and Yurchenco, 2004; Yurchenco, 2011). LM-511 is a prototypical laminin capable of network formation through interactions of its three LN or polymerization domains (Yurchenco, 2011). LM-332, on the other hand, has truncated subunits, with the predominant form found in MDCK cells lacking two of the three LN domains. Given that our results suggest possible cooperation between laminin isoforms, it is tempting to speculate that truncated laminin isoforms such as LM-332 act by disrupting the LM-511 network and thereby modulating laminin signaling to the cell. Recent model experiments conducted with purified laminins and visualized by atomic force microscopy are consistent with this interpretation (Chiang et al., 2011). As discussed in Results, we cannot completely rule out the assembly and secretion of alternative laminin trimers when expression of LMα5 is suppressed, most notably LM-311. However, because LM-311 would also contain the truncated α3 subunit and therefore be incapable of network formation, its presence together with LM-332 would not invalidate our overall proposal.
MATERIALS AND METHODS
Cell culture
Stock cultures of MDCK cells (type II, Heidelberg isolate, passages 7–35), cultured as described previously (Matlin et al., 1981), were adapted to minimal essential medium (MEM; Invitrogen, Carlsbad, CA) supplemented with 5% (vol/vol) fetal bovine serum (Hyclone-Fisher, Rockford, IL), 2 mM l-glutamine, and 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid–KOH, pH 7.3 (MEM growth medium). All experiments, except where indicated, were done under serum-free conditions using ExCell hormone-supplemented MDCK cell growth medium (M3803; Sigma-Aldrich, St. Louis, MO) as previously described (Moyano et al., 2010). Similar results were obtained with OptiPRO SFM (12309-019; Invitrogen), an alternative serum-free medium.
For all the experiments, unless otherwise stated, culture dishes or glass coverslips were precoated with 10 μg/cm2 of collagen-I (5005-B; PureCol, Advance BioMatrix, San Diego, CA) in PBS without divalent cations.
Antibodies
Antibodies used in this study are listed in Table 1.
TABLE 1:
Antibodies used in this study.
| Antigen | Name | Species | Working concentration | Source/catalogue no. |
|---|---|---|---|---|
| Mouse LM-111 | Anti-laminin (detects β1 and γ1 of LM-511) | Rabbit polyclonal | IP: 2 μg; IF: 1:100 | Sigma-Aldrich/L9393 |
| Human LM-332 | 8LN5 | Rabbit polyclonal | IP: 4 μg | M. Koch |
| Human laminin β3 | Anti–kalinin B1 | Mouse monoclonal | IF: 1:50 | BD Biosciences (San Diego, CA)/610423 (discontinued) |
| Canine podocalyxin | GP135 (3F21D8) | Mouse monoclonal supernatant | IF: 1:2 | Ojakian and Schwimmer (1988) |
| Human E-cadherin C-terminal | Purified mouse anti–E-cadherin (clone 36/E-cadherin) | Mouse monoclonal | IF: 1:100 | BD Transduction Laboratories (Lexington, KY)/610182 |
| Human Cdc42 | CDC42 | Mouse monoclonal | WB: 1:1500 | BD Transduction Laboratories/610928 |
| Human Rac1 | Rac1 | Mouse monoclonal | WB: 1:1000 | BD Transduction Laboratories/610650 |
| Human RhoA (aa 120–150) | RhoA (26C4) | Mouse monoclonal | WB: 1:500 | Santa Cruz Biotechnology (Santa Cruz, CA)/sc418 |
| Cortactin | Anti-cortactin (p80/85), clone 4F11 | Mouse monoclonal | IF: 1:100 | Millipore/05-180 |
| Mouse Akt (residues surrounding Ser-473) | Phospho-Akt (Ser-473) | Rabbit polyclonal | WB: 1:1000 | Cell Signaling (Beverly, MA)/9271 |
| Mouse Akt (C-terminus) | Akt | Rabbit polyclonal | WB: 1:1000 | Cell Signaling/9272 |
| Human PKCζ (residues surrounding Thr-410) | Phospho-PKCζ/λ (Thr-410/403) | Rabbit polyclonal | WB: 1:2000 | Cell Signaling/9378 |
| Rat PKCζ (C-terminus, aa 577–592) | Anti-PKCζ | Rabbit polyclonal | WB: 1:1000; IF: 1:100 | Upstate, Millipore/07-264 |
| Human giantin (N-terminus aa 1–469) | Rabbit polyclonal to giantin-Golgi marker | Rabbit polyclonal | IF: 1:500 | Abcam (Cambridge, MA)/ab24586 |
| Human β1 integrin | Human β1 integrin TS2/16 | Mouse monoclonal | IF: 1:100 | American Type Culture Collection |
| Human α6 integrin (extracellular epitope) | Purified Rat anti–human CD49f (clone GoH3) | Rat monoclonal | IF: 1:50 | BD Biosciences PharMingen (San Diego, CA)/555734 |
| Human α6 integrin (isoform α6XIA) (aa 526–803) | Rabbit polyclonal to integrin α6 | Rabbit polyclonal | WB: 1:1000 | Abcam/ab97760 |
| Mouse VLA-3α (N-terminus aa 110–325) | Integrin α3/VLA-3α | Mouse monoclonal | WB: 1:500 | BD Transduction Laboratories/611044 |
| Human GAPDH (C-terminus) | Anti-GAPDH | Rabbit polyclonal | WB: 1:10,000 | Millipore/ABS16 |
| Mouse IgG | Peroxidase-conjugated AffiniPure donkey anti–mouse IgG (H+L) | Donkey polyclonal | WB: 1:1000 | Jackson ImmunoResearch (West Grove, PA)/715-035-151 |
| Rabbit IgG | Peroxidase-conjugated AffiniPure donkey anti–rabbit IgG (H+L) | Donkey polyclonal | WB: 1:1000 | Jackson ImmunoResearch/711-035-152 |
| Mouse IgG | Alexa Fluor 488 goat anti–mouse IgG (H+L) | Goat polyclonal | IF: 1:250 | Invitrogen/A11029 |
| Mouse IgG | Alexa Fluor 555 goat anti–mouse IgG (H+L) | Goat polyclonal | IF: 1:250 | Invitrogen/A21424 |
| Rabbit IgG | Alexa Fluor 488 goat anti–rabbit IgG (H+L) | Goat polyclonal | IF: 1:250 | Invitrogen/A11034 |
| Rat IgG | Alexa Fluor 488 goat anti–rat IgG (H+L) | Goat polyclonal | IF: 1:250 | Invitrogen/A11006 |
aa, amino acids; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IF, immunofluorescence microscopy; IgG, immunoglobulin G; IP, immunoprecipitation; WB, Western blot.
DNA constructs and transient transfection
The plasmid GEX2TK-PAK-Crib was a gift from John G. Collard (Netherlands Cancer Institute, Amsterdam, Netherlands). The plasmid coding for the GFP-PH-Akt fusion protein (Varnai and Balla, 1998), a kind gift from Tamás Balla (National Institutes of Health, Bethesda, MD), was transiently transfected using the AMAXA system (VCA-1005; Lonza AG, Cologne, Germany) with the program L-005.
Generation of laminin and integrin knockdowns
Target sequences to knock down LMα5 (KD2: GCAAGUACGUGGAUCUCAA; KD4: GAGAGGAUCAACGUGUUCA) or LMα3 (GGAACCGCAACUUUGGAAA) synthesis were chosen using algorithms at the siDESIGN Center (www.dharmacon.com/designcenter) and the Whitehead Institute (siRNA-help@wi.mit.edu). Oligonucleotides encoding the corresponding shRNA sequences for LMα5 and a scrambled sequence (GGAGAGUUCAUCGUAAGCA) were cloned into the pSUPER.retro.puro vector (Oligoengine, Seattle, WA). Retroviruses were generated by transfecting 24 μg of each vector and 2.4 μg of pVSV-G into the Phoenix gag-pol packaging cell line (American Type Culture Collection, Manassas, VA) using Lipofectamine 2000 (Invitrogen) as described (Schuck et al., 2004). MDCK cells plated the day before at 2.5 × 104 cells/cm2 in 35-mm dishes were infected with 1 ml of filtered, virus-containing media supplemented with 4 μg/ml polybrene (Sigma-Aldrich) and reinfected the next day. Selection of infected cells was done in the presence of 2 μg/ml puromycin (Sigma-Aldrich). For clonal selection, cells were plated at 10 cells/cm2, and individual colonies isolated and expanded.
To transiently knock down the synthesis of LMα3, cells were transfected with siRNAs against the LMα3 subunit, or luciferase (GUGCGUUGCUAGUACCAAC) as control, as described.
For the generation of the integrin α3 and α6 knock down, cell lines were generated as described before using the corresponding shRNA sequences cloned in the pRVHpuro vector (Myllymaki et al., 2011).
qPCR and standard RT-PCR
Expression of the α5 and α3 subunits of LM-511 and LM-332, respectively, was determined as described before (Moyano et al., 2010) using the canine primers Cf_LAMA5_1_SG and Cf_LAMA3_1_SG QuantiTect Primer Assay (QT01450638 and QT01465975, respectively; Qiagen, Valencia, CA). To determine α4 expression, the canine primer Cf_LAMA4_1_SG (QT00903308; Qiagen) was used in standard RT-PCR using the same program as before.
Preparation of endogenous LM-511–enriched extracellular matrix
Parental MDCK cells were plated at a density of 7.5 × 104 cells/cm2 on collagen-coated surfaces for 4 d to deposit an ECM enriched in LM-511. Cells were then washed with PBS− three times and once with double-distilled H2O, and the monolayer was removed by incubating ∼10 min in 20 mM NH4OH at room temperature, followed by extensive washes with PBS−. All solutions were supplemented with 1 mM phenylmethanesulfonyl fluoride (PMSF) and protease inhibitor cocktail (PIC; Roche Applied Science, Indianapolis, IN). Finally, the ECM was washed with PBS+ three times for 5 min before plating cells.
Analysis of protein synthesis and deposition by metabolic labeling
For biosynthesis analysis, cells were pulse labeled for 30 min with [35S]Met/Cys as previously described (Moyano et al., 2010). Protein was extracted with RIPA buffer (10 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1% [wt/vol] IGEPAL CA-630, 0.5% [wt/vol] SDS, 1% [wt/vol] sodium deoxycholate) with 1 mM PMSF, PIC, and phosphatase inhibitor cocktail (PhIC; Sigma-Aldrich), and LM-511 and LM-332 were immunoprecipitated with either anti–LM-111 or 8LN5 pAbs, respectively (Table 1). Radiolabeled proteins were resolved by SDS–PAGE (see later discussion) and detected by autoradiography.
For the analysis of ECM protein deposition, 4 × 104 cells/cm2 were plated on collagen in ExCell, pulsed with [35S]Met/Cys for 40 min, and then chased for 3 h in ExCell supplemented with 15 μg/ml of unlabeled Met/Cys. The ECM was prepared as described and then extracted in 10 mM Tris-Cl, pH 7.5, 150 mM NaCl, and 1% (wt/vol) SDS with 1 mM PMSF, PIC/PhIC, and 100 μM idoacetamide (IAA). Samples were diluted 1:1 (vol/vol) in 10 mM Tris-Cl, pH 7.5, 150 mM NaCl, 2% (wt/vol) IGEPAL CA-630, 2% (wt/vol) sodium deoxycholate with 1 mM PMSF, PIC/PhIC, and 100 μM IAA. Radiolabeled LM-511 or LM-332 was then immunoprecipitated and analyzed by SDS–PAGE and autoradiography.
Immunoprecipitation, small RhoGTPase pull-down assays, and Western blotting
For immunoprecipitation, equal amounts of protein extracted in RIPA buffer, determined by the bicinchoninic acid assay (Thermo Scientific, Rockford, IL), were incubated overnight at 4°C with the appropriate concentration of antibody. Immune complexes were captured with 60 μl of 1:1 (vol/vol) slurry of protein A–Trisacryl (20338; Thermo Scientific) for 2 h at 4°C and washed three times with RIPA buffer and once with 10 mM Tris-Cl, pH 8.6. Bound proteins were solubilized in 2× Laemmli buffer with 20 mM dithiothreitol (DTT), heated at 95°C for 3 min, and alkylated with IAA before SDS–PAGE.
Cdc42 and Rac1 pull-down assays were performed as described previously (Sander et al., 1999). Briefly, protein was extracted in glutathione-S-transferase (GST)–Fish buffer (10% [wt/vol] glycerol, 50 mM Tris-Cl, pH 7.4, 100 mM NaCl, 1% [vol/vol] IGEPAL CA-630, 2 mM MgCl2) with 1 mM PMSF and PIC/PhIC and incubated with 60 μl of 1:1 (vol/vol) slurry of GST-PAK-Crib glutathione agarose beads for 45 min at 4°C. The beads were then washed three times with GST-Fish buffer, and proteins were solubilized in 2× Laemmli buffer with 20 mM DTT, and heated at 95°C for 3 min. For Rho pull-down assays, Rhotekin-RBD-agarose beads (14-383; Millipore, Billerica, MA) were used following the manufacturer's instructions.
For Western blotting, cells were washed twice with PBS− and extracted in RIPA buffer supplemented with 1 mM PMSF and PIC/PhIC. Samples were resolved by SDS–PAGE and analyzed by Western blotting as described previously (Moyano et al., 2010).
Immunofluorescence
Cells grown on collagen-coated coverslips were washed twice with PBS+, fixed with 3% paraformaldehyde (PFA) in PBS+ for 30 min at room temperature, and then washed three times with PBS−. Unreacted PFA was quenched with 0.1% (wt/vol) NaBH4 in PBS− for 15 min, followed by three washes with PBS−. In most cases, cells were permeabilized with cold 0.1% (wt/vol) Triton X-100 in PBS− for 4 min and washed twice with PBS−. Samples were blocked with 10% (vol/vol) goat serum or 0.2% (wt/vol) fish skin gelatin in PBS− (blocking solution) for 30 min at room temperature. Primary antibodies were diluted in blocking solution and incubated for 1 h at room temperature. Samples were then washed with blocking solution and incubated with secondary antibodies conjugated to Alexa fluorochromes (Invitrogen) for 30 min at room temperature. After washing, coverslips were mounted with Mowiol-based mounting. Transwell permeable supports (3412; Corning Life Sciences, Lowell, MA) were fixed and stained for immunofluorescence as described (Moyano et al., 2010). Digital images were collected on a Zeiss LSM510 confocal fluorescence microscope with AIM software (Carl Zeiss, Jena, Germany) and optimized with ImageJ (National Institutes of Health, Bethesda, MS) and Photoshop (Adobe Systems, San Jose, CA).
Live-cell time-lapse videomicroscopy
Cells were seeded at 2.2 × 104 cells/cm2 on eight-well Lab-Tek chamber slides (155411; Thermo Scientific) precoated with collagen I and incubated for 1 h at 37°C. The slides were then observed with a laser confocal microscope (Zeiss LSM-510) equipped with a heated stage inside of an environmental chamber (37°C, 5% CO2). Differential interference contrast (DIC) images for the different experimental groups were acquired with a 20× Plan Achromat objective (numerical aperture 0.8) every 10 min for up to 13 h using an automated stage.
Cell trajectory was quantified by individually tracking frame by frame 6–10 cells within an area of ∼0.5 mm2 (field) using the Manual Tracking plug-in for ImageJ (F. Cordelieres, Institute Curie, Paris, France). An average of five fields per group were analyzed, and each experiment was repeated at least three times. The vector data generated for each field were then loaded onto the Chemotaxis and Migration Tool plug-in (G. Trapp and E. Horn, Ibidi, Munich, Germany), and accumulated distance, Euclidean distance, directionality index (Euclidean/accumulated distance ratio), and cell velocity were calculated. The number of events in which cells within a field persistently migrated at a particular angle, divided in 10º sectors, was represented as a circular histogram or windrose plot. To quantify the specificity of persistent migration as a function of circular distribution, the Rayleigh statistical test for the vectorial data corresponding to each field was applied, and the p value was calculated (see later discussion).
To quantify the stability of nascent cell–cell contacts, individual single cells within fields were manually tracked and initial cell–cell contacts scored for the ability to either remain adherent (stable contact) or detach (broken contact) over the length of the time-lapse analysis.
Raw time-lapse files were exported as QuickTime movies using the Sorenson codec set at 8 frames/s using ImageJ and compiled using iMovie'11 (Apple, Cupertino, CA).
Wound-healing assays
Cells plated at 2.5 × 105 cells/cm2 on collagen-coated coverslips or chamber slides were grown to confluency in ExCell. Monolayers were then wounded with either a pipette tip or a razor blade and washed twice with PBS+. The cultures were then incubated in ExCell and recorded using time-lapse videomicroscopy. For immunofluorescence, cells were fixed and stained 12 h after wounding.
Hanging-drop assays
Hanging-drop assays of cell–cell adhesion stability were conducted essentially as described (Ehrlich et al., 2002). Trypsinized cells were suspended at a final density of 5 × 105 cells/ml in ExCell, and 20 μl of each cell suspension was incubated as a hanging drop from the lid of 60-mm dishes filled with PBS−. After 24 h, cell aggregates were collected, triturated by pipetting up and down 15 times with a P20 Pipetman, transferred to a Neubauer chamber, and photographed. Cell area of the aggregates was measured using ImageJ.
Statistical analysis
Statistical significance was determined by one-way analysis of variance with the Student's t Newman–Keuls post hoc test for multiple comparisons and two-tailed unpaired t test for single comparisons using the InStat application (GraphPad, La Jolla, CA). The p values are indicated, considering 0.05 or less as statistically significant. To assess whether the distribution of the preferred direction of each cell was unimodal, the Rayleigh test for vectorial data was applied, and p < 0.15 was chosen as the criterion for rejecting the null hypothesis of random directionality.
Supplementary Material
Acknowledgments
We are grateful to Joel Collier (University of Chicago, Chicago, IL) and Jonathan Jones (Northwestern University, Chicago, IL) for critically reading the manuscript. This work was supported by Grant RO1 DK068568 from the National Institutes of Health.
Abbreviations used:
- aPKC
atypical protein kinase C
- ECM
extracellular matrix
- GFP
green fluorescent protein
- KD
knockdown
- LM
laminin
- MDCK
Madin-Darby canine kidney epithelial cells
- PH
plecsktrin homology domain
- PIP3
phosphatidylinositol (3,4,5)-trisphosphate
- Sc
scramble
- SEM
standard error of the mean
- shRNA
short hairpin RNA
- siRNA
small interference RNA
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-08-0718) on October 26, 2011.
REFERENCES
- Aumailley M, Rousselle P. Laminins of the dermo-epidermal junction. Matrix Biol. 1999;18:19–28. doi: 10.1016/s0945-053x(98)00004-3. [DOI] [PubMed] [Google Scholar]
- Brakeman PR, Liu KD, Shimizu K, Takai Y, Mostov KE. Nectin proteins are expressed at early stages of nephrogenesis and play a role in renal epithelial cell morphogenesis. Am J Physiol Renal Physiol. 2009;296:F564–F574. doi: 10.1152/ajprenal.90328.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryce N, Clark E, Leysath J, Currie J, Webb D, Weaver A. Cortactin promotes cell motility by enhancing lamellipodial persistence. Curr Biol. 2005;15:1276–1285. doi: 10.1016/j.cub.2005.06.043. [DOI] [PubMed] [Google Scholar]
- Calaluce R, Beck S, Bair E, Pandey R, Greer K, Hoying A, Hoying J, Mount D, Nagle R. Human laminin-5 and laminin-10 mediated gene expression of prostate carcinoma cells. Prostate. 2006;66:1381–1390. doi: 10.1002/pros.20393. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Champliaud M, Burgeson R, Marinkovich M, Yurchenco P. Self-assembly of laminin isoforms. J Biol Chem. 1997;272:31525–31532. doi: 10.1074/jbc.272.50.31525. [DOI] [PubMed] [Google Scholar]
- Chiang LY, Poole K, Oliveira BE, Duarte N, Sierra YA, Bruckner-Tuderman L, Koch M, Hu J, Lewin GR. Laminin-332 coordinates mechanotransduction and growth cone bifurcation in sensory neurons. Nat Neurosci. 2011;14:993–1000. doi: 10.1038/nn.2873. [DOI] [PubMed] [Google Scholar]
- Chiharu T, Daisuke T, Koji S, Katsutoshi Y, Hisayoshi I, Kazuyo N, Masamitsu I, Hiromi K. Spatial and temporal control of laminin-511 and -332 expressions during catagen. J Dermatol Sci. 2010;58:55–63. doi: 10.1016/j.jdermsci.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Choma D, Pumiglia K, DiPersio C. Integrin a3b1 directs the stabilization of a polarized lamellipodium in epithelial cells through activation of Rac1. J Cell Sci. 2004;117:3947–3959. doi: 10.1242/jcs.01251. [DOI] [PubMed] [Google Scholar]
- deHart G, Healy K, Jones J. The role of alpha3beta1 integrin in determining the supramolecular organization of laminin-5 in the extracellular matrix of keratinocytes. Exp Cell Res. 2003;283:67–79. doi: 10.1016/s0014-4827(02)00028-9. [DOI] [PubMed] [Google Scholar]
- DiPersio C, van der Neut R, Georges-Labouesse E, Kreidberg J, Sonnenberg A, Hynes R. alpha3beta1 and alpha6beta4 integrin receptors for laminin-5 are not essential for epidermal morphogenesis and homeostasis during skin development. J Cell Sci. 2000;113:3051–3062. doi: 10.1242/jcs.113.17.3051. [DOI] [PubMed] [Google Scholar]
- Drubin DG, Nelson WJ. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- Ehrlich J, Hansen M, Nelson W. Spatio-temporal regulation of Rac1 localization and lamellipodia dynamics during epithelial cell-cell adhesion. Dev Cell. 2002;3:259–270. doi: 10.1016/s1534-5807(02)00216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell. 2001;106:489–498. doi: 10.1016/s0092-8674(01)00471-8. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A. Cdc42 regulates GSK-3beta and adenomatous polyposis coli to control cell polarity. Nature. 2003;421:753–756. doi: 10.1038/nature01423. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S, Manneville J-B, Nicholls S, Ferenczi MA, Hall A. Cdc42 and Par6-PKCzeta regulate the spatially localized association of Dlg1 and APC to control cell polarization. J Cell Biol. 2005;170:895–901. doi: 10.1083/jcb.200412172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D, Carter W. Laminin 5 deposition regulates keratinocyte polarization and persistent migration. J Cell Sci. 2004;117:1351–1363. doi: 10.1242/jcs.01003. [DOI] [PubMed] [Google Scholar]
- Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10:445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- Goldfinger LE, Hopkinson SB, deHart GW, Collawn S, Couchman JR, Jones JC. The alpha3 laminin subunit, alpha6beta4 and alpha3beta1 integrin coordinately regulate wound healing in cultured epithelial cells and in the skin. J Cell Sci. 1999;112:2615–2629. doi: 10.1242/jcs.112.16.2615. [DOI] [PubMed] [Google Scholar]
- Hamill K, Kligys K, Hopkinson S, Jones J. Laminin deposition in the extracellular matrix: a complex picture emerges. J Cell Sci. 2009;122:4409–4417. doi: 10.1242/jcs.041095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen K, Abrass CK. Laminin-8/9 is synthesized by rat glomerular mesangial cells and is required for PDGF-induced mesangial cell migration. Kidney Int. 2003;64:110–118. doi: 10.1046/j.1523-1755.2003.00039.x. [DOI] [PubMed] [Google Scholar]
- Hartwig B, Borm B, Schneider H, Arin M, Kirfel G, Herzog V. Laminin-5-deficient human keratinocytes: defective adhesion results in a saltatory and inefficient mode of migration. Exp Cell Res. 2007;313:1575–1587. doi: 10.1016/j.yexcr.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Hintermann E, Bilban M, Sharabi A, Quaranta V. Inhibitory role of a6b4-associated Erbb-2 and phosphoinositide 3-kinase in keratinocyte haptotactic migration dependent on a3b1 integrin. J Cell Biol. 2001;153:465–478. doi: 10.1083/jcb.153.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iden S, Collard JG. Crosstalk between small GTPases and polarity proteins in cell polarization. Nat Rev Mol Cell Biol. 2008;9:846–859. doi: 10.1038/nrm2521. [DOI] [PubMed] [Google Scholar]
- Kitt KN, Nelson WJ. Rapid suppression of activated Rac1 by cadherins and nectins during de novo cell-cell adhesion. PLoS One. 2011;6:e17841. doi: 10.1371/journal.pone.0017841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kligys K, Claiborne J, DeBiase P, Hopkinson S, Wu Y, Mizuno K, Jones J. The slingshot family of phosphatases mediates Rac1 regulation of cofilin phosphorylation, laminin-332 organization, and motility behavior of keratinocytes. J Biol Chem. 2007;282:32520–32528. doi: 10.1074/jbc.M707041200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komura H, Ogita H, Ikeda W, Mizoguchi A, Miyoshi J, Takai Y. Establishment of cell polarity by afadin during the formation of embryoid bodies. Genes Cells. 2008;13:79–90. doi: 10.1111/j.1365-2443.2007.01150.x. [DOI] [PubMed] [Google Scholar]
- Li R. Symmetry breaking in biology. Cold Spring Harb Perspect Biol. 2009;2:a003475. doi: 10.1101/cshperspect.a003475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak G, Kavanaugh G, Buschmann M, Stickley S, Koch M, Goss K, Waechter H, Zuk A, Matlin K. Regulated synthesis and functions of laminin 5 in polarized Madin-Darby canine kidney epithelial cells. Mol Biol Cell. 2006;17:3664–3677. doi: 10.1091/mbc.E05-11-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margadant C, Raymond K, Kreft M, Sachs N, Janssen H, Sonnenberg A. Integrin a3b1 inhibits directional migration and wound re-epithelialization in the skin. J Cell Sci. 2009;122:278–288. doi: 10.1242/jcs.029108. [DOI] [PubMed] [Google Scholar]
- Marinkovich M. Tumour microenvironment: laminin 332 in squamous-cell carcinoma. Nat Rev Cancer. 2007;7:370–380. doi: 10.1038/nrc2089. [DOI] [PubMed] [Google Scholar]
- McMillan J, Akiyama M, Nakamura H, Shimizu H. Colocalization of multiple laminin isoforms predominantly beneath hemidesmosomes in the upper lamina densa of the epidermal basement membrane. J Histochem Cytochem. 2006;54:109–118. doi: 10.1369/jhc.5A6701.2005. [DOI] [PubMed] [Google Scholar]
- Matlin KS, Reggio H, Helenius A, Simons K. Infectious entry pathway of influenza virus in a canine kidney cell line. J Cell Biol. 1981;91:601–613. doi: 10.1083/jcb.91.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner J, Patton B, Lentz S, Gilbert D, Snider W, Jenkins N, Copeland N, Sanes J. The laminin alpha chains: expression, developmental transitions, and chromosomal locations of alpha1–5, identification of heterotrimeric laminins 8–11, and cloning of a novel alpha3 isoform. J Cell Biol. 1997;137:685–701. doi: 10.1083/jcb.137.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner JH, Yurchenco PD. Laminin functions in tissue morphogenesis. Annu Rev Cell Dev Biol. 2004;20:255–284. doi: 10.1146/annurev.cellbio.20.010403.094555. [DOI] [PubMed] [Google Scholar]
- Moyano JV, Greciano PG, Buschmann MM, Koch M, Matlin KS. Autocrine transforming growth factor-b1 activation mediated by integrin aVb3 regulates transcriptional expression of laminin-332 in Madin–Darby canine kidney epithelial cells. Mol Biol Cell. 2010;21:3654–3668. doi: 10.1091/mbc.E10-06-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myllymaki SM, Teravainen TP, Manninen A. Two distinct integrin-mediated mechanisms contribute to apical lumen formation in epithelial cells. PLoS One. 2011;6:e19453. doi: 10.1371/journal.pone.0019453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WJ. Remodeling epithelial cell organization: transitions between front-rear and apical-basal polarity. Cold Spring Harb Perspect Biol. 2009;1:a000513. doi: 10.1101/cshperspect.a000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiuchi R, Murayama O, Fujiwara H, Gu J, Kawakami T, Aimoto S, Wada Y, Sekiguchi K. Characterization of the ligand-binding specificities of integrin a3b1 and a6b1 using a panel of purified laminin isoforms containing distinct a chains. J Biochem. 2003;134:497–504. doi: 10.1093/jb/mvg185. [DOI] [PubMed] [Google Scholar]
- Nishiuchi R, Takagi J, Hayashi M, Ido H, Yagi Y, Sanzen N, Tsuji T, Yamada M, Sekiguchi K. Ligand-binding specificities of laminin-binding integrins: a comprehensive survey of laminin-integrin interactions using recombinant alpha3beta1, alpha6beta1, alpha7beta1 and alpha6beta4 integrins. Matrix Biol. 2006;25:189–197. doi: 10.1016/j.matbio.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Ojakian GK, Schwimmer R. The polarized distribution of an apical cell surface glycoprotein is maintained by interactions with the cytoskeleton of Madin-Darby canine kidney cells. J Cell Biol. 1988;107:2377–2387. doi: 10.1083/jcb.107.6.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani N, Peglion F, Chavrier P, Etienne-Manneville S. Cdc42 localization and cell polarity depend on membrane traffic. J Cell Biol. 2010;191:1261–1269. doi: 10.1083/jcb.201003091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie R, Doyle A, Yamada K. Random versus directionally persistent cell migration. Nat Rev Mol Cell Biol. 2009;10:538–549. doi: 10.1038/nrm2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plopper G, Domanico S, Cirulli V, Kiosses W, Quaranta V. Migration of breast epithelial cells on laminin-5: differential role of integrins in normal and transformed cell types. Breast Cancer Res Treat. 1998;51:57–69. doi: 10.1023/a:1006086218174. [DOI] [PubMed] [Google Scholar]
- Qin Y, Capaldo C, Gumbiner BM, Macara IG. The mammalian Scribble polarity protein regulates epithelial cell adhesion and migration through E-cadherin. J Cell Biol. 2005;171:E1061–E1071. doi: 10.1083/jcb.200506094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander EE, ten Klooster JP, van Delft S, van der Kammen RA, Collard JG. Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J Cell Biol. 1999;147:1009–1022. doi: 10.1083/jcb.147.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S, Manninen A, Honsho M, Fullekrug J, Simons K. Generation of single and double knockdowns in polarized epithelial cells by retrovirus-mediated RNA interference. Proc Natl Acad Sci USA. 2004;101:4912–4917. doi: 10.1073/pnas.0401285101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal B, DeBiase P, Matzno S, Chew T, Claiborne J, Hopkinson S, Russell A, Marinkovich M, Jones J. Integrin beta4 regulates migratory behavior of keratinocytes by determining laminin-332 organization. J Biol Chem. 2006;281:35487–35498. doi: 10.1074/jbc.M606317200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin K, Wang Q, Margolis B. PATJ regulates directional migration of mammalian epithelial cells. EMBO Rep. 2007;8:158–164. doi: 10.1038/sj.embor.7400890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin L, Pausch F, Durbeej M, Ekblom P. Differential expression of five laminin alpha (1–5) chains in developing and adult mouse kidney. Dev Dyn. 1997;210:446–462. doi: 10.1002/(SICI)1097-0177(199712)210:4<446::AID-AJA8>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Stipp CS. Laminin-binding integrins and their tetraspanin partners as potential antimetastatic targets. Expert Rev Mol Med. 2010;12:e3. doi: 10.1017/S1462399409001355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara K, Tsuruta D, Kobayashi H, Ikeda K, Hopkinson S, Jones J, Ishii M. Spatial and temporal control of laminin-332 (5) and -511 (10) expression during induction of anagen hair growth. J Histochem Cytochem. 2007;55:43–55. doi: 10.1369/jhc.6A6920.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai Y, Miyoshi J, Ikeda W, Ogita H. Nectins and nectin-like molecules: roles in contact inhibition of cell movement and proliferation. Nat Rev Mol Cell Biol. 2008;9:603–615. doi: 10.1038/nrm2457. [DOI] [PubMed] [Google Scholar]
- Varnai P, Balla T. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J Cell Biol. 1998;143:501–510. doi: 10.1083/jcb.143.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen T, Zhang Z, Yu Y, Qu H, Koch M, Aumailley M. Integrin α3 subunit regulates events linked to epithelial repair, including keratinocyte migration and protein expression. Wound Repair Regen. 2010;18:325–334. doi: 10.1111/j.1524-475X.2010.00590.x. [DOI] [PubMed] [Google Scholar]
- Yu W, Datta A, Leroy P, O'Brien L, Mak G, Jou T, Matlin K, Mostov K, Zegers M. Beta1-integrin orients epithelial polarity via Rac1 and laminin. Mol Biol Cell. 2005;16:433–445. doi: 10.1091/mbc.E04-05-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurchenco PD. Basement membranes: cell scaffoldings and signaling platforms. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Kramer R. Laminin 5 deposition promotes keratinocyte motility. Exp Cell Res. 1996;227:309–322. doi: 10.1006/excr.1996.0280. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.