Sequestration of Cdc14 from the cytoplasm ensures Chs2 ER retention after MEN activation. The interdependence of chromosome segregation, MEN activation, decrease in mitotic CDK activity, and Cdc14 dispersal provides an effective mechanism for cells to order late mitotic events.
Abstract
Cytokinesis, which leads to the physical separation of two dividing cells, is normally restrained until after nuclear division. In Saccharomyces cerevisiae, chitin synthase 2 (Chs2), which lays down the primary septum at the mother–daughter neck, also ensures proper actomyosin ring constriction during cytokinesis. During the metaphase-to-anaphase transition, phosphorylation of Chs2 by the mitotic cyclin-dependent kinase (Cdk1) retains Chs2 at the endoplasmic reticulum (ER), thereby preventing its translocation to the neck. Upon Cdk1 inactivation at the end of mitosis, Chs2 is exported from the ER and targeted to the neck. The mechanism for triggering Chs2 ER export thus far is unknown. We show here that Chs2 ER export requires the direct reversal of the inhibitory Cdk1 phosphorylation sites by Cdc14 phosphatase, the ultimate effector of the mitotic exit network (MEN). We further show that only Cdc14 liberated by the MEN after completion of chromosome segregation, and not Cdc14 released in early anaphase by the Cdc fourteen early anaphase release pathway, triggers Chs2 ER exit. Presumably, the reduced Cdk1 activity in late mitosis further favors dephosphorylation of Chs2 by Cdc14. Thus, by requiring declining Cdk1 activity and Cdc14 nuclear release for Chs2 ER export, cells ensure that septum formation is contingent upon chromosome separation and exit from mitosis.
INTRODUCTION
Cytokinesis, a key event at the end of mitosis in the budding yeast, is the physical separation of mother and daughter cells through the deposition of a septum as the actomyosin ring undergoes constriction. Actomyosin ring constriction and septum formation are interdependent (Schmidt et al., 2002; VerPlank and Li, 2005) and require the proper assembly of various neck components and the actomyosin ring throughout the cell division cycle (Balasubramanian et al., 2004), as well as the timely arrival of Chs2 to the mother–daughter neck at the end of mitosis (VerPlank and Li, 2005; Zhang et al., 2006; Teh et al., 2009). More important, mechanisms must exist in dividing cells to coordinate sister chromatid segregation, mitotic exit, and targeted secretion so as to ensure the execution of cytokinesis and septation only after nuclear division.
Chitin synthase 2 (Chs2) lays down the septum at the mother–daughter neck (Walther and Wendland, 2003; Cabib, 2004) as the plasma membrane is pulled inward during the constriction of the actomyosin ring in cytokinesis (Schmidt et al., 2002; VerPlank and Li, 2005). CHS2 is expressed in metaphase (Pammer et al., 1992; Choi et al., 1994; Spellman et al., 1998) but is retained in the endoplasmic reticulum (ER; Zhang et al., 2006) by the phosphorylation of a cluster of N-terminal Ser-Pro sites by the yeast cyclin-dependent kinase (Cdk1) Cdc28 (Martinez-Rucobo et al., 2009; Teh et al., 2009). Only after sister chromatid segregation, when the destruction of the mitotic cyclins such as Clb2 leads to loss of mitotic Cdk1 activity, is Chs2 exported from the ER and targeted to the neck, where it deposits the primary septum (VerPlank and Li, 2005; Zhang et al., 2006; Meitinger et al., 2010).
The destruction of mitotic cyclins, resulting in the termination of Cdk1 activity, is known as mitotic exit. Mitotic exit depends upon the ubiquitin-mediated destruction of the mitotic cyclins by the E3 ligase known as the anaphase-promoting complex/cyclosome (APC/C). The activation of the APC/C requires the activator proteins Cdc20 and Cdh1. Cdc20 acts from metaphase to anaphase to cause the ubiquitination of Clb2, whereas Cdh1 is functional midway in anaphase through G1 to ensure the complete destruction of Clb2 (reviewed in Sullivan and Morgan, 2007). A pathway known as the mitotic exit network (MEN) is needed to activate Cdh1. The MEN consists of Tem1 (a GTPase), Lte1 (a GTP/GDP exchange factor), Cdc15, Cdc5, Dbf2, and Dbf20 (Ser/Thr kinases), Mob1, and Cdc14 (a phosphatase; Jaspersen et al., 1998; Morgan, 1999; Lee et al., 2001). The primary function of the MEN is to prevent spindle disassembly, mitotic exit, and cytokinesis from occurring until nuclear division has been properly executed, thereby ensuring the equivalent distribution of chromosomes to the mother and daughter cells (Fraschini et al., 2008).
The MEN cascade ends with Cdc14, a dual-specificity phosphatase that has been genetically and biochemically implicated in both the inactivation of Cdk1 and the reversal of Cdk1-catalyzed phosphorylation events (Visintin et al., 1998). Cdc14 is sequestered in the nucleolus for most of the cell cycle (reviewed in Stegmeier and Amon, 2004). During sister chromatid separation, Cdc14 is released into the nucleoplasm in response to a separase-dependent signaling pathway termed the Cdc fourteen early anaphase release (FEAR) network, where it helps regulate anaphase spindle function and segregation of rDNA loci (D'Amours and Amon, 2004). Cdc14 release in response to the FEAR pathway is transient, and Cdc14 is returned to the nucleolus shortly afterward. Subsequently, in telophase, Cdc14 is dispersed much more broadly from the nucleolus to the cytoplasm to promote complete Cdk1 inactivation, which leads to exit from mitosis.
Despite the essential role of Cdc14 in mitotic exit in budding yeast, regulation of cytokinesis may actually be one of the most highly conserved functions for the Cdc14 family in eukaryotes (Clifford et al., 2008). Surprisingly, substrates of Cdc14 involved in cytokinesis remain mostly unknown (Mocciaro and Schiebel, 2010). Although it is clear that Chs2 exits the ER only upon MEN activation and destruction of Cdk1 activity (VerPlank and Li, 2005; Zhang et al., 2006; Teh et al., 2009; Meitinger et al., 2010), the molecular mechanism by which its release from the ER is triggered has been unclear. Given the timing of Cdc14 cytoplasmic release and Chs2 neck localization, one likely possibility is that Chs2 is directly dephosphorylated by Cdc14, thus relieving the inhibitory phosphorylation that retains it in the ER. However, other possibilities exist as well, including dephosphorylation by other phosphatases after Cdk1 inactivation, phosphorylation by one of the MEN kinases, or other regulatory events initiated by, but downstream of, the MEN (Meitinger et al., 2010).
In this study, we show that the Cdc14 specifically liberated from the nucleolus to the cytoplasm after sister chromatid segregation and MEN activation promotes the Chs2 ER export by directly reversing Cdk1-catalyzed phosphorylation of the Chs2 N-terminus. Chs2 is therefore one of the first identified Cdc14 targets that is a cargo of the secretory pathway needed for septation and that also plays a role in cytokinesis. By coupling Chs2 ER export to Cdc14 cytoplasmic localization, cells ensure that septum formation during cytokinesis depends on MEN activation and mitotic exit, thereby preventing premature partitioning of the mother and daughter cells.
RESULTS
Chs2–green fluorescent protein (GFP) fails to exit from the ER in the MEN cdc14-3 mutant when mitotic Cdk1 is reduced
As part of the MEN, the primary functions of Cdc14 are promoting mitotic cyclin destruction by dephosphorylating Cdh1, and Cdk1 inhibition by dephosphorylating the Cdk1 inhibitor Sic1 and its transcription factor Swi5 (Visintin et al., 1998; Sullivan and Morgan, 2007). However, Cdc14 is believed to act more broadly by reversing phosphorylation on many other Cdk1 substrates (Stegmeier and Amon, 2004).
We examined the possible requirement for Cdc14 in Chs2 ER export independent of its role in promoting mitotic exit. To this end, we constructed a cdc14-3 CHS2-GFP GAL-sic1-NTΔ strain in which the overexpression of the truncated form of Sic1, sic1-NTΔ, allows the bypass of the mitotic exit defects in the cdc14-3 cells (Mendenhall and Hodge, 1998; Noton and Diffley, 2000).
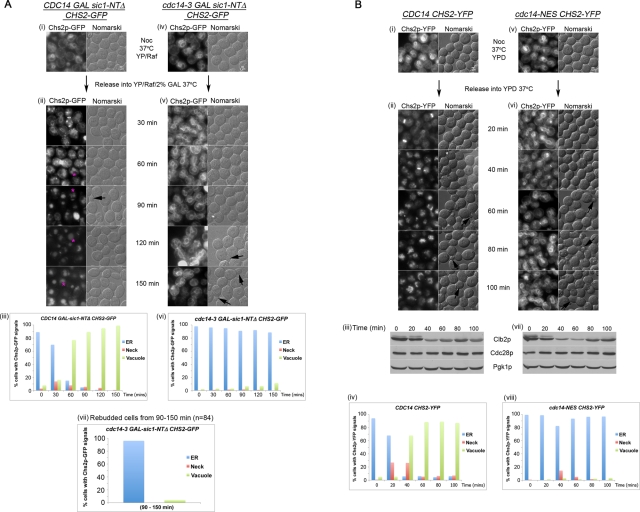
Of interest, in cdc14-3 CHS2-GFP GAL-sic1-NTΔ cells released from a nocodazole (Noc) arrest at the restrictive temperature (37°C) in galactose (Gal), Chs2-GFP signals failed to appear at the bud neck in a significant number of the cells (Figure 1A, v, 30 min onward). Instead, Chs2-GFP ER signals persisted in 89% (Figure 1A, vi) of the cells at the 150-min time point after Noc wash-off, when cells had exited from mitosis, as indicated by the rebudding of the cells (Figure 1A, v, 120–150 min, arrows). A count of the rebudded cdc14-3 cells revealed that 96% (Figure 1A, vii) of these cells retained Chs2-GFP in the ER, confirming a failure in Chs2 ER export.
FIGURE 1:
(A) Chs2-GFP fails to exit from the ER in cdc14-3 cells when Cdk1 activity is reduced. CDC14 (i) or cdc14-3 (iv) cells carrying CHS2-GFP GAL-SIC1-NTΔ were arrested at 24°C in YP/Raff + Noc for 4 h (i, iv). Cells were then shifted up to 37°C for 1 h, after which they were washed into YP/Raff/2% Gal at 37°C (ii, v). (iii, vi) Plots of CDC14 and cdc14-3 cells at 30-min intervals showing Chs2-GFP ER signals or Chs2-GFP neck and spot signals, indicating endocytosed Chs2-GFP (pink asterisks) (ii). Based on the timing and size, the smaller, intense spots observed are likely to be Chs2-YFP traversing through Golgi compartments (Teh et al., 2009), whereas the larger spots (pink asterisks) indicate Chs2-GFP internalized from the neck and transported to endosomes/vacuoles (Supplemental Figure S1). One hundred cells were counted per time point. ER counts refer to clearly visible ER signals. For neck counts, any cells with Chs2 at the neck were classified as having neck signals. Endosomal/vacuolar counts refer to cells with signals in the cell body without ER or neck signals. (vii) Plots show percentages of Chs2-GFP localization in rebudded cdc14-3 cells from the 90- to 150-min time points. (B) Chs2-YFP exit from the ER is defective in cdc14 nuclear export mutant. (i) CDC14 CHS2-YFP and (v) cdc14-NES CHS2-YFP cells were arrested at metaphase in YPD containing Noc at 24°C. After 4.75 h, the cells were shifted to 37°C for 15 min (ii, vi) before they were released from Noc into YPD at 37°C. Western blots show cells releasing from Noc arrest (iii, vii). (iv, viii) Chs2-YFP signals were enumerated as in A, iii and vi.
CDC14 CHS2-GFP GAL-sic1-NTΔ cells treated similarly showed relatively quick exit of Chs2-GFP from the ER (Figure 1A, ii, 60–150 min), as evident from the complete loss of ER signals in the CDC14 cells by the 150-min time point after release from Noc. Because Chs2p neck localization was transient due to its endocytosis upon cytokinesis (Roh et al., 2002; Teh et al., 2009), we observed only a peak of ∼14% of the cells with neck signals (Figure 1A, iii).
However, the decrease in ER signals indicating transport of Chs2-GFP to the neck in 99% of the cells (Figure 1A, iii, 150 min) was accompanied by the appearance of Chs2-GFP signals in the endosomes/vacuole (Bryant and Stevens, 1998; Figure 1A, ii, 60–150 min, pink asterisks) due to endocytosis of Chs2 (Chuang and Schekman, 1996) during cytokinesis (Roh et al., 2002; Teh et al., 2009). Indeed, these endosomal/vacuolar signals were specifically absent from the endocytosis mutant sla2Δ when cells exited mitosis (Supplemental Figure S1). The endosomal/vacuolar signals therefore served as an indication of the passage of Chs2 from the ER to the neck, where Chs2 is endocytosed and transported to the vacuole. Our data imply that Cdc14 is needed for Chs2 ER export in addition to its role in promoting Cdk1 inactivation.
Chs2–yellow fluorescent protein (YFP) exit from the ER is defective in cdc14 nuclear export mutant
We also examined Chs2 trafficking in a temperature-sensitive cdc14 nuclear export mutant (referred to as cdc14-NES) that fails to export Cdc14-NES to the cytoplasm normally. These cells however, are able to trigger the destruction of the mitotic cyclin Clb2 and enter the subsequent G1 (Bembenek et al., 2005).
In CDC14 CHS2-YFP cells released from Noc at 37°C, Chs2-YFP exited the ER with an increase in fluorescence signals at the neck and vacuole (Figure 1B, ii, 20 min onward). After Noc release, Chs2-YFP neck signals were seen in 27% of the cells at 30 min and 26% at 60 min (Figure 1B, iv). Moreover, after 100 min, 87% of the cells showing vacuolar signals could be observed with a corresponding loss of ER signals.
In the cdc14-NES CHS2-YFP cells, although 23% of the total cells (Figure 1B, viii) showed detectable Chs2-YFP neck signals (Figure 1B, vi, 20–100 min), these cells generally still contained visible ER signals, indicating inefficient ER export. Furthermore, vacuolar signals were seen in only 11% of total cells, implying that Chs2-YFP failed to be exported from the ER to the neck. Consistent with this, at least 82% of the cdc14-NES cells exhibited Chs2-YFP ER signals at every time point, confirming a failure in Chs2-YFP ER export.
The cdc14-NES cells showed a decrease in mitotic cyclin Clb2 levels comparable to the wild-type cells (Figure 1B, iii and vii), indicating that the failure of Chs2-YFP ER exit was not due to a mitotic exit defect. Similar to CDC14 cells (Figure 1B, ii, 60–100 min, arrows), the cdc14-NES cells were also able enter the next G1, as seen by rebudding (Figure 1B, vi, 60–100 min, arrows). The defect in Chs2 ER export in the mutant was therefore likely due to the altered dynamics of nuclear export of the Cdc14-NES mutant at the end of mitosis (Bembenek et al., 2005). When Cdc14 function was restored in the cdc14-NES strain by overexpressing CDC14 under the GAL promoter, the defect in Chs2p ER export was rescued (data not shown). Collectively, the data confirmed that Chs2 exit out of the ER depends upon Cdc14.
Failure to export Chs2 from the ER in the cdc14-NES mutant can be bypassed by abolishing the Cdk1 sites on Chs2
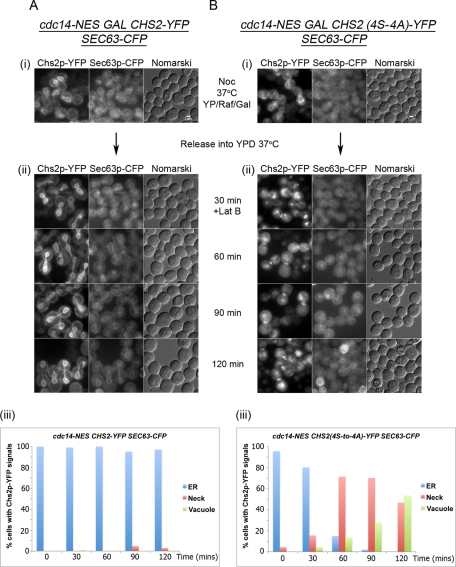
To test whether the defective Chs2 ER export was due to a failure in Chs2 dephosphorylation in the cdc14 mutants (Figure 1), we observed the localization of the phosphorylation-deficient Chs2(4S-to-4A)-YFP mutant (Teh et al., 2009) in the cdc14-NES mutant. The Chs2(4S-to-4A)-YFP constitutively exits the ER in metaphase cells due to the loss of four consensus Cdk1 sites at the N-terminus (Teh et al., 2009). Because of the rapid dynamics of Chs2 endocytosis from the neck as cells were released from metaphase (Roh et al., 2002; Teh et al., 2009), we enriched for the neck signals using the actin inhibitor latrunculin (Lat) B, a variant of Lat A (Ayscough et al., 1997), to slow down internalization of Chs2 in the cells, given that endocytosis is actin dependent (Moseley and Goode, 2006). Lat B was used to retard endocytosis of Chs2-FP, as sla2Δ in the cdc14-NES strain reduced the fitness of the cells in our hands (data not shown).
Wild-type Chs2-YFP, when induced in the cdc14-NES GAL-CHS2-YFP SEC63-CFP strain, was retained in the ER for up to 120 min following release from metaphase at 37°C (Figure 2A, ii) in almost 100% of the cells (Figure 2A, iii), as judged by the ER marker Sec63-CFP, consistent with the foregoing observations (Figure 1B). Overall, 8% of the cells showed Chs2-YFP neck signals, whereas vacuolar signals were mostly absent, confirming a defect in ER export of Chs2-YFP.
FIGURE 2:
Failure to export Chs2 from the ER in the cdc14-NES mutant can be bypassed by abolishing the Cdk1 sites on Chs2. cdc14-NES SEC63-CFP containing GAL-CHS2-YFP (A) or GAL-CHS2(4S-to-A)-YFP (B) cells was arrested in YP/Raff with Noc for 5 h, following which CHS2 was induced with 2% Gal for 45 min. The cells were then shifted to 37°C for 15 min before release into YPD prewarmed at 37°C (ii). At 30 min after release into YPD, 32 μM Lat B was added into both cultures and cells further incubated at 37°C for an additional 90 min. (A, iii; B, iii) Chs2-YFP signals were enumerated as in Figure 1.
In contrast, Chs2(4S-to4A)-YFP in cdc14-NES GAL-CHS2(4S-to-4A)-YFP SEC63-CFP cells exited the ER relatively quickly under the same conditions, with the depletion of Chs2(4S-to-4A)-YFP ER signals (Figure 2B, ii, 60–120 min) occurring in 100% of the cells by 120 min after Noc release (Figure 2B, iii). Chs2(4S-to-4A)-YFP neck signals were found in ∼70% of the cells at the 60- and 90-min time points due to the reduced rate of endocytosis in the presence of Lat B. In addition, because of the slower internalization, the percentages of cells showing vacuolar signals were correspondingly lower.
Taken together, our data imply that the failure of Chs2-YFP ER export in the cdc14 mutants (Figures 1 and 2) is due to a defect in the dephosphorylation of Chs2 at the Cdk1 sites when cells exit from mitosis. Because Chs2 depends upon the secretory pathway for trafficking to the neck (Chuang and Schekman, 1996; Zhang et al., 2006), the results with the cdc14-NES mutant also excluded the possibility that the defective Chs2-YFP ER export in the absence of Cdc14 function and mitotic exit is due to a general shutdown of the secretory pathway.
Chs2 is a novel substrate of Cdc14
Several in vivo Cdk1 phosphorylation sites near the Chs2 N-terminus have been directly identified in mass spectrometry (MS)–based phosphoproteomic studies (Smolka et al., 2007; Holt et al., 2009; Bodenmiller et al., 2010). However, many Cdk1 substrates are dephosphorylated by phosphatases prior to mitotic exit (Sullivan and Morgan, 2007), and the phosphorylation status of Chs2 at mitotic exit has not been directly examined. Considering the results from Figures 1 and 2 and the fact that Cdc14 shows specificity for Cdk1 substrates (Stegmeier and Amon, 2004), we tested whether Chs2 phosphorylation status at mitotic exit is directly controlled by Cdc14 and whether phosphorylated Chs2 is an efficient Cdc14 substrate.
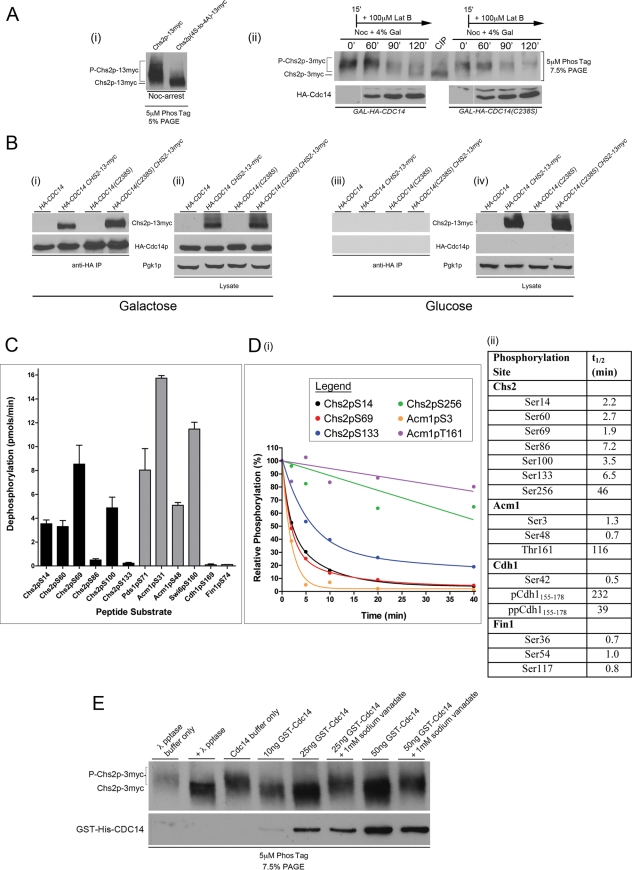
First, we examined whether Chs2 remained heavily phosphorylated on its N-terminal Cdk1 sites in MEN mutants that fail to activate Cdc14. We arrested cdc15-2 CHS2-13myc cells at 37°C, isolated Chs2-13myc using myc antibody-coupled beads, and analyzed its phosphorylation status by tandem MS. We detected phosphorylation on Cdk1 consensus sites at Ser-14, Ser-69, Ser-86, Ser-100, and Ser-133 (Supplemental Figure S2). The average stoichiometry for these sites was 35% based on simple comparison of extracted ion signals for the unmodified and phosphorylated forms of each peptide (data not shown). Thus Chs2 remains heavily phosphorylated on Cdc28 phosphorylation sites in late anaphase, immediately prior to Cdc14 activation by the MEN.
We next tested whether the overexpression of CDC14 in metaphase cells results in Chs2 dephosphorylation. Using the Phos-tag gel system (Kinoshita et al., 2009), we first established the mobility patterns of Chs2-13myc or Chs2(4S-to4A)-13myc immunoprecipitated from Noc-arrested cells. The ER-retained and phosphorylated Chs2-13myc migrated more slowly than the phosphorylation-deficient Chs2(4S-to-4A)-13myc mutant that is constitutively exported from the ER (Teh et al., 2009; Figure 3A, i).
FIGURE 3:
Chs2 is a novel substrate of Cdc14. (Ai) GAL-CHS2-13myc and GAL-CHS2(4S-to-4A)-13myc cells were arrested in YPD + Noc and anti-myc IPs performed. Immunoprecipitates were resolved on 5% SDS–PAGE + 5 μM Phos-tag, and anti-myc antibodies were used for detection of Chs2-13myc and its phosphorylated forms (denoted with a P). (A, ii) GAL-HA-CDC14 CHS2-3myc and GAL-HA-CDC14(C283S) CHS2-3myc cells were released from α-factor into YP/Raff + Noc. On Noc-arrest (0′), 4% Gal was added to the cultures. At 15 min after Noc was introduced, 100 μM Lat B was added to both cultures. Samples harvested at indicated time points were lysed and anti-myc IPs performed. As a control for the migration of dephosphorylated Chs2-3myc, 10 U of calf intestinal phosphatase (CIP) was incubated with an IP performed on an additional sample harvested from the 0′ time point. CIP treatment was for 1 h at 37°C. Immunoprecipitates were resolved on 7.5% SDS–PAGE + 5 μM Phos-tag, and anti-myc antibodies were used for detection of Chs2-3myc and its phosphorylated forms (top). HA-Cdc14 and HA-Cdc14(C283S) were detected in Western blot analysis of lysates using anti-HA antibodies (bottom). (B) GAL-HA-CDC14 CHS2-13myc and GAL-HA-CDC14(C283S) CHS2-13myc cells were arrested in YP/Raff + Noc for 5 h. Gal 2% was added to half of the culture (i, ii) and 2% Glu to the other half (iii, iv). Cells were harvested and lysed, and Co-IPs were performed using anti-HA beads, followed by Western blot analysis of Chs2-13myc and HA-Cdc14 (i, iii). As controls, 80 μg of total lysates was analyzed directly on Western blots (ii, iv). (C) Rates of dephosphorylation of all indicated phosphopeptide substrates by Cdc14 were determined at a single substrate concentration of 200 μM. Data are averages of three trials with SDs and are normalized for Cdc14 concentration (see Materials and Methods for peptide sequences). (D) Representative plots of phosphopeptide abundance from recombinant protein substrates during Cdc14 phosphatase reactions, monitored by label-free MS as described in Materials and Methods (i). Data were fitted with exponential decay equations or by linear regression using GraphPad Prism and are averages from two independent experiments. To compare relative rates of Cdc14-catalyzed dephosphorylation at individual Cdk1 phosphorylation sites on intact protein substrates, half-life values (t1/2) were determined from the fit lines of phosphorylation decay plots (ii). Smaller t1/2 values indicate faster dephosphorylation rates. The Cdh1155-178 peptide has four potential Cdk1 phosphorylation sites (data not shown), and the overall dephosphorylation rates for the singly phosphorylated (p) and doubly phosphorylated (pp) species are shown. (E) Chs2-3myc immunoprecipitated from TCA lysates prepared from Noc-arrested cells was treated with phosphatase buffer (lane 3) or the indicated amount of affinity-purified GST-6His-Cdc14 at 30°C for 60 min (lanes 4–8). As a positive control, λ protein phosphatase was used (lane 2).
In vivo Cdc14 phosphatase assays were then performed using GAL-HA-CDC14 CHS2-3MYC and GAL-HA-CDC14(C283S) CHS2-3MYC cells released from G1 into YP/Raff/Noc. On metaphase arrest, expression of HA-CDC14 and HA-CDC14(C283S), an active-site mutant that lacks phosphatase activity but retains high-affinity interaction with substrates (Hall et al., 2008), was induced with Gal. Lat B (100 μM) was added after 15 min. In GAL-HA-CDC14 CHS2-3MYC cells, the phosphorylated form of Chs2-3myc at Noc-arrest (Figure 3A, ii) was converted to faster-migrating bands, consistent with dephosphorylation of Chs2. In the GAL-CDC14(C283S) CHS2-3MYC cells, however, the phosphorylated form of Chs2 persisted (Figure 3A, ii). The ectopic expression of CDC14 in metaphase therefore promotes the dephosphorylation of Chs2.
We also tested whether Cdc14 physically associates with Chs2. Coimmunoprecipitation (Co-IP) experiments were performed using GAL-HA-CDC14 CHS2-13myc and GAL-HA-CDC14(C283S) CHS2-13myc cells arrested with Noc to enrich for Chs2-13myc. Chs2-13myc coprecipitated with both hemagglutinin (HA)-Cdc14 and the substrate trap mutant HA-Cdc14(C283S) (Figure 3B, i). Western blot analysis of the lysates used for the Co-IPs showed similar expression levels of epitope-tagged proteins (Figure 3B, ii). Control experiments performed in parallel without Gal induction of the GAL-HA-CDC14 CHS2-13myc and GAL-HA-CDC14(C283S) CHS2-13myc cells indicated that the coprecipitated Chs2 was specific (Figure 3B, iii and iv).
To determine whether Cdc14 can efficiently dephosphorylate the N-terminal Cdk1 phosphorylation sites on Chs2, we compared dephosphorylation of Chs2 to other known Cdc14 substrates using synthetic phosphopeptides and in vitro phosphorylated recombinant proteins. Although Cdc14 is believed to possess general specificity for Cdk1-type phosphorylation sites (S/T-P; Gray et al., 2003; Stegmeier and Amon, 2004), it has become apparent that many Cdk1 substrates are poor Cdc14 substrates (Sullivan and Morgan, 2007; Bloom et al., 2011). We first compared the ability of purified recombinant Cdc14 to dephosphorylate phosphopeptide sequences derived from the Chs2 N-terminus and several known Cdc14 substrates. Four Chs2 phosphopeptides (containing S14, S60, S69, and S100) were dephosphorylated with rates comparable to phosphopeptides from Acm1 (including the most efficient in vitro Cdc14 peptide substrate we have identified, Acm1pS31) and Swi6pS160 and Pds1pS71 (representing two of the few mapped in vivo Cdk1 sites targeted by Cdc14; Geymonat et al., 2004; Holt et al., 2008). In contrast, other Cdk1 site–containing Chs2 phosphopeptides (S86 and S133) and other Cdk phosphorylation sequences from known Cdc14 substrates (Fin1pS74 and Cdh1pS169) were dephosphorylated, but at much lower rates (Figure 3C).
Using an MS assay based on label-free quantification that allows independent measurement of dephosphorylation at multiple phosphorylation sites, we further compared the Cdc14-catalyzed dephosphorylation of an intact recombinant N-terminal domain of Chs2, glutathione S-transferase (GST)–Chs2(1-300), to that of the known Cdc14 substrates GST-Acm1, GST-Cdh1(1-211), and GST-Fin1 (Figure 3D). Recombinant proteins were phosphorylated in vitro with purified Clb2-Cdc28. Protein substrate concentrations were kept constant, and the average starting phosphorylation stoichiometry was similar for all protein substrates (data not shown). Tandem MS analysis confirmed phosphorylation at all six N-terminal Ser-Pro phosphorylation sites, plus an additional Ser-Pro site at Ser-256 (not shown). All six N-terminal Chs2 phosphorylation sites were rapidly dephosphorylated by Cdc14 with rates only slightly slower than the best sites on Acm1, Cdh1, and Fin1 (Figure 3D, i and ii). In contrast, Chs2 pSer256 was a poor Cdc14 substrate, as were other Cdk1 sites on Acm1 and Cdh1.
To ascertain whether Cdc14 could directly dephosphorylate full-length endogenous Chs2 isolated from metaphase cells, in vitro phosphatase assays using recombinant purified GST-6His-Cdc14 and Chs2-3myc immunoprecipitated from TCA extracts of Noc-arrested cells were performed. The dephosphorylation of Chs2-3myc was apparent in the assay using only 10 ng (∼3.8 nM) of recombinant Cdc14, as judged by the increased electrophoretic mobility of Chs2-3myc (Figure 3E). The level of dephosphorylation by the GST-6His-Cdc14 was comparable to that using the general λ phosphatase. The phosphatase activity of the GST-6His-Cdc14 against Chs2-3myc was inhibited by the protein tyrosine phosphatase family inhibitor sodium vanadate, supporting the specificity of the activity.
Collectively, the data in Figure 3 show that Cdc14 associates with phosphorylated Chs2 in mitosis, promotes Chs2 dephosphorylation in vivo, and has an intrinsic capacity to dephosphorylate the Cdk1-phosphorylated Chs2 N-terminus with high efficiency, comparable to other well-characterized Cdc14 substrates. We conclude that Chs2 is a bona fide in vivo Cdc14 substrate and represents the first identified secretory cargo substrate of Cdc14 in any organism.
Cdc14 nucleolar release precedes Chs2 neck localization
Our observations thus far imply that Chs2 requires the nuclear export of Cdc14 into the cytoplasm (Figure 1B) to reverse the phosphorylation on its N-terminal Cdk1 sites (Figures 2 and 3) to promote its ER export. However, the relative timings of Cdc14 dispersal from the nucleolus and Chs2 ER export are unclear.
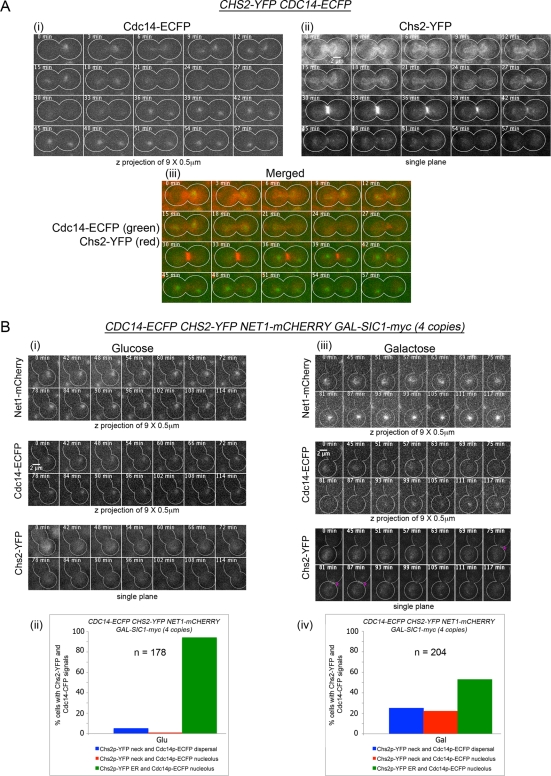
To establish the physiological relevance of our genetic and biochemical data showing Cdc14 dependence for Chs2 ER export, we compared the temporal localization of Cdc14 and Chs2 in CDC14-ECFP CHS2-YFP cells released from Noc arrest by time-lapsed microscopy (n = 48). Cdc14-enhanced cyan fluorescent protein (ECFP) in the nucleolus (Figure 4A, i, 0 min) was dispersed completely by 24–30 min in all cells prior to Chs2-YFP arrival at the neck (Figure 4A, ii, 30–42 min). The time interval between Cdc14-ECFP nucleolar export and Chs2-YFP neck localization was 7 ± 2 min, with Cdc14-ECFP dispersal to the cytoplasm preceding Chs2-YFP neck localization in all 48 cells observed. The duration of Cdc14 export to the cytoplasm likely allows sufficient time for Cdc14 to act on Chs2 as Cdk1 activity declines.
FIGURE 4:
(A) Cdc14 nucleolar release precedes Chs2 neck localization. CHS2-YFP CDC14-ECFP cells were arrested in YPD/Noc at 24°C. After 4 h, the cells were then released from Noc arrest and mounted onto an agarose pad for time-lapsed imaging. Images were acquired at 3-min intervals, with nine Z-planes at 0.5 μm between each plane for Cdc14-ECFP (i) and single planes for Chs2-YFP (ii). Merged images are shown in (iii). (B) Premature Cdc14 dispersal and Chs2 ER export from nucleolus at metaphase can be triggered by Sic1 overexpression. CDC14-ECFP CHS2-YFP NET1-mCHERRY GAL-SIC1-myc(4copies) cells were arrested in Noc in YP/Raff for 5 h at 24°C. At Noc arrest, Glu (i) or Gal (iii) was added to the cells to a final concentration of 2%, and cells were mounted onto agarose pads containing Noc and examined by time-lapsed microscopy. Images were acquired as in A but only selected time points are shown. In addition, Net1-mCherry signals were captured similar to Cdc14-ECFP. Percentages of cells with Cdc14-ECFP nucleolar localization or dispersal and Chs2-YFP neck localization were plotted (ii, v).
Premature Cdc14 dispersal and Chs2 ER export at metaphase can be triggered by Sic1 overexpression
We previously showed that Chs2 could be forced to localize at the neck at metaphase (Zhang et al., 2006) upon induction of the Cdk1 inhibitor SIC1. This observation brings to question whether Cdc14, normally sequestered in the nucleolus at metaphase, can indeed be released from the nucleolus upon SIC1 induction, thereby accounting for the Chs2 ER exit as observed.
CDC14-ECFP CHS2-YFP NET1-mCHERRY GAL-SIC1-myc(4copies) cells arrested in Noc with or without Gal induction of SIC1 were examined by time-lapsed imaging. That the cells maintained arrest in metaphase was judged by the presence of a single Cdc14-ECFP nucleolar region in each cell, indicating that no nuclear division had occurred (Figure 4B, i). An mCherry fusion with Net1, which is needed to sequester Cdc14 in the nucleolus (Shou et al., 1999; Visintin et al., 1999), was used as an indication of nucleolar position. In cells without SIC1 induction, only 5% showed Cdc14-ECFP nucleolar dispersal with Chs2-YFP neck localization (Figure 4B, ii). Consistently, we noted that in the absence of SIC1 induction, when Cdc14-ECFP remained nucleolar, Chs2-YFP was retained in the ER 94% of the time.
In SIC1-induced cells, 25% showed a transient but complete disappearance of the characteristic nucleolar Cdc14-ECFP signal (Figure 4B, iii, 69–93 min, middle), consistent with its liberation from the nucleolus into the cytoplasm. The cells subsequently were able to resequester Cdc14 back into the nucleolus during 99–117 min. Net1-mCherry signals were present throughout the duration of the time-lapsed experiment. The transient Cdc14 release was possibly due to the inability of the cells to maintain a Cdc14 cytoplasmic localization in the absence of full MEN activity in a Noc-induced arrest (Stegmeier et al., 2002; Mohl et al., 2009). In all cells in which Cdc14-ECFP was dispersed, Chs2-YFP was seen at the neck (Figure 4B, iii, 75–87 min, pink asterisks, bottom).
In addition, it is noteworthy that in 22% (Figure 4B, iv) of cells in which Cdc14-ECFP was visible in the nucleolus throughout the time-lapsed experiment (Supplemental Figure S3, middle), Chs2-YFP neck signals could be seen (Supplemental Figure S3, 9–69 min, pink asterisks, bottom). This could simply be due to a lower level of SIC1 expression in these cells leading to a slow to moderate release of Cdc14-ECFP that was not easily detected but that still caused dephosphorylation and ER export of Chs2-YFP. In these cells, the Chs2-YFP neck signal intensity was lower, although the duration of visible neck signals was longer. This indicated a low but sustained release of Cdc14, leading to a gradual dephosphorylation and ER export of the Chs2 to the neck. Nonetheless, our data support the notion that SIC1 induction in metaphase can cause the premature export of Chs2 from the ER due to the forced release of Cdc14 to the cytoplasm.
That SIC1 overexpression in Noc-arrested cells caused Cdc14 nucleolar exit was rather intriguing, given that Cdk1 activity is needed for the FEAR-dependent nucleolar export during early anaphase (Azzam et al., 2004). It is unlikely that the Cdc14 release was a result of FEAR pathway activation via the liberation of Esp1p (e.g., Stegmeier et al., 2002; Sullivan and Uhlmann, 2003) due to an escape from the Noc arrest, as only cells maintaining a Noc arrest were enumerated in the experiments.
However, our observations should not be too surprising, given that SIC1 overexpression rescues most MEN mutants except cdc14 (Fitzpatrick et al., 1998; Jaspersen et al., 1998). Presumably, the rescue of such MEN mutants by ectopic expression of SIC1 requires at some point dephosphorylation of the Cdk1 substrates so that cells can exit mitosis in the absence of MEN function. The presence of Sic1 in these mutants leading to a decline in Cdk1 activity could have led to the release of Cdc14 from the nucleolus and the dephosphorylation of the Cdk1 substrates by Cdc14. Indeed, previous studies provided clear genetic and biochemical data showing that the overexpression of SIC1 in metaphase or telophase can result in the dephosphorylation of Cdc14 substrates (e.g., Bloom and Cross, 2007; Jin et al., 2008; Zhai et al., 2010), although no direct evidence for the cellular distribution of Cdc14 in relation to substrate localization documented in these reports.
We posit that the second wave of MEN-triggered nucleolar export of Cdc14 to the cytoplasm during a normal telophase might also require a lowering of Cdk1 activity to help promote Cdc14 nucleolar dispersal. This idea is consistent with a previous report showing that lowering of Cdk1 activity contributes to mitotic exit (Lu and Cross, 2009). In addition, a previous study showed that ectopic expression of SIC1 can give rise to supernumerary Cdc14 release in 29% of cells without nucleolar separation, albeit in a bub2Δ 2XGAL-SIC1 background (Lu and Cross, 2010). Moreover, it was observed that in Cdc20p-depleted pMET3-CDC20 pds1Δ cdh1Δ pGAL-SIC1 cells, induction of SIC1 did not prevent Cdc14 export out of the nucleolus (Visintin et al., 2008). Although it is unclear how SIC1 overexpression leads to Cdc14 nucleolar dispersal at metaphase, our data nonetheless underscore the significance of having a complex network regulating mitotic exit and the release of Cdc14 (Stegmeier and Amon, 2004) during transition through mitosis.
Premature MEN but not FEAR activation in Noc-treated cells can cause Chs2 ER export
We further tested whether the premature release of Cdc14 in cdc55Δ or bub2Δ cells could trigger ER export of Chs2. Deletion of CDC55, which encodes for the regulatory B subunit of the protein phosphatase 2A (Jiang, 2006), can cause premature release of Cdc14 by the FEAR pathway (Wang and Ng, 2006; Yellman and Burke, 2006). The MEN pathway can be triggered in metaphase (Wang et al., 2000; Wang and Ng, 2006) by the deletion of BUB2 that encodes for a spindle-positioning checkpoint component (Gardner and Burke, 2000).
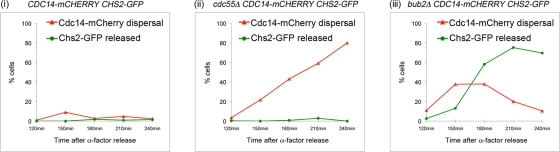
Wild-type, cdc55Δ, and bub2Δ strains each carrying CDC14-mCHERRY CHS2-GFP were released into Noc-containing medium from a G1 arrest. At Noc-induced arrest, Cdc14-mCherry distribution and Chs2-GFP localization were observed. Wild-type cells maintained a tight nucleolar localization of Cdc14-mCherry and Chs2-GFP in the ER (data not shown and Figure 5A, i). In the cdc55Δ mutant, we observed Cdc14-mCherry to be diffused in the nucleus that is surrounded by the perinuclear ER signal of Chs2-GFP (data not shown). Very low percentages of cells with diffused Cdc14-mCherry signals in the cdc55Δ cells showed Chs2-GFP neck signals (Figure 5A, ii), suggesting that premature FEAR activation did not trigger ER export of Chs2-GFP. In the bub2Δ strain, a relatively high proportion of cells (Figure 5A, iii) with both Cdc14-mCherry dispersal and Chs2-GFP neck signal were observed (data not shown), indicating that ER export had occurred.
FIGURE 5:
Premature MEN but not FEAR activation in Noc-treated cells can cause Chs2 ER export. Wild-type (i), cdc55Δ (ii), or bub2Δ (iii) cells harboring CDC14-mCHERRY CHS2-GFP were released from α-factor–induced G1 into YPD + Noc. Cells were observed every 30 min from 120 min after G1 release when cells were in metaphase. Each cell counted was evaluated for Chs2-GFP and Cdc14-mCherry localization. Chs2-GFP released refers to absence of ER signals. Cdc14-mCherry dispersal was taken as loss of distinct nucleolar signals. Percentages of cells showing Cdc14 cytoplasmic dispersal and Chs2 neck localization are plotted.
These results show that in metaphase, premature activation of the MEN leading to cytoplasmic Cdc14 release, but not the FEAR pathway leading to nuclear Cdc14 release, causes Chs2 exit from the ER. This finding is consistent with the fact that the N-terminus of Chs2 that is phosphorylated by Clb2-Cdc28 (Ubersax et al., 2003; Loog and Morgan, 2005) faces the cytoplasm (Martinez-Rucobo et al., 2009; Teh et al., 2009).
In addition to Noc-arrested cells in which Cdc14 is prematurely exported from the nucleolus, we also examined Chs2 ER export in cycling cells to ascertain whether its regulation was similar during unperturbed cell cycle progression. We found that phospho-mimetic Chs2(4S-to-4E)-YFP was constitutively localized in the ER in cycling cells (data not shown), whereas the Chs2(4S-to-4A)-YFP was located at the neck prior to mitotic exit and wild-type Chs2-YFP was localized to the neck largely in cells exiting mitosis (Supplemental Figure S4A). For instance, in a cycling culture, Chs2-YFP neck localization in large-budded cells was well correlated with Inn1-mCherry, a marker for mitotic exit (Meitinger et al., 2010; Supplemental Figure S4, i and ii), but Chs2(4S-to-4A)-YFP was observed at the neck even while Inn1-mCherry was absent.
Moreover, in cells released from α-factor, wild-type Chs2-YFP observed at the neck was tightly correlated with mitotic exit (Supplemental Figure S4B). We previously showed that Chs2 phosphorylation by Clb2-Cdc28 leads to its retention in the ER at metaphase (Zhang et al., 2006). Collectively, our data here indicate that the dephosphorylation of Chs2 by Cdc14 released to the cytoplasm during mitotic exit ensures that Chs2 export from the ER occurs only after mitotic kinase destruction.
DISCUSSION
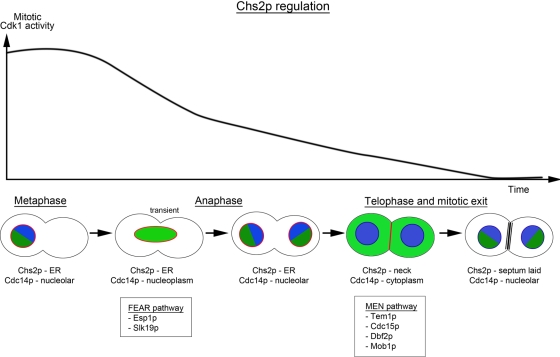
Taken together, our data provide a model by which the timely export of Chs2 from the ER to the bud neck for septum formation can be explained (Figure 6). CHS2 that is expressed during metaphase (Pammer et al., 1992; Choi et al., 1994; Spellman et al., 1998) is retained in the ER prior to sister chromatid separation by Clb2-Cdc28 phosphorylation at the N-terminus (VerPlank and Li, 2005; Zhang et al., 2006; Teh et al., 2009). On APCCdc20 activation, sister chromatids separate and the destruction of the mitotic cyclin Clb2 occurs, leading to the initiation of the first phase of mitotic exit (Yeong et al., 2000; Wasch and Cross, 2002). At this point, Cdc14 is released throughout the nucleus by the FEAR pathway, and Chs2 that is phosphorylated continues to be retained in the ER. This is consistent with previous observations that MEN mutants defective in reducing Cdk1 activity are unable to export Chs2 from the ER (VerPlank and Li, 2005; Zhang et al., 2006; Teh et al., 2009; Meitinger et al., 2010) as Chs2 remains phosphorylated at telophase (Supplemental Figure S2).
FIGURE 6:
Schematic showing Cdc14 (green) and Chs2 (red) distribution during mitotic exit. See the text for details.
As sister chromatids separate and the spindles elongate, the migration of one spindle pole body into the daughter cell results in the activation of the MEN that promotes the complete inactivation of Cdk1 (Stegmeier and Amon, 2004). At the same time, Cdc14 released from the nucleolus to the cytoplasm (Figure 4) during MEN activation now has access to the N-terminal of Chs2 and acts efficiently to relieve the inhibitory phosphorylation on Chs2 (Figure 3). In the absence of competing Cdk1 activity, dephosphorylated Chs2 is exported from the ER via the secretory pathway to the bud neck (Chuang and Schekman, 1996; Zhang et al., 2006) in time to deposit the primary septum and stabilize the actomyosin ring as it undergoes constriction (VerPlank and Li, 2005).
The dispersal of Cdc14 to the nucleoplasm in early anaphase by FEAR and to the cytoplasm in late mitosis by MEN has been suggested to provide a means by which cells regulate the dephosphorylation of distinct groups of substrates of Cdc14 at different times in mitosis (Sullivan and Morgan, 2007). However, in addition to Cdc14 dispersal, the Cdk1 activity could also contribute to the differential dephosphorylation of Cdc14 substrates. For instance, in early anaphase, substrates including Ase1 (Khmelinskii et al., 2007, 2009), Sli15 (Pereira and Schiebel, 2003), and Ask1 (Higuchi and Uhlmann, 2005) are dephosphorylated by the FEAR-released Cdc14 in the nucleoplasm even though the Cdk1 activity is still relatively high. This could reflect the higher efficiency that Cdc14 has in dephosphorylating these substrates. Substrates such as Cdh1 (Zachariae et al., 1998; Jaspersen et al., 1999), Swi5 (Moll et al., 1991; Visintin et al., 1998). and Sic1 (Visintin et al., 1998) are dephosphorylated in late anaphase by Cdc14 released into the cytoplasm when APCCdc20 is active and the Cdk1 activity likely has decreased to 50% of its peak level. In line with our data, Chs2 likely represents a member of the group of late Cdc14 substrates that are dephosphorylated in telophase when Cdk1 activity has been reduced to a very low level (Figure 6). As such, the different levels of Cdk1 activity at early anaphase, late anaphase, and telophase, coupled to the nucleoplasmic or cytoplasmic localization of Cdc14, together contribute toward ordering mitotic events.
Consistent with this notion, we observed that the export of Chs2 was effectively triggered when Cdk1 was inhibited by overexpression of SIC1 in metaphase-arrested cells (Figure 4B). Presumably, the Cdc14 that is forced out of the nucleolus under these conditions acts unopposed in the absence of Cdk1 and efficiently dephosphorylates its substrates, including Chs2. These observations are in line with the findings that cdc55Δ, leading to FEAR activation, did not cause premature Chs2 ER export in metaphase (Figure 5A, ii), but, rather, bub2Δ that leads to activation of the MEN and Cdc14 cytoplasmic localization caused Chs2 ER export (Figure 5A, iii).
Besides promoting mitotic exit, the MEN components such as Tem1p, Cdc15, Dbf2, Dbf20, and Cdc14 are known to localize to the neck at the end of mitosis to promote events needed for cytokinesis and septation (Yeong et al., 2002; Balasubramanian et al., 2004). The exact function(s) of the MEN components in cytokinesis and septum formation is unclear. It was recently suggested that the MEN plays a role in the neck localization of cytokinesis components such as Inn1, Cyk3, and Chs2 (Meitinger et al., 2010). In dbf2-2 dbf20Δ cells in which SIC1 was overexpressed to inhibit the mitotic Cdk1 activity, Chs2 translocation to the neck was inefficient. Consequently, the authors proposed that the MEN is needed for targeting Chs2 to the neck. However, these observations could be due to the fact that Dbf2 is needed for promoting Cdc14 cytoplasmic retention during exit from mitosis (Mohl et al., 2009) and that in the dbf2-2 dbf20Δ GAL-SIC1 cells, Cdc14 cytoplasmic localization was not efficient even in the presence of Sic1. As a result, Chs2 remained at the ER and failed to translocate to the neck.
It should also be noted that the cdc14-NES mutant shows a cytokinesis defect (Bembenek et al., 2005). Observation of the GFP-tagged Cdc14-NES protein revealed that the mutant protein is defective in neck localization, suggesting that Cdc14 neck localization might be important for septum formation and/or cytokinesis. However, in the cdc14-NES mutant, phosphorylation-deficient Chs2(4S-to-4A)-YFP was localized to the neck even in metaphase (Figure 2). This indicates that Chs2 localization at the neck per se might not normally depend upon Cdc14 function at the neck, but, rather, that Cdc14 acts on Chs2 at the ER to promote its export and translocation to the neck. The cytokinesis defect in the cdc14-NES mutant is therefore likely to be a failure of Chs2 ER export.
It remains to be seen how the dephosphorylation of Chs2 by Cdc14 during mitotic exit allows for its incorporation into COP II vesicles (Lee and Miller, 2007; Sato and Nakano, 2007) for its transport to the neck. It would be of interest to determine whether the COP II coat protein Sec24 interacts with the dephosphorylated form of Chs2 and, if so, what the underlying basis is that allows for the association. Furthermore, it is unclear how the interaction with COP II complex enables the selection of Chs2 into ER exit sites (Budnik and Stephens, 2009) in a timely manner for targeting to the bud neck to aid in actomyosin ring constriction, and this requires further study. Given that several other cargoes of the secretory pathway studied are constitutively exported out of the ER (Makarow, 1988; Nevalainen et al., 1989) unlike Chs2, understanding the differences in the mechanisms by which the COP II coat proteins and other components of the early secretory pathway interacts with distinct cargoes is an important future goal.
Cdc14 belongs to a family of highly conserved dual-specificity phosphatases (Mocciaro and Schiebel, 2010), with several novel substrates in mitosis and cytokinesis identified recently (Bloom et al., 2011). Our finding that Chs2, a cargo of the secretory pathway (Chuang and Schekman, 1996; VerPlank and Li, 2005; Zhang et al., 2006), is a novel Cdc14 substrate highlights an important link between the cell division machinery and the protein-trafficking pathway in the coordination of cell cycle events. More than transporting Chs2, the secretory vesicles carrying Chs2 that are targeted to the neck might also be required for the delivery of other proteins and, more critically, membranes that are needed during cytokinesis (McKay and Burgess, 2011). It is intriguing that Cdc14A in mammalian cells (Lanzetti et al., 2007) has been shown to interact with Rab5, a small GTPase implicated in the secretory and the endocytic pathways (Zerial and McBride, 2001), whereas Cdc14C has been found to localize to the ER (Rosso et al., 2008). The implications of these findings are unknown, although the findings point to possible roles of Cdc14 in higher eukaryotes in membrane trafficking.
Although questions remain regarding how dephosphorylation of Chs2 affects its selection into COP II vesicles and how Cdc14 might further contribute to cytokinesis through its role in promoting vesicular transport, our data nonetheless highlight the significance of the combined effects of low Cdk1 activity and cytoplasmic localization of Cdc14 in promoting the dephosphorylation of telophase substrates such as Chs2 (Figure 6). The constitutive neck localization of Chs2p(4S-to-4A)-YFP in cycling cells even when mitotic exit has not occurred (Supplemental Figure S4) further suggests that the presence of phosphorylation at the N-terminal Cdk1 sites normally prevents untimely localization of Chs2 during a regular cell division. Conversely, it also implies that the dephosphorylation of Chs2 is needed to alleviate the restraint on its ER export. More important, the untimely dephosphorylation of Chs2 can result in premature transport to the neck in an unperturbed cell division cycle.
The tight regulation of Chs2 ER export at the end of mitosis is critical, as premature localization of Chs2 to the neck in mitosis can result in aberrant septation (Zhang et al., 2006; Meitinger et al., 2010), whereas the phosphorylation-deficient Chs2(4S-to-4A) kills cells when overexpressed (Teh et al., 2009). Moreover, a transient release of Cdc14 from the nucleolus in metaphase upon SIC1 induction (Figure 4B) was sufficient to cause Chs2 ER export, indicating that the sequestration of Cdc14 from the cytoplasm normally ensures Chs2 ER retention until after the activation of the MEN. The observation further raises the possibility that the lowering of Cdk1 during mitotic exit could contribute to Cdc14 cytoplasmic localization. Such an interdependence of chromosome segregation, MEN activation, decrease in mitotic Cdk1 activity, and Cdc14 dispersal is likely to play a critical role in the execution of late mitotic events. Indeed, the coupling of these events in promoting late mitotic processes such as the export of Chs2 from the ER provides a simple yet effective mechanism for cells to order sister chromatid separation, mitotic exit, and cytokinesis in the proper temporal sequence.
MATERIALS AND METHODS
Yeast culture reagents
Supplemental Table S1 lists the strains used in this study. Cells were routinely grown in yeast extract peptone (YP) or selective medium supplemented with 2% dextrose at 24°C. For experiments requiring Gal induction, cells were grown in YP supplemented with 2% raffinose (Raff), followed by addition of Gal to a final concentration of 2% unless otherwise stated. Each experiment was performed at least three times, and 100 cells were counted for each time point unless otherwise stated. Plots shown are typical representations of three experiments.
Strains and plasmids
A combination of standard molecular biology and molecular genetics techniques such as PCR-based tagging of endogenous genes and tetrad dissection was used to construct plasmids and strains (Supplemental Table S1). The plasmids for the CFP and YFP cassettes were obtained from the European Saccharomyces cerevisiae Archive for Functional Analysis. Further information regarding strain and plasmid constructions will be provided upon request.
Cell synchrony
For experiments requiring synchronized cultures, exponential-phase cells were diluted to 107 cells/ml in growth medium at 24°C. For G1 arrest, cells were treated with α-factor (US Biological, Swampscott, MA) at 0.4 μg/ml for 3 h. After the cells were arrested, they were washed by filtration and resuspended in media at the required conditions as described in the various sections. For a typical Noc arrest, cells were arrested with 7.5 μg/ml Noc (US Biological) for 2.5 h at 24°C, followed by the further addition of 7.5 μg/ml for another 2.5 h at 24 or 37°C, depending on the strain. The drug was washed off by centrifugation of the cells. Cells were then released and sampled at intervals as described in the relevant sections.
Western blot analysis
Samples were taken at the time points indicated and proteins extracted as described in Zhang et al. (2006). Anti-Cdc28, anti-Myc, and anti-HA antibodies (all from Santa Cruz Biotechnology, Santa Cruz, CA) were used at a 1:1000 dilution, and anti-Clb2 antibodies (Santa Cruz Biotechnology) were used at a 1:5000 dilution. The enhanced chemiluminescence kit (Pierce, Thermo Fisher Scientific, Rockford, IL) was used according to the manufacturer's recommendations.
Coimmunoprecipitation
Yeast pellets were resuspended in IP-lyse buffer (1% NP-40, 9% glycerol, 18 mM Tris-Cl, pH 8.0, 121 mM NaCl) supplemented with 1× protease inhibitor and 1× phosphatase inhibitor (Sigma-Aldrich, St. Louis, MO). Zirconia beads were then added to one-fourth total of the total liquid volume, and cells were agitated in vortexer at 4°C until most of the cells were lysed. Extracts were cleared by centrifugation force at 13,200 rpm at 4°C. Protein concentrations of the lysates were determined using the bicinchoninic acid assay (Pierce). Anti-HA beads (Santa Cruz Biotechnology) were added to 2 mg of total lysate in each IP and incubated on a rotating platform for 2 h. Beads were washed six times with IP wash buffer (10% glycerol, 20 mM Tris-Cl, pH 8.0, 135 mM NaCl). Bound proteins were eluted by boiling beads in standard 2× SDS dye.
In vitro phosphatase assay with full-length Chs2
Recombinant GST-6His-Cdc14 was expressed in Escherichia coli and purified as described previously (Hall et al., 2008) with some modifications. GST-6His-Cdc14 was affinity purified using glutathione agarose resin (Pierce), followed by further purification using Ni2+ beads (Sigma-Aldrich) and elution using imidazole as recommended by the manufacturer. Diafiltration of GST-6His-Cdc14 into storage buffer (Hall et al., 2008) was carried out using Amicon Ultra-15 centrifugation tubes as recommended by the manufacturer (Millipore, Billerica, MA). GST-6His-Cdc14 concentration was judged by running an aliquot against known amounts of bovine serum albumin as a standard on an SDS–PAGE with Coomassie blue staining.
For the phosphatase assay, Chs2p-3myc was first immunoprecipitated from trichloroacetic acid (TCA) lysates of yeast cells arrested in Noc. Chs2p-3myc beads were incubated in 30 μl of phosphatase buffer (Traverso et al., 2001) with or without purified GST-6His-Cdc14 for 1 h at 30°C. The beads were then washed three times with IP wash buffer, eluted with 2× SDS dye, and subjected to Phos-tag gel analysis (Kinoshita et al., 2009). Sodium orthovanadate 1 mM (Sigma-Aldrich) was used as an inhibitor of GST-6His-Cdc14.
Peptide synthesis
Phosphopeptide substrates were from New England Peptide (Gardner, MA) and were purified in house by standard reverse-phase high-performance liquid chromatography, lyophilized, and reconstituted in water. Phosphopeptide concentrations were determined by an ashing procedure and malachite green–based spectrometric assay described previously (Buss and Stull, 1983). Sequences of phosphopeptides used in this study are as follows (pS in parentheses is the phosphorylated residue): Chs2pS14, MVEPSNG(pS)PNRRGASN; Chs2pS60, VFQGLPA(pS)PSRAALRY; Chs2pS69, SRAALRY(pS)PDRRHRTQ; Chs2pS86, YRDSAHN(pS)PVAPNRYA; Chs2pS100, YAANLQE(pS)PKRAGEAV; Chs2pS133, PVDPYHL(pS)PQQQPSNN; Chs2pS256, DTLPRRN(pS)PEFTEMRY; Acm1pS31, VKGNELR(pS)PSKRRSQI; Acm1pS48, TDYALRR(pS)PIKTIQIS; Pds1pS71, IQGGKEV(pS)PTKRLHTH; Swi6pS160, DAHRELG(pS)PLKKLKID; Cdh1pS169, AAGLEEF(pS)PHSTPVTP; Fin1pS74, SIQVTPRRIM(pS)PECLK.
Protein purification
The recombinant proteins GST-Acm1, GST-Fin1, GST-Chs2(1-300), and GST-Cdh1(1-211) were overexpressed from pGEX6P-1 (GE Healthcare, Piscataway, NJ) in log-phase E. coli culture by addition of 0.5 mM isopropyl-β-d-thiogalactoside and incubation for ∼8 h at 23°C and purified on glutathione agarose resin (EMD Biosciences, San Diego, CA) in 50 m Tris-HCl (pH 7.8), 250 mM NaCl, 10% glycerol, 1% Triton X-100, 1 mM EDTA, and 1 mM dithiothreitol. GST fusion proteins were eluted by addition of 10 mM reduced glutathione, dialyzed into 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES; pH 7.5), 100 mM NaCl, 10 mM MgCl2, 0.5 mM dithiothreitol, and 40% glycerol and stored directly in small aliquots at −80°C. Purity and concentration were assessed by SDS–PAGE using known amounts of bovine serum albumin as a standard.
Cdk1 (Clb2-Cdc28) was purified by expressing a Clb2-protein A fusion protein from the GAL1 promoter on the 2-μm plasmid BG1805 (Open Biosystems, Thermo Biosystems, Huntsville, AL) in a sic1Δ strain by addition of 2% galactose to 4 l of log-phase culture and incubation for 4 h at 30°C. Whole-cell extracts were generated by glass bead lysis in 50 mM Tris-HCl (pH 8.0), 250 mM NaCl, 10% glycerol, 0.1% Triton X-100, 20 mM sodium fluoride, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 μM pepstatin, and 100 μM leupeptin, and Cdk1 was isolated by incubation with 100 μl of immunoglobulin G–coupled agarose resin (Sigma-Aldrich) for 2 h at 4°C. Beads were washed extensively with the same buffer lacking protease inhibitors, washed once with kinase buffer (10 mM HEPES, pH 7.5, 10 mM MgCl2, 50 mM NaCl, 10% glycerol, 0.5 mM dithiothreitol, 0.005% Triton X-100), and stored at −20°C until use.
Recombinant protein phosphorylation
To phosphorylate GST fusion proteins with purified Cdk1, each protein was diluted to ∼5 μM with kinase buffer, supplemented with 1 mM ATP, mixed with ∼25 μl of Cdk1 beads, and incubated at 30°C for 45 min with gentle agitation to keep beads suspended. Cdk1 beads were pelleted by centrifugation, and phosphorylated proteins in the supernatant were removed, analyzed by MS to evaluate phosphorylation, and used for phosphatase assays.
Peptide phosphatase assays
Cdc14-catalyzed dephosphorylation of phosphopeptides was measured by absorbance detection of the released Pi in a spectrophotometer at 650 nm after reaction with BIOMOL Green reagent (Enzo Life Sciences, Plymouth, PA). Enzyme assays were performed in 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, and 3 mM β-mercaptoethanol at 30°C for varying times and stopped with BIOMOL Green as directed by the supplier. Standard curves were generated using defined concentrations of Na2PO4 under identical solution conditions.
Dephosphorylation of recombinant protein substrates
Purified GST-Cdc14, 250 nM, was added to Cdk1-phosphorylated GST-Fin1, GST-Acm1, GST-Chs2(1-300), or GST-Cdh1(1-211) and the reaction allowed to proceed at 30°C for the indicated times. Aliquots of 20 μl were removed at each time point and added to 7 μl of 4× SDS–PAGE loading dye and heated to 95°C. Proteins were processed by SDS–PAGE, stained with Coomassie blue, and subjected to in-gel digestion with either Lys-C (for Acm1 and Fin1) or trypsin (for Chs2 and Cdh1) as described (Hall et al., 2008).
Liquid chromatography–mass spectrometry analysis and label-free quantification
Peptide samples from recombinant protein reactions were resuspended in 0.1% formic acid and analyzed by liquid chromatography–mass spectrometry on an LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific, Waltham, MA) using a 45-min gradient of 3–40% acetonitrile in 0.1% formic acid at 300 nl/min and collecting cycles of MS and eight data-dependent MSMS scans. Samples were loaded directly into 75-μm–inner diameter fused silica capillary (Biotaq, Gaithersburg, MD) electrospray ionization tips packed with Magic C18 resin (Bruker-Microm, Auburn, CA).
Extracted ion chromatograms for individual phosphopeptides and nonphosphorylated standard peptides used for normalization were generated from the XCalibur software (Thermo Fisher) and exported into GraphPad Prism (GraphPad Software, La Jolla, CA) for peak integration and data processing. To generate graphs of relative phosphorylation over time, the raw signals for phosphopeptides at each time point were first normalized using the average percentage deviation of at least three standard peptides (not containing Cdk1 phosphorylation sequences) generated from the same protein relative to the zero time point. Subsequently, the normalized phosphopeptide signals were converted to percentages of the zero–time point signals.
Chs2-13myc IP and phosphorylation-site mapping
A 2-l culture of cdc15-2 CHS2-13myc cells was shifted to 37°C in mid-log phase to arrest cells in telophase with inactive Cdc14. Cells were lysed by blending with glass beads in 20 mM Tris-HCl (pH 8), 400 mM NaCl, 10% glycerol, 1% NP-40, and 1 mM dithiothreitol supplemented with protease inhibitors (5 mM EDTA, 100 μM leupeptidin, 1 μM pepstatin, and 1 mM PMSF) at 4°C. Extracts were cleared by centrifugation at 17,000 × g for 30 min and incubated with 100 μl of EZview anti-myc antibody resin (Sigma-Aldrich) for 3 h at 4°C with gentle mixing. The resin was washed four times with the same buffer lacking protease inhibitors and then boiled in 1× SDS–PAGE loading dye. Proteins were separated by SDS–PAGE and detected with Coomassie blue, and the Chs2-13myc band was excised from the gel and digested with trypsin. Tryptic peptides were analyzed on the LTQ-Orbitrap Velos exactly as described earlier for recombinant proteins. Phosphorylated peptides were first identified by database search of the yeast protein database using Proteome Discoverer software (Thermo Scientific) and then by manual inspection of the raw MS data.
Fluorescence microscopy
Samples for fluorescence microscopy were taken at time points indicated in the relevant sections. Images were captured and processed as described previously (Zhang et al., 2006; Teh et al., 2009). ImageJ (National Institutes of Health, Bethesda, MA) and Photoshop (Adobe, San Jose, CA) were used for the production of the figures.
Time-lapsed microscopy
For time-lapsed microscopy, cells released from a Noc arrest were resuspended in complete synthetic media containing glucose (Glu) or Gal as required and mounted onto 5% agarose pads on slides. Time-lapsed images were obtained using an IX81 wide-field fluorescence microscope (Olympus, Center Valley, PA). Typically, the exposure time for the acquisition of the images was ∼180–200 ms for YFP, 20 ms per plane for CFP, and 18 ms per plane for RFP. Nine optical Z-sections at 0.5-μm intervals were obtained for each time point. Images shown were either maximal projection of the Z-stacks or images taken at a single plane, as indicated in the relevant sections. ImageJ and Adobe Photoshop were used for the production of the montages and figures.
Supplementary Material
Acknowledgments
We thank Harry Charbonneau, Hong Tao Yu, and Chun Liang for various strains and constructs. We are grateful to Yuan Yuan Chew, Jesse Murphy, Christie Eissler, and Juan Martinez for technical assistance. We also thank Anton Iliuk and W. Andy Tao for access to and assistance with operating the LTQ-Orbitrap Velos. We also acknowledge the European Saccharomyces cerevisiae Archive for Functional Analysis and the Yeast Resource Center for various GFP and epitope-tagging constructs. We thank the anonymous reviewers for their comments and suggestions for improving the manuscript. M.C.H. is supported by National Science Foundation Grant MCB 0841748 and by the Purdue Center for Cancer Research Small Grants Program. F.M.Y. is funded by Singapore Ministry of Education Grant R183-000-246-112.
Abbreviations used:
- FEAR
Cdc fourteen early anaphase release
- Gal
galactose
- GFP
green fluorescence protein
- Glu
glucose
- GST
glutathione S-transferase
- MEN
mitotic exit network
- MS
mass spectrometry
- Noc
nocodazole
- YP
yeast extract peptone
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-05-0434) on November 9, 2011.
REFERENCES
- Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DG. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzam R, Chen SL, Shou W, Mah AS, Alexandru G, Nasmyth K, Annan RS, Carr SA, Deshaies RJ. Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus. Science. 2004;305:516–519. doi: 10.1126/science.1099402. [DOI] [PubMed] [Google Scholar]
- Balasubramanian MK, Bi E, Glotzer M. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr Biol. 2004;14:R806–R818. doi: 10.1016/j.cub.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Bembenek J, Kang J, Kurischko C, Li B, Raab JR, Belanger KD, Luca FC, Yu H. Crm1-mediated nuclear export of Cdc14 is required for the completion of cytokinesis in budding yeast. Cell Cycle. 2005;4:961–971. doi: 10.4161/cc.4.7.1798. [DOI] [PubMed] [Google Scholar]
- Bloom J, Cristea IM, Procko AL, Lubkov V, Chait BT, Snyder M, Cross FR. Global analysis of cdc14 phosphatase reveals diverse roles in mitotic processes. J Biol Chem. 2011;286:5434–5445. doi: 10.1074/jbc.M110.205054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J, Cross FR. Novel role for Cdc14 sequestration: Cdc14 dephosphorylates factors that promote DNA replication. Mol Cell Biol. 2007;27:842–853. doi: 10.1128/MCB.01069-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant NJ, Stevens TH. Vacuole biogenesis in Saccharomyces cerevisiae: protein transport pathways to the yeast vacuole. Microbiol Mol Biol Rev. 1998;62:230–247. doi: 10.1128/mmbr.62.1.230-247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B, et al. Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci Signal. 2010;3:rs4. doi: 10.1126/scisignal.2001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budnik A, Stephens DJ. ER exit sites—localization and control of COPII vesicle formation. FEBS Lett. 2009;583:3796–3803. doi: 10.1016/j.febslet.2009.10.038. [DOI] [PubMed] [Google Scholar]
- Buss JE, Stull JT. Measurement of chemical phosphate in proteins. Methods Enzymol. 1983;99:7–14. doi: 10.1016/0076-6879(83)99035-3. [DOI] [PubMed] [Google Scholar]
- Cabib E. The septation apparatus, a chitin-requiring machine in budding yeast. Arch Biochem Biophys. 2004;426:201–207. doi: 10.1016/j.abb.2004.02.030. [DOI] [PubMed] [Google Scholar]
- Choi WJ, Santos B, Duran A, Cabib E. Are yeast chitin synthases regulated at the transcriptional or the posttranslational level? Mol Cell Biol. 1994;14:7685–7694. doi: 10.1128/mcb.14.12.7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang JS, Schekman RW. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. J Cell Biol. 1996;135:597–610. doi: 10.1083/jcb.135.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford DM, Chen CT, Roberts RH, Feoktistova A, Wolfe BA, Chen JS, McCollum D, Gould KL. The role of Cdc14 phosphatases in the control of cell division. Biochem Soc Trans. 2008;36:436–438. doi: 10.1042/BST0360436. [DOI] [PubMed] [Google Scholar]
- D'Amours D, Amon A. At the interface between signaling and executing anaphase—Cdc14 and the FEAR network. Genes Dev. 2004;18:2581–2595. doi: 10.1101/gad.1247304. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick PJ, Toyn JH, Millar JB, Johnston LH. DNA replication is completed in Saccharomyces cerevisiae cells that lack functional Cdc14, a dual-specificity protein phosphatase. Mol Gen Genet. 1998;258:437–441. doi: 10.1007/s004380050753. [DOI] [PubMed] [Google Scholar]
- Fraschini R, Venturetti M, Chiroli E, Piatti S. The spindle position checkpoint: how to deal with spindle misalignment during asymmetric cell division in budding yeast. Biochem Soc Trans. 2008;36:416–420. doi: 10.1042/BST0360416. [DOI] [PubMed] [Google Scholar]
- Gardner RD, Burke DJ. The spindle checkpoint: two transitions, two pathways. Trends Cell Biol. 2000;10:154–158. doi: 10.1016/s0962-8924(00)01727-x. [DOI] [PubMed] [Google Scholar]
- Geymonat M, Spanos A, Wells GP, Smerdon SJ, Sedgwick SG. Clb6/Cdc28 and Cdc14 regulate phosphorylation status and cellular localization of Swi6. Mol Cell Biol. 2004;24:2277–2285. doi: 10.1128/MCB.24.6.2277-2285.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray CH, Good VM, Tonks NK, Barford D. The structure of the cell cycle protein Cdc14 reveals a proline-directed protein phosphatase. EMBO J. 2003;22:3524–3535. doi: 10.1093/emboj/cdg348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MC, Jeong DE, Henderson JT, Choi E, Bremmer SC, Iliuk AB, Charbonneau H. Cdc28 and Cdc14 control stability of the anaphase-promoting complex inhibitor Acm1. J Biol Chem. 2008;283:10396–10407. doi: 10.1074/jbc.M710011200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi T, Uhlmann F. Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature. 2005;433:171–176. doi: 10.1038/nature03240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt LJ, Krutchinsky AN, Morgan DO. Positive feedback sharpens the anaphase switch. Nature. 2008;454:353–357. doi: 10.1038/nature07050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Charles JF, Morgan DO. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr Biol. 1999;9:227–236. doi: 10.1016/s0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Charles JF, Tinker-Kulberg RL, Morgan DO. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:2803–2817. doi: 10.1091/mbc.9.10.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y. Regulation of the cell cycle by protein phosphatase 2A in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2006;70:440–449. doi: 10.1128/MMBR.00049-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Liu H, Liang F, Rizkallah R, Hurt MM, Wang Y. Temporal control of the dephosphorylation of Cdk substrates by mitotic exit pathways in budding yeast. Proc Natl Acad Sci USA. 2008;105:16177–16182. doi: 10.1073/pnas.0808719105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A, Lawrence C, Roostalu J, Schiebel E. Cdc14-regulated midzone assembly controls anaphase B. J Cell Biol. 2007;177:981–993. doi: 10.1083/jcb.200702145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A, Roostalu J, Roque H, Antony C, Schiebel E. Phosphorylation-dependent protein interactions at the spindle midzone mediate cell cycle regulation of spindle elongation. Dev Cell. 2009;17:244–256. doi: 10.1016/j.devcel.2009.06.011. [DOI] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Koike T. Separation and detection of large phosphoproteins using Phos-tag SDS–PAGE. Nat Protoc. 2009;4:1513–1521. doi: 10.1038/nprot.2009.154. [DOI] [PubMed] [Google Scholar]
- Lanzetti L, Margaria V, Melander F, Virgili L, Lee MH, Bartek J, Jensen S. Regulation of the Rab5 GTPase-activating protein RN-tre by the dual specificity phosphatase Cdc14A in human cells. J Biol Chem. 2007;282:15258–15270. doi: 10.1074/jbc.M700914200. [DOI] [PubMed] [Google Scholar]
- Lee MC, Miller EA. Molecular mechanisms of COPII vesicle formation. Semin Cell Dev Biol. 2007;18:424–434. doi: 10.1016/j.semcdb.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Lee SE, Frenz LM, Wells NJ, Johnson AL, Johnston LH. Order of function of the budding-yeast mitotic exit-network proteins Tem1, Cdc15, Mob1, Dbf2, Cdc5. Curr Biol. 2001;11:784–788. doi: 10.1016/s0960-9822(01)00228-7. [DOI] [PubMed] [Google Scholar]
- Loog M, Morgan DO. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–108. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- Lu Y, Cross F. Mitotic exit in the absence of separase activity. Mol Biol Cell. 2009;20:1576–1591. doi: 10.1091/mbc.E08-10-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Cross FR. Periodic cyclin-Cdk activity entrains an autonomous Cdc14 release oscillator. Cell. 2010;141:268–279. doi: 10.1016/j.cell.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarow M. Secretion of invertase in mitotic yeast cells. EMBO J. 1988;7:1475–1482. doi: 10.1002/j.1460-2075.1988.tb02965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Rucobo FW, Eckhardt-Strelau L, Terwisscha van Scheltinga AC. Yeast chitin synthase 2 activity is modulated by proteolysis and phosphorylation. Biochem J. 2009;417:547–554. doi: 10.1042/BJ20081475. [DOI] [PubMed] [Google Scholar]
- McKay HF, Burgess DR. “Life is a highway”: membrane trafficking during cytokinesis. Traffic. 2011;12:247–251. doi: 10.1111/j.1600-0854.2010.01139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F, Petrova B, Lombardi IM, Bertazzi DT, Hub B, Zentgraf H, Pereira G. Targeted localization of Inn1, Cyk3 and Chs2 by the mitotic-exit network regulates cytokinesis in budding yeast. J Cell Sci. 2010;123:1851–1861. doi: 10.1242/jcs.063891. [DOI] [PubMed] [Google Scholar]
- Mendenhall MD, Hodge AE. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1998;62:1191–1243. doi: 10.1128/mmbr.62.4.1191-1243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocciaro A, Schiebel E. Cdc14: a highly conserved family of phosphatases with non-conserved functions? J Cell Sci. 2010;123:2867–2876. doi: 10.1242/jcs.074815. [DOI] [PubMed] [Google Scholar]
- Mohl DA, Huddleston MJ, Collingwood TS, Annan RS, Deshaies RJ. Dbf2-Mob1 drives relocalization of protein phosphatase Cdc14 to the cytoplasm during exit from mitosis. J Cell Biol. 2009;184:527–539. doi: 10.1083/jcb.200812022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll T, Tebb G, Surana U, Robitsch H, Nasmyth K. The role of phosphorylation and the CDC28 protein kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor SWI5. Cell. 1991;66:743–758. doi: 10.1016/0092-8674(91)90118-i. [DOI] [PubMed] [Google Scholar]
- Morgan DO. Regulation of the APC and the exit from mitosis. Nat Cell Biol. 1999;1:E47–E53. doi: 10.1038/10039. [DOI] [PubMed] [Google Scholar]
- Moseley JB, Goode BL. The yeast actin cytoskeleton: from cellular function to biochemical mechanism. Microbiol Mol Biol Rev. 2006;70:605–645. doi: 10.1128/MMBR.00013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevalainen LT, Louhelainen J, Makarow M. Post-translational modifications in mitotic yeast cells. Eur J Biochem. 1989;184:165–172. doi: 10.1111/j.1432-1033.1989.tb15003.x. [DOI] [PubMed] [Google Scholar]
- Noton E, Diffley JF. CDK inactivation is the only essential function of the APC/C and the mitotic exit network proteins for origin resetting during mitosis. Mol Cell. 2000;5:85–95. doi: 10.1016/s1097-2765(00)80405-0. [DOI] [PubMed] [Google Scholar]
- Pammer M, Briza P, Ellinger A, Schuster T, Stucka R, Feldmann H, Breitenbach M. DIT101 (CSD2, CAL1), a cell cycle-regulated yeast gene required for synthesis of chitin in cell walls and chitosan in spore walls. Yeast. 1992;8:1089–1099. doi: 10.1002/yea.320081211. [DOI] [PubMed] [Google Scholar]
- Pereira G, Schiebel E. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science. 2003;302:2120–2124. doi: 10.1126/science.1091936. [DOI] [PubMed] [Google Scholar]
- Roh DH, Bowers B, Schmidt M, Cabib E. The septation apparatus, an autonomous system in budding yeast. Mol Biol Cell. 2002;13:2747–2759. doi: 10.1091/mbc.E02-03-0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso L, Marques AC, Weier M, Lambert N, Lambot MA, Vanderhaeghen P, Kaessmann H. Birth and rapid subcellular adaptation of a hominoid-specific CDC14 protein. PLoS Biol. 2008;6:e140. doi: 10.1371/journal.pbio.0060140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Nakano A. Mechanisms of COPII vesicle formation and protein sorting. FEBS Lett. 2007;581:2076–2082. doi: 10.1016/j.febslet.2007.01.091. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Bowers B, Varma A, Roh DH, Cabib E. In budding yeast, contraction of the actomyosin ring and formation of the primary septum at cytokinesis depend on each other. J Cell Sci. 2002;115:293–302. doi: 10.1242/jcs.115.2.293. [DOI] [PubMed] [Google Scholar]
- Shou W, Seol JH, Shevchenko A, Baskerville C, Moazed D, Chen ZW, Jang J, Shevchenko A, Charbonneau H, Deshaies RJ. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- Smolka MB, Albuquerque CP, Chen SH, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci USA. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Amon A. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu Rev Genet. 2004;38:203–232. doi: 10.1146/annurev.genet.38.072902.093051. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Visintin R, Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- Sullivan M, Morgan DO. Finishing mitosis, one step at a time. Nat Rev Mol Cell Biol. 2007;8:894–903. doi: 10.1038/nrm2276. [DOI] [PubMed] [Google Scholar]
- Sullivan M, Uhlmann F. A non-proteolytic function of separase links the onset of anaphase to mitotic exit. Nat Cell Biol. 2003;5:249–254. doi: 10.1038/ncb940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh EM, Chai CC, Yeong FM. Retention of Chs2p in the ER requires N-terminal CDK1-phosphorylation sites. Cell Cycle. 2009;8:2964–2974. [PubMed] [Google Scholar]
- Traverso EE, Baskerville C, Liu Y, Shou W, James P, Deshaies RJ, Charbonneau H. Characterization of the Net1 cell cycle-dependent regulator of the Cdc14 phosphatase from budding yeast. J Biol Chem. 2001;276:21924–21931. doi: 10.1074/jbc.M011689200. [DOI] [PubMed] [Google Scholar]
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- VerPlank L, Li R. Cell cycle-regulated trafficking of Chs2 controls actomyosin ring stability during cytokinesis. Mol Biol Cell. 2005;16:2529–2543. doi: 10.1091/mbc.E04-12-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin C, Tomson BN, Rahal R, Paulson J, Cohen M, Taunton J, Amon A, Visintin R. APC/C-Cdh1-mediated degradation of the Polo kinase Cdc5 promotes the return of Cdc14 into the nucleolus. Genes Dev. 2008;22:79–90. doi: 10.1101/gad.1601308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- Visintin R, Hwang ES, Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- Walther A, Wendland J. Septation and cytokinesis in fungi. Fungal Genet Biol. 2003;40:187–196. doi: 10.1016/j.fgb.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Wang Y, Hu F, Elledge SJ. The Bfa1/Bub2 GAP complex comprises a universal checkpoint required to prevent mitotic exit. Curr Biol. 2000;10:1379–1382. doi: 10.1016/s0960-9822(00)00779-x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ng TY. Phosphatase 2A negatively regulates mitotic exit in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:80–89. doi: 10.1091/mbc.E04-12-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasch R, Cross FR. APC-dependent proteolysis of the mitotic cyclin Clb2 is essential for mitotic exit. Nature. 2002;418:556–562. doi: 10.1038/nature00856. [DOI] [PubMed] [Google Scholar]
- Yellman CM, Burke DJ. The role of Cdc55 in the spindle checkpoint is through regulation of mitotic exit in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:658–666. doi: 10.1091/mbc.E05-04-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeong FM, Lim HH, Padmashree CG, Surana U. Exit from mitosis in budding yeast: biphasic inactivation of the Cdc28-Clb2 mitotic kinase and the role of Cdc20. Mol Cell. 2000;5:501–511. doi: 10.1016/s1097-2765(00)80444-x. [DOI] [PubMed] [Google Scholar]
- Yeong FM, Lim HH, Surana U. MEN, destruction and separation: mechanistic links between mitotic exit and cytokinesis in budding yeast. Bioessays. 2002;24:659–666. doi: 10.1002/bies.10106. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Schwab M, Nasmyth K, Seufert W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 1998;282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- Zhai Y, Yung PY, Huo L, Liang C. Cdc14p resets the competency of replication licensing by dephosphorylating multiple initiation proteins during mitotic exit in budding yeast. J Cell Sci. 2010;123:3933–3943. doi: 10.1242/jcs.075366. [DOI] [PubMed] [Google Scholar]
- Zhang G, Kashimshetty R, Ng KE, Tan HB, Yeong FM. Exit from mitosis triggers Chs2p transport from the endoplasmic reticulum to mother-daughter neck via the secretory pathway in budding yeast. J Cell Biol. 2006;174:207–220. doi: 10.1083/jcb.200604094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.