Global translation is inhibited in Saccharomyces cerevisiae cells under osmotic stress; nonetheless, osmostress-protective proteins are synthesized. We found that translation mediated by the mRNA cap-binding protein Cbc1 is stress-resistant and necessary for the rapid translation of osmostress-protective proteins under osmotic stress.
Abstract
In response to osmotic stress, global translation is inhibited, but the mRNAs encoding stress-protective proteins are selectively translated to allow cell survival. To date, the mechanisms and factors involved in the specific translation of osmostress-responsive genes in Saccharomyces cerevisiae are unknown. We find that the mRNA cap-binding protein Cbc1 is important for yeast survival under osmotic stress. Our results provide new evidence supporting a role of Cbc1 in translation initiation. Cbc1 associates with polysomes, while the deletion of the CBC1 gene causes hypersensitivity to the translation inhibitor cycloheximide and yields synthetic “sickness” in cells with limiting amounts of translation initiator factor eIF4E. In cbc1Δ mutants, translation drops sharply under osmotic stress, the subsequent reinitiation of translation is retarded, and “processing bodies” containing untranslating mRNAs remain for long periods. Furthermore, osmostress-responsive mRNAs are transcriptionally induced after osmotic stress in cbc1Δ cells, but their rapid association with polysomes is delayed. However, in cells containing a thermosensitive eIF4E allele, their inability to grow at 37ºC is suppressed by hyperosmosis, and Cbc1 relocalizes from nucleus to cytoplasm. These data support a model in which eIF4E-translation could be stress-sensitive, while Cbc1-mediated translation is necessary for the rapid translation of osmostress-protective proteins under osmotic stress.
INTRODUCTION
One important response of eukaryotic cells to stress is the change in gene expression patterns through a general shutdown and reprogramming of protein synthesis. Stress signals inhibit the translation of “housekeeping” proteins and promote the translation of stress-protective proteins (Yamasaki and Anderson, 2008; Spriggs et al., 2010). Defects in translation reprogramming in response to stress have been associated with disease, including cancer, diabetes, and inflammatory processes (Keene, 2007). How specific mRNAs become translated under stress, when general translation is down-regulated, is an important issue for understanding the mechanisms regulating gene expression.
Eukaryotic translation is a complex process controlled by a wide range of regulatory factors. Stress signals usually inhibit the process in the initiation step. Normal translation takes place through a cap-dependent initiation mechanism; the 40S ribosome subunit is recruited to a modified base at the 5′ end of the mRNA (the m7G cap). This process is coordinated by a complex with eukaryotic initiation factors (eIFs) called eIF4F, in which eIF4G is a scaffold protein that facilitates the interactions between proteins and the small ribosomal subunit, and eIF4E recognizes and binds the cap structure of mRNA. Once Met-tRNA, the 40S subunit, and the eukaryotic initiation factors form a complex on capped mRNA, the start codon is recognized, and the large ribosomal subunit attaches. One key target for the control of translation in the initiation step is eIF4E, which can be bound by inhibitory proteins that subsequently impede mRNA binding (Wang and Proud, 2007; Lackner and Bähler, 2008; Van Der Kelen et al., 2009).
Whereas global translation is inhibited during stress, the mRNAs encoding stress-response proteins must be able to engage with ribosomes and be translated. To achieve this, several mechanisms have evolved in eukaryotic cells. For some of them, the recruitment of ribosomes is regulated by elements in the 5′ and 3′ untranslated regions (UTRs) of mRNAs, including the internal ribosome entry sites (IRES) that bypass the cap-dependent recruitment of mRNA, upstream open reading frames, and microRNA target sites (Yamasaki and Anderson, 2008; Van Der Kelen et al., 2009; Spriggs et al., 2010). However, for most stress conditions in which translation reprogramming has been described, the selective translation mechanism of stress-specific response mRNAs remains unknown. The relocalization upon stress of “housekeeping” mRNAs to cytoplasmic foci, where they are stored in a translationally silenced state, would allow the dedication of a basal translation to synthesize protective proteins (Bond, 2006; Buchan and Parker, 2009; Lui et al., 2010).
One of the experimental models for studying the regulation of gene expression by stress is the response of Saccharomyces cerevisiae cells to hyperosmotic shock. A sudden increase in external osmolarity produces several rapid effects on yeast cells, such as a rapid decrease in protein synthesis and activation of the HOG mitogen-activated protein kinase (MAPK) pathway. Activation by phosphorylation of MAPK Hog1, which is homologous to mammalian p38 MAPK, is necessary for the control of different cellular processes that allow cells to adapt and to survive the stress condition (reviewed in de Nadal et al., 2002; de Nadal and Posas, 2010; Hohmann, 2002). One of the main effects of Hog1 activation is a quick, massive change in gene expression. Thus the levels of around 600 mRNAs (representing 10% of all yeast genes) significantly change after a few minutes of stress, as a result of changes in their transcription rates and stability (Posas et al., 2000; Rep et al., 2000; Molin et al., 2009; Romero-Santacreu et al., 2009; Miller et al., 2011). The quick and transient general inhibition of translation observed under osmotic stress occurs in the initiation step by an unidentified mechanism that is apparently independent of Hog1. However, this MAPK is involved in the later activation of translation, possibly through Rck2, this being a yeast kinase that resembles mammalian MAPK-activated protein kinases (Bilsland-Marchesan et al., 2000; Teige et al., 2001). Rck2 could affect global translation by directly regulating elongation factor EF-2, but an effect on initiation factors, which occurs in the homologous mammalian p38 MAPK pathway, cannot be ruled out. It is important to note that recent studies have shown that a Hog1-dependent, short-term regulation of mRNA translation rates takes place after hyperosmotic stress. Therefore, during the global inhibition of translation, important genes for cell survival under this stress condition are translationally up-regulated, and their protein synthesis increases (Melamed et al., 2008; Halbeisen and Gerber, 2009; Warringer et al., 2010). In this scenario, the connections among translation, mRNA degradation and the formation of RNA containing granules, for example, stress granules and “processing bodies” (P-bodies), seem to be important, because, parallel to the regulation of translation rates, the stability of functionally related groups of mRNAs is selectively modified and P-bodies are seen to temporarily increase upon hyperosmotic shock (Greatrix and van Vuuren, 2006; Hilgers et al., 2006; Molin et al., 2009; Romero-Santacreu et al., 2009; Miller et al., 2011). Translationally repressed mRNAs can be degraded in P-bodies by the main mRNA decay pathway, which includes the decapping enzyme Dcp1/Dcp2 and the 5′–3′ exonuclease Xrn1. Under stress, however, mRNAs can be stored together with some translation initiation factors, such as eIF4E, in P-bodies, which allows their return to translation after adaptation (Hoyle et al., 2007; Parker and Sheth, 2007; Buchan and Parker, 2009; Romero-Santacreu et al., 2009). To date, the precise mechanisms and factors modulating the changes in mRNA translation and fate under osmotic stress remain unknown.
The nuclear cap-binding complex (CBC) in S. cerevisiae, composed of Cbc1 and Cbc2, is recruited cotranscriptionally to the cap structure of mRNAs (Görlich et al., 1996; Wong et al., 2007). The CBC complex is mainly nuclear, but can shuttle between the nucleus and the cytoplasm using the importin α/β system (Görlich et al., 1996). Individual CBC subunits are not essential in yeast, but deletions of the corresponding genes diminish cell growth in glucose media, while mutants show different phenotypes related to different aspects of RNA metabolism (Das et al., 2000); consequently, CBC has been proposed as a participant in a wide variety of mRNA-associated processes. Nuclear CBC is involved in splicing (Colot et al., 1996; Fortes et al., 1999; Zhang and Rosbash, 1999; Görnemann et al., 2005; Hossain et al., 2009), the formation of the preinitiation complex at active genes (Lahudkar et al., 2011), transcription termination (Das et al., 2000; Wong et al., 2007), and nuclear mRNA degradation (Das et al., 2003; Kuai et al., 2005). Moreover, in mammalian cells, CBC plays a role in exporting capped RNA, but its role is not clear in yeast (Izaurralde et al., 1995; Görlich et al., 1996; Wong et al., 2007).
Besides the nuclear roles of Cbc1, its mammalian homologue CBP80 has been proposed as forming part of the pioneer translation initiation complex. The bulk of cellular translation initiation is mediated by cytoplasmic cap-binding initiation factor eIF4E, but CBP80-bound mRNA, which is a precursor to eIF4E-bound mRNA, can also be translated during a pioneer round of translation, in which aberrant mRNAs are eliminated by the nonsense-mediated mRNA decay (NMD) pathway (Ishigaki et al., 2001; Lejeune et al., 2002; Chiu et al., 2004). Supporting this model, CBP80 can be found associated with subpolysomal fractions, and the translation initiation factors eIF4GI, eIF3, eIF4I, and eIF2A and poly(A)-binding protein (PAB)1, to coimmunopurify with both CBP80 and eIF4E, which suggests that each factor functions in both translation modes.
In yeast, Cbc1 has also been proposed as a mediator for a pioneer round of translation; nonetheless, contradictory data provide an unclear view of Cbc1's role in translation. On the one hand, Gao et al. (2005) showed that NMD occurs in Cbc1-bound mRNAs and also in eIF4E-mRNAs in yeast. However, Kuperwasser et al. (2004) argued against a Cbc1/Cbc2-mediated pioneer round of translation, since they found that NMD occurs minimally in Cbc2-bound mRNA. Moreover, and remarkably, deletion in CBC1 resulted in synthetic lethality with an eIF4GI version mutated in the sites of interaction with eIF4E and Pab1 (Fortes et al., 2000). These authors' work showed that eIF4GI can simultaneously interact competitively with Cbc1 and eIF4E and that Cbc1 stimulates a 2.5-fold translation in extracts of yeast strains harboring a mutated eIF4GI that interacts only weakly with eIF4E. Nevertheless, later results have revealed that the cells expressing eIF4GI, with a point mutation that abolishes binding to Cbc1, fail to manifest a detectable defect in either the growth or composition of the transcriptome (Baron-Benhamou et al., 2003). Collectively, all these results suggest a connection between yeast Cbc1 and translation, but they do not explain the functional significance of this connection for yeast cells.
The importance of a mammalian CBP80-dependent pioneer round of translation in response to stress conditions has been recently documented. In mammalian cells under prolonged hypoxia or during serum starvation, a steady-state translation dependent on eIF4E is dramatically inhibited; however, NMD, which in mammalian cells restricts only the pioneer round of translation via CBP80, occurs efficiently (Oh et al., 2007a, 2007b). Moreover, CBP80 translation differs from the steady-state translation, and a specific translation initiation factor, CTIF, is preferentially involved in CBP80-translation (Lee et al., 2008; Kim et al., 2009).
For the purpose of identifying those proteins involved in the posttranscriptional regulation of gene expression in S. cerevisiae cells under osmotic stress, we performed an osmotic stress sensitivity screening using 182 mutants in yeast genes encoding for RNA-binding proteins. One of the most sensitive strains carried a deletion in CBC1. Our results show that Cbc1 is directly involved in translation under normal conditions and that Cbc1-mediated translation ensures the rapid synthesis of osmostress-responsive genes upon hyperosmotic shock.
RESULTS
Deletion of the CBC1 gene results in growth reduction and loss of viability under osmotic stress
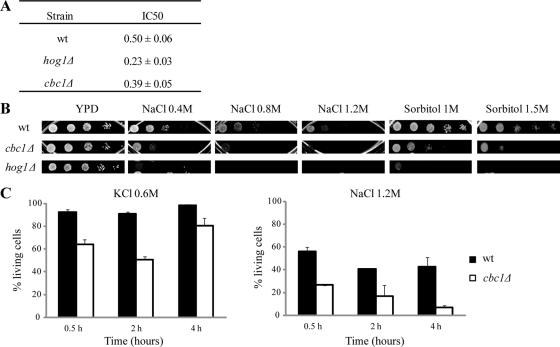
For the purpose of discovering the yeast proteins involved in the regulation of gene expression in response to osmotic stress at the co- and posttranscriptional levels, we checked the tolerance to NaCl of the mutants in those genes encoding for the proteins described to be involved in RNA metabolism. To select mutants, we searched in the Saccharomyces Genome Database and found 182 genes for which deletions were viable, and whose products were described to be involved in mRNA export, translation, degradation, or splicing; we also considered genes of unknown functions whose corresponding proteins contained RNA-binding domains. The NaCl tolerance of these 182 mutants (from the Euroscarf mutant collection, http://web.uni-frankfurt.de/fb15/mikro/euroscarf) was assayed by determining the NaCl concentration that causes a growth reduction of 50% (IC50 index; Supplemental Table S1). One of the mutants that showed reduced tolerance to NaCl contained a deletion of the CBC1 gene (Figure 1A), which encodes the nuclear cap-binding protein Cbc1, homologous to the mammalian CBP80 protein (Lewis et al., 1996). The sensitivity of the cbc1Δ mutant to NaCl was due to a reduced tolerance to osmotic stress, since the cbc1Δ mutant also displayed hypersensitivity to high concentrations of sorbitol (Figure 1B). Moreover, cbc1Δ cells showed a strong reduction in viability after treatments with 0.6 M KCl or 1.2 M NaCl, when compared with wild-type (wt) cells (Figure 1C; between 40% in 0.6 M KCl and 30% in 1.2 M NaCl).
FIGURE 1:
The cbc1Δ mutant shows growth defects and loss of viability under osmotic stress. (A) The NaCl concentration required to inhibit 50% of growth (IC50) as a measure of the osmotic stress sensitivity of the indicated strains. The data are the mean of at least three independent experiments and the SE is indicated. (B) Sensitivity to different osmotic stress conditions of the cbc1Δ and hog1Δ mutant cells. Exponentially growing cells were spotted on YPD plates supplemented with NaCl or sorbitol. Plates were incubated at 30°C. (C) Viability analysis of the cbc1Δ mutant during osmotic stress. Exponentially growing wt and cbc1Δ cells in YPD were subjected to the indicated osmotic stress conditions or kept in YPD without stress. Samples were taken at the indicated times and dead cells were determined by flow cytometry using PI. The percentage of living cells for each condition is compared with the cells in an equivalent aliquot of the same strain incubated in YPD. The data are the mean and the SE of two independent experiments.
Osmotic stress–activated mRNAs are induced in cbc1Δ mutants upon hyperosmotic shock
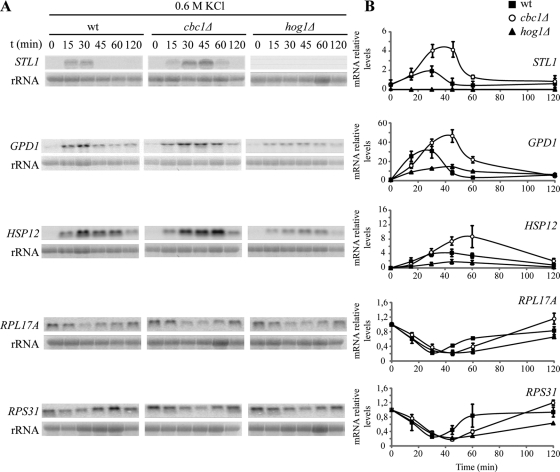
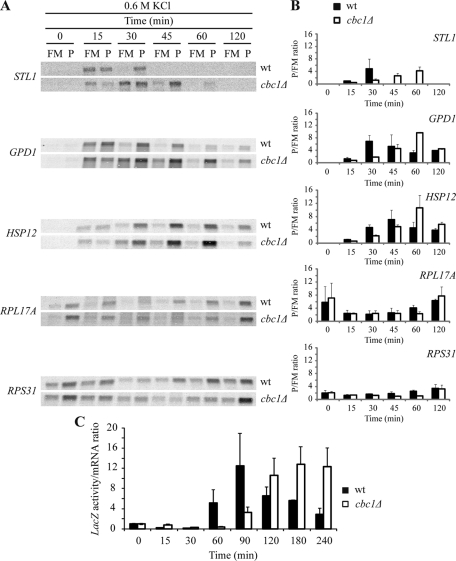
Gene expression patterns are massive, rapid, and transiently changed in response to hyperosmotic shock. These changes take place at the transcriptional and posttranscriptional levels and yield, among other things, a variation in mRNA amounts of around 10% of all the yeast genes, mostly controlled by Hog1 MAPK (reviewed in de Nadal and Posas, 2010). The Cbc1 protein is cotranscriptionally bound to mRNAs through their cap structures, and cells in which the CBC1 gene has been deleted are hypersensitive to osmotic stress, we therefore reasoned that regulation of gene expression could be defective in a cbc1Δ mutant, which is the case in many osmostress-hypersensitive mutants, including mutations in HOG pathway components (O'Rourke and Herskowitz, 2004). To test this possibility, we first investigated the induction of the total mRNA levels of STL1, GPD1, and HSP12, which are involved in glycerol transport, glycerol production, and maintenance of membrane organization, respectively, and whose corresponding mRNAs are strongly and transiently induced by osmotic stress (Posas et al., 2000; Romero-Santacreu et al., 2009). As previously reported (Rep et al., 1999; Posas et al., 2000), the osmostress induction of STL1 was fully dependent on Hog1, while the inductions of GPD1 and HSP12 were partially dependent on this MAPK (Figure 2). In the cbc1Δ mutant, the three genes were induced after treatment with 0.6 M KCl. The increase in the levels of STL1, GPD1, and HSP12 during the first part of the time course (the first 15 min for STL1; the first 30 min for GPD1 and HSP12) was similar in cbc1Δ and wt cells, with a slight delay noted in the mutant (Figure 2B). However, the kinetics of mRNA induction showed differences between cbc1Δ and wt at the maximal levels reached and in the length of response. In the cbc1Δ mutant, the increase in the STL1, GPD1, and HSP12 mRNA amounts was approximately twofold higher and peaked 10–20 min later than in wt (Figure 2). The amount of mRNA encoding ribosomal proteins (RPs), such as RPL17A and RPS31, has also been described as reduced under osmotic stress (Posas et al., 2000; Romero-Santacreu et al., 2009). Our results reveal that variation in the mRNA levels of these two genes during osmotic stress treatment with 0.6 M KCl was similar in cbc1Δ and wt cells, and only a delay in the recovery of the basal level of RPS31 mRNA was observed in the cbc1Δ mutant cells (Figure 2). In summary, our results indicate that the growth defect and loss of viability of the cbc1Δ mutant under hyperosmotic stress do not correlate with an inability to induce the mRNA levels of the osmotic stress-responsive genes. On the contrary, our results indicate that this induction in the cbc1Δ mutant is stronger and lasts longer than in the wt strain.
FIGURE 2:
Increase in the levels of osmostress-responsive mRNAs is delayed, but not inhibited, in the cbc1Δ mutants. The wt, hog1Δ, and cbc1Δ strains were grown exponentially in YPD, and then cells were subjected to osmotic stress by addition of 0.6 M KCl. Samples were taken at the indicated times and the expression of the indicated genes was analyzed by Northern blotting (A). (B) Quantifications of the mRNA levels normalized to rRNA. The data are the mean and the SE of three independent experiments.
The cbc1Δ mutant shows a defect in the reinitiation of translation after hyperosmotic shock
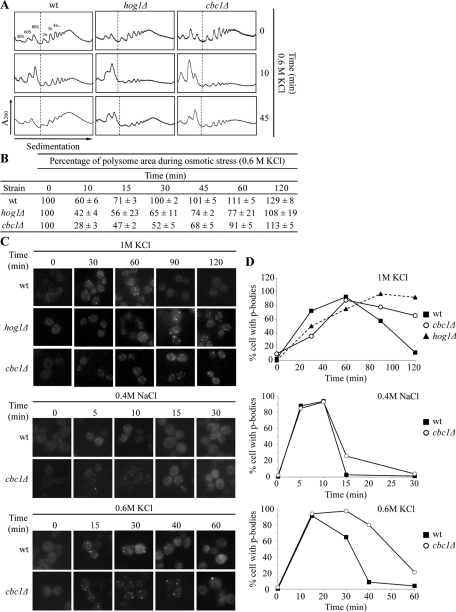
Gene expression also changed at the translation level upon osmotic shock. In yeast, increased extracellular osmolarity leads to a transient inhibition of translation in the initiation step. After adaptation, protein synthesis is restored through an unknown Hog1-dependent mechanism (Uesono and Toh-e, 2002). This stop in global translation occurs parallel to a transient increase in the P-bodies due to the incorporation of mRNAs from polysomes, which is delayed and lasts longer in the hog1Δ mutant cells (Teixeira et al., 2005; Romero-Santacreu et al., 2009). Thus, as a second step in the study of gene expression under osmotic stress in the cbc1Δ mutant, we analyzed changes in global translation by obtaining polysome profiles and by following the P-bodies' status. Under nonstress conditions, the cbc1Δ and hog1Δ mutants showed a similar translation profile to that of the wt strain (Figure 3A). After 10 min of osmotic stress treatment with 0.6 M KCl, global translation dropped in all three strains, as expected. The cbc1Δ mutant displayed a more marked decrease than the wt or even the hog1Δ mutants (Figure 3, A and B). Global translation recovered after 30 min of stress in wt cells, while the polysome levels did not recover after 60 min in the cbc1Δ mutant. At this time, it was possible to see that global translation in the hog1Δ mutant had not completely recovered (Figure 3B). The P-bodies' status under osmotic stress was followed by visualizing a green fluorescent protein (GFP)-tagged version of the decapping enzyme Dcp2 (Sheth and Parker, 2003). As previously described (Teixeira et al., 2005; Romero-Santacreu et al., 2009), under treatment with 1 M KCl, P-bodies could be visualized in wt cells after 30 min of stress and remained until 60 min; meanwhile, in hog1Δ cells, an assembly of P-bodies was observed at 60 min, while bright foci were still visible after 2 h of stress (Figure 3, C and D). In the cbc1Δ mutant, P-body assembly/disassembly kinetics were similar to those of the hog1Δ mutant: P-bodies were clearly visible after 60 min of stress and were still observed with a similar brightness after 2 h of treatment (Figure 3C). Mild osmotic stress conditions, such as treatment with 0.6 M KCl or 0.4 M NaCl, cause a quicker and more transitory formation of P-bodies (Romero-Santacreu et al., 2009; Figure 3C). Under both conditions, P-bodies were also visualized for longer periods in cbc1Δ cells compared with wt cells (Figure 3, C and D). These results indicate that Cbc1 is not essential for P-body formation. However, delay in the recovery of global translation and the prolonged permanence of P-bodies in the cbc1Δ mutant suggest a role for the cap-binding protein in P-body disassembly, the return of mRNA to polysomes, and the reinitiation of translation. A similar role in the control of mRNA cytoplasmic fate in response to osmotic stress has also been suggested for MAPK Hog1 (Romero-Santacreu et al., 2009; Warringer et al., 2010). Altogether, our results indicate that the cbc1Δ mutant is not defective in the induction of osmostress mRNAs but is defective in the reinitiation of translation during adaptation. Thus the cbc1Δ mutant resembles only part of the defects in the regulation of gene expression caused by HOG1 deletion.
FIGURE 3:
The cbc1Δ mutant shows a defect in translation recovery after hyperosmotic stress. (A) Reinitiation of translation after adaptation to osmotic stress is delayed in the cbc1Δ mutant. Polysome profiles show translation inhibition and recovery during osmotic stress in the wt, hog1Δ, and cbc1Δ strains. Cells grown exponentially in YPD were treated with 0.6 M KCl at the indicated times. Polysomal profiles were obtained as described in Materials and Methods. The relative amount of rRNA in each fraction, measured by online UV detection at 260 nm, shows the ribosomal light subunits (40S), heavy subunits (60S), monosomes (80S), and polysomes (2n, 3n, 4n, …). Representative absorbance curves (out of five repetitions) are displayed. (B) Percentage of the polysomal area during osmotic stress (0.6 M KCl) in wt, hog1Δ, and cbc1Δ. The polysome percentage is relative to the polysomal area of each strain at time 0. The data are given as the mean and SE of three independent experiments. (C) P-body assembly and disassembly under osmotic shock are delayed in the cbc1Δ mutant. The wt, hog1Δ, and cbc1Δ strains expressing a GFP-tagged version of the decapping enzyme Dcp2 were grown until the exponential phase, and then treated with 1 M KCl, 0.6 M KCl, or with 0.4 M NaCl. Dcp2-GFP allowed P-body visualization by fluorescence microscopy. (D) The quantification of the percentage of cells with P-bodies is indicated in the graphs. At least 100 cells were quantified for each condition.
cbc1Δ cells are hypersensitive to the translation inhibitor cycloheximide and show a genetic interaction with translation initiation factor eIF4E
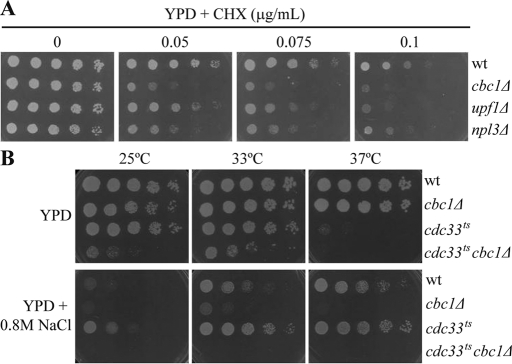
Previously, it was proposed that Cbc1 could mediate translation (Fortes et al., 2000; Gao et al., 2005), yet other results have argued against this role (Baron-Benhamou et al., 2003; Kuperwasser et al., 2004). Our results, which revealed defects of the cbc1Δ mutants in the recovery of translation and P-body disassembly after hyperosmotic shock, prompted us to further investigate the involvement of Cbc1 in translation. With this aim in mind, we examined the sensitivity of the cbc1 mutant to the inhibitor of translation elongation cycloheximide (CHX; McKeehan and Hardesty, 1969). It has been reported that yeast strains with translational defects are hypersensitive to the growth inhibitory effect of CHX (Gross et al., 2007). This is the case for mutants in ribonucleoprotein Npl3, which promotes translation termination accuracy, and in Upf1, a component of the NMD RNA surveillance pathway with a role in translation (Leeds et al., 1991; Estrella et al., 2009). As Figure 4A depicts, the cbc1Δ strain is hypersensitive to CHX; in fact, it is more sensitive than the upf1Δ and npl3Δ mutants (Figure 4A).
FIGURE 4:
Cbc1 is involved in translation under nonstress and osmotic stress conditions. (A) Sensitivity to CHX of the cbc1Δ mutant. The wt, cbc1Δ mutant, and strains carrying deletions in the UPF1 and NPL3 genes were grown exponentially in YPD. An aliquot of each culture was diluted and 8 μl of fivefold serial dilutions were spotted on plates containing YPD or YPD supplemented with 0.05, 0.075, and 0.1 μg/ml CHX. Plates were grown for 2 d at 30°C. The results in this figure are representative of at least two independent experiments. (B) Synthetic sickness of the double mutant cdc33tscbc1Δ and resistance to osmotic stress of the single mutant cdc33ts. Cultures of the wt, cbc1Δ, cdc33ts, and cdc33tscbc1Δ strains were treated as in (A), and growth was tested in YPD and YPD supplemented with 0.8 M NaCl. Plates were left to grow for 4 d at the indicated temperatures. The results in this figure are representative of at least two independent experiments.
Translation initiation factor eIF4E replaces Cbc1 in the cap structure of cytoplasmic mRNAs. Nevertheless, it has also been proposed that both eIF4E and Cbc1 conduct pioneer rounds of translation in yeast (Gao et al., 2005). However, while eIF4E is essential for cell survival, deletion of CBC1 yields only a slight growth defect under optimal conditions (Figure 4, A and B), indicating that bulk translation is initiated by eIF4E (Gao et al., 2005). Our results of hypersensitivity to CHX support the hypothesis of a role for Cbc1 in translation under nonstress conditions. We hypothesized that Cbc1's role in translation would become relevant for survival under conditions in which the eIF4E function was compromised. To test this, we introduced a CBC1 deletion into yeast cells carrying a thermosensitive mutation in eIF4E (cdc33ts; Schwartz and Parker, 1999). Yeast cells carrying the cdc33ts allele (cdc33-42) have previously been shown to perform almost 80% of the wt translation at 24°C, and this ratio lowered to 19% at 38°C (Schwartz and Parker, 1999). Accordingly, cdc33ts cells did not show important growth defects on the yeast extract–peptone–dextrose (YPD) plates incubated at 25°C or 33°C and were almost unable to grow at 37°C (Figure 4B). It is interesting to note that the cdc33ts cbc1Δ double mutant clearly grew less well than the cdc33ts mutant at 33°C (Figure 4B). Additionally, we tested sensitivity to CHX in plates incubated for a longer period to allow the growth of the synthetic sick cdc33ts cbc1Δ strain. We observed that the hypersensitive defect of the cdc33ts and cbc1Δ single mutant strains was exacerbated in the double mutant (Supplemental Figure S1A). Collectively, these results suggest that Cbc1, together with eIF4E, functions in the initiation of translation under optimal conditions, which is consistent with the hypothesis of Cbc1's involvement in a cytoplasmic pioneer round of translation.
As mentioned earlier, our results indicate that the cbc1Δ mutant is hypersensitive to osmotic stress (Figure 1). If both proteins eIF4E and Cbc1 are involved in translation under normal conditions, and Cbc1 is required for the reinitiation of translation after the global translation inhibition caused by osmotic stress (Figure 2), there is every reason to investigate the osmotic stress sensitivity of both the double mutant cdc33ts cbc1Δ and the single mutant cdc33ts. As Figure 4B depicts, the cdc33ts cbc1Δ strain was unable to grow in media with 0.8 M NaCl at any temperature. On the other hand, growth of cdc33ts cells at a permissive temperature, at which both eIF4E and Cbc1 are functional, was considerably reduced when 0.8 M NaCl was added. This suggests that NaCl retains the ability to affect growth independently of both eIF4E and Cbc1. Unexpectedly, thermosensitive mutant cdc33ts grew even better than a wt strain under hyperosmotic media, and even at a nonpermissive temperature (Figures 4B and S1B). The viability of cdc33ts at 37°C in hyperosmotic media was not explained by the recovery of the wt levels of protein eIF4E (Figure S1C). In summary, these results suggest that Cbc1 and eIF4E play redundant roles in translation, and that the roles of eIF4E under nonstress and of Cbc1 under osmotic stress conditions are prevalent. Depletion of most of the eIF4E protein is not deleterious for yeast cells growing under osmotic stress, instead making yeast cells more osmotolerant, and this suppression phenotype is lost when the CBC1 gene is deleted, which suggests a potentiation of Cbc1's role in translation when the amount of protein eIF4E is limited. This behavior could result from a more stable interaction between Cbc1 and eIF4G for this condition (Fortes et al., 2000).
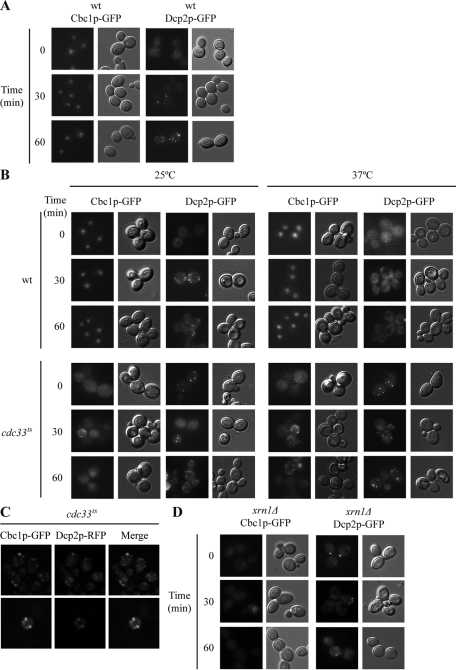
Cbc1 is located in the nucleus under nonstress and osmotic stress, and redistributes to the cytoplasm in the cells lacking either translation initiation factor eIF4E or exonuclease Xrn1
We have shown that the reinitiation of translation and P-body disassembly after adaptation to osmotic stress depend on Hog1 and Cbc1 (Romero-Santacreu et al., 2009; Figure 3). One possibility is that the Cbc1 expression or function could be regulated by osmotic stress; this regulation may, or may not, be Hog1-dependent. We observed no change in the amount of Cbc1 protein or in the modification of the protein that could change its electrophoretic mobility during osmotic stress (Figure S2). Previous studies have demonstrated how Cbc1 accumulates in the nucleus under optimal growing conditions (Görlich et al. 1996, Shen et al., 2000). We checked whether the Cbc1 protein changed its cellular localization in hyperosmotic media. To test this possibility, we followed the distribution of a Cbc1-GFP fusion during the treatment with 1 M KCl at the times when global translation stopped and P-bodies appeared (Figure 5A; Teixeira et al., 2005; Romero-Santacreu et al., 2009). We observed that Cbc1-GFP was enriched in the nucleus, regardless of osmotic stress treatment (Figure 5A). Thus all these results provide no indication of Cbc1 regulation by osmotic stress.
FIGURE 5:
Cbc1 accumulates in the nucleus in wt cells under nonstress and osmotic stress conditions, but is distributed all over the cell in mutants in either translation initiation factor eIF4E or mRNA exonuclease Xrn1. (A and D) The wt and xrn1Δ strains expressing Cbc1-GFP or Dcp2-GFP (P-body marker) were exponentially grown in YPD. Cultures were then incubated with 1 M KCl at 30°C. Cbc1-GFP localization and P-bodies (Dcp2-GFP) were visualized at 0, 30, and 60 min of osmotic stress treatment by fluorescence microscopy (left panels). Cells were also photographed by Nomarski optics (right panels). (B) The wt and cdc33ts mutant strains expressing Cbc1-GFP or Dcp2-GFP were exponentially grown in YPD at 25°C. One aliquot of each culture was treated with 1 M KCl and incubated at a permissive temperature (25°C). Another aliquot was incubated at a restrictive temperature (37°C) for 30 min and was then treated with 1M KCl. Cbc1-GFP localization, P-bodies (Dcp2-GFP), and cells were visualized as described in (A and D). (C) Cbc1 partially colocalizes with P-bodies. The cdc33ts cells coexpressing Cbc1-GFP and Dcp2-RFP (P-body marker) were exponentially grown in YPD at 25°C. Afterward the culture was incubated at a restrictive temperature (37°C) for 30 min and was then treated with 1 M KCl for 60 min. Cbc1-GFP and Dcp2-RFP were visualized by fluorescence microscopy.
Next we tested whether the Cbc1 protein changed localization in the cdc33ts strain, since our results in preceding section(s) suggest that Cbc1's role in the initiation of translation becomes relevant in this strain, given the limitation in the eIF4E-driven initiation of translation (Schwartz and Parker, 1999). As previously described, P-bodies were observed in cdc33ts cells at any temperature (Figures 5B and S3; Teixeira et al., 2005; Brengues and Parker, 2007). It is noteworthy that Cbc1 was distributed throughout the cell in the cdc33ts mutant strain under both nonstress and osmotic stress conditions when cells were incubated at 25°C (Figure 5C). At a restrictive temperature (37°C), Cbc1 was also distributed all over the cell, but when cdc33ts cells were additionally stressed with 1M KCl for 30 and 60 min, part of the Cbc1 protein was found in cytoplasmic foci, which partially colocalized with P-bodies (Figure 5, B and C).
The inhibition of mRNA degradation by a mutation of the 5′ to 3′ exonuclease Xrn1 has been described as increasing the number of P-bodies in yeast cells under normal conditions (Sheth and Parker, 2003). A role for eIF4E in targeting messenger ribonucleoproteins (mRNPs) to mammalian P-bodies has been suggested (Andrei et al., 2005; Ferraiuolo et al., 2005), and eIF4E has been detected in P-bodies in yeast cells (Brengues and Parker, 2007). We hypothesize that the amount of eIF4E protein dedicated to active translation would be limited in xrn1Δ cells, because of a possible stall in the degradation of the eIF4E-containing mRNPs in P-bodies, and if this were the case, that the localization of Cbc1 in xrn1Δ cells would change. As Figure 5D illustrates, Cbc1 lost the nuclear accumulation pattern in xrn1Δ cells and was localized throughout the cell. This distribution of Cbc1 in the cell was also observed in xrn1Δ cells under osmotic stress (Figure 5D).
In summary, our results indicate no change in Cbc1 nuclear localization during osmotic stress treatment, when global translation is transiently inhibited and P-bodies appear due to the incorporation of bulk mRNAs (Uesono and Toh, 2002; Teixeira et al., 2005). Conversely, in cells in which eIF4E is limited, as in the cdc33ts and xrn1Δ strains, most Cbc1 relocalizes to the cytoplasm under both nonstress and osmotic stress conditions. One simple interpretation of this result is that the recycling of Cbc1 to the nucleus needs the replacement of Cbc1 with eIF4E in the cap structure of cytoplasmic mRNA. However, an alternative and nonexclusive interpretation is that more Cbc1 is needed in the cytoplasm to replace the eIF4E functions in translation when the amount of eIF4E is limited. Indeed, resistance of cdc33ts to osmotic stress, even at a restrictive temperature (Figures 4B and S1B), suggests that an increase in Cbc1 in the cytoplasm offers cells an advantage in osmotic stress conditions, possibly through Cbc1's competence in translation. Finally, localization of part of the Cbc1 protein in the cytoplasmic foci in cdc33ts cells when simultaneously treated with a high temperature and osmotic stress resembles the visualization of the eIF4E initiation factor in P-bodies when global translation is inhibited (Brengues and Parker, 2007), and could also be indicative of the replacement of the eIF4E function in translation with Cbc1 under this condition.
Cbc1 associates with polysomes
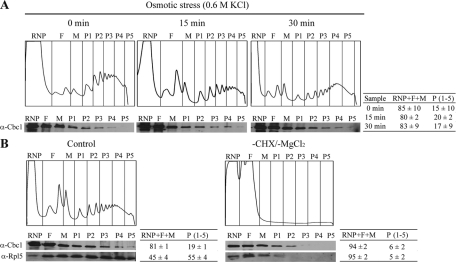
Cbc1's role in translation initiation could be performed through a direct association of Cbc1 with the active translation machinery. In mammalian cells, CBP80 associates with polysomal fractions to carry out the pioneer round of translation (Chiu et al., 2004; Kim et al., 2009). In a study into the Npl3 function in translation in yeast, Cbc1 was not detected in the polysomal fractions (Windgassen et al., 2004). However, we decided to do an in-depth analysis of Cbc1's ability to cosediment with polysomes under both optimal and osmotic stress conditions. rRNA profiles, measured at A260, indicated the position of ribonucleoproteins, ribosomal subunits, monosomes, and polysomes (Figure 6). The Western blot analysis of all fractions revealed that significant amounts of Cbc1 protein (between 15 and 20% of total Cbc1) were detectable in polysome-containing fractions of the wt cells grown under optimal conditions (Figure 6A). The association of Cbc1 with polysomes was dependent on blocking translation elongation by CHX (Figures 6B and S4), indicating that Cbc1 associates with translationally active ribosomes (Moldave, 1985; Ramírez et al., 1991; Harel-Sharvit et al., 2010). Similar to CBP80 in mammalian cells, Cbc1 was detected in all the polysomal fractions, but was concentrated in the subpolysomal fractions (Figure 6), while eIF4E was mainly detected in higher polysomal fractions (Chiu et al., 2004; Hoyle et al., 2007). These results suggest that Cbc1-bound mRNAs are less efficiently translated than eIF4E-bound mRNAs.
FIGURE 6:
Cbc1p associates with polysomes under nonstress and osmotic stress conditions. (A) The wt culture growing exponentially in YPD was treated with 0.6M KCl and polysomal profiles were determined at the indicated times. mRNP, FM, and P (P1–P5) fractions were collected and analyzed by Western blotting with the anti-Cbc1 antibody. (B) wt cells were treated with 0.6 M KCl for 15 min. Polysomal profiles were obtained from one aliquot of the culture (Control), as described in Materials and Methods. Another aliquot of the culture was treated with NaN3 for 30 min at 30°C, and CHX and MgCl2 were omitted in the cell extract and sucrose gradient to disrupt polysomes (−CHX/−MgCl2). An analysis of the Cbc1 protein in the fractions was performed as in (A). Detection of RP Rpl5 with a specific antibody was used as a control of polysome disassembly. The polysome profiles and the Western blot analyses shown in this figure correspond to a representative experiment. The percentage of Cbc1 (A and B) and of Rpl5 (B) in nonpolysomal (RNP + F + M) and in polysomal fractions (P1–P5) is indicated. The data are provided as the mean and SE of at least two independent experiments.
Association with polysomes has also been described for other yeast nucleus–cytoplasm shuttling mRNA-bound proteins, such as Npl3, Gbp2, and Rpb4. For all these proteins, as for Cbc1, their fusions with GFP were only visualized in the nucleus, due to the higher concentration of the proteins in this compartment (Windgassen et al., 2004; Harel-Sharvit et al., 2010). In mammalian cells, CBP80 associated with polysomes and was also highly enriched in the nucleus (Kim et al., 2009).
Next we examined the association of Cbc1 with polysomes under osmotic stress. The proportion of polysome-associated Cbc1 did not significantly change after 15 or 30 min of addition of 0.6 M KCl (Figure 6A). Thus, similar to the nuclear accumulation of Cbc1, and regardless of osmotic stress, the proportion of the Cbc1 associated with polysomes remained unchanged by stress. However, it should be noted that the proportion of the Cbc1 associated with polysomes was constant, independent of the reduction of polysomes due to the translation inhibition provoked by osmotic stress (Figure 6A); thus Cbc1 constituted a larger fraction of the polysome-associated proteins.
We also examined the association of Cbc1 with polysomes in the cdc33ts and xrn1Δ mutants. Our results indicate that the cytoplasmic localization of Cbc1 protein in cdc33ts and xrn1Δ (Figure 5) does not lead to an increased association of the protein with polysomes (Figure S5, A and B).
Together, all these data suggest that: 1) Association of Cbc1 with polysomes is independent of osmotic stress; however, the proportion of Cbc1-dependent translation increases under osmotic stress, because bulk translation is inhibited under this condition, although the proportion of the Cbc1 protein associated with polysomes remains constant. 2) Association of Cbc1 with polysomes remains constant despite the abrogation of Xrn1 or eIF4E; thus this result suggests that the proportion of Cbc1-bound mRNA in relation to the total mRNA in active translation increases when the eIF4E translation initiation factor amount is limited in cdc33ts and putatively in the xrn1Δ mutants.
Cbc1 is required for a rapid translation of osmotic-induced mRNAs under osmotic stress
All the above results enabled us to hypothesize that Cbc1 is directly involved in translation initiation under osmotic stress, when bulk translation is inhibited. Given this stress condition, Cbc1-dependent translation would still work efficiently and be relevant for cell adaptation. Recently it has been reported that specific functional groups of mRNAs increase their association with polysomes under osmotic stress conditions in which global translation is inhibited (Melamed et al., 2008; Halbeisen and Gerber, 2009; Warringer et al., 2010). Some of the genes that are translationally up-regulated are also up-regulated at the transcriptional level, including genes essential for survival of osmotic stress, such as GPD1 (Melamed et al., 2008; Warringer et al., 2010). One prediction of our model of Cbc1's role in translation under osmotic stress is that the association of the osmostress-regulated mRNAs with polysomes would be affected in those yeast cells lacking the CBC1 gene. To test this possibility, we examined the association of several osmostress-regulated mRNAs with polysomes during an osmotic stress–treatment time course (Figure 7). For GPD1, STL1, and HSP12, mRNA amounts increased after 15 min of treatment with 0.6 M KCl (Figures 2 and 7A). At this time, an important proportion of the three mRNAs was detected in the polysomal fraction in wt cells (Warringer et al., 2010), whereas the polysomal association of these osmostress-regulated mRNAs was clearly lower in cbc1Δ cells (Figure 7A). It is noteworthy that a significant proportion of the mRNAs remained associated with the monosomal and the free fraction in cbc1Δ cells at 30 min of stress, whereas most mRNAs were associated with polysomes in wt cells (Figure 7A). As described earlier (Figure 2), the STL1, GPD1, and HSP12 mRNA levels were still high in the cbc1Δ strain at times of osmotic stress when these levels dropped in the wt strain (Figure 7A). The analysis of the polysomal (P) to the free and monosomal (FM) ratios indicates that most of STL1, GPD1, and HSP12 mRNAs were actively translated at 60 min of stress in the cbc1Δ strain (Figure 7B). We also analyzed the translational status of two mRNAs, RPL17A and RPS31, that encode RPs. It has been reported that a decrease in the levels of the mRNAs encoding for RPs occurs quickly after osmotic stress (Posas et al., 2000; Molin et al., 2009; Romero-Santacreu et al., 2009) and this takes place parallel to a translational down-regulation for many of the RP-encoding mRNAs (Melamed et al., 2008; Halbeisen and Gerber, 2009; Warringer et al., 2010). Accordingly, the levels and the P/FM ratios for RPL17 and RPS31 mRNAs were lower after treatment with 0.6 M KCl in a wt strain (Figure 7, A and B). Similar results were obtained with the cbc1Δ mutant strain during the first 30 min of stress; however, the recovery in the polysomal association of both the RP-encoding mRNAs was delayed in cbc1Δ, compared with wt cells (Figure 7, A and B; see the differences in P/FM ratios at 60 min).
FIGURE 7:
Rapid translation reprogramming under hyperosmotic stress is delayed in the cbc1Δ mutant. (A) Association of osmostress response mRNAs and RP mRNAs with polysomes in wt and cbc1Δ under osmotic stress. The wt and cbc1Δ cultures growing exponentially in YPD were treated with 0.6 M KCl, and polysomal profiles were determined at the indicated times. The fractions corresponding to FM and to P were collected and analyzed by Northern blotting using specific probes that recognize the indicated mRNAs. The RNP fraction was excluded because it never gave a signal for the mRNAs tested here (unpublished data). (B) The quantified signals from three independent replicates were used to determine the average change in the P/FM ratios for the indicated genes. (C) Translation efficiency of the STL1-LacZ fusion in wt and cbc1Δ under osmotic stress. The wt cells expressing fusion STL1-LacZ from a plasmid (the STL1 promoter fused to LacZ, LEU2+ [2 µm]) were grown in selective media at 30°C until the exponential phase, and were then transferred to YPD and incubated at 30°C for 4 h. Cells were then subjected to osmotic stress by adding 0.6 M KCl, while samples were taken at the indicated times. β-galactosidase activity was measured as described in Materials and Methods, and the LacZ mRNA level was determined by Northern blotting. Translation efficiency was calculated as the β-galactosidase activity/LacZ mRNA ratio, and the results were normalized to efficiency at time 0. The data shown are the mean and SE of three independent experiments.
To independently confirm Cbc1's role in the rapid translation of transcripts after osmotic stress, we monitored translation efficiency in cbc1Δ cells by checking the expression of an osmotic stress-inducible reporter gene. We used a fusion gene between the STL1 promoter and the LacZ reporter gene (STL1-LacZ), and measured the β-galactosidase activity of the corresponding protein in relation to the increase in the LacZ mRNA level during osmotic stress treatment of cells with 0.6 M KCl. As Figure 7C shows, the efficiency of the translation of STL1-LacZ mRNA (proportion of reporter protein activity in relation to the mRNA amount) increased significantly faster in wt than in cbc1Δ cells. According to our previous data regarding the association of osmostress-induced mRNAs with polysomes (Figure 7B), the translation efficiency of STL1-LacZ needed more time to recover in a cbc1Δ mutant after osmotic shock (Figure 7C). The importance of 3′-UTRs for the proper up-regulation of mRNA translation under osmotic stress has been indicated (Warringer et al., 2010). Our result obtained with STL1-LacZ indicates an impaired reporter-RNA translation in cbc1Δ cells that is independent of the native 3′UTR of STL1 mRNA. One likely interpretation is that STL1-LacZ translation is regulated through its STL1 5′-UTR.
Collectively, all these results indicate that Cbc1 is necessary for the rapid translation of mRNAs induced by osmotic stress, but that the down-regulation of the translation of those mRNAs encoding for RPs is independent of Cbc1. In summary, these results suggest that Cbc1-dependent translation is not inhibited by osmotic stress, but is required for rapid translation reprogramming under this stress condition.
DISCUSSION
In response to osmotic stress, global translation is inhibited in S. cerevisiae cells. Nonetheless, many osmostress-responsive mRNAs engage with polysomes, and their corresponding stress-protective proteins are efficiently synthesized (Uesono and Toh, 2002; Melamed et al., 2008; Halbeisen and Gerber, 2009; Warringer et al., 2010). The results presented herein identify, for the first time, an mRNA-binding protein, yeast cap-binding protein Cbc1, involved in this rapid translation reprogramming upon osmotic stress.
The involvement of yeast Cbc1 in translation has been a matter of debate in the past few years (see Introduction for details). Our present results favor a direct role of Cbc1 in translation initiation, which is not essential for cell viability under normal conditions, in which Cbc1 is replaced with eIF4E in the cap of most transcripts in the cytoplasm (Gao et al., 2005). First, cbc1 mutants are sensitive to translation inhibitor CHX (Figure 4A). Second, the deletion of the CBC1 gene in cells carrying a thermosensitive allele of eIF4E (cdc33ts) confers synthetic sickness at any temperature (Figure 4B). A functional interaction between eIF4E and Cbc1 is also demonstrated by the synergistic CHX hypersensibility of the cdc33ts cbc1Δ double mutant (Figure S1A). Third, Cbc1 cosediments with polysomes in a CHX-dependent (i.e., translation-dependent) manner. Ribosome-associated Cbc1 is concentrated in subpolysomal fractions and involves only a small fraction of the Cbc1 protein (15–20%; Figures 6 and S4).
Our data highlight the important role played by Cbc1-dependent translation under osmotic stress conditions. First, the cbc1Δ mutant shows low viability in hyperosmotic media (Figure 1). Second, the decrease in the polysome content in response to osmotic stress is more extensive in the cbc1Δ mutant than in wt cells, and the recovery of normal translation profiles is delayed in the cbc1Δ mutant (Figure 3, A and B). Third, the P-bodies formed when global translation is inhibited upon osmotic shock remain for longer periods in cbc1Δ mutants than in wt cells (Figure 3C), indicating a defect in recovery. Fourth, osmostress-responsive mRNAs are induced in a cbc1Δ mutant in response to osmotic stress (Figure 2); however, their rapid association with polysomes, as observed in wt cells, is significantly delayed in the mutant (Figure 7A). Moreover, the increase in translation efficiency upon osmotic stress of the reporter gene STL1-LacZ and the subsequent decline are also delayed in the cbc1Δ mutant, compared with wt cells (Figure 7B). Fifth, the growth inability of cdc33ts mutants at 37°C is suppressed by hyperosmosis and, in those cells with limiting amounts of eIF4E, Cbc1 relocalizes to the cytoplasm. Sixth, the Cbc1 protein fraction associated with polysomes remains constant under osmotic stress, when global translation is inhibited; this indicates that the proportion of Cbc1-bound mRNA in active translation increases upon stress (Figure 6A). All these phenotypes of the cbc1Δ mutant in translation under osmotic stress are not merely the result of Cbc1's role in mRNA metabolism, since other mutants in genes involved in splicing, transcription, or mRNA export do not show similar effects (Figure S6).
We suggest a model in which the bulk translation driven by eIF4E is stress-sensitive and is mostly inhibited by osmotic stress, while Cbc1-mediated translation is stress-resistant. Thus the newly synthesized mRNA encoding osmostress-protective proteins will be bound by Cbc1 in the nucleus and will undergo Cbc1-dependent translation in the cytoplasm. At the same time, the pool of housekeeping mRNAs, which was actively translated mostly by cytoplasmic eIF4E before stress, will be kept in the P-bodies until adaptation. A similar scenario has been proposed for CBP80-mediated translation in mammalian cells under stress conditions provoked by prolonged hypoxia and serum starvation (Oh et al., 2007a, 2007b). Under such stresses, the steady-state translation mediated by eIF4E is drastically abolished, whereas CBP80-mediated translation efficiently occurs. According to our model, the level of yeast Cbc1 protein remains constant, and the protein does not seem to be modified by osmotic stress (Figure S2).
One open question is whether Cbc1-mediated translation mechanistically differs from eIF4E-mediated translation. As stated before, Cbc1 can interact with eIF4GI (Fortes et al., 2000), in a manner similar to the interaction between mammalian CBP80 and eIF4GI (McKendrick et al., 2001; Chiu et al., 2004). However, in mammalian cells, the CBP80-eIF4GI interaction is of little functional relevance, and a new translation factor, CTIF, is specifically involved in CBP80-dependent translation (Chiu et al., 2004; Lee et al., 2008; Kim et al., 2009). In yeast, additional studies should be conducted to determine the Cbc1-mediated translation mechanism.
Another unresolved issue is whether Cbc1 mediates the translation of any mRNA or whether Cbc1-dependent translation is restricted to specific transcripts, such as osmostress-responsive mRNAs. We have shown that the rapid translation of newly synthesized endogenous osmoresponsive mRNAs under osmotic stress conditions depends on Cbc1, as does the translation of the osmotic stress–induced STL1-LacZ fusion (Figure 7). Finally, the modulation of translation under osmotic stress by Cbc1 could also imply a direct role in the movement of mRNAs from P-bodies to polysomes. Consistently, P-bodies remain longer and fewer polysomes are observed in the cbc1Δ mutant cells under osmotic stress (Figure 2). Moreover, Cbc1 is localized in cytoplasmic foci that partially colocalize with the P-bodies in cdc33ts cells when translation is abolished by a simultaneous treatment with high temperature and osmotic stress (Figure 5, B and C).
The hypersensitivity and loss of viability of cbc1Δ mutant cells under osmotic stress could relate to Cbc1's role in translation reprogramming. However, we cannot rule out other Cbc1 functions in response of yeast to osmotic stress. Recently a cytoplasmic membrane-anchored version of Hog1 that is unable to induce the transcription of osmostress-responsive genes showed no growth defects in hyperosmotic media, which has led to the proposal that the regulation of transcription by Hog1 is not essential for yeast survival under osmotic stress (Westfall et al., 2008). However, cytoplasmic Hog1 could still be able to regulate translation; therefore, we do not know how important this regulation is for cell survival.
The translation mediated by mammalian CBP80 seems to be resistant to several stress conditions, for example, heat shock, prolonged hypoxia, or serum starvation (Maquat and Li, 2001; Marín-Vinader et al., 2006; Oh et al., 2007a, 2007b). Whether yeast Cbc1 is involved in translation reprogramming under other stress conditions should be addressed.
The fact that osmo-mRNA levels exhibit a greater and prolonged increase in cbc1Δ than do those in wt under osmotic stress (Figure 2) suggests a defect in the transcription shutdown and/or mRNA destabilization that occurs after an initial increase in transcription and stability (Molin et al., 2009; Romero-Santacreu et al., 2009). Future studies will reveal whether Cbc1 plays a role in cytoplasmic mRNA decay and/or whether there is cross-talk between the translation and transcription mediated by Cbc1. Recently the Rpb4/7 subunits of polymerase II have been postulated as mRNA coordinators that couple transcription, mRNA decay, and translation (Goler-Baron et al., 2008; Harel-Sharvit et al., 2010). The involvement of nuclear Cbc1 in transcription (Das et al. 2000; Wong et al., 2007; Lahudkar et al., 2011), mRNA splicing (Lewis et al., 1996), and nuclear degradation (Das et al., 2003), and of cytoplasmic Cbc1 in translation (Gao et al., 2005; this work) and perhaps in mRNA decay, suggest that Cbc1 could also be one of the mRNA coordinators connecting the fate of mRNA to cellular circumstances.
MATERIALS AND METHODS
Yeast strains and growth conditions
The genotypes of the strains used in this study are listed in Table 1. The strains containing the CBC1 gene deletion and those expressing Cbc1 protein C-terminally tagged with GFP were constructed following the PCR-based gene modification method described by Longtine et al. (1998). Purified PCR products were used for yeast transformation by the lithium acetate method (Gietz et al., 1995), and transformants were selected by plating on appropriate selective-medium plates.
TABLE 1:
The S. cerevisiae strains used in this work.
| Strain | Relevant genotype | Source |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Euroscarf |
| PAY611a | CBC1-GFP::KanMX6 | This study |
| 6566a | cbc1Δ::KanMX4 | Euroscarf |
| 2724a | hog1Δ::KanMX4 | Euroscarf |
| 6214a | upf1Δ::KanMX4 | Euroscarf |
| 4268a | npl3Δ::KanMX4 | Euroscarf |
| 4540a | xrn1Δ::KanMX4 | Euroscarf |
| PAY664a | xrn1Δ CBC1-GFP::His3MX6 | This study |
| yRP840 | MATa his4, leu2, trp1, ura3, cup1::LEU2/PGK1pG/MFA2pG | Schwartz and Parker (1999) |
| PAY671b | CBC1-GFP::KanMX6 | This study |
| yRP1321b | cdc33::LEU2 (cdc33-42/TRP1) | Schwartz and Parker (1999) |
| PAY673b | cdc33::LEU2 (cdc33-42/TRP1) CBC1-GFP::KanMX6 | This study |
| PAY678b | cbc1Δ::KanMX6 | This study |
| PAY679b | cdc33::LEU2 (cdc33-42/TRP1) cbc1Δ::KanMX6 | This study |
a Strain derivative of the EUROSCARF wt BY4741 strain.
b Strain derivative of the wt yRP840 strain.
NaCl tolerance screening was done by growing yeast strains in 220 μl of YPD medium (1% yeast extract, 2% peptone, 2% glucose) in 96-well microtiter plates for 16 h. Cultures were then diluted 5000 times in eight cultures of 215 μl of YPD plus NaCl at 0, 0.15, 0.25, 0.4, 0.55, 0.75, 1, and 1.25 M. Plates were incubated at 30°C for 17–24 h, and OD620 (optical density) was measured by an absorbance microplate reader (SpectraFluor; Tecan, Männedorf, Switzerland). IC50 (concentration of NaCl required to inhibit 50% of growth in relation to the maximal growth observed for the same strain in YPD without NaCl) was calculated using the OriginLab software (Northampton, MA).
Thermosensitive mutants were grown at 25°C and were then transferred to a permissive (25°C or 33°C, as indicated) or a restrictive (37°C) temperature. Hyperosmotic stress was provoked in a liquid medium by adding KCl or NaCl to cultures to obtain the indicated final salt concentration. To test osmotic stress sensitivity in a solid medium, YPD was supplemented with NaCl (0.4, 0.8, or 1.2 M) or with sorbitol (1 or 1.5 M). CHX sensitivity tests were performed on solid YPD containing CHX at the indicated concentration. All the tests for growth on solid media were done with exponentially growing cultures (OD600 = 0.5), then 5- or 10-fold serial dilutions were made, and 8 μl of each dilution was spotted on the indicated plates. The incubation times and temperature are indicated in each case.
Flow cytometry
Yeast cells were grown until the exponential phase in YPD and were subjected to osmotic stress by adding 0.6 M KCl or 1.2 M NaCl, or they were kept in YPD as the control condition. After stress incubation, 100 μl of the culture was harvested by centrifugation at 13,000 rpm for 30 s, and cells were resuspended in 400 μl of PBS buffer (140 mM NaCl, 40 mM Na2HPO4, 2.7 mM KCl, 1.8 mM KH2PO4). To identify dead cells, propidium iodide 15 μM (PI) dye was added, and fluorescence was measured in a flow cytometer (EPICS XL-MCL; Beckman Coulter, Brea, CA) after 5 min of incubation at room temperature. The yeast cells kept in YPD were used as a 100% viability control. The fluorescence of the cells under the control conditions without PI was used as a baseline.
Polysomal fractionation and analysis of the specific RNAs and proteins from the fractions
Cells grown exponentially in YPD media to an OD600 of 0.5 were subjected to 0.6 M KCl stress for the indicated times. A portion of the culture (80 ml) was recovered and chilled for 5 min on ice in the presence of 0.1 mg/ml CHX. Cells were harvested by centrifugation at 6000 × g for 4 min at 4°C and resuspended in lysis buffer (20 mM Tris-HCl, pH 8, 140 mM KCl, 5 mM MgCl2, 0.5 mM dithiothreitol, 1% Triton X-100, 0.1 mg/ml CHX, and 0.5 mg/ml heparin). After being washed, cells were resuspended in 700 μl of lysis buffer, a 0.3-ml volume of glass beads was added, and cells were disrupted by vortexing eight times for 30 s. Lysates were cleared by being centrifuged at 5000 rpm for 5 min, after which the supernatant was recovered and was centrifuged at 8000 rpm for 5 min. Finally, glycerol was added to the supernatant at a final concentration of 5% before the extracts were stored at −70°C. Samples of 10–20 A260 units were loaded onto 10–50% sucrose gradients and were separated by ultracentrifugation for 2 h and 40 min at 35,000 rpm in a Beckman SW41 rotor at 4°C. Gradients were then fractionated using isotonic pumping of 60% sucrose from the bottom; this was followed by a recording of the polysomal profiles by online UV detection at 260 nm (Density Gradient Fractionation System; Teledyne Isco, Lincoln, NE). For the runoff experiments, cultures were incubated for 20 min with 1 mM NaN3 before cells were collected, and CHX and MgCl2 were omitted in both the lysis buffer and the gradient solutions. For analysis of the RNA of the polysomal fractions, 10 mM Tris-HCl, 10 mM EDTA and 0.5% SDS were added to the fractions. Afterward, two extractions with phenol (pH 7.9):chloroform (1:1) were prepared, and RNA was precipitated with ethanol-sodium acetate. Specific RNAs were analyzed by Northern blotting. For analysis of the protein content of the fractions, proteins were precipitated by 10% TCA and twice washed with cold anhydrous acetone. Dry pellets were resuspended in SB 1X (360 mM Tris-HCl, 18% SDS, 60% glycerol, 24% β-mercaptoethanol, 0.12% bromophenol blue). The extracted proteins were analyzed by Western blotting as described in Western blot analysis.
Total RNA extraction and Northern blotting
For the analysis of the cellular mRNA levels, exponentially growing cultures were incubated at the indicated conditions and times, and the cells from the 20-ml culture were collected by centrifugation and frozen at −20°C. Total RNA was extracted from the cell pellets using a FastPrep device (FastPrep 120, Bio101 ThermoSavant; Qbiogene, Illkirch, France) in 0.5 ml LETS buffer (0.1 M LiCl, 0.01 M EDTA, pH 8.0, 0.01 M Tris-HCl, pH 7.4, and 0.2% SDS [wt/vol]), 0.5 ml phenol (pH 4.5):chloroform:isoamylic alcohol (25/24/1; vol/vol/vol), and 0.3 ml glass beads. Supernatants were extracted with phenol:chloroform:isoamylic alcohol (25/24/1; vol/vol/vol) and chloroform:isoamylic alcohol (24/1; vol/vol). RNA was precipitated by adding two volumes of 96% ethanol and 0.1 volumes of 5 M LiCl and by incubating at −20°C for at least 3 h. Isolated RNA from the polysomal fractions and total RNA were separated by electrophoresis in denaturing 1% (wt/vol) agarose gel (2.2 M formaldehyde, 1X MOPS [20 mM 3-(N-morpholino)-propanesulfonic acid, 8 mM NaAc, 1 mM EDTA, pH 7.0]), and were blotted on Hybond N+ membranes (Amersham, GE Healthcare, Waukesha, WI; Sambrook and Russell, 2001). Ethidium bromide-stained gels were read in a phosphorimager scanner (FLA-3000; Fujifilm, Tokyo, Japan) to quantify the signal of 18S or 25S rRNA (rRNA). The membrane was then hybridized for 24 h at 65°C in 0.5 M sodium phosphate (pH 7.2), 7% SDS (wt/vol), and 1 mM EDTA (pH 7.0). DNA probes were obtained by PCR and were labeled by random priming using the Ready-to-Go kit (Amersham) and [33P]dCTP. After hybridization, membranes were washed once for 10 min at 25°C in 1X saline sodium citrate (SSC), 0.1% SDS, and once for 15 min at 65°C in 0.5X SSC , 0.1% SDS. Membranes were exposed to an imaging plate (BAS-MP; Fujifilm), which was read in a phosphorimager scanner (FLA-3000; Fujifilm).
Western blot analysis
To analyze the total cellular content of the Cbc1 protein under nonstress and osmostress conditions, we followed the protein extraction protocol detailed in Alepuz et al. (2001). The total protein extracts and the protein extracts from polysomal fractions were resolved by 10% SDS–PAGE. Gels were then transferred to a nitrocellulose membrane (Protran; Whatman, Piscataway, NJ) and blocked in 5% nonfat milk overnight at 4°C. The antibodies utilized were α-Cbc1 (1:20,000 dilution; kindly provided by D. Görlich) and α-Rpl5 (1:5000 dilution; kindly provided by J. L. Woolford). The secondary antibody was anti-rabbit (1:25,000 dilutions; Amersham). Membranes were developed using an ECL Advance and Typhoon scanner (GE Healthcare). Images were quantified using Image Gauge software (Fujifilm).
β-Galactosidase assays
The wt and cbc1Δ strains containing STL1-LacZ fusion in a multicopy plasmid (STL1 promoter fused to LacZ, LEU2+ [2 μm]; a kind gift from E. de Nadal) were grown until the exponential phase in selective media. For the β-galactosidase assays, 3 ml of each culture, either untreated or subjected to the indicated conditions, was recovered by centrifugation. Cells were washed once and resuspended in 500 μl Z buffer (16.1 g/l Na2HPO4·7H2O, 5.5 g/l NaH2PO4·H2O, 0.75 g/l KCl, 0.246 g/l MgSO4·7H2O, pH 7.0). To permeabilize cells, 50 μl of sample was kept in liquid N2 for 1 min and then heated at 37°C for 1 min. To start the reaction, 600 μl of Z buffer containing 37.4 mM β-mercaptoethanol and 160 μl of 4 mg/ml ortho-nitrophenyl-β-galactoside were added to each permeabilized cell sample. Samples were incubated at 30°C until the appearance of a yellow color, and the reaction was then stopped by adding 400 μl 1M Na2CO3. The absorbance at 420 nm was measured in the supernatants. The β-galactosidase units were calculated as previously described (Miller, 1972). The results shown are the mean and SE of at least three independent experiments.
Fluorescence microscopy
Cells expressing Dcp2-GFP or Dcp2-RFP from plasmids (Sheth and Parker, 2003) or Cbc1-GFP from the CBC1 genomic locus were grown overnight in YPD or in synthetic complete (SC) medium supplemented with the appropriate amino acids, respectively. Cells were then transferred to YPD and grown to an OD600 of 0.5–0.6. Cells were washed and resuspended in SC plus amino acids, without salt or with 1M KCl, as indicated. Observations were made at the indicated times using an Axio Imager Z1 fluorescence microscope (Zeiss, Tokyo, Japan), an FV1000 system, and a FV10-ASW 2.1 Viewer software for the colocalization experiments.
Supplementary Material
Acknowledgments
We thank D. Görlich for the anti-Cbc1 antibody, J. L. Woolford for the anti-Rpl5 antibody, R. Parker for the cdc33ts strain and the Dcp2-GFP plasmid, and E. de Nadal for the STL1-LacZ plasmid. We thank F. Carrasco for lab technical assistance, A. Flores from the Flow Cytometry Service SCSIE for help with the yeast viability experiments, and J. Warringer for helping with the initial polysome experiments. We also thank J. E. Pérez-Ortín for helpful discussions and for critical reading of the manuscript. This work was supported by grants BFU2008-02114 (Ministerio Español de Ciencia cofinanciado con Fondos FEDER) and GV05/051 and Prometeo 2011/088 (Generalitat Valenciana–Regional Valencian Government) to P.A. and 2010-4645 (Swedish Research Council) and UNICELLSYS, LSHG-CT2007-201142 (European Commission FP7) to P.S. A visit by P.A. to P.S.'s lab was supported by a grant from the European Science Foundation, “Frontiers of Functional Genomics” Activity. L.R.-S. was supported by an F.P.I. fellowship from the Spanish Ministry of Science and N.D.C. by a Santiago Grisolía Fellowship from the Generalitat Valenciana.
Abbreviations used:
- CBC
cap-binding complex
- CHX
cycloheximide
- eIFs
eukaryotic initiation factors
- FM
free and monosomal
- GFP
green fluorescent protein
- IRES
internal ribosome entry sites
- MAPK
mitogen-activated protein kinase
- MOPS
3-(N-morpholino)-propanesulfonic acid
- mRNP
messenger ribonucleoproteins
- NMD
nonsense-mediated mRNA decay
- P
polysomal
- PAB1
poly(A)-binding protein 1
- P-bodies
processing bodies
- PI
propidium iodide
- RP
ribosomal protein
- SC
synthetic complete
- SSC
saline sodium citrate
- UTR
untranslated region
- wt
wild type
- YPD
yeast extract–peptone–dextrose
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-05-0419) on November 9, 2011.
REFERENCES
- Alepuz PM, Jovanovic A, Reiser V, Ammerer G. Stress-induced map kinase Hog1 is part of transcription activation complexes. Mol Cell. 2001;7:767–777. doi: 10.1016/s1097-2765(01)00221-0. [DOI] [PubMed] [Google Scholar]
- Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Luhrmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron-Benhamou J, Fortes P, Inada T, Preiss T, Hentze MW. The interaction of the cap-binding complex (CBC) with eIF4G is dispensable for translation in yeast. RNA. 2003;9:654–662. doi: 10.1261/rna.5100903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilsland-Marchesan E, Arino J, Saito H, Sunnerhagen P, Posas F. Rck2 kinase is a substrate for the osmotic stress-activated mitogen-activated protein kinase Hog1. Mol Cell Biol. 2000;20:3887–3895. doi: 10.1128/mcb.20.11.3887-3895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond U. Stressed out! Effects of environmental stress on mRNA metabolism. FEMS Yeast Res. 2006;6:160–170. doi: 10.1111/j.1567-1364.2006.00032.x. [DOI] [PubMed] [Google Scholar]
- Brengues M, Parker R. Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2592–2602. doi: 10.1091/mbc.E06-12-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu SY, Lejeune F, Ranganathan AC, Maquat LE. The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes Dev. 2004;18:745–754. doi: 10.1101/gad.1170204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot HV, Stutz F, Rosbash M. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes Dev. 1996;10:1699–1708. doi: 10.1101/gad.10.13.1699. [DOI] [PubMed] [Google Scholar]
- Das B, Guo Z, Russo P, Chartrand P, Sherman F. The role of nuclear cap binding protein Cbc1p of yeast in mRNA termination and degradation. Mol Cell Biol. 2000;20:2827–2838. doi: 10.1128/mcb.20.8.2827-2838.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B, Butler JS, Sherman F. Degradation of normal mRNA in the nucleus of Saccharomyces cerevisiae. Mol Cell Biol. 2003;23:5502–5515. doi: 10.1128/MCB.23.16.5502-5515.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nadal E, Alepuz PM, Posas F. Dealing with osmostress through MAP kinase activation. EMBO Rep. 2002;3:735–740. doi: 10.1093/embo-reports/kvf158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nadal E, Posas F. Multilayered control of gene expression by stress-activated protein kinases. EMBO J. 2010;29:4–13. doi: 10.1038/emboj.2009.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrella LA, Wilkinson MF, Gonzalez CI. The shuttling protein Npl3 promotes translation termination accuracy in Saccharomyces cerevisiae. J Mol Biol. 2009;394:410–422. doi: 10.1016/j.jmb.2009.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraiuolo MA, Basak S, Dostie J, Murray EL, Schoenberg DR, Sonenberg N. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J Cell Biol. 2005;170:913–924. doi: 10.1083/jcb.200504039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes P, Inada T, Preiss T, Hentze MW, Mattaj IW, Sachs AB. The yeast nuclear cap binding complex can interact with translation factor eIF4G and mediate translation initiation. Mol Cell. 2000;6:191–196. [PubMed] [Google Scholar]
- Fortes P, Kufel J, Fornerod M, Polycarpou-Schwarz M, Lafontaine D, Tollervey D, Mattaj IW. Genetic and physical interactions involving the yeast nuclear cap-binding complex. Mol Cell Biol. 1999;19:6543–6553. doi: 10.1128/mcb.19.10.6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Das B, Sherman F, Maquat LE. Cap-binding protein 1-mediated and eukaryotic translation initiation factor 4E-mediated pioneer rounds of translation in yeast. Proc Natl Acad Sci USA. 2005;102:4258–4263. doi: 10.1073/pnas.0500684102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Goler-Baron V, Selitrennik M, Barkai O, Haimovich G, Lotan R, Choder M. Transcription in the nucleus and mRNA decay in the cytoplasm are coupled processes. Genes Dev. 2008;22:2022–2027. doi: 10.1101/gad.473608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Kraft R, Kostka S, Vogel F, Hartmann E, Laskey RA, Mattaj IW, Izaurralde E. Importin provides a link between nuclear protein import and U snRNA export. Cell. 1996;87:21–32. doi: 10.1016/s0092-8674(00)81319-7. [DOI] [PubMed] [Google Scholar]
- Görnemann J, Kotovic KM, Hujer K, Neugebauer KM. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell. 2005;19:53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Greatrix BW, van Vuuren HJ. Expression of the HXT13, HXT15 and HXT17 genes in Saccharomyces cerevisiae and stabilization of the HXT1 gene transcript by sugar-induced osmotic stress. Curr Genet. 2006;49:205–217. doi: 10.1007/s00294-005-0046-x. [DOI] [PubMed] [Google Scholar]
- Gross T, Siepmann A, Sturm D, Windgassen M, Scarcelli JJ, Seedorf M, Cole CN, Krebber H. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science. 2007;315:646–649. doi: 10.1126/science.1134641. [DOI] [PubMed] [Google Scholar]
- Halbeisen RE, Gerber AP. Stress-dependent coordination of transcriptome and translatome in yeast. PLoS Biol. 2009;7:e105. doi: 10.1371/journal.pbio.1000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel-Sharvit L, Eldad N, Haimovich G, Barkai O, Duek L, Choder M. RNA polymerase II subunits link transcription and mRNA decay to translation. Cell. 2010;143:552–563. doi: 10.1016/j.cell.2010.10.033. [DOI] [PubMed] [Google Scholar]
- Hilgers V, Teixeira D, Parker R. Translation-independent inhibition of mRNA deadenylation during stress in Saccharomyces cerevisiae. RNA. 2006;12:1835–1845. doi: 10.1261/rna.241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MA, Claggett JM, Nguyen T, Johnson TL. The cap binding complex influences H2B ubiquitination by facilitating splicing of the SUS1 pre-mRNA. RNA. 2009;15:1515–1527. doi: 10.1261/rna.1540409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyle NP, Castelli LM, Campbell SG, Holmes LE, Ashe MP. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J Cell Biol. 2007;179:65–74. doi: 10.1083/jcb.200707010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki Y, Li X, Serin G, Maquat LE. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, McGuigan C, Mattaj IW. A cap-binding protein complex mediating U snRNA export. Nature. 1995;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Kim KM, Cho H, Choi K, Kim J, Kim BW, Ko YG, Jang SK, Kim YK. A new MIF4G domain-containing protein, CTIF, directs nuclear cap-binding protein CBP80/20-dependent translation. Genes Dev. 2009;23:2033–2045. doi: 10.1101/gad.1823409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuai L, Das B, Sherman F. A nuclear degradation pathway controls the abundance of normal mRNAs in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2005;102:13962–13967. doi: 10.1073/pnas.0506518102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuperwasser N, Brogna S, Dower K, Rosbash M. Nonsense-mediated decay does not occur within the yeast nucleus. RNA. 2004;10:1907–1915. doi: 10.1261/rna.7132504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner DH, Bähler J. Translational control of gene expression from transcripts to transcriptomes. Int Rev Cell Mol Biol. 2008;271:199–251. doi: 10.1016/S1937-6448(08)01205-7. [DOI] [PubMed] [Google Scholar]
- Lahudkar S, Shukla A, Bajwa P, Durairaj G, Stanojevic N, Bhaumik SR. The mRNA cap-binding complex stimulates the formation of pre-initiation complex at the promoter via its interaction with Mot1p in vivo. Nucleic Acids Res. 2011;39:2188–2209. doi: 10.1093/nar/gkq1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Cho H, Kim YK. Ectopic expression of eIF4E-transporter triggers the movement of eIF4E into P-bodies, inhibiting steady-state translation but not the pioneer round of translation. Biochem Biophys Res Commun. 2008;369:1160–1165. doi: 10.1016/j.bbrc.2008.03.017. [DOI] [PubMed] [Google Scholar]
- Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- Lejeune F, Ishigaki Y, Li X, Maquat LE. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J. 2002;21:3536–3545. doi: 10.1093/emboj/cdf345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Gorlich D, Mattaj IW. A yeast cap binding protein complex (yCBC) acts at an early step in pre-mRNA splicing. Nucleic Acids Res. 1996;24:3332–3336. doi: 10.1093/nar/24.17.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lui J, Campbell SG, Ashe MP. Inhibition of translation initiation following glucose depletion in yeast facilitates a rationalization of mRNA content. Biochem Soc Trans. 2010;38:1131–1136. doi: 10.1042/BST0381131. [DOI] [PubMed] [Google Scholar]
- Maquat LE, Li X. Mammalian heat shock p70 and histone H4 transcripts, which derive from naturally intronless genes, are immune to nonsense-mediated decay. RNA. 2001;7:445–456. doi: 10.1017/s1355838201002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín-Vinader L, van Genesen ST, Lubsen NH. mRNA made during heat shock enters the first round of translation. Biochim Biophys Acta. 2006;1759:535–542. doi: 10.1016/j.bbaexp.2006.10.003. [DOI] [PubMed] [Google Scholar]
- McKeehan W, Hardesty B. The mechanism of cycloheximide inhibition of protein synthesis in rabbit reticulocytes. Biochem Biophys Res Commun. 1969;36:625–630. doi: 10.1016/0006-291x(69)90351-9. [DOI] [PubMed] [Google Scholar]
- McKendrick L, Thompson E, Ferreira J, Morley SJ, Lewis JD. Interaction of eukaryotic translation initiation factor 4G with the nuclear cap-binding complex provides a link between nuclear and cytoplasmic functions of the m(7) guanosine cap. Mol Cell Biol. 2001;21:3632–3641. doi: 10.1128/MCB.21.11.3632-3641.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed D, Pnueli L, Arava Y. Yeast translational response to high salinity: global analysis reveals regulation at multiple levels. RNA. 2008;14:1337–1351. doi: 10.1261/rna.864908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C, et al. Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol Syst Biol. 2011;7:458. doi: 10.1038/msb.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. Experiments in Molecular Genetics. New York: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Moldave K. Eukaryotic protein synthesis. Annu Rev Biochem. 1985;54:1109–1149. doi: 10.1146/annurev.bi.54.070185.005333. [DOI] [PubMed] [Google Scholar]
- Molin C, Jauhiainen A, Warringer J, Nerman O, Sunnerhagen P. mRNA stability changes precede changes in steady-state mRNA amounts during hyperosmotic stress. RNA. 2009;15:600–614. doi: 10.1261/rna.1403509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke SM, Herskowitz I. Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol Biol Cell. 2004;15:532–542. doi: 10.1091/mbc.E03-07-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh N, Kim KM, Cho H, Choe J, Kim YK. Pioneer round of translation occurs during serum starvation. Biochem Biophys Res Commun. 2007a;362:145–151. doi: 10.1016/j.bbrc.2007.07.169. [DOI] [PubMed] [Google Scholar]
- Oh N, Kim KM, Choe J, Kim YK. Pioneer round of translation mediated by nuclear cap-binding proteins CBP80/20 occurs during prolonged hypoxia. FEBS Lett. 2007b;581:5158–5164. doi: 10.1016/j.febslet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Posas F, Chambers JR, Heyman JA, Hoeffler JP, de Nadal E, Ariño J. The transcriptional response of yeast to saline stress. J Biol Chem. 2000;275:17249–17255. doi: 10.1074/jbc.M910016199. [DOI] [PubMed] [Google Scholar]
- Ramírez M, Wek RC, Hinnebusch AG. Ribosome association of GCN2 protein kinase, a translational activator of the GCN4 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:3027–3036. doi: 10.1128/mcb.11.6.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M, Albertyn J, Thevelein JM, Prior BA, Hohmann S. Different signalling pathways contribute to the control of GPD1 gene expression by osmotic stress in Saccharomyces cerevisiae. Microbiology. 1999;145:715–727. doi: 10.1099/13500872-145-3-715. [DOI] [PubMed] [Google Scholar]
- Rep M, Krantz M, Thevelein JM, Hohmann S. The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J Biol Chem. 2000;275:8290–8300. doi: 10.1074/jbc.275.12.8290. [DOI] [PubMed] [Google Scholar]
- Romero-Santacreu L, Moreno J, Pérez-Ortín JE, Alepuz P. Specific and global regulation of mRNA stability during osmotic stress in Saccharomyces cerevisiae. RNA. 2009;15:1110–1120. doi: 10.1261/rna.1435709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory; 2001. [Google Scholar]
- Schwartz DC, Parker R. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:5247–5256. doi: 10.1128/mcb.19.8.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen EC, Stage-Zimmermann T, Chui P, Silver PA. 7The yeast mRNA-binding protein Npl3p interacts with the cap-binding complex. J Biol Chem. 2000;275:23718–23724. doi: 10.1074/jbc.M002312200. [DOI] [PubMed] [Google Scholar]
- Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010;40:228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- Teige M, Scheikl E, Reiser V, Ruis H, Ammerer G. Rck2, a member of the calmodulin-protein kinase family, links protein synthesis to high osmolarity MAP kinase signaling in budding yeast. Proc Natl Acad Sci USA. 2001;98:5625–5630. doi: 10.1073/pnas.091610798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D, Sheth U, Valencia-Sánchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uesono Y, Toh-e A. Transient inhibition of translation initiation by osmotic stress. J Biol Chem. 2002;277:13848–13855. doi: 10.1074/jbc.M108848200. [DOI] [PubMed] [Google Scholar]
- Van Der Kelen K, Beyaert R, Inze D, De VL. Translational control of eukaryotic gene expression. Crit Rev Biochem Mol Biol. 2009;44:143–168. doi: 10.1080/10409230902882090. [DOI] [PubMed] [Google Scholar]
- Wang X, Proud CG. Methods for studying signal-dependent regulation of translation factor activity. Methods Enzymol. 2007;431:113–142. doi: 10.1016/S0076-6879(07)31007-0. [DOI] [PubMed] [Google Scholar]
- Warringer J, Hult M, Regot S, Posas F, Sunnerhagen P. The HOG pathway dictates the short-term translational response after hyperosmotic shock. Mol Biol Cell. 2010;21:3080–3092. doi: 10.1091/mbc.E10-01-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall PJ, Patterson JC, Chen RE, Thorner J. Stress resistance and signal fidelity independent of nuclear MAPK function. Proc Natl Acad Sci USA. 2008;105:12212–12217. doi: 10.1073/pnas.0805797105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windgassen M, Sturm D, Cajigas IJ, Gonzalez CI, Seedorf M, Bastians H, Krebber H. Yeast shuttling SR proteins Npl3p, Gbp2p, and Hrb1p are part of the translating mRNPs, and Npl3p can function as a translational repressor. Mol Cell Biol. 2004;24:10479–10491. doi: 10.1128/MCB.24.23.10479-10491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CM, Qiu H, Hu C, Dong J, Hinnebusch AG. Yeast cap binding complex impedes recruitment of cleavage factor IA to weak termination sites. Mol Cell Biol. 2007;27:6520–6531. doi: 10.1128/MCB.00733-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki S, Anderson P. Reprogramming mRNA translation during stress. Curr Opin Cell Biol. 2008;20:222–226. doi: 10.1016/j.ceb.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Rosbash M. Identification of eight proteins that cross-link to pre-mRNA in the yeast commitment complex. Genes Dev. 1999;13:581–592. doi: 10.1101/gad.13.5.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.