The functions of eight Tetrahymena thermophila genes encoding RNase T2 family proteins (Rnt2 proteins) are explored in strains lacking one RNT2 gene or combinations of genes. At least three Tetrahymena RNase T2 enzymes are involved in the conditionally induced turnover of tRNA and rRNA.
Abstract
RNase T2 enzymes are produced by a wide range of organisms and have been implicated to function in diverse cellular processes, including stress-induced anticodon loop cleavage of mature tRNAs to generate tRNA halves. Here we describe a family of eight RNase T2 genes (RNT2A–RNT2H) in the ciliate Tetrahymena thermophila. We constructed strains lacking individual or combinations of these RNT2 genes that were viable but had distinct cellular and molecular phenotypes. In strains lacking only one Rnt2 protein or lacking a subfamily of three catalytically inactive Rnt2 proteins, starvation-induced tRNA fragments continued to accumulate, with only a minor change in fragment profile in one strain. We therefore generated strains lacking pairwise combinations of the top three candidates for Rnt2 tRNases. Each of these strains showed a distinct starvation-specific profile of tRNA and rRNA fragment accumulation. These results, the delineation of a broadened range of conditions that induce the accumulation of tRNA halves, and the demonstration of a predominantly ribonucleoprotein-free state of tRNA halves in cell extract suggest that ciliate tRNA halves are degradation intermediates in an autophagy pathway induced by growth arrest that functions to recycle idle protein synthesis machinery.
INTRODUCTION
RNase T2 enzymes are single-strand–specific endoribonucleases with an acidic optimum for activity around pH 4–5 and no strong cleavage preference for any base (Deshpande and Shankar, 2002; Luhtala and Parker, 2010). RNase T2 proteins are broadly conserved, with family members encoded in genomes of animals, plants, protozoans, prokaryotes, and some viruses (Irie, 1999). Most organisms produce one or two RNase T2 enzymes, but evolutionary expansion of the RNase T2 family in plants generated more numerous paralogues (five in Arabidopsis and eight in rice) that can be divided into subgroups based on sequence, expression profile, and biological function (MacIntosh et al., 2010). Eukaryotic RNase T2s are generally targeted to the secretory pathway for extracellular secretion or are intracellularly directed to lysosomes. Specific RNase T2 enzymes have also been reported to be vacuolar, cytoplasmic, or nuclear in localization (Irie, 1999; Deshpande and Shankar, 2002; Luhtala and Parker, 2010).
Cellular roles of RNase T2 enzymes have been defined in plants, where they contribute to phosphate and nucleotide scavenging, gametophytic self-incompatibility, pathogen defense, and rRNA decay (MacIntosh et al., 2010; Hillwig et al., 2011). An RNase T2 function in rRNA turnover has also been proposed for the animal enzymes, based on the enzyme depletion phenotype of rRNA overaccumulation in zebrafish neural tissue (Haud et al., 2011). This same phenotype is proposed to underlie the familial cystic leukoencephalopathy associated with human RNase T2 (RNASET2) loss-of-function gene mutations (Henneke et al., 2009). RNase T2 enzymes also can have functions independent of catalytic activity (Luhtala and Parker, 2010). A subgroup of plant RNase T2 paralogues that do not retain catalytically critical enzyme active-site residues has been suggested to play a role in pathogen defense (MacIntosh et al., 2010). Human RNASET2 has tumor-suppressor function independent of catalytic activity (Acquati et al., 2005; Smirnoff et al., 2006), and the influence of yeast RNase T2 (Rny1p) on cell viability is likewise independent of catalytic activity (Thompson and Parker, 2009a).
Yeast Rny1p has a catalytic role in the stress-induced production of tRNA fragments by endonucleolytic cleavage of mature tRNAs in the anticodon loop region (Thompson and Parker, 2009a). Diverse sizes and sequences of tRNA fragments have been uncovered by deep-sequencing projects across a broad range of organisms (Thompson and Parker, 2009b; Phizicky and Hopper, 2010; Pederson, 2010). Some types of ∼20-nucleotide (nt) tRNA fragments have been implicated in cellular regulation of translation or proliferation (Lee et al., 2009; Zhang et al., 2009). In the ciliate Tetrahymena thermophila, specifically processed ∼20-nt mature tRNA 3′-end fragments are bound by the Piwi-family protein Twi12 for a growth-essential ribonucleoprotein (RNP) function (Couvillion et al., 2010). In contrast, the tRNA halves produced by endonucleolytic cleavage in the anticodon loop region are substantially larger than 20 nt and conditional in their accumulation, thus representing a distinct class of tRNA fragments. Accumulation of tRNA halves was initially described in cultures of T. thermophila starved for essential amino acid(s) under conditions that promote developmental progression (Lee and Collins, 2005). Other single-celled organisms also conditionally accumulate tRNA halves, including Saccharomyces cerevisiae, Aspergillus fumigates, and Giardia lamblia (Jochl et al., 2008; Li et al., 2008; Thompson et al., 2008). Cultured mammalian cells also produce tRNA fragments when subject to some forms of cellular stress, suggesting that regulated cleavage of mature tRNAs in the anticodon loop region may be a general eukaryotic phenomenon (Thompson and Parker, 2009b). However, human tRNA halves are proposed to result from action of the RNase A family member angiogenin rather than RNASET2 (Fu et al., 2009; Yamasaki et al., 2009).
In this work we sought to address the endonuclease(s) responsible for the stress-induced production of tRNA fragments in T. thermophila. Although this ciliated protozoan lacks any predicted protein homologous RNase A, we identified eight predicted paralogous members of an expanded RNase T2 family (designated Rnt2 A–H). By characterization of RNT2 gene disruption strains and by molecular and biochemical characterization of inducibly accumulated tRNA and rRNA fragments in wild-type and gene-disruption strain backgrounds, we demonstrate that several T. thermophila RNase T2 family members function in regulated tRNA and rRNA turnover.
RESULTS
The T. thermophila genome encodes eight putative Rnt2 family proteins
We searched the expressed macronuclear T. thermophila genome (Eisen et al., 2006) and genome database (Tetrahymena Genome Database Wiki) by annotation and by BLAST using human, yeast, and zebrafish RNase T2s (RNASET2, Rny1p, Dre1, and Dre2) and also using human angiogenin. By this approach we were unable to find any credible homologues of the RNase A family member angiogenin, whereas we identified eight RNase T2 homologues designated Rnt2 A–H (Table 1 and Figure 1). T. thermophila Rnt2 A, B, and C showed the highest homology (E < 5.7 × 10−5) with yeast Rny1p, and Rnt2 A, B, C, G, and H had reliable homology (E < 8.2 × 10−7) with human RNASET2.
TABLE 1:
Properties of the eight predicted RNase T2 paralogues in Tetrahymena (Rnt2 A–H).
| Gene name | TTHERMa | Size (amino acids)b | Molecular mass (kDa)b | pIb | Rny1p homologue rank | RNASET2 homologue rank | Signal peptide | Conserved active-site motif | Expression profilec | Expression level maximumd |
|---|---|---|---|---|---|---|---|---|---|---|
| Rnt2 A | 00222300 | 224 | 25.1 | 8.37 | 3 | 1 | Y | Y | stat/starv/conj | 16,181 |
| Rnt2 B | 00784550 | 203 | 23.1 | 4.86 | 1 | 3 | Y | Y | conj | 80 |
| Rnt2 C | 00264790 | 235 | 27.0 | 4.88 | 2 | 5 | Y | Y | stat/starv/conj | 1,598 |
| Rnt2 D | 00815150 | 213 | 25.4 | 5.67 | 5 | 9 | Y | N | stat | 77 |
| Rnt2 E | 00815180 | 213 | 25.3 | 5.95 | 9 | 8 | Y | N | conj | 68 |
| Rnt2 F | 00815210 | 213 | 25.2 | 6.06 | 7 | 12 | Y | N | gen | 47 |
| Rnt2 G | 00444290 | 207 | 23.7 | 5.60 | 4 | 2 | Y | Y | gen | 42 |
| Rnt2 H | 00125450 | 201 | 22.9 | 5.16 | 6 | 4 | Y | Y | gen | 47 |
aTetrahymena Genome Database Wiki identification number.
bCalculated for mature proteins lacking the signal peptide.
cAt least double expression level in stationary phase (stat), starvation (starv), or conjugation (conj) compared with lowest level of expression, or expressed at similar levels across all conditions (gen).
dRelative value.
FIGURE 1:
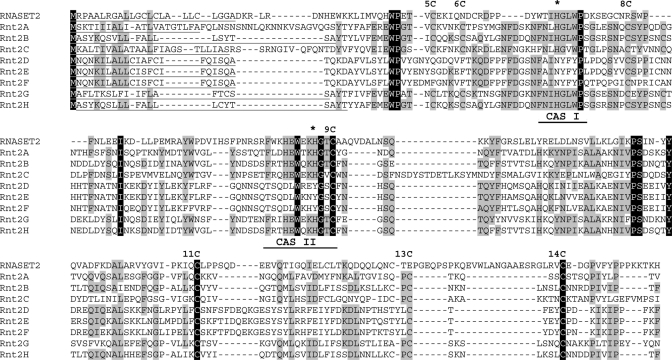
Amino acid sequence alignment of T. thermophila Rnt2 family members. Absolutely conserved residues are indicated with black background, and residues conserved in five to eight sequences are indicated with gray background. The two active-site motifs CAS I and CAS II are indicated below the sequence alignment, and the two catalytic histidines are marked with an asterisk above. The conserved cysteines involved in disulfide bridges are also indicated above the sequence alignment and are numbered according to previous annotation (Irie, 1999). The N-terminal signal peptides predicted by SignalP are underlined.
The automated prediction of an open reading frame (ORF) for Rnt2 C gave a significantly longer ORF than encoded by the other RNT2 genes, but a closer inspection of the gene suggested that an ATG codon downstream of the initially predicted start codon is likely to be the correct site of translation initiation. With this correction Rnt2 C becomes close in length to all of the other predicted T. thermophila Rnt2 proteins (201–235 amino acids, 22.9–27.0 kDa; Table 1) and to RNase T2 family members in other eukaryotes (Deshpande and Shankar, 2002). In addition, only the corrected Rnt2 C ORF encodes an N-terminal signal peptide, which is present in all of the other T. thermophila Rnt2 ORFs (Table 1 and Figure 1) and RNase T2 protein ORFs in other eukaryotes. Of note, despite their predicted signal peptides, Rnt2 proteins were not found in a recent proteomic study of the T. thermophila secretome (Madinger et al., 2010), suggesting that they could be predominantly trafficked to a cell-internal compartment rather than to extracellular secretion.
Variation in isoelectric point (pI) has been associated with specific subgroups of RNase T2 proteins with distinct biological properties and roles (MacIntosh et al., 2010). Of note, Rnt2 A is predicted to be significantly more basic than the other T. thermophila Rnt2 proteins, with a pI of 8.37, compared with other Rnt2 pI values between 4.86 and 6.06 (Table 1). Alignment of the eight predicted Rnt2 paralogues with human RNASET2 reveals that the T. thermophila Rnt2 paralogues also differ in their conservation of amino acid residues important for catalytic activity (Figure 1). Rnt2 A, B, C, G, and H conserve the RNase T2 active-site motifs (CAS I and CAS II), including the two histidines essential for catalysis (Irie, 1999). In contrast, the Rnt2 D, E, and F paralogues, which are similar in sequence to each other and are encoded by uninterrupted tandem genes, have not retained consensus CAS I and CAS II motifs. In these three proteins the histidines directly involved in catalysis have been replaced with asparagine and tyrosine (Figure 1), likely rendering them catalytically inactive. Nonetheless, Rnt2 D, E, and F do conserve the cysteines involved in two structurally critical RNase T2 disulfide bridges, 8C–9C and 11C–14C (Figure 1). In addition to the aforementioned cysteines, Rnt2 A, B, C, G, and H have the potential to form an additional disulfide bridge between 5C and 6C that is present in RNase T2s of plants and animals but not fungi (Irie, 1999).
Data from microarray studies of mRNA expression in growing, early stationary phase, starving, and sexually conjugating cells (Miao et al., 2009) suggest that most RNT2 mRNAs have low abundance throughout the T. thermophila life cycle, with the exception of the mRNAs encoding Rnt2 A and Rnt2 C (Table 1). Of note, these two mRNAs have enhanced levels under the starvation conditions that induce the accumulation of tRNA halves, making them particularly interesting candidates for a tRNA cleavage nuclease.
Individual losses of RNT2 loci impose modest phenotypes
To study the biological function(s) of T. thermophila Rnt2 proteins, we targeted RNT2 loci in the expressed macronucleus for replacement with a neo2 or bsr2 cassette, which confers resistance to paromomycin or blasticidin, respectively. Initial transformation replaces only a few of the 45 macronuclear gene copies with the resistance cassette, but continuous selection pressure yields complete replacement of nonessential genes (Gaertig and Kapler, 2000). The genes encoding Rnt2 A, B, C, G, and H were targeted individually, whereas the genes encoding Rnt2 D, E, and F were targeted in combination due to their tandem arrangement in the macronuclear genome.
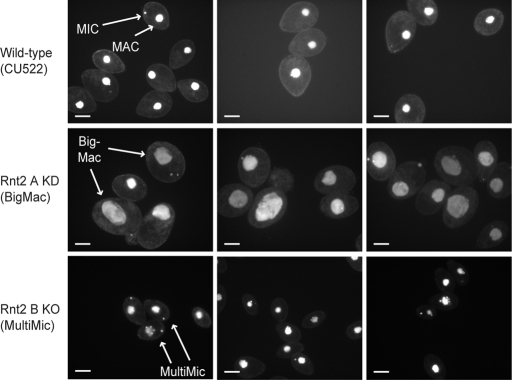
Cells undergoing depletion of Rnt2 A showed considerable difficulties in growth during selection and were often were bigger than wild-type cells. To investigate whether an increased cellular content of DNA or RNA was linked to the increased cell size, we stained wild-type and RNT2 loci targeted cells with the general nucleic acid 4′,6-diamidino-2-phenylindole (DAPI) stain and the RNA-specific SYTO RNASelect stain. With SYTO RNASelect, only weak general staining of the cytosol was observed in any strain background. Of interest, DAPI staining revealed that cells undergoing depletion of Rnt2 A had enlarged macronuclei that correlated with increased cell size (BigMac phenotype; Figure 2). DAPI staining of cells undergoing depletion of Rnt2 B instead showed an apparent increase in the number of micronuclei in many of the cells (MultiMic phenotype; Figure 2). T. thermophila BigMac and MultiMic phenotypes are consequences of aberrant or aborted cell division, suggesting that depletion of Rnt2 A or Rnt2 B can impose a cell cycle defect. However, these cellular phenotypes disappeared over the course of ∼200–400 cell generations in cultures propagated in rich growth medium.
FIGURE 2:
Cellular phenotypes associated with Rnt2 depletion. DAPI staining of macronuclei and micronuclei in wild-type CU522 compared with Rnt2 A KD or Rnt2 B KO cells with BigMac or MultiMic phenotypes, respectively (arrows). Scale bars, 20 μm.
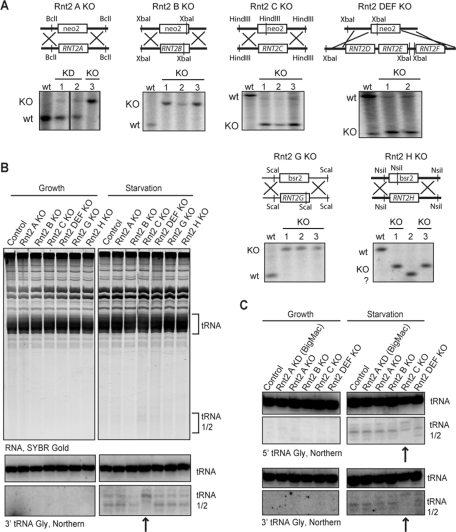
After an interval of drug selection, cells targeted for disruption of RNT2 loci were then grown in rich medium without drug to allow back-assortment of incompletely replaced loci. Individual cells were isolated and expanded as clonal cell lines prior to genomic DNA extraction. Southern blot hybridization was used to query the extent of replacement of macronuclear RNT2 loci with the drug resistance cassette (note that the micronuclear copies of the gene will not be replaced but are not expressed). Based on the extent of locus assortment, we obtained complete macronuclear gene knockout (KO) strains for Rnt2 A, B, C, G, H, and the triple D–F combination. For most strains, at least three of the four clonal cell lines tested for genotype were complete KOs (Figure 3A). However, only one of 11 clonal cell lines tested for RNT2A locus replacement was a complete KO, whereas the rest were partial gene copy number knockdowns (KDs). This is likely due to the compromised growth of cells depleted for Rnt2 A as noted, which resulted in fewer population doublings during the selection interval.
FIGURE 3:
Profiles of tRNA fragments in Rnt2 KO strains. (A) Southern blot analysis confirming complete replacement of macronuclear RNT2 genes with the resistance cassette neo2 or bsr2. Genomic DNA was digested with the indicated restriction enzymes to resolve wild-type (wt) from KO chromosomes as schematized above each blot. Two or three independent clonal cell lines are compared with wt for each RNT2 locus targeting. (B) Total RNA from control and KO strains grown in defined complete synthetic medium without or with transfer to starvation medium for a subsequent 3 h. RNA was analyzed by SYBR Gold stain (top) and by Northern blot hybridization with a probe against the 3′ end of tRNA Gly (bottom; only tRNA-relevant blot sections are shown). The positions of mature tRNA and tRNA halves are indicated. An arrow highlights the Rnt2 C KO lane with an altered size of tRNA halves. (C) RNA from cells grown in complex, rich medium NEFF with or without subsequent transfer to starvation medium. RNA was analyzed by Northern blot hybridization with probes against tRNA Gly 5′ and 3′ ends.
We chose to eliminate RNT2 loci in an epitope-tagged Twi12 strain background. The Piwi-family protein Twi12 carries ∼20-nt tRNA 3′-end fragments specifically processed from mature tRNAs, unlike the Dicer-processed small RNAs carried by other T. thermophila Piwi-family proteins (Couvillion et al., 2009, 2010). The use of this strain allowed us to isolate Twi12 RNPs by protein affinity purification and assess any difference in the Twi12-bound tRNA fragment profile. Consistent with the essential function of Twi12 RNPs (Couvillion et al., 2010), Twi12-bound tRNA 3′ fragments appeared unaffected in the RNT2 KO strains (data not shown).
Rnt2 C KO alters the size profile of starvation-induced tRNA halves
We examined the Rnt2 KO strains for potentially altered production of tRNA halves. Cells were grown in defined complete synthetic medium, precluding the ingestion of exogenous tRNA or other noncoding RNA fragments, and then transferred to starvation medium. No accumulation of tRNA halves was observed in growing cells of wild-type or KO genotype, and wild-type and all of the KO strains did accumulate tRNA halves following 3 h of starvation (Figure 3B). Curiously, the pattern of the tRNA cleavage products detected by Northern blot hybridization was shifted by ∼2 nt in total RNA from the Rnt2 C KO strain (Figure 3B, arrow). Similar results were observed for cells grown in the typical rich medium containing protease peptone and yeast extract and for Rnt2 A KD cells, with almost exclusively the BigMac phenotype, as well as the Rnt2 A KO cells, passaged over more generations (Figure 3C).
Double-KO strains accumulate high levels of starvation-induced tRNA and rRNA fragments
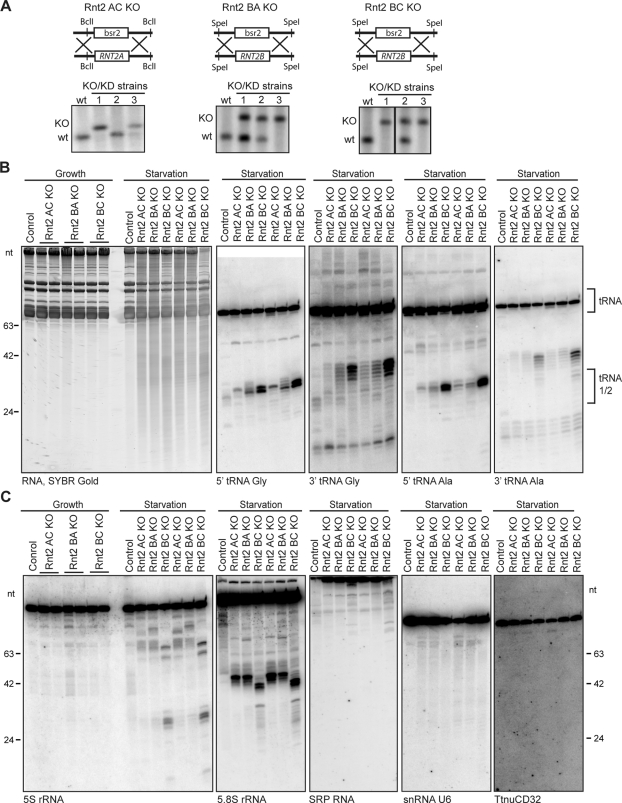
To investigate functional redundancy among the T. thermophila Rnt2 proteins, we generated strains with double-gene KOs that eliminated each pairwise combination of the three Rnt2 proteins that are most homologous to yeast Rny1p, clearly expressed as mRNA, and predicted to be catalytically active (Table 1; Rnt2 AC, BA, and BC KO strains). The second, sequentially targeted RNT2 locus was sometimes completely and sometimes partially assorted in the clonal cell lines produced, as described for the initial locus KO (Figure 4A). This demonstrates that despite some compromise of cell growth rate during selection, pairwise combinations of T. thermophila Rnt2 A, B, and C proteins can be eliminated without loss of strain viability.
FIGURE 4:
Elevated levels of tRNA and rRNA fragments in Rnt2 double-KO strains. (A) Southern blot analysis confirming complete replacement of a second RNT2 locus with the bsr2 resistance cassette. Genomic DNA from wild-type (wt) cells and three independent double-targeted clonal cell lines was digested with appropriate restriction enzymes to resolve wild-type and KO chromosomes as schematized above each blot. (B) RNA harvested from growing cells or cells starved for 3.5 h from two independent cell lines for each Rnt2 double KO, stained with SYBR Gold (left). Results are shown from Northern blot hybridization to detect 5′ and 3′ fragments of tRNA Gly and tRNA Ala (right). (C) Northern blot hybridization analysis of RNA as earlier described. Results from probes complementary to 5S rRNA, 5.8S rRNA, SRP RNA, small nuclear RNA U6, and small nucleolar RNATtnuCD32 are shown.
We purified total RNA from growing and starving Rnt2 double-KO strains, as well as from control cell cultures. SYBR Gold staining revealed no obvious difference in the RNA profiles of growing cells, but in starving cells a heterogeneous smear of RNA fragments was much more prominent in the total RNA of each double-KO strain compared with wild-type (Figure 4B, left). Northern blot hybridization with probes complementary to several different tRNAs confirmed a dramatic increase in tRNA fragments in all of the double-KO strains and for Rnt2 BC KO cells in particular (Figure 4B). Cells of each double-KO genotype had a distinct tRNA fragment pattern reproducible across two independently selected clonal cell lines of each double-KO genotype (Figure 4B, shown as two sets of the three double-KO genotypes). It is striking that the tRNA fragments that accumulated in each double-KO strain were predominantly larger than the wild-type tRNA halves for both the tRNA 5′ and 3′ pieces (Figure 4B, see labels under each Northern blot). The combined longer 5′ and 3′ fragment lengths approach the length of a mature tRNA, as if the fragments that accumulate in wild-type cells have been trimmed of the single-stranded portions of the anticodon loop, whereas in the double-KO cells fewer nucleotides have been removed after an initial endonuclease cleavage event.
RNA fragments detected by SYBR Gold staining in the Rnt2 double-KO starved cells (Figure 4B, left) include a prominent distribution of ∼35- to 40-nt RNAs matching the migration of the oversized tRNA halves. In addition, RNA fragments extend to well above the maximum size of intact tRNAs, suggesting that tRNA degradation does not account for the entire increase in RNA fragments in the double-KO starved cells compared with wild type. We therefore tested whether fragments from other structured noncoding RNAs could be detected, using Northern blot probes complementary to 5S and 5.8S rRNAs and other functional noncoding RNAs (Figure 4C). Fragments from the RNA Pol III–transcribed 5S and RNA Pol I–transcribed 5.8S rRNAs accumulated specifically in starved cells, dependent on the double-KO genotype. Fragments from 5S rRNA were particularly evident in total RNA from Rnt2 BC KO cells, and a distinct pattern of 5.8S rRNA fragments was produced in Rnt2 BC KO cells compared with Rnt2 AC or BA KO cells (Figure 4C, see labels under each Northern blot). In comparison, more limited if any accumulation of fragments from the predominantly cytoplasmic signal recognition particle (SRP) RNA, the nuclear U6 RNA, or the nucleolar TtnuCD32 RNA were detected (Figure 4C, right). We conclude that Rnt2 AC, BA, and BC double-KO strains overaccumulate both tRNA and rRNA fragments, suggesting that the T. thermophila RNase T2 enzymes have starvation-induced roles in turnover of the RNA machinery for protein translation.
Of note, following ∼500 cell generations of culture growth, the severity of the Rnt2 double-KO strain phenotypes abated. The only persistent molecular phenotype was a shift from wild-type sized tRNA halves to slightly larger tRNA halves in Rnt2 AC and BC KO cells, reflecting the phenotype persistent in cells with Rnt2 C KO alone (Figure 3, B and C). Southern blot hybridization confirmed that this reduced phenotypic severity was not accompanied by genotype reversal (data not shown). Thus it seems as if T. thermophila adapts to the loss of Rnt2 A and Rnt2 B by bypassing the induction of RNA degradation, by up-regulating other RNA degradation pathways, and/or by trafficking RNA fragments to extracellular secretion rather than intracellular retention. A large number of possible genetic and epigenetic changes could underlie this adaptation.
Growth arrest culture conditions induce the production of tRNA halves
The accumulation of T. thermophila tRNA halves was detected originally (Lee and Collins, 2005) and in the work described here as a consequence of shifting growing cells to nutrient starvation. Data from genome-wide microarray analysis of mRNA expression (Miao et al., 2009) suggest that Rnt2 A and Rnt2 C mRNA levels increase in stationary phase as well as in starvation. Therefore, if these nucleases are generally involved in tRNA (fragment) turnover, we would predict that tRNA halves accumulate transiently in wild-type cells in stationary phase as well as starvation. Total RNA was prepared from wild-type cells in starvation, stationary phase, and also mild or severe heat shock or cold shock. All of these stress conditions are physiologically relevant for a free-living, pond-dwelling, single-celled organism such as Tetrahymena. As expected, tRNA halves were not detectable in rapidly dividing cells in log-phase growth but accumulated at readily detectable levels within the first 6 h of starvation (Figure 5). If growing cells were allowed to reach maximum density and then kept shaking for an additional 16–18 h of stationary-phase culture, tRNA halves did indeed accumulate to a similar level as in starved cells (Figure 5, Stat. phase).
FIGURE 5:
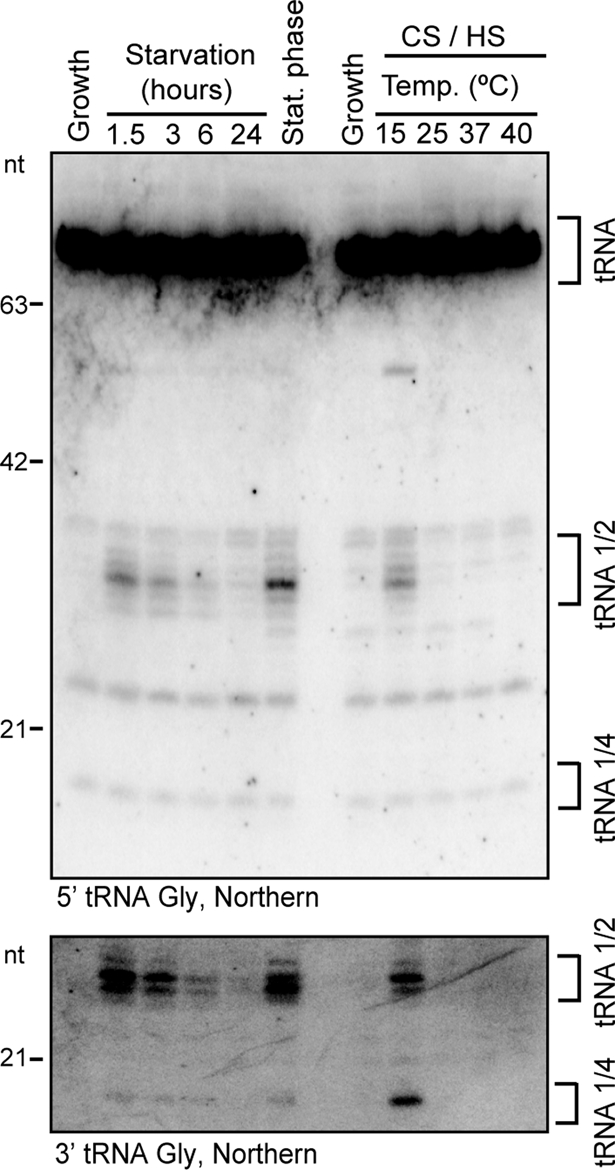
Accumulation levels of tRNA fragments compared across culture conditions. The accumulation of 5′ (top) and 3′ (bottom) tRNA Gly fragments upon various stresses: starvation, stationary phase, mild or severe cold shock (CS), and mild or severe heat shock (HS). The positions of mature tRNAs, tRNA halves, and tRNA quarters, as well as RNA size markers, are indicated.
T. thermophila cells are routinely grown in the laboratory at a favorable temperature of 30°C. Slight expression of heat shock factors is observed when growth temperatures are raised or lowered within the range of 25 or 37°C, but a more drastic cold-shock or heat-shock response ensues at a temperature of 15 or 40°C, respectively. We observed no tRNA fragment accumulation with even severe heat shock, but following severe cold shock tRNA halves did accumulate (Figure 5; additional time course data not shown). Of interest, cells at 40°C can divide, whereas starving cells, stationary-phase cells, and cold-shocked cells kept at 15°C showed little if any cell division. These findings suggest that tRNA halves are produced in response to cell growth arrest rather than as a more general response to stress.
Evidence for a bulk population of RNP-free tRNA halves
It remains unclear whether tRNA halves have a specific biological function or fortuitously accumulate as degradation intermediates, with tRNA turnover as the biologically significant process. To approach this question, because most noncoding RNAs function as RNP complexes (Hogg and Collins, 2008), we investigated tRNA fragment association with cellular proteins. To evaluate whether the tRNA halves are incorporated into RNPs, we analyzed cell extracts from starved cells (not shown) or cold-shocked cells (Figure 6) by glycerol gradient sedimentation. We were not able to detect tRNA halves in any fractions containing high–molecular weight complexes. Instead, the tRNA halves sedimented with free tRNAs in the gradient top fractions. As an important control, the ∼20-nt tRNA 3′-end fragments bound to Twi12 did demonstrate a sedimentation profile slightly shifted from free tRNA (Figure 6, bottom), consistent with their biological stabilization in the form of Twi12 RNP (Couvillion et al., 2010). These results suggest that the bulk population of tRNA halves is present as free RNA rather than RNP.
FIGURE 6:
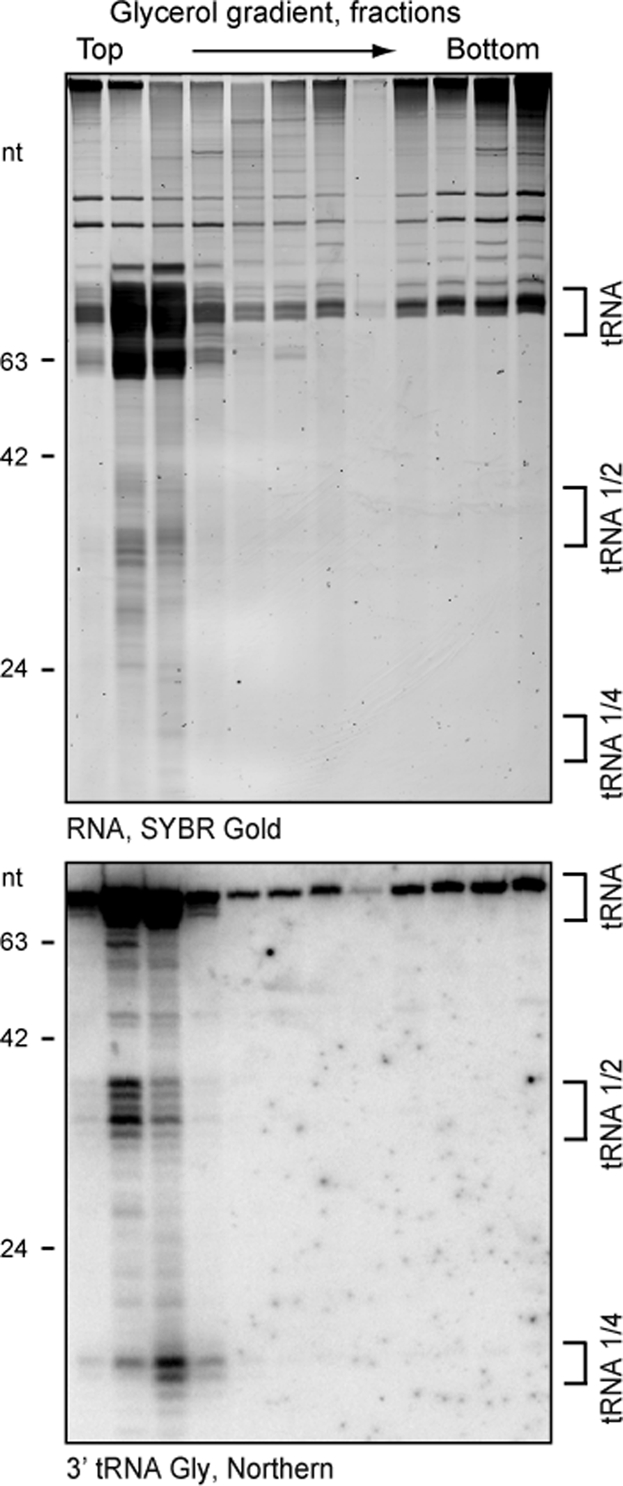
Evidence for an RNP-free state of tRNA halves in cell extract. RNA was harvested from every second fraction of a glycerol gradient fractioning cell extract from cold-shocked cells. SYBR Gold–stained RNA (top) and Northern blot hybridization of the same RNA transferred to a membrane (bottom) probed with an oligonucleotide complementary to the tRNA Gly 3′ end are shown. Size markers and tRNA species are indicated.
DISCUSSION
We identified eight RNase T2 homologues in T. thermophila, an expansion from the one or sometimes two encoded in animal and yeast genomes and comparable to the number of family members characterized in plant species. Subgroups of the plant RNase T2 proteins have distinct biological functions, which could be the case for ciliate Rnt2 proteins as well. Biochemical assays have suggested at least three distinct Tetrahymena RNase T2 enzyme activities (Maouri and Georgatsos, 1987; McKee and Prescott, 1991). Our work supports the physiological function of at least three T. thermophila Rnt2 proteins: Rnt2 A, B, and C. For these three paralogues, each single-gene KO produced a cellular or molecular phenotype and each double-KO combination was strongly synthetic in phenotype. On the other hand, any biological function(s) of the three T. thermophila Rnt2 paralogues predicted to lack catalytic activity remain a mystery: even the triple-KO elimination of Rnt2 D, E, and F had no cellular or molecular phenotype that we could detect across the various assays described.
Formation of tRNA halves by cleavage(s) in the anticodon loop region may be a conserved outcome of eukaryotic cell exposure to certain stresses (Thompson and Parker, 2009b). In T. thermophila it was demonstrated that deprivation of even a single essential amino acid was sufficient to induce tRNA half accumulation (Lee and Collins, 2005). We expanded the previous study by showing that stationary-phase and cold-shocked but not heat-shocked cells accumulate tRNA halves, with induction of tRNA fragment accumulation correlated with conditions that arrest cell growth and division. However, because cycloheximide treatment abolished the starvation-induced formation of tRNA halves (Lee and Collins, 2005), growth arrest is not sufficient to induce tRNA halves in the absence of translation. Synthesis of Rnt2 A and Rnt2 C could account at least in part for this translation requirement, as their mRNA levels increase after the onset of starvation.
Of note, no single Rnt2 KO abolished tRNA half accumulation. Rnt2 C KO did affect the size of the accumulating tRNA fragments. In addition, KO combinations of Rnt2 A, B, and C produced tRNA fragments with slower gel mobility and much greater accumulation than the wild-type-sized tRNA halves. Double-KO-strain phenotypes indicate that Rnt2 A, B, and C are at least partly redundant in biological function. Nonetheless, because each Rnt2 double KO had a distinct pattern of tRNA fragment accumulation, the substrate specificities of each enzyme may differ. Technical challenges in the complete macronuclear locus assortment of a third selectable marker make the construction of a triple-targeted KO strain beyond the scope of the present study. We speculate that a strain with quintuple KO of all five T. thermophila Rnt2 paralogues predicted to be catalytically active would show decreased rather than increased accumulation of tRNA halves, due to the lack of any starvation-induced tRNA anticodon-region endonucleolytic cleavage activity.
Zebrafish and plant RNase T2 enzymes have been proposed to function within an intracellular compartment, acting selectively on rRNAs targeted for ribophagy (Haud et al., 2011; Hillwig et al., 2011). Yeast Rny1p could also have this role, since ribophagy in yeast degrades ribosomes targeted to the vacuole (Kraft et al., 2008). T. thermophila Rnt2 enzymes could serve this role as well, based on the observed increase in accumulation of 5S and 5.8S rRNA fragments in Rnt2 double-KO strains. As one model, tRNAs could be degraded as part of the ribophagy pathway. Alternatively, because multiple autophagy pathways have been distinguished in eukaryotic cells (Beau et al., 2008), a tRNA-specific recycling pathway could operate in parallel to ribophagy. As a further alternative, because yeast Rny1p is proposed to cleave tRNAs in the cytoplasm after stress-enabled escape from the vacuole (Thompson and Parker, 2009a), T. thermophila tRNA halves and/or rRNA fragments could be generated without their encapsulation within a membrane-bound compartment. We favor the hypothesis that tRNA turnover occurs subsequent to targeting of these RNAs, or tRNA-containing RNP complexes, for destruction by autophagy. Cytological studies decades ago described the starvation-induced appearance of Tetrahymena autophagosomes with kinetics that can account for the appearance of tRNA halves (Nilsson, 1984; Lee and Collins, 2005). Combining these results and others described here, it seems likely that T. thermophila Rnt2 enzymes play a role in rRNA and tRNA turnover by acting in autophagy pathway(s) that preferentially recycle idle protein translation machinery.
MATERIALS AND METHODS
Sequence analysis
The search for angiogenin and RNase T2 homologues in the T. thermophila macronuclear genome was done using the Tetrahymena Genome Database Wiki. Sequence searching was done by BLASTP with human angiogenin (AAA51678.1), human RNASET2 (NP_003721.2), yeast Rny1p (NP_015202.1), and zebrafish Dre1a and Dre2 (ACK38069.1, ACK38071.1). Isoelectric point and molecular mass were determined using ExPASy proteomic tools. Signal peptides were predicted using SignalP (Bendtsen et al., 2004) with default settings. Multiple alignment was carried out using T-Coffee (Notredame et al., 2000) with default settings.
Strains and cell culture
Clonal KO cell lines were made in strain MZZFT12L, which has a transgene-encoded, epitope-tagged Twi12 protein under expression control of the MTT1 promoter at the BTU1 locus in CU522 strain background. Cells were grown shaking at 30°C in NEFF (0.25% proteose peptone, 0.25% yeast extract, 0.5% glucose, 30 μM FeCl3), SUPP (2% Proteose Peptone, 0.2% yeast extract, 0.2% glucose, 30 μM FeCl3), or defined synthetic complete medium (Szablewski et al., 1991) and starved by washing cells twice before resuspending in 10 mM Tris (pH 7.5). For heat shock and cold shock, cells were harvested, resuspended in an equal volume of preheated/precooled NEFF medium and incubated at the desired temperature. For stationary phase, cells were cultured 16–18 h after reaching maximum density ([1–3] × 106 cells/ml).
Gene disruption constructs and transformation
Strains with single-locus disruptions of Rnt2 A, B, or C or the triple KO of Rnt2 D, E, and F were made by targeted replacement of the RNT2 ORFs by the neo2 paromomycin resistance cassette (Gaertig et al., 1994). Similarly, locus disruptions to eliminate Rnt2 G or H or the second gene KO to generate Rnt2 AC, BA, and BC double-KO strains were obtained by targeted replacement of RNT2 ORFs with the bsr2 blasticidin S resistance cassette (Brown et al., 1999; Mary Couvillion, University of California, Berkeley, unpublished data). PCR primers to amplify and clone genomic sequences are listed in Supplemental Table S1. Transformed cells were selected for maximal locus assortment to the recombinant chromosome using paromomycin or blasticidin S. Cells were then isolated to obtain clonal cell lines, and KD or KO was determined by Southern blot hybridization as follows: 2 ml of cell culture was harvested and the cell pellet lysed by resuspending in 250 μl of lysis buffer (10 mM Tris-HCl, pH 9.5, 0.5 M EDTA, 1% SDS). Then 500 μl of water was added, and the lysed cells were incubated at 60°C for 2–4 h before cooling to room temperature and proteinase K treatment (50 μg/ml) at 37°C overnight. Following phenol:chloroform:isoamyl alcohol 25:24:1 (PCI) extraction and isopropanol precipitation, the genomic DNA was RNase A treated (100 μg/ml) for 2 h at 37°C, reextracted, and precipitated. The DNA was subsequently digested with an appropriate restriction enzyme to resolve wild-type from KO chromosomes (Figures 3A and 4A), PCI extracted, and precipitated. About 5–10 μg DNA was resolved on a 0.7% agarose gel and transferred to a Hybond-N+ membrane (GE Healthcare, Piscataway, NJ). The membrane was hybridized with a 32P-labeled probe made by random priming with hexamers using either the 5′ or 3′ flanking genomic region of the KO construct as template.
RNA purification, fractionation, and detection
Total RNA was isolated from cells using TRIzol (Invitrogen, Carlsbad, CA). SYBR Gold (Molecular Probes, Invitrogen) was used to visualize RNA following separation by denaturing 12–15% acrylamide–urea 0.6× Tris borate–EDTA gels. To detect specific RNAs by Northern blot hybridization, RNA was transferred to Hybond N+ membranes and hybridized to an appropriate 5′ end–radiolabeled DNA oligonucleotide in Denhardt's hybridization buffer (4× Denhardt's, 6× standard saline citrate, 0.1% SDS). For tRNA detection, complementary 20- to 22-nt oligonucleotides with an endpoint at the mature tRNA 5′ or 3′ end (excluding the 3′ CCA) were used (oligonucleotides are listed in Supplemental Table S1). For cell extract sedimentation through an 11-ml 15–45% glycerol gradient, cells were lysed by rotating for 10 min at 4°C in 20 mM Tris (pH 7.5), 5 mM MgCl2, 200 mM NaCl, and 10% glycerol supplemented with 0.2% NP40, 0.1% Triton X-100, 0.1 mM phenylmethylsulfonyl fluoride, and 10 mM β-mercaptoethanol. Cell lysate was cleared by centrifuging at 14,000 × g for 10 min and loaded onto the gradient. Sedimentation was done for 16–18 h at 197,000 × g at 4°C. RNA from fractions was purified by PCI, precipitated, and resolved by denaturing acrylamide gel.
Microscopy
For live-cell microscopy, cells from 1 ml of culture were washed once in 10 mM Tris (pH 7.5), resuspended in a small volume, and mounted in 2% low-melt agarose in 10 mM Tris (pH 7.5) for immobilization. For DAPI staining, cells were fixed for 1 h at room temperature in 2% paraformaldehyde in PHEM buffer (60 mM 1,4-piperazinediethanesulfonic acid, pH 6.9, 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 10 mM ethylene glycol tetraacetic acid [EGTA], 2 mM MgCl2), washed in modified phosphate-buffered saline (PBS; 130 mM NaCl, 2 mM KCl, 8 mM Na2HPO4, 2 mM KH2PO4, 10 mM EGTA, 2 mM MgCl2, pH 7.2), and stained with DAPI (0.1 μg/ml final concentration) for 10 min at room temperature in modified PBS. Residual DAPI was removed by washing in modified PBS for 5 min at room temperature. Cells were mounted in 90% glycerol.
Supplementary Material
Acknowledgments
We thank Mary Couvillion for supplying the epitope-tagged Twi12 strain and the bsr2 cassette and George Katibah and Mary Couvillion for discussions regarding tRNA fragments and ribonucleases. K.L.A. was partly funded by a gift from Agilent Technologies Foundation.
Abbreviations used:
- KD
knockdown
- KO
knockout
- nt
nucleotide
- ORF
open reading frame
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-08-0689) on November 2, 2011.
REFERENCES
- Acquati F, et al. Tumor and metastasis suppression by the human RNASET2 gene. Int J Oncol. 2005;26:1159–1168. [PubMed] [Google Scholar]
- Beau I, Esclatine A, Codogno P. Lost to translation: when autophagy targets mature ribosomes. Trends Cell Biol. 2008;18:311–314. doi: 10.1016/j.tcb.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von HG, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Brown JM, Marsala C, Kosoy R, Gaertig J. Kinesin-II is preferentially targeted to assembling cilia and is required for ciliogenesis and normal cytokinesis in Tetrahymena. Mol Biol Cell. 1999;10:3081–3096. doi: 10.1091/mbc.10.10.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvillion MT, Lee SR, Hogstad B, Malone CD, Tonkin LA, Sachidanandam R, Hannon GJ, Collins K. Sequence, biogenesis, and function of diverse small RNA classes bound to the Piwi family proteins of Tetrahymena thermophila. Genes Dev. 2009;23:2016–2032. doi: 10.1101/gad.1821209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvillion MT, Sachidanandam R, Collins K. A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. Genes Dev. 2010;24:2742–2747. doi: 10.1101/gad.1996210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande RA, Shankar V. Ribonucleases from T2 family. Crit Rev Microbiol. 2002;28:79–122. doi: 10.1080/1040-840291046704. [DOI] [PubMed] [Google Scholar]
- Eisen JA, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583:437–442. doi: 10.1016/j.febslet.2008.12.043. [DOI] [PubMed] [Google Scholar]
- Gaertig J, Gu L, Hai B, Gorovsky MA. High frequency vector-mediated transformation and gene replacement in Tetrahymena. Nucleic Acids Res. 1994;22:5391–5398. doi: 10.1093/nar/22.24.5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertig J, Kapler G. Transient and stable DNA transformation of Tetrahymena thermophila by electroporation. Methods Cell Biol. 2000;62:485–500. doi: 10.1016/s0091-679x(08)61552-6. [DOI] [PubMed] [Google Scholar]
- Haud N, et al. rnaset2 mutant zebrafish model familial cystic leukoencephalopathy and reveal a role for RNase T2 in degrading ribosomal RNA. Proc Natl Acad Sci USA. 2011;108:1099–1103. doi: 10.1073/pnas.1009811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneke M, et al. RNASET2-deficient cystic leukoencephalopathy resembles congenital cytomegalovirus brain infection. Nat Genet. 2009;41:773–775. doi: 10.1038/ng.398. [DOI] [PubMed] [Google Scholar]
- Hillwig MS, Contento AL, Meyer A, Ebany D, Bassham DC, MacIntosh GC. RNS2, a conserved member of the RNase T2 family, is necessary for ribosomal RNA decay in plants. Proc Natl Acad Sci USA. 2011;108:1093–1098. doi: 10.1073/pnas.1009809108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg JR, Collins K. Structured non-coding RNAs and the RNP renaissance. Curr Opin Chem Biol. 2008;12:684–689. doi: 10.1016/j.cbpa.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie M. Structure-function relationships of acid ribonucleases: lysosomal, vacuolar, and periplasmic enzymes. Pharmacol Ther. 1999;81:77–89. doi: 10.1016/s0163-7258(98)00035-7. [DOI] [PubMed] [Google Scholar]
- Jochl C, Rederstorff M, Hertel J, Stadler PF, Hofacker IL, Schrettl M, Haas H, Huttenhofer A. Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic Acids Res. 2008;36:2677–2689. doi: 10.1093/nar/gkn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- Lee SR, Collins K. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J Biol Chem. 2005;280:42744–42749. doi: 10.1074/jbc.M510356200. [DOI] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Luo J, Zhou H, Liao JY, Ma LM, Chen YQ, Qu LH. Stress-induced tRNA-derived RNAs: a novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res. 2008;36:6048–6055. doi: 10.1093/nar/gkn596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhtala N, Parker R. T2 Family ribonucleases: ancient enzymes with diverse roles. Trends Biochem Sci. 2010;35:253–259. doi: 10.1016/j.tibs.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntosh GC, Hillwig MS, Meyer A, Flagel L. RNase T2 genes from rice and the evolution of secretory ribonucleases in plants. Mol Genet Genomics. 2010;283:381–396. doi: 10.1007/s00438-010-0524-9. [DOI] [PubMed] [Google Scholar]
- Madinger CL, Collins K, Fields LG, Taron CH, Benner JS. Constitutive secretion in Tetrahymena thermophila. Eukaryot Cell. 2010;9:674–681. doi: 10.1128/EC.00024-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maouri A, Georgatsos JG. Specificity and other properties of three ribonucleases of Tetrahymena pyriformis. Eur J Biochem. 1987;168:523–528. doi: 10.1111/j.1432-1033.1987.tb13449.x. [DOI] [PubMed] [Google Scholar]
- McKee T, Prescott DJ. Characterization of the extracellular ribonuclease of Tetrahymena pyriformis. W. J Protozool. 1991;38:465–471. doi: 10.1111/j.1550-7408.1991.tb04818.x. [DOI] [PubMed] [Google Scholar]
- Miao W, Xiong J, Bowen J, Wang W, Liu Y, Braguinets O, Grigull J, Pearlman RE, Orias E, Gorovsky MA. Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. PLoS One. 2009;4:e4429. doi: 10.1371/journal.pone.0004429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson JT. On starvation-induced autophagy in Tetrahymena. Carlsberg Res Commun. 1984;49:323–340. [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Pederson T. Regulatory RNAs derived from transfer RNA? RNA. 2010;16:1865–1869. doi: 10.1261/rna.2266510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnoff P, Roiz L, Angelkovitch B, Schwartz B, Shoseyov O. A recombinant human RNASET2 glycoprotein with antitumorigenic and antiangiogenic characteristics: expression, purification, and characterization. Cancer. 2006;107:2760–2769. doi: 10.1002/cncr.22327. [DOI] [PubMed] [Google Scholar]
- Szablewski L, Andreasen PH, Tiedtke A, Florin-Christensen J, Florin-Christensen M, Rasmussen L. Tetrahymena thermophila: growth in synthetic nutrient medium in the presence and absence of glucose. J Eukaryot Microbiol. 1991;38:62–65. [Google Scholar]
- Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Parker R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol. 2009a;185:43–50. doi: 10.1083/jcb.200811119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell. 2009b;138:215–219. doi: 10.1016/j.cell.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Sun L, Kragler F. The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol. 2009;150:378–387. doi: 10.1104/pp.108.134767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.