hRio1 is an atypical protein kinase of the conserved RIO family. Depletion of hRio1 affects the last step of 18S rRNA maturation and causes defects in recycling of trans-acting factors from pre-40S subunits in the cytoplasm. The kinase activity of hRio1 is essential for recycling of the endonuclease hNob1 and its binding partner hDim2 from pre-40S.
Abstract
RIO proteins form a conserved family of atypical protein kinases. Humans possess three distinct RIO kinases—hRio1, hRio2, and hRio3, of which only hRio2 has been characterized with respect to its role in ribosomal biogenesis. Here we show that both hRio1 and hRio3, like hRio2, are associated with precursors of 40S ribosomal subunits in human cells. Furthermore, we demonstrate that depletion of hRio1 by RNA interference affects the last step of 18S rRNA maturation and causes defects in the recycling of several trans-acting factors (hEnp1, hRio2, hLtv1, hDim2/PNO1, and hNob1) from pre-40S subunits in the cytoplasm. Although the effects of hRio1 and hRio2 depletion are similar, we show that the two kinases are not fully interchangeable. Moreover, rescue experiments with a kinase-dead mutant of hRio1 revealed that the kinase activity of hRio1 is essential for the recycling of the endonuclease hNob1 and its binding partner hDim2 from cytoplasmic pre-40S. Kinase-dead hRio1 is trapped on pre-40S particles containing hDim2 and hNob1 but devoid of hEnp1, hLtv1, and hRio2. These data reveal a role of hRio1 in the final stages of cytoplasmic pre-40S maturation.
INTRODUCTION
The synthesis of ribosomal subunits in eukaryotic cells depends on more than 200 accessory proteins, so-called trans-acting factors, that promote rRNA processing, modification and folding, assembly of ribosomal proteins, and transport of preribosomal subunits across the nuclear envelope (Fatica and Tollervey, 2002; Tschochner and Hurt, 2003; Zemp and Kutay, 2007; Panse and Johnson, 2010). Many of these trans-acting factors possess enzymatic activities, including various types of nucleoside triphosphatases (NTPases) such as GTPases, DEAD-box helicases, AAA-ATPases, and protein kinases. These enzymes play key roles at different stages during the multistep pathway along which ribosomal subunits are assembled. Revealing the mode of action of these enzymes is important for a mechanistic understanding of the ribosome synthesis pathway.
The use of trans-acting factors as baits in tandem affinity purification (TAP) has been instrumental to determine the composition of preribosomal subunits and to sketch a map of the ribosome assembly pathway. A large number of distinct preribosomal particles has been purified from yeast cells and analyzed by mass spectrometry (Bassler et al., 2001; Harnpicharnchai et al., 2001; Dragon et al., 2002; Nissan et al., 2002; Milkereit et al., 2003; Schäfer et al., 2003). Ribosomal subunit biogenesis starts in the nucleolus, where a long precursor rRNA (pre-rRNA) is transcribed. There, the first ribosomal proteins and trans-acting factors are deposited on this pre-rRNA to form a large preribosomal particle that serves as precursor for both 40S and 60S subunits. An important step during nucleolar maturation is the endonucleolytic cleavage of the pre-rRNA, splitting the large precursor particle into pre-40S and pre-60S subunits. The subunit precursors are further matured in the nucleus with respect to both rRNA processing and protein composition. Many trans-acting factors join the ribosomal precursors at early stages of subunit assembly and are released during subsequent nuclear maturation steps. Others accompany preribosomal subunits to the cytoplasm, where they are released during the last cytoplasmic steps of subunit maturation and then shuttled back to the nucleus to serve another round of subunit assembly (Fatica and Tollervey, 2002; Tschochner and Hurt, 2003; Zemp and Kutay, 2007; Panse and Johnson, 2010). Release of trans-acting factors during cytoplasmic maturation not only allows subunits to gain competence for translation but might also contribute to the functional proofreading of the emerging subunits.
Cytoplasmic maturation of pre-40S subunits is dependent on atypical protein kinases of the RIO (right open reading frame) family (Vanrobays et al., 2001, 2003; Geerlings et al., 2003; Schäfer et al., 2003; Rouquette et al., 2005; Zemp et al., 2009). RIO kinases are conserved throughout all kingdoms of life (Angermayr et al., 2002; LaRonde-LeBlanc and Wlodawer, 2005). However, for all RIO kinases studied to date, substrates have remained elusive. Three subfamilies of eukaryotic RIO kinases can be distinguished, namely Rio1, Rio2, and Rio3. Whereas members of the Rio1 and Rio2 subfamilies are conserved from archaea to humans, Rio3 is only found in higher eukaryotes (Angermayr et al., 2002; LaRonde-LeBlanc and Wlodawer, 2005). RIO kinases share a conserved RIO kinase domain, but the subfamilies differ in their N- and C-terminal extensions. The RIO domain adopts the typical bilobed structure characteristic for protein kinases (LaRonde-LeBlanc and Wlodawer, 2004; Laronde-Leblanc et al., 2005). At the primary sequence level, the RIO domain harbors a “kinase signature” but otherwise does not show high sequence conservation with conventional eukaryotic protein kinases (ePKs; Angermayr and Bandlow, 2002). Furthermore, some subdomains of the RIO kinase domain are quite different when compared with ePKs, and other subdomains, including the activation loop and the putative substrate-binding site, are absent. Instead, RIO kinases contain a flexible loop insertion that harbors the mapped autophosphorylation sites of archaea Rio1 and Rio2 (LaRonde-LeBlanc and Wlodawer, 2004; Laronde-Leblanc et al., 2005).
The yeast kinase Rio1/Rrp10 is required for the last, cytoplasmic processing step of the 18S rRNA in yeast, namely cleavage of 20S pre-rRNA at site D (Vanrobays et al., 2001). Depletion of Rio1 causes a strong cytoplasmic accumulation of 20S pre-rRNA and a severe decrease in 18S rRNA levels, whereas biogenesis of large ribosomal subunits remains unaffected (Vanrobays et al., 2001; Soudet et al., 2011). Rio1 is a cytoplasmic protein that cosediments with 40S subunits on sucrose gradients (Vanrobays et al., 2001, 2003). Yet Rio1 has not been isolated as part of any 40S precursors purified by TAP, suggesting a transient interaction of Rio1 with the pre-40S particle. Although no other 40S trans-acting factor has been identified to interact with Rio1, 20S pre-rRNA has been found enriched in Rio1 pull-down experiments from yeast cell extracts (Vanrobays et al., 2001), suggesting that yeast Rio1 can associate with pre-40S particles. It is unclear whether kinase activity of Rio1, which is promoted by casein kinase 2, is required for its role in 40S production (Angermayr et al., 2007).
In contrast to Rio1, Rio2 is a core component of 40S precursors (Schäfer et al., 2003). Like Rio1, Rio2 is required for D-site cleavage in the cytoplasm (Geerlings et al., 2003; Vanrobays et al., 2003). However, Rio2 has also been proposed to support nuclear export of pre-40S subunits since the 40S export reporter ribosomal protein of the small subunit 2 (Rps2)–green fluorescent protein (GFP) accumulates in the nucleus in cells carrying a temperature-sensitive mutant allele of RIO2 (rio2-1) (Schäfer et al., 2003). Of note, Rio1 and Rio2 are both encoded by essential genes in yeast, indicating that they harbor, at least in part, different functions.
Among the human RIO kinases, only human Rio2 (hRio2) has been functionally linked to ribosome biogenesis (Rouquette et al., 2005; Zemp et al., 2009). Like its yeast counterpart, hRio2 is a component of late 40S precursors, and it promotes the final maturation of 40S subunits in the cytoplasm (Rouquette et al., 2005; Zemp et al., 2009). Depletion of hRio2 causes cytoplasmic accumulation of 18S-E rRNA, a 3′ extended 18S rRNA precursor. This rRNA processing defect might be the consequence of severe cytoplasmic particle remodeling defects, since hRio2 is required for the release of several 40S trans-acting factors from cytoplasmic pre-40S, including hEnp1/bystin, hDim2/PNO1, hRrp12, hLtv1, and hNob1 (Zemp et al., 2009; Wyler et al., 2011). The recycling of all of these factors, with the exception of hEnp1, depends on the kinase activity of hRio2. Here we set out to characterize the role of human Rio1 (hRio1) and hRio3 in ribosome synthesis in mammalian cells.
RESULTS
hRio1 and hRio3 are components of late 40S precursors
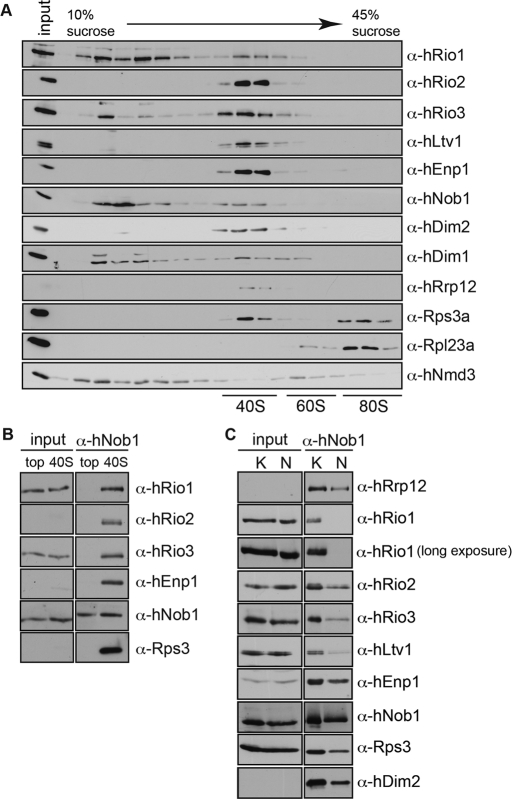
The atypical protein kinase hRio2 acts as a trans-acting factor in the biogenesis of 40S ribosomal subunits. hRio2 is required for maturation of 40S subunits in the cytoplasm and a stable component of pre-40S particles (Schäfer et al., 2003; Vanrobays et al., 2003; Zemp et al., 2009). To investigate whether the homologous human protein kinases Rio1 (hRio1) and Rio3 (hRio3) could likewise be involved in 40S biogenesis, we first analyzed the association of both kinases with 40S-sized particles. HeLa cell cytosol was fractionated on linear sucrose gradients (Figure 1A). The gradient fractions were analyzed for their content of Rio kinases, ribosomal proteins, and trans-acting factors by Western blotting. hRio1 was mainly found in the top fractions of the gradient. In addition, a pool of hRio1 cosedimented with pre-40S particles together with the established pre-40S components hRio2, hEnp1/bystin, hNob1, and hDim2/PNO1 (Zemp et al., 2009; Wyler et al., 2011). In contrast to hRio1, only little free hRio3 was found at the top of the gradient, and most of hRio3 was detected in (pre)-40S–containing fractions.
FIGURE 1:
hRio1 and hRio3 are components of 40S precursors. (A) All three Rio kinases cosediment with (pre)-40S ribosomal subunits. A hypotonic extract of HeLa cells was fractionated on a 10–45% linear sucrose gradient (monovalent ions: 100 mM KCl). Gradient fractions were precipitated and analyzed by Western blotting using the indicated antibodies. (B) hRio1 and hRio3 coprecipitate with pre-40S particles. hNob1 was precipitated from either pooled 40S fractions or pooled fractions from the top of the gradient (monovalent ions: 100 mM KCl). Precipitates were analyzed by Western blotting for the presence of hRio1 and hRio3, as well as for the established pre-40S components hNob1, hEnp1, hRio2, and Rps3. (C) hNob1 was immunoprecipitated from total HeLa cell extract under either low (150 mM K acetate; K) or high (250 mM NaCl; N) salt conditions to select for weak or strong protein–protein interactions. Eluates were analyzed by Western blotting for the indicated proteins. Whereas hRio3 is a stable component of 40S precursors, hRio1 was found to interact with α-hNob1 particles only under low-salt conditions.
To verify that hRio1 and hRio3 are associated with precursors of the small ribosomal subunit, we immunoprecipitated pre-40S from 40S–containing sucrose gradient fractions using an antibody directed against the pre-40S component hNob1. hNob1 is the human homologue of the yeast endonuclease Nob1 that promotes the final processing of 20S to 18S rRNA in the cytoplasm (Pertschy et al., 2009). Western blot analysis revealed that several known pre-40S components, including hRio2, hEnp1, and Rps3, were precipitated by the anti-hNob1 antibody from 40S-containing fractions (Figure 1B). Both hRio1 and hRio3 kinases were indeed part of these pre-40S particles. Neither hRio1 nor hRio3 coprecipitated with hNob1 from the top fractions of the gradient (Figure 1B). hRio1 and hRio3 were also found associated with pre-40S isolated from total HeLa cell extract (Figure 1C). Of note, the association of hRio1 with pre-40S was lost in buffers of higher ionic strength, indicating a weak interaction of hRio1 with the subunit precursors. Sucrose gradient analysis combined with isolation of pre-40S particles provides evidence that both hRio1 and hRio3, like hRio2, are components of pre-40S particles. In comparison to hRio2 and hRio3, hRio1 appears less stably associated with pre-40S.
hRio1 is associated with components of the 20S methylosome
Next we generated epitope-tagged hRio1 or hRio3 for tandem affinity purification (TAP) to isolate associated factors. Both kinases were tagged at either their N- or C-terminus with a tandem affinity tag (HASt-tag and StHA-tag, respectively; Wyler et al., 2011). Like endogenous hRio1 and hRio3, tagged hRio1 and hRio3 fusion proteins localized predominantly to the cytoplasm (Supplemental Figure S1). Of note, some hRio1 was also localized to the nucleus, which was confirmed by cell fractionation.
The tagged fusion proteins were then used to generate inducible HEK293 cell lines for TAP. All four cell lines expressed the fusion proteins upon induction by doxycycline. However, hRio3 TAP experiments were not successful. For hRio3-StHA, only the bait protein was retrieved, whereas HASt-hRio3 could not be purified (data not shown). Because both fusion proteins were at least in part incorporated into 40S-sized particles as analyzed by sucrose gradients, we assume that the tags are not accessible to allow for pre-40S purification.
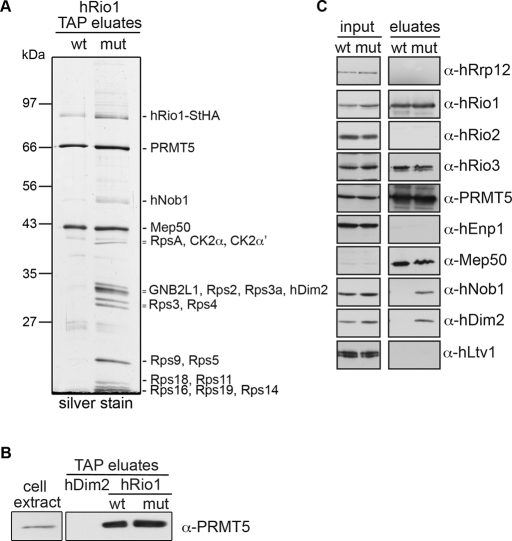
In contrast, both HASt-hRio1 (data not shown) and hRio1-StHA (Figure 2A) copurified two binding partners that were identified as PRMT5 and MEP50 by mass spectrometry. Both PRMT5, a protein arginine N-methyltransferase, and MEP50, a WD repeat protein, are part of the so-called methylosome that methylates Sm proteins during small nuclear ribonucleoprotein biogenesis (Guderian et al., 2011). Binding of PRMT5 to hRio1 is specific, as PRMT5 was not retrieved on the 40S trans-acting factor hDim2 (Figure 2B). In agreement with our interaction data, hRio1 was recently identified as binding partner of PRMT5 and MEP50 and suggested to function as adapter for PRMT5-mediated methylation of nucleolin (Guderian et al., 2011).
FIGURE 2:
Kinase-dead hRio1 copurifies a pre-40S particle. (A) Cell extracts of HEK293 cells expressing either hRio1(wt)-StHA or the kinase-inactive hRio1(D324A)-StHA mutant (mut) were subjected to tandem affinity purification. Protein eluates were analyzed by SDS–PAGE followed by either silver (gel shown) or Coomassie staining for subsequent analysis by mass spectrometry to identify proteins in the indicated bands. Note that background binding to either beads or the TAP tag alone is negligible (Wyler et al., 2011). (B) Cell extracts of HEK293 cells expressing HASt-Dim2, hRio1(wt)-StHA, or hRio1(mut)-StHA were subjected to one-step purification on StrepTactin Sepharose. Protein eluates were analyzed by SDS–PAGE, followed by immunoblotting using anti-PRMT5 antibodies. (C) The experiment in A was repeated, and the input and eluate samples were subjected to Western blot analysis using the indicated antibodies. Note that hRio1(wt)-StHA only purified a complex consisting of hRio1, PRMT5, and MEP50, as well as hRio3, whereas a subset of pre-40S components additionally copurified with hRio1(D324A)-StHA.
Kinase-dead hRio1 is more strongly associated with a pre-40S particle than wild-type hRio1
Because hRio1 was found to be associated with pre-40S particles under conditions of low but not high ionic strength (Figure 1) and no pre-40S was copurified with hRio1 by TAP (Figure 2A), we reasoned that the association of hRio1 with pre-40S might be too transient or unstable to isolate an associated 40S precursor. Therefore we next tested whether a kinase-inactive hRio1 mutant could be used to pull down pre-40S particles. On the basis of sequence alignments of the kinase domain of Rio1 from different species with protein kinase A, we determined Asp-324 as the catalytic base for the kinase reaction (Supplemental Figure S2). Mutation of Asp-324 to Ala (D324A) led to loss of hRio1's autophosphorylation activity in in vitro kinase assays (Supplemental Figure S2).
Next we introduced the D324A mutation into the hRio1-StHA fusion protein. When we used hRio1(D324A)-StHA stably expressed in HEK293 cells as bait in TAP, we not only precipitated PRMT5 and MEP50, but we also recovered a pre-40S particle as judged by both mass spectrometric analysis of copurifying proteins and immunoblotting for pre-40S components (Figure 2). Among the identified pre-40S–associated trans-acting factors were hNob1 and hDim2. In contrast, hEnp1 and hLtv1 were not enriched, suggesting that a late cytoplasmic pre-40S particle from which hEnp1/hLtv1 had been released was copurified. These data indicate that the kinase-dead form of hRio1 binds to 40S precursors more stably than wild type (wt), suggesting that active hRio1 is more rapidly dissociated from 40S precursors. In support of this conclusion, sucrose gradient analysis of cell extracts from hRio1(wt)-StHA– and hRio1(D324A)-StHA–expressing cells demonstrated that a fraction of the mutant protein comigrated with (pre)-40S–sized particles (Supplemental Figure S3).
hRio1 is required for cytoplasmic pre-40S maturation
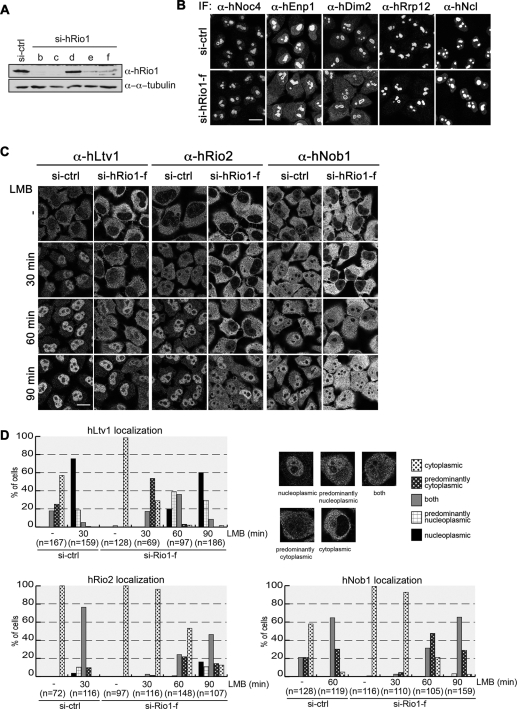
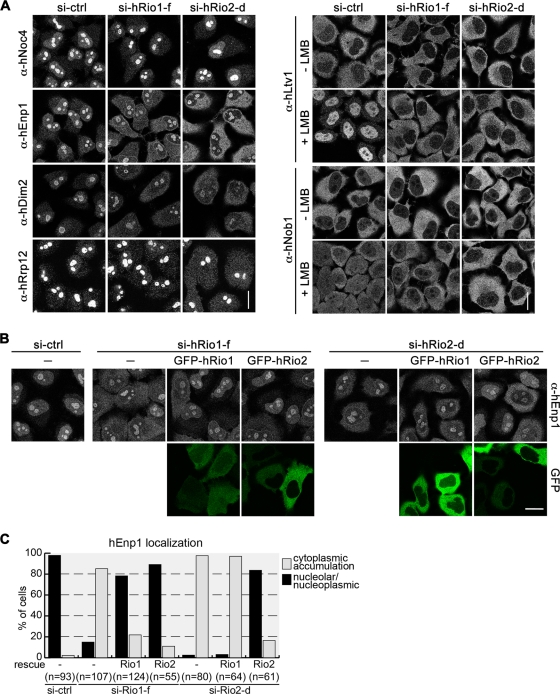
To study the role of hRio1 and hRio3 in 40S biogenesis, we depleted cells of hRio1 or hRio3 by RNA interference (RNAi). Down-regulation of the kinases was efficient, as judged by immunoblotting (Figure 3A and Supplemental Figure S1A). Effects on 40S biogenesis were determined by analyzing the localization of different trans-acting factors involved in 40S synthesis. The nucleolar proteins hEnp1 and hDim2, which join precursors of the 40S subunit early during nucleolar maturation and accompany them out of the nucleus, accumulated in the cytoplasm after hRio1 knockdown, suggesting that hRio1 is required for cytoplasmic maturation of 40S subunits. Other early nuclear trans-acting factors, such as hNoc4, hRrp12, and nucleolin, which also localize to the nucleolus at steady state, were not mislocalized upon depletion of hRio1 (Figure 3B). Consistent with the observation that nuclear steps in 40S biogenesis are not disturbed by hRio1 depletion, we did not detect any defect using an inducible Rps2–yellow fluorescent protein (YFP) HeLa reporter cell line that allows for scoring of nuclear defects in 40S biogenesis (Wild et al., 2010; B. Widmann, unpublished data).
FIGURE 3:
hRio1 is required for the cytoplasmic release of 40S trans-acting factors. (A) HeLa cells were transfected with five different siRNAs (17.5 nM) targeting hRio1. Cells were harvested after 3 d, and down-regulation of hRio1 was analyzed by Western blotting. For further analysis, the siRNA Rio1-f was chosen, as it targets the 3′UTR of the mRNA and allows for rescue experiments. (B) hRio1 was depleted from HeLa cells using si-Rio1-f as in A. Cells were fixed, and the localization of different nucleolar 40S trans-acting factors was analyzed by immunofluorescence. Whereas hDim2 and hEnp1 accumulated in the cytoplasm after depletion of hRio1, localization of nucleolin (hNcl), hNoc4, and hRrp12 was not altered. Scale bar, 20 μm. (C) Cells were depleted of hRio1 as in B. Before fixation, cells were treated with 20 nM LMB or ethanol (-) for the indicated times. Localization of hLtv1, hRio2, and hNob1 was analyzed by immunofluorescence. Scale bar, 20 μm. (D) Quantification of experiment shown in C. Based on the localization of the trans-acting factors, cells were assigned to the following phenotypic classes: cytoplasmic, predominantly cytoplasmic, both (equal cytoplasmic and nuclear localization), predominantly nucleoplasmic, and nucleoplasmic. Example cells are shown for hLtv1 localization. hLtv1 and hRio2 accumulate in the nucleus 30 and 60 min after inhibition of hCrm1, respectively. However, upon depletion of hRio1, their shuttling kinetics are delayed, indicating defects in recycling of both factors from pre-40S. hNob1 shuttling, although slower than shuttling of hLtv1 and hRio2 in control cells, is further delayed upon knockdown of hRio1.
To substantiate the role of hRio1 in cytoplasmic steps of 40S biogenesis, we analyzed whether hRio1 knockdown interferes with the localization of the cytoplasmic trans-acting factors hRio2, hNob1, and hLtv1. Indeed, all three factors, which are almost exclusively cytoplasmic in control cells, showed an even stronger cytoplasmic signal upon hRio1 knockdown (Figure 3C, top). Sucrose gradient analysis of cell extracts from control and hRio1-depleted cells showed that all the factors that accumulate in the cytoplasm after hRio1 down-regulation, namely hEnp1, hDim2, hRio2, hLtv1, and hNob1, cosedimented with 40S-sized particles in hRio1-depleted cells, indicating that these factors might be trapped as part of cytoplasmic 40S precursors after knockdown of hRio1 (Supplemental Figure S4).
The cytoplasmic steady-state localization of hRio2, hLtv1, and hNob1 is known to be dependent on CRM1-mediated nuclear export. Inhibition of CRM1 with the small-molecule inhibitor leptomycin B (LMB) leads to a nuclear accumulation of all three factors (Zemp et al., 2009; Figure 3, C and D). Thus responsiveness to LMB treatment can be used as tool to analyze defects in their nucleocytoplasmic shuttling caused by failures in release from cytoplasmic 40S precursors. Cells were treated with either control or hRio1 small interfering RNAs (siRNAs) and then incubated with LMB for 30, 60, or 90 min. The fast shuttling protein hLtv1 accumulated in the nucleus of control cells after only 30 min of LMB treatment. However, in hRio1-knockdown cells, shuttling of hLtv1 was strongly impaired (Figure 3, C and D). Quantification of the data showed that after 30 min of LMB treatment, hLtv1 was found in the nucleus in 75% of control cells, whereas in roughly 80% of hRio1-depleted cells hLtv1 was predominantly or exclusively cytoplasmic. hRio2 was equally distributed between the cytoplasm and the nucleus in 75% of control cells after 30 min of LMB (Figure 3, C and D). It is striking that in >90% of the hRio1-depleted cells, hRio2 localized still completely to the cytoplasm after 30 min of LMB treatment. After 90 min of LMB treatment, both hLtv1 and hRio2 showed almost the same level of nuclear accumulation in hRio1-depleted cells and in control cells (Figure 3C, bottom), indicating that shuttling of hLtv1 and hRio2 is not blocked but kinetically impaired. Furthermore, the shuttling behavior of hNob1, which is per se slower than that of hRio2 and hLtv1, was also delayed after down-regulation of hRio1. These data demonstrate that several 40S trans-acting factors are transiently trapped on 40S precursors in the cytoplasm in hRio1-depleted cells, indicative of defects in cytoplasmic 40S maturation.
Because hRio1 is associated with PRMT5 (Guderian et al., 2011; Figure 2), we tested whether the observed 40S maturation defects after hRio1 depletion could be mediated by PRMT5. Therefore we addressed whether PRMT5 is important for 40S synthesis. To analyze potential defects in cytoplasmic steps of pre-40S maturation, we determined the localization of hEnp1, hDim2, hLtv1, and hRio2 by indirect immunofluorescence after down-regulation of PRMT5. None of these 40S trans-acting factors was mislocalized upon PRMT5 knockdown (Supplemental Figure S5). We also depleted PRMT5 in the inducible Rps2-YFP HeLa reporter cell line. Although depletion of PRMT5 was efficient as judged by immunoblotting, there was no detectable defect in 40S biogenesis, and Rps2-YFP showed its normal, mostly cytoplasmic localization. Thus our analysis did not reveal any influence of PRMT5 on 40S biogenesis.
In addition, the down-regulation of hRio3 by RNAi led neither to defects in 40S biogenesis as scored by the Rps2-YFP reporter (Wild et al., 2010) nor to detectable changes in the localization of any of the analyzed factors (hDim2, hEnp1, hNob1, or hRio2 [Supplemental Figure 1E] and hLtv1, hNoc4, hRrp12, or nucleolin [not shown]). However, we cannot exclude that RNAi was too inefficient to pinpoint a contribution of PRMT5 or hRio3 to 40S biogenesis. In addition, it is also possible that these factors perform a function that is not detectable by our assays.
The kinase activity of hRio1 is required for recycling of hNob1 and hDim2 during cytoplasmic pre-40S maturation
To test whether kinase activity of hRio1 is required for the late cytoplasmic maturation steps in 40S biogenesis, we performed RNAi-rescue experiments using StHA-tagged wild-type or kinase-dead hRio1 (Figure 4). Cells were treated with an siRNA targeting the 3′-untranslated region (UTR) of hRio1 for 48 h and then transiently transfected with the rescue constructs. Twenty-four hours later, cells were fixed and analyzed by immunofluorescence. First, we investigated effects on hEnp1 localization. The cytoplasmic accumulation of hEnp1 caused by hRio1 depletion could be rescued by expression of both hRio1(wt)-StHA and hRio1(D324A)-StHA such that nucleolar localization of hEnp1 was restored (Figure 4A). Apparently, the recycling of hEnp1 only depends on the physical presence of hRio1 but not on its kinase activity. Of note, compared with hRio1(wt)-StHA, kinase-inactive hRio1(D324A)-StHA was less nuclear.
FIGURE 4:
Kinase activity of hRio1 is not required for cytoplasmic release of the 40S trans-acting factor hEnp1. HeLa cells were treated with either control or Rio1-f siRNAs as in Figure 3. For rescue analysis, cells were transfected with plasmids expressing either hRio1(wt)-StHA or hRio1(D324A)-StHA. (A) Cells were analyzed for localization of hEnp1 and the StHA-tagged rescue constructs by immunostaining and data quantified by assigning cells to one of two categories (cytoplasmic vs. nucleolar/nucleoplasmic). For the rescue experiments, only cells transfected with the rescue constructs were analyzed. Both hRio1(wt) and hRio1(D324A) rescue hEnp1 mislocalization observed upon hRio1 depletion. Scale bar, 20 μm. (B) Cells were treated with LMB for 30 min prior to fixation as indicated. Cells were analyzed for localization of hLtv1 or hRio2 and the rescue constructs by immunostaining. The experiment was quantified as in Figure 3D. For the rescue experiments, only cells transfected with the rescue constructs were analyzed. Scale bar, 20 μm.
Moreover, we tested whether the kinase activity of hRio1 is required for release of hLtv1 and hRio2 from cytoplasmic pre-40S by analyzing the response of hLtv1 and hRio2 localization to LMB treatment. We found that expression of hRio1(wt)-StHA almost completely rescued the shuttling of both factors to the nucleus in the presence of LMB after down-regulation of hRio1. The hRio1(D324A)-StHA mutant rescued the localization of hLtv1 partially, such that hLtv1 was equally distributed between the nucleus and the cytoplasm in most cells. In addition, for hRio2, the rescue by hRio1(D324A)-StHA was partial such that hRio2 still displayed a predominantly yet not exclusively cytoplasmic localization after 30 min of LMB treatment (Figure 4B). These data suggest that the kinase activity of hRio1 is not essential for the release of hLtv1 and hRio2 from cytoplasmic 40S precursors but is required for these steps to occur efficiently.
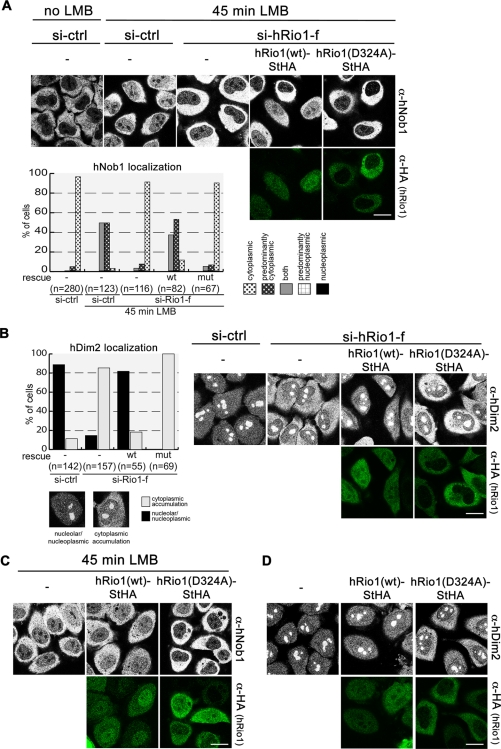
Next we tested whether kinase-dead hRio1 can rescue the recycling defects observed for hNob1 and hDim2 upon depletion of hRio1. For hNob1, we again monitored its ability to enter the nucleus after treatment of cells with LMB. Whereas hRio1(wt) restored the LMB-induced relocalization of hNob1 to the nucleus, kinase-inactive hRio1(D324A) enhanced the nuclear exclusion of hNob1 caused by down-regulation of hRio1 (Figure 5A). Similarly, hRio1(wt) rescued the phenotype of hRio1 depletion for hDim2 such that cytoplasmic localization of hDim2 was strongly reduced (Figure 5B). The kinase-inactive version of hRio1, in contrast, enhanced cytoplasmic localization of hDim2. These data suggested that kinase-dead hRio1 might even exert a dominant-negative effect on the localization of hNob1 and hDim2. Therefore we repeated the experiment without hRio1 down-regulation. Indeed, we observed that expression of hRio1(D324A)-StHA but not hRio1(wt)-StHA had a dominant-negative effect since hDim2 accumulated in the cytoplasm and the LMB-induced relocalization of hNob1 to the nucleus was decreased (Figure 5, C and D). Both hNob1 and hDim2 were trapped in the cytoplasm upon transfection of the mutant kinase. Of note, this dominant-negative effect hRio1(D324A)-StHA was only observed for these two factors but not for hLtv1, hRio2, and hEnp1 (data not shown).
FIGURE 5:
hRio1 kinase activity is required for recycling of hNob1 and hDim2. (A) HeLa cells were treated with either control or Rio1-f siRNAs and transfected with rescue constructs as in Figure 4. For the analysis of hNob1 localization, cells were treated with LMB 45 min prior to fixation. The localization of hNob1 was analyzed by immunostaining and quantified as in Figure 3D. Note that hNob1, which is trapped in the cytoplasm after hRio1 knockdown, can shuttle back into the nucleus in cells expressing hRio1(wt) but not in cells expressing hRio1(D324A). Scale bar, 20 μm. (B) Localization of Dim2 and the StHA-tagged rescue constructs was analyzed by immunostaining and quantified as for hEnp1 in Figure 4A. Scale bar, 20 μm. (C) HeLa cells were treated with control siRNAs and then transfected with either hRio1(wt)-StHA or hRio1(D324A)-StHA. Forty-five minutes prior to fixation, cells were treated with 20 nM LMB. The localization of hNob1 was analyzed by immunostaining. Note that hRio1(D324A) exerts a dominant-negative effect on hNob1 such that it is trapped in the cytoplasm. Scale bar, 20 μm. (D) HeLa cells were transfected as in C and analyzed for localization of hDim2 by immunostaining. Expression of hRio1(D324A) exerts a dominant-negative effect on hDim2 leading to its accumulation in the cytoplasm. Scale bar, 20 μm.
hRio1 and hRio2 are not functionally interchangeable
Recently, we demonstrated that the related kinase hRio2 is required for cytoplasmic recycling steps of 40S trans-acting factors. In fact, the defects observed upon depletion of both kinases are similar but not identical (Figure 6A and Table 1; Zemp et al., 2009). Downregulation of either hRio1 or hRio2 causes hNob1 and hDim2 to accumulate on cytoplasmic pre-40S particles. These defects can be rescued by overexpression of the respective wild-type kinase, whereas the kinase activities of both hRio1 and hRio2 are required for proper recycling of hNob1 and hDim2. However, whereas overexpression of kinase-dead hRio1 exerts a dominant negative effect on hDim2 and hNob1 localization, kinase-dead hRio2 does not (Zemp et al., 2009). For recycling of hLtv1, the kinase activity of hRio2 is indispensable, whereas the physical presence of inactive hRio1 is sufficient for hLtv1 recycling to occur partially. Moreover, whereas hRio1 depletion affected the LMB-induced nuclear relocalization of hRio2 (Figure 3), hRio2 depletion did not influence the localization of hRio1 (Supplemental Figure S6).
FIGURE 6:
hRio1 and hRio2 are not functionally interchangeable. (A) HeLa cells were depleted of hRio1 or hRio2 for 72 h and the localization of the indicated nucleolar (left) and cytoplasmic factors (right) was determined by immunofluorescence. Where indicated, cells were treated with LMB for 30 min prior to fixation. Note that hRio1 and hRio2 are required for recycling of hDim2, hEnp1, hLtv1, and hNob1. Recycling of hRrp12 only depends on hRio2. Scale bar, 20 μm. (B) HeLa cells were depleted of hRio1 or hRio2 by RNAi using siRNAs targeting the mRNA-UTRs. After 48 h, GFP-tagged, wild-type hRio1 or hRio2 was transfected, and cells were fixed after further 24 h. Note that expression of GFP-hRio2(wt) can rescue the recycling defect of hEnp1 caused by hRio1 depletion. However, GFP-hRio1 cannot rescue the mislocalization of hEnp1 caused by hRio2 knockdown. Scale bar, 20 μm. (C) Data from B were quantified as in Figure 4A.
TABLE 1:
Comparison of the requirements for hRio1 and hRio2 with respect to the cytoplasmic release of 40S trans-acting factors.
| 40S trans-acting factor | Relocalization after depletion of hRio1 | Rescue by hRio1 wt | Rescue by hRio1 kinase dead |
|---|---|---|---|
| hNoc4 | Noa | - | - |
| hRrp12 | Noa | - | - |
| hEnp1 | Yesa | Yesa | Yesa |
| hLtv1 | Yesa | Yesa | Partiala |
| hRio2 | Yesa | Yesa | Partiala |
| hNob1 | Yesa | Yesa | Noa |
| hDim2 | Yesa | Yesa | Noa |
| 40S trans-acting factor | Relocalization after depletion of hRio2 | Rescue by hRio2 wt | Rescue by hRio2 kinase dead |
| hNoc4 | Nob | - | - |
| hRio1 | Noc | - | - |
| hRrp12 | Yesb | Yesb | Nob |
| hEnp1 | Yesd | Yesd | Yesd |
| hLtv1 | Yesd | Yesd | Nod |
| hNob1 | Yesd | Yesd | Nod |
| hDim2 | Yesd | Yesd | Nod |
Of all the trans-acting factors analyzed, only the release of hEnp1 from the cytoplasmic pre-40S particle was independent of the kinase activities of both hRio1 and hRio2. Therefore we used hEnp1 as readout to test whether hRio1 and hRio2 are partially interchangeable with respect to their function in ribosome biogenesis. hEnp1 accumulates on cytoplasmic 40S precursors upon knockdown of either hRio1 or hRio2. It is striking that expression of either GFP-hRio1(wt) or GFP-hRio2(wt) rescued the aberrant cytoplasmic localization of hEnp1 in hRio1-depleted cells (Figure 6, B and C). In contrast, when we did the experiment the other way around, we observed that expression of GFP-hRio1(wt) could not rescue the recycling defect of hEnp1 after hRio2 depletion (Figure 6, B and C). On the basis of these observations, we conclude that the two Rio kinases hRio1 and hRio2 play at least in part distinct roles during the last maturation steps of the small ribosomal subunit and are not fully functionally interchangeable.
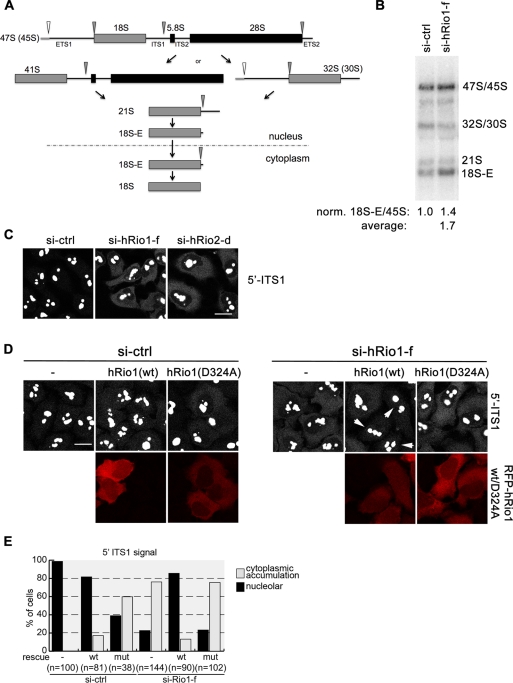
hRio1 kinase activity supports processing of 18S-E rRNA
Depletion of hRio2 from HeLa cells by RNAi leads to defects in the final step of 18S rRNA maturation, leading to the cytoplasmic accumulation of 18S-E rRNA (Rouquette et al., 2005; Zemp et al., 2009). Because cytoplasmic 40S maturation is stalled in the absence of hRio1, we wondered whether hRio1 is required for processing of the 18S-E pre-rRNA to mature 18S (for a scheme, see Figure 7A). To analyze 18S rRNA processing in hRio1-depleted cells, we analyzed the levels of 18S rRNA precursors by Northern blotting. Quantification showed that relative to the amount of 45′S/45S pre-rRNA, 18S-E levels were increased by a factor of ∼1.4 upon hRio1 knockdown, revealing a mild 18S-E rRNA processing defect caused by hRio1 depletion (Figure 7B). To substantiate these data, we also performed fluorescence in situ hybridization (FISH) analysis after hRio1 RNAi using a Cy5-labeled oligonucleotide complementary to the 5′-end of ITS1 as probe (Rouquette et al., 2005). In control cells, the FISH signal was observed in nucleoli, reflecting early, nucleolar precursors of 18S rRNA, whereas only a very weak signal was detected in the cytoplasm. However, after down-regulation of hRio1, the cytoplasmic signal increased (Figure 7C). The observed cytoplasmic increase of the 5′ITS signal was weak but highly reproducible. Again, we tested whether the kinase activity of hRio1 is involved in the last step of 18S pre-rRNA processing. Only wild-type hRio1 could rescue the pre-rRNA processing defect caused by hRio1 depletion, whereas the mutant kinase was not sufficient to rescue the defect in 18S-E rRNA cleavage (Figure 7, D and E).
FIGURE 7:
The kinase activity of hRio1 is required for 18S-E to 18S rRNA processing in the cytoplasm. (A) Simplified, schematic representation of the rRNA processing pathways generating 18S rRNA in human cells. The primary transcript pre-rRNA is processed along at least two different pathways, generating a 21S rRNA intermediate. 21S rRNA is converted to 18S-E rRNA, which still contains the 5′ region of ITS1, in the nucleus. After nuclear export of pre-40S, 18S-E rRNA is matured to 18S rRNA in the cytoplasm. (B) HeLa cells were depleted for hRio1 as in Figure 6. Total RNA was extracted and analyzed by Northern blotting using a probe specific for the 5′ end of ITS1. Depletion of hRio1 leads to increased accumulation of 18S-E relative to 45S rRNA level by a factor of 1.4 for the experiment represented. The average 18S-E/45S rRNA ratio derived from five independent experiments was 1.7. (C) Cells were depleted of hRio1 or hRio2 (as in Figure 6). After 72 h, cells were fixed and analyzed by FISH using a Cy5-labeled 5′ ITS1 probe. Note that this probe detects cytoplasmic 18S-E rRNA and all nuclear precursors thereof. Down-regulation of either hRio1 or hRio2 leads to the accumulation of 18S-E pre-rRNA in the cytoplasm. Scale bar, 20 μm. (D) HeLa cells were treated with either control or hRio1-f siRNAs for 72 h. At 24 h prior to fixation, RFP-hRio1(wt) or RFP-hRio1(D324A) rescue constructs were transfected. Cells were analyzed by FISH as in C. Note that only hRio1(wt) and not the kinase-dead mutant can rescue the 18S-E prerRNA processing defect caused by hRio1 depletion (right). White arrows point at cells transfected with RFP-hRio1(wt) in which the effect of hRio1 depletion is rescued. Scale bar, 20 μm. (E) Data from D were quantified assigning cells to one of two categories (cytoplasmic, cells displaying cytoplasmic 5′-ITS1 FISH signal in addition to the nucleolar signal; nucleolar, only nucleolar 5′-ITS1 FISH signal).
Given the dominant-negative effect of overexpression of kinase-dead hRio1 on recycling of hDim2 and hNob1, we wondered whether overexpression of kinase-dead hRio1 would interfere with 18S-E pre-rRNA processing. Indeed, hRio1(mut) caused accumulation of 18S-E rRNA in the cytoplasm (Figure 7, D and E). Of note, although 60% of cells showed the phenotype (Figure 7, D and E), the intensity of cytoplasmic signal of 18S-E rRNA was not as strong as observed after hRio1 RNAi, suggesting that endogenous hRio1 can partially fulfill the role of hRio1 in 18S-E pre-rRNA processing in presence of overexpressed hRio1(D324A). Taken together, the kinase activity of hRio1 is important for final steps of cytoplasmic 40S maturation, including pre-rRNA processing and recycling of hNob1 and hDim2 from the late 40S precursor.
DISCUSSION
Immature, cytoplasmic 40S subunits contain a number of trans-acting factors that need to be released from the subunit precursors during final steps of maturation. This maturation process also involves a last cleavage step at the 3′ end of the rRNA precursor mediated by the endonuclease hNob1 that generates mature 18S rRNA. It was previously found that both final rRNA processing and the recycling of trans-acting factors depend on the atypical protein kinase hRio2, which is a component of pre-40S subunits (Rouquette et al., 2005; Zemp et al., 2009). Here we demonstrated that both hRio1 and hRio3 are also associated with pre-40S subunits and characterized their potential function in 40S synthesis.
hRio1 is involved in the recycling of trans-acting factors and rRNA maturation
RNAi experiments showed that hRio1 is involved in the release of at least five different 40S trans-acting factors from pre-40S particles in the cytoplasm, namely hEnp1, hDim2, hRio2, hLtv1, and hNob1. In addition, hRio1 promotes efficient 18S-E to 18S rRNA processing. RNAi-rescue experiments using either wild-type or kinase-dead hRio1 revealed that differences exist with respect to the requirement of hRio1 kinase activity for the release of distinct trans-acting factors. The recycling of both hNob1 and hDim2 could not be rescued by the expression of kinase-dead hRio1, which even exerted a dominant-negative effect on the release of both trans-acting factors. In contrast, the recycling of hEnp1 required only the physical presence of hRio1 but did not depend on the kinase activity of hRio1. Similarly, release of hLtv1 could be partially rescued by the kinase-dead hRio1 mutant. Of note, yeast Enp1 and Ltv1 are dissociated together from pre-40S subunits in a step that is dependent on the activity of Hrr25, a yeast homologue of casein kinase 1 (Schäfer et al., 2006). The fact that kinase activity of hRio1 is not essential for recycling of hEnp1 and hLtv1 is thus in line with the model that dissociation of a complex of both factors is mediated by the activity of another, yet-unidentified factor in human cells. Still, in analogy to what we previously observed for hRio2, the release of hEnp1 requires the physical presence of hRio1. The molecular basis for these observations is unclear, but possibly the absence of either kinase might affect 1) particle structure, 2) the incorporation of a ribosomal protein, 3) the recruitment of another trans-acting factor required for maturation, or 4) the dissociation of a factor that inhibits further subunit remodeling. So far, we have not observed any obvious changes in the composition of pre-40S particles isolated by hDim2-TAP from cells in which hRio1 was down-regulated (data not shown).
Our data provide a strong link between hRio1 kinase activity and the recycling of hNob1 and hDim2, indicating that the enzymatic activity of hRio1 is required very late during 40S maturation. In support for such a late role of hRio1 kinase activity, we observed that the pre-40S particle purified by TAP using the kinase-dead hRio1 mutant contained hNob1 and hDim2, whereas hEnp1, hLtv1, and hRio2 were absent, suggesting that the release of the latter factors had been completed. Thus, consistent with the results of the RNAi-rescue experiments only hDim2 and hNob1, the recycling of which is strictly dependent on hRio1 kinase activity, are present in a hRio(D324A)-associated particle. The strong association of hRio(D324A) with pre-40S might indicate that the kinase-dead version of hRio1 behaves like a kinase-substrate trap, bringing hDim2 and hNob1 as well as other factors associated with this precursor into the spotlight for the hRio1 substrate hunt. Yeast Dim2 and Nob1 bind each other directly (Campbell and Karbstein, 2011), and the same holds true for their mammalian homologues (Supplemental Figure S7). Thus both factors might be kept together on pre-40S and be dissociated in a step dependent on Rio1 kinase activity. Therefore we also tested binding of recombinant hRio1 to hDim2 and/or hNob1 but could not detect any significant interaction (B. Widmann, F. Wandrey, and U. Kutay, unpublished results). Of note, hDim2 and hNob1 can be phosphorylated by hRio1 purified from baculovirus-infected insect cells but not by hRio1 purified from Escherichia coli (unpublished observations). The caveat, however, is that hRio1 from insect cells might contain additional, copurifiying protein kinases, and despite many attempts, we have been unable to purify the kinase-inactive version as control.
hRio1 and its association with PRMT5 and MEP50
Consistent with recently published data (Guderian et al., 2011), we found hRio1 in a stable complex with PRMT5 and MEP50, two components of the methylosome (Friesen et al., 2001). This observation raised the possibility that PRMT5 possesses a function along with hRio1 in 40S biogenesis. However, depletion of PRMT5 did not phenocopy the effect of depletion of hRio1 on 40S biosynthesis, even though RNAi against PRMT5 was very efficient. Furthermore, only very small amounts of PRMT5 could be detected on late pre-40S subunits immunoprecipitated with an anti-hNob1 antibody (Supplemental Figure S5). Therefore we consider it unlikely that the PRMT5-MEP50-hRio1 complex plays a decisive role in late steps of 40S production. Using the Rps2-YFP reporter cell line, we also failed to observe defects in nuclear steps of 40S biogenesis in absence of PRMT5. Therefore the PRMT5-mediated methylation of the early rRNA processing factor nucleolin (Guderian et al., 2011) seems not to be essential for nuclear 40S maturation. Still, it is possible that PRMT5-mediated methylation of ribosome biogenesis factors assists ribosome production in a way that is not measurable by our assays. In addition, the function of PRMT5 could be redundant with that of other PRMTs. Of interest, it was shown recently that the small ribosomal protein Rps10 is a substrate of PRMT5 (Ren et al., 2010). Rps10 is required for cytoplasmic steps of 40S biogenesis, and its absence causes defects in hEnp1 recycling (Wild et al., 2010). Lack of Rps10 methylation causes Rps10 instability and is accompanied by slight defects in protein synthesis, as well as by a small drop in the ratio of free 40S to 60S subunits (Ren et al., 2010), indicating that methylation of Rps10 might somehow promote either 40S stability or synthesis.
hRio1 and hRio2 have similar but distinguishable roles in 40S biogenesis
Both yeast Rio kinases Rio1 and Rio2 are essential, indicating that they cannot have entirely redundant roles (Vanrobays et al., 2001, 2003; Geerlings et al., 2003). Yet the defects in human 40S subunit biogenesis caused by depletion of either hRio1 or hRio2 are at first sight highly similar. Whereas localization of hNoc4 or hDim1 is not affected by hRio1 or hRio2 knockdown, hEnp1, hDim2, hLtv1, and hNob1 are trapped on cytoplasmic 40S precursors (Table 1). However, there are also clear differences in the requirements of hRio1 and hRio2 in late 40S maturation, indicating that different functions can indeed be attributed to both kinases. For instance, the cytoplasmic recycling of the nucleolar trans-acting factor hRrp12 only depends on the presence of hRio2 (Wyler et al., 2011) and not on hRio1. Furthermore, whereas kinase-dead hRio1 has a dominant-negative effect on recycling of hDim2 and hNob1 (Figure 5), kinase-dead hRio2 does not exacerbate the recycling defects of hDim2 and hNob1 after hRio2 RNAi (Zemp et al., 2009). Finally, defects in hEnp1 recycling caused by hRio1 depletion can be rescued by the overexpression of hRio2, but the converse experiment failed. This indicates that at least one aspect of hRio2 function, namely the release of hEnp1, cannot be complemented by hRio1. The rescue of hRio1 depletion by hRio2 overexpression could suggest that both kinases might in principle be able to occupy the same binding site on pre-40S particles. This raises the question as to whether both kinases ever simultaneously bind to the same pre-40S particle. Although hRio1 and hRio2 are both associated with pre-40S subunits isolated by immunoprecipitation of hNob1, these likely represent a mixture of particles of different maturation stages. Thus it is possible that the two kinases are bound to different subsets of pre-40S. In support of this notion, hRio2 is absent from pre-40S subunits that are associated with the kinase-dead mutant of hRio1, and we did not identify hRio1 on pre-40S isolated by immunoprecipitation of hRio2 (Zemp et al., 2009). This favors a model of mutually exclusive binding of both kinases to cytoplasmic pre-40S particles. Whether this reflects binding to the same binding pocket or to structurally distinct particles will require the identification of the binding sites for both proteins on pre-40S subunits. The hRio1(D324A) mutant that stably interacts with 40S precursors might serve as valuable tool for such future analysis.
Studies in yeast have yielded a map of cytoplasmic 60S maturation ordering the function of trans-acting factors in this pathway (Lo et al., 2010). In comparison, our understanding of cytoplasmic 40S maturation is still too incomplete to establish a fixed order of events in a similar way for the small subunit. Our observations that hRio1 depletion compromises hRio2 recycling, whereas loss of hRio2 does not appear to affect hRio1 release from pre-40S, could suggest that hRio1 functions upstream of hRio2. However, the fact that kinase activity of hRio1 is required for the release of hNob1 and hDim2, together with the observation that kinase-dead hRio1 is associated with a particle from which hEnp1, hLtv1, and hRio2 have been dissociated, would place the requirement for hRio1's kinase activity downstream of hRio2 function and is thus at odds with a simple, linear model. Instead of a fixed, linear order of events for cytoplasmic 40S maturation, it is also possible that release of different 40S trans-acting factors in the cytoplasm can occur, at least to some extent, in a parallel or noninterdependent manner.
hRio3 is a component of cytoplasmic pre-40S subunits
The third Rio kinase, hRio3, was also identified as stable component of pre-40S subunits, both by immunoprecipitation of hNob1 and as part of a pre-40S particle isolated by TAP using hRio1(D324A) as bait. This demonstrates that all three human RIO kinases are components of 40S biogenesis intermediates. Although we were unable to detect defects in 40S biogenesis upon down-regulation of hRio3 by RNAi, it is interesting that hRio3 was coisolated along with Rio1 in TAP experiments (Figure 2). This observation suggests that hRio1 and hRio3 are in the same protein complex.
Of interest, hRio3 was identified as a polyubiquitin-binding protein that modulates NF-κB signaling. hRio3 interacts with both K63- and K48-linked polyubiquitin chains via its N-terminal domain (Fenner et al., 2009). These previous observations, together with our identification of hRio3 as a pre-40S component, open the possibility of hRio3 serving as a pre-40S–associated factor that safeguards 40S biogenesis and might transmit defects in pre-40S maturation into the NF-κB pathway. Such a safeguarding function could explain why we failed to detect defects in 40S biogenesis upon depletion of hRio3. However, it is also possible that 1) depletion of hRio3 was too inefficient, 2) the function of hRio3 is redundant with the function of hRio2 or hRio1, or 3) our assays are inadequate to detect a function of hRio3 in 40S synthesis.
Outlook
Clearly, many questions remain to be answered for all members of the RIO family. Key to understanding their molecular function in cytoplasmic 40S maturation will be the identification of phosphorylation substrates and the unraveling of how phosphorylation of these factors explains the cellular function of RIO kinases. Furthermore, it is also unclear which cellular role the autophosphorylation activity of RIO kinases plays. Autophosphorylation is shared by all RIO kinases but is not required for kinase activity of Archaeoglobus fulgidus Rio1 (LaRonde-LeBlanc et al., 2005). The single autophosphorylation site of AfRio1 is not conserved in yeast and human Rio1, and the identification of such site(s) will be a prerequisite to define the biological function of RIO autophosphorylation. Taking into account the lack of success in the field in identifying RIO phosphorylation substrates, a provocative assumption could be that RIO kinases have no other substrates but themselves. Yet it is also possible that RIO substrates are only phosphorylated on pre-40S subunits of specific composition or conformation. Another important aspect that is only poorly understood is the question of how the activities of RIO kinases are regulated. In this context, another level of complexity is added by the control of yeast Rio1 kinase activity and levels through casein kinase II–mediated phosphorylation of its C-terminal domain (Angermayr et al., 2007). It remains to be established whether a similar regulation of RIO kinase activity exists for other RIO family members and in other organisms. Thus RIO kinases remain a challenging subject for future studies on how 40S subunits are matured in the cytoplasm to gain functionality for protein translation.
MATERIALS AND METHODS
Molecular cloning, protein expression, and purification
HASt-hRio1/HASt-hRio3 and hRio1-StHA(wt)/hRio1-StHA(D324A)/hRio3-StHA constructs were cloned into the pcDNA5/FRT/TO/nHASt-TAP and pcDNA5/FRT/TO/cStHA-TAP vectors, respectively, using KpnI/NotI as described (Wyler et al., 2011). The D324A point mutation was introduced into hRio1 using the QuikChange Kit (Stratagene, Santa Clara, CA).
For expression of zz-hRio1 in E. coli, the hRio1 coding sequence was inserted into pQE70-zz. For transient transfection of HeLa cells, hRio1 was cloned into a monomeric red fluorescent protein–encoding vector derived from pEGFP-C1 (Clontech, Mountain View, CA).
zz-hRio1(wt) and zz-hRio1(D324A) were expressed in BLR(pRep4) cells at 20°C. Cells were pelleted and lysed by sonication in 50 mM Tris/HCl (pH 7.6), 700 mM NaCl, 5 mM MgCl2, and 5% (vol/vol) glycerol. The lysate was cleared by ultracentrifugation and purified using nickel-nitrilotriacetic acid agarose (Qiagen, Valencia, CA). Purified proteins were rebuffered to 50 mM Tris/HCl (pH 7.6), 200 mM NaCl, and 5 mM MgCl2.
Generation of cell lines
hRio1-HASt cell lines were established as described (Wyler et al., 2011). In short, 0.1 μg of plasmid containing hRio1-StHA(wt) or (D324A) was transfected with 0.9 μg of pOG44 into HEK293 FlpIn TRex cells (Invitrogen, Carlsbad, CA). Cells were selected using 100 μg/ml hygromycin B and 15 μg/ml blasticidin. All cell foci were pooled, yielding polyclonal TAP cell lines.
Antibodies
Antibodies against hRio1, hRio3, and PRMT5 were raised against purified recombinant glutathione S-transferase–tagged (hRio1) or histidine-tagged (hRio3, PRMT5) proteins and affinity purified using the respective antigens coupled to SulfoLink (Thermo Fisher Scientific, Waltham, MA). All other antibodies were as described (Zemp and Kutay, 2007; Wyler et al., 2011). Anti–α-tubulin and anti-actin antibodies were purchased from Sigma-Aldrich (St. Louis, MO), and α-MEP50 was a kind gift from Utz Fischer (University of Würzburg, Würzburg, Germany). Secondary antibodies for immunofluorescence experiments were purchased from Invitrogen.
Transient transfections and RNAi
Cells were transiently transfected using the X-tremeGENE 9 reagent (Roche, Indianapolis, IN) and fixed in 4% paraformaldehyde (PFA). For most RNAi experiments, cells were transfected with 17.5 nM siRNA using Interferin (Polyplus-transfection, Illkirch, France) for 72 h and fixed in 4% PFA or harvested for Western blot analysis, RNA extraction, or cell extract preparation. For hRio2 depletion, the siRNA concentration was 9 nM. AllStars (Qiagen) served as negative control. The following siRNA oligonucleotides were used (sense strand):
si-hRio1-b r(UGAGUUUCAUCGGUAAAGA)d(TT), open reading frame (ORF)
si-hRio1-c r(GCCCAACAAGATAATATTCTA)d(TT), ORF
si-hRio1-d r(CAGAGGCACTCATGGGAAA)d(TT), 3′UTR
si-hRio1-e r(GCCACUGUUGGCUUCUGAA)d(TT), 5′UTR
si-hRio1-f r(GCAUCUGGAAGAUGGCUUA)d(TT), 3′UTR
si-hRio2-d r(GGAUCUUGGAUAUGUUUAA)d(TT), 3′UTR
si-hRio3-a r(AUCACUGGCUGUAUUAGUA)d(TT), ORF
si-hRio3-b r(AGGAGUCUGUUGUCUUUCA)d(TT), ORF
si-hRio3-UII r(CAAAUGAAGUUAUGGGUGA)d(TT), 3′UTR
si-hRio3-UIII r(GAUACUGGCUUUACAGAAA)d(TT), 3′UTR
si-PRMT5 r(AGGGCUCAAGCCACCAAUCUA)d(TT), 3′UTR
In rescue experiments, cells were additionally transiently transfected with hRio1-StHA(wt) or hRio1-StHA(D324A) 24 h prior to fixation.
Preparation of cell extracts
Cell extracts for TAP were prepared as described (Wyler et al., 2011). Cells were lysed on ice in 10 mM Tris (pH 7.6), 100 mM KCl, 2 mM MgCl2, 1 mM dithiothreitol (DTT), 0.5% NP-40 substitute (Fluka, Sigma-Aldrich), 0.5 mM NaF, 0.1 mM Na3VO4, 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 10 μM pepstatin using a douncer. The extract was cleared by centrifugation at 5000 × g for 12 min at 4°C.
Hypotonic cell extracts for sucrose gradient analysis of RNAi experiments (Supplemental Figure S4) were prepared as described (Zemp et al., 2009), but without addition of Triton X-100.
HeLa cell low-salt extract used for immunoprecipitations was prepared using digitonin. HeLa cell pellets of 5 × 109 cells were resuspended in 50 mM Tris (pH 7.6), 150 mM KOAc, 2 mM MgCl2, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 10 μg/ml pepstatin, and 0.15% (wt/vol) digitonin and lysed by douncing on ice. Cell debris was pelleted by centrifugation at 4500 rpm for 15 min at 4°C.
Sucrose gradient analysis
For sucrose gradient analysis of low-salt extract (Figure 1), extract (1 mg of protein) was loaded on a linear 10–45% sucrose gradient in 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)/KOH, pH 7.5, 100 mM KCl, and 3 mM MgCl2 and centrifuged for 3 h at 40,000 rpm in an SW41 rotor (Beckman Coulter, Brea, CA). For analysis of RNAi experiments (Supplemental Figures S3 and S4), hypotonic cell extract (400 μg of protein) was loaded onto a linear 10–45% sucrose gradient (50 mM HEPES/KOH, pH 7.5, 100 mM KCl, 3 mM MgCl2) and centrifuged for 2 h at 55,000 rpm at 4°C in a TLS55 rotor (Beckman Coulter). Gradient fractions were precipitated with trichloroacetic acid (TCA), followed by Western blot analysis.
To prepare pooled 40S fractions for immunoprecipitation experiments (Figure 1), low-salt extract was fractionated on a linear sucrose gradient using a SW32 rotor (Beckman Coulter) as described (Zemp et al., 2009).
Tandem affinity purification
TAP was performed as described (Wyler et al., 2011). Briefly, cell lysates were incubated with StrepTactin Sepharose (IBA, Göttingen, Germany) for 30 min and washed three times with TAP buffer (10 mM Tris/HCl, pH 7.6, 100 mM KCl, 2 mM MgCl2, 0.5 mM NaF, 0.1 mM Na3VO4, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 0.1 μg/ml pepstatin). Protein elution was performed three times with 300 μl of TAP buffer supplemented with 2.5 mM desthio-biotin (Sigma-Aldrich). Eluates were pooled and incubated with hemagglutinin-agarose (Sigma-Aldrich) for 1 h. After washing twice with TAP buffer and once with 10 mM Tris/HCl (pH 7.6) and 2 mM MgCl2, purified protein complexes were eluted with SDS sample buffer without DTT. Mass spectrometry analysis was performed as described (Wyler et al., 2011).
Immunoprecipitation
hNob1-specific antibodies coupled to a 9:1 mixture of protein A:protein G Sepharose (GE Healthcare, Piscataway, NJ) were incubated with low-salt extract containing 0.001% (vol/vol) Triton X-100. After washing, beads were eluted with 200 mM glycine/HCl (pH 2.2). Eluates were precipitated with TCA and analyzed by SDS–PAGE and Western blotting. For immunoprecipitations from the pooled top or 40S-containing fractions of sucrose gradients, fractions were diluted 1:2 with 50 mM Tris/HCl, pH 7.5, 100 mM KCl, and 5 mM MgCl2 prior to incubation with beads.
Northern blotting and FISH
Northern blotting was performed as described (Wyler et al., 2011). FISH was performed as described previously (Rouquette et al., 2005) using a Cy5-labeled ITS1-specific probe (5′-CCTCGCCCTCCGGGCTCCGTTAATGATC-3′). FISH images were processed in parallel as described (Rouquette et al., 2005; Zemp et al., 2009).
Immunofluorescence analysis
Cells were fixed in 4% PFA for 15 min and permeabilized in 0.02% SDS and 0.1% Triton X-100 in phosphate-buffered saline (PBS) for 5 min. After blocking in 10% goat serum (Sigma-Aldrich) and 2% bovine serum albumin (BSA) in PBS, primary antibodies were applied for 1 h in blocking solution. Next cells were washed three times for 5 min with 2% BSA in PBS and incubated with secondary antibody (Alexa Fluor 488 or Alexa Fluor 561 labeled; Invitrogen) diluted in blocking solution for 30 min. Cells were washed again three times and mounted in VectaShield (Vector Laboratories, Burlingame, CA).
Kinase assays
A total of 1 μg of zz-hRio1(wt) or zz-hRio1(D324A) was incubated with 1 μCi of γ-32P-ATP in a total volume of 10 μl using 50 mM Tris (pH 7.6), 200 mM NaCl, and 5 mM MgCl2. After incubation at 30°C for 20 min, the reaction was stopped by addition of 30 μl of SDS–PAGE sample buffer. Samples were analyzed by SDS–PAGE, and autoradiography was detected using phosphoimaging.
Supplementary Material
Acknowledgments
We thank C. Ashiono for excellent technical assistance. Imaging was performed on instruments of the ETH Zurich Light Microscopy Centre facility. This work was supported by the Swiss National Science Foundation and an intramural ETH Zurich grant (both to U.K.).
Abbreviations used:
- HASt-tag
hemagglutin-epitope/streptavidin-binding peptide tag
- LMB
leptomycin B
- Rps
ribosomal protein of the small subunit
- StHA-tag
streptavidin-binding peptide/hemagglutinin-epitope tag
- TAP
tandem affinity purification.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-07-0639) on November 9, 2011.
REFERENCES
- Angermayr M, Bandlow W. RIO1, an extraordinary novel protein kinase. FEBS Lett. 2002;524:31–36. doi: 10.1016/s0014-5793(02)02993-9. [DOI] [PubMed] [Google Scholar]
- Angermayr M, Hochleitner E, Lottspeich F, Bandlow W. Protein kinase CK2 activates the atypical Rio1p kinase and promotes its cell-cycle phase-dependent degradation in yeast. FEBS J. 2007;274:4654–4667. doi: 10.1111/j.1742-4658.2007.05993.x. [DOI] [PubMed] [Google Scholar]
- Angermayr M, Roidl A, Bandlow W. Yeast Rio1p is the founding member of a novel subfamily of protein serine kinases involved in the control of cell cycle progression. Mol Microbiol. 2002;44:309–324. doi: 10.1046/j.1365-2958.2002.02881.x. [DOI] [PubMed] [Google Scholar]
- Bassler J, Grandi P, Gadal O, Lessmann T, Petfalski E, Tollervey D, Lechner J, Hurt E. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol Cell. 2001;8:517–529. doi: 10.1016/s1097-2765(01)00342-2. [DOI] [PubMed] [Google Scholar]
- Campbell MG, Karbstein K. Protein-protein interactions within late pre-40S ribosomes. PLoS One. 2011;6:e16194. doi: 10.1371/journal.pone.0016194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragon F, et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PubMed] [Google Scholar]
- Fatica A, Tollervey D. Making ribosomes. Curr Opin Cell Biol. 2002;14:313–318. doi: 10.1016/s0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- Fenner BJ, Scannell M, Prehn JH. Identification of polyubiquitin binding proteins involved in NF-kappaB signaling using protein arrays. Biochim Biophys Acta. 2009;1794:1010–1016. doi: 10.1016/j.bbapap.2009.02.013. [DOI] [PubMed] [Google Scholar]
- Friesen WJ, Paushkin S, Wyce A, Massenet S, Pesiridis GS, Van Duyne G, Rappsilber J, Mann M, Dreyfuss G. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol Cell Biol. 2001;21:8289–8300. doi: 10.1128/MCB.21.24.8289-8300.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerlings TH, Faber AW, Bister MD, Vos JC, Raue HA. Rio2p, an evolutionarily conserved, low abundant protein kinase essential for processing of 20 S Pre-rRNA in Saccharomyces cerevisiae. J Biol Chem. 2003;278:22537–22545. doi: 10.1074/jbc.M300759200. [DOI] [PubMed] [Google Scholar]
- Guderian G, Peter C, Wiesner J, Sickmann A, Schulze-Osthoff K, Fischer U, Grimmler M. RioK1, a new interactor of protein arginine methyltransferase 5 (PRMT5), competes with pICln for binding and modulates PRMT5 complex composition and substrate specificity. J Biol Chem. 2011;286:1976–1986. doi: 10.1074/jbc.M110.148486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harnpicharnchai P, et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- Laronde-Leblanc N, Guszczynski T, Copeland T, Wlodawer A. Structure and activity of the atypical serine kinase Rio1. FEBS J. 2005;272:3698–3713. doi: 10.1111/j.1742-4658.2005.04796.x. [DOI] [PubMed] [Google Scholar]
- LaRonde-LeBlanc N, Wlodawer A. Crystal structure of A. fulgidus Rio2 defines a new family of serine protein kinases. Structure. 2004;12:1585–1594. doi: 10.1016/j.str.2004.06.016. [DOI] [PubMed] [Google Scholar]
- LaRonde-LeBlanc N, Wlodawer A. The RIO kinases: an atypical protein kinase family required for ribosome biogenesis and cell cycle progression. Biochim Biophys Acta. 2005;1754:14–24. doi: 10.1016/j.bbapap.2005.07.037. [DOI] [PubMed] [Google Scholar]
- Lo KY, Li Z, Bussiere C, Bresson S, Marcotte EM, Johnson AW. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol Cell. 2010;39:196–208. doi: 10.1016/j.molcel.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit P, Strauss D, Bassler J, Gadal O, Kuhn H, Schutz S, Gas N, Lechner J, Hurt E, Tschochner H. A Noc complex specifically involved in the formation and nuclear export of ribosomal 40 S subunits. J Biol Chem. 2003;278:4072–4081. doi: 10.1074/jbc.M208898200. [DOI] [PubMed] [Google Scholar]
- Nissan TA, Bassler J, Petfalski E, Tollervey D, Hurt E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002;21:5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse VG, Johnson AW. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem Sci. 2010;35:260–266. doi: 10.1016/j.tibs.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertschy B, Schneider C, Gnadig M, Schäfer T, Tollervey D, Hurt E. RNA helicase Prp43 and its co-factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J Biol Chem. 2009;284:35079–35091. doi: 10.1074/jbc.M109.040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Wang Y, Liang Y, Zhang Y, Bao S, Xu Z. Methylation of ribosomal protein S10 by protein-arginine methyltransferase 5 regulates ribosome biogenesis. J Biol Chem. 2010;285:12695–12705. doi: 10.1074/jbc.M110.103911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquette J, Choesmel V, Gleizes PE. Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J. 2005;24:2862–2872. doi: 10.1038/sj.emboj.7600752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer T, Maco B, Petfalski E, Tollervey D, Bottcher B, Aebi U, Hurt E. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature. 2006;441:651–655. doi: 10.1038/nature04840. [DOI] [PubMed] [Google Scholar]
- Schäfer T, Strauss D, Petfalski E, Tollervey D, Hurt E. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 2003;22:1370–1380. doi: 10.1093/emboj/cdg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soudet J, Gelugne JP, Belhabich-Baumas K, Caizergues-Ferrer M, Mougin A. Immature small ribosomal subunits can engage in translation initiation in Saccharomyces cerevisiae. EMBO J. 2011;29:80–92. doi: 10.1038/emboj.2009.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschochner H, Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13:255–263. doi: 10.1016/s0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- Vanrobays E, Gelugne JP, Gleizes PE, Caizergues-Ferrer M. Late cytoplasmic maturation of the small ribosomal subunit requires RIO proteins in Saccharomyces cerevisiae. Mol Cell Biol. 2003;23:2083–2095. doi: 10.1128/MCB.23.6.2083-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanrobays E, Gleizes PE, Bousquet-Antonelli C, Noaillac-Depeyre J, Caizergues-Ferrer M, Gelugne JP. Processing of 20S pre-rRNA to 18S ribosomal RNA in yeast requires Rrp10p, an essential non-ribosomal cytoplasmic protein. EMBO J. 2001;20:4204–4213. doi: 10.1093/emboj/20.15.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild T, Horvath P, Wyler E, Widmann B, Badertscher L, Zemp I, Kozak K, Csucs G, Lund E, Kutay U. A protein inventory of human ribosome biogenesis reveals an essential function of exportin 5 in 60S subunit export. PLoS Biol. 2010;8:e1000522. doi: 10.1371/journal.pbio.1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyler E, Zimmermann M, Widmann B, Gstaiger M, Pfannstiel J, Kutay U, Zemp I. Tandem affinity purification combined with inducible shRNA expression as a tool to study the maturation of macromolecular assemblies. RNA. 2011;17:189–200. doi: 10.1261/rna.2325911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemp I, Kutay U. Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett. 2007;581:2783–2793. doi: 10.1016/j.febslet.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Zemp I, Wild T, O'Donohue MF, Wandrey F, Widmann B, Gleizes PE, Kutay U. Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J Cell Biol. 2009;185:1167–1180. doi: 10.1083/jcb.200904048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.