Although UHRF1 is essential for many epigenetic marks, the mechanism that regulates UHRF1 is not understood. This study shows that a key component of the cell cycle machinery—cyclin-dependent kinase 2/cyclin A2—phosphorylates UHRF1 and that this phosphorylation is essential for early zebrafish development.
Abstract
Ubiquitin-like, containing PHD and RING finger domains 1 (uhrf1) is regulated at the transcriptional level during the cell cycle and in developing zebrafish embryos. We identify phosphorylation as a novel means of regulating UHRF1 and demonstrate that Uhrf1 phosphorylation is required for gastrulation in zebrafish. Human UHRF1 contains a conserved cyclin-dependent kinase 2 (CDK2) phosphorylation site at Ser-661 that is phosphorylated in vitro by CDK2 partnered with cyclin A2 (CCNA2), but not cyclin E. An antibody specific for phospho-Ser-661 recognizes UHRF1 in both mammalian cancer cells and in nontransformed zebrafish cells, but not in zebrafish bearing a mutation in ccna2. Depleting Uhrf1 from zebrafish embryos by morpholino injection causes arrest before gastrulation and early embryonic death. This phenotype is rescued by wild-type UHRF1, but not by UHRF1 in which the phospho-acceptor site is mutated, demonstrating that UHRF1 phosphorylation is essential for embryogenesis. UHRF1 was detected in the nucleus and cytoplasm, whereas nonphosphorylatable UHRF1 is unable to localize to the cytoplasm, suggesting the importance of localization in UHRF1 function. Together, these data point to an essential role for UHRF1 phosphorylation by CDK/CCNA2 during early vertebrate development.
INTRODUCTION
UHRF1 (also called Np95 in mice and ICBP90 in humans; Unoki et al., 2008) is a multi-domain protein implicated in many aspects of chromatin modification and epigenetic regulation of gene expression. Moreover, changes in UHRF1 levels may influence cell cycle progression, apoptosis, and response to DNA damage (Jeanblanc et al., 2005; Kim et al., 2009; Tien et al., 2011). It is not clear how these multiple functions of UHRF1 are coordinated, as few studies have explored the possibility that UHRF1 is regulated by posttranslational modifications.
Zebrafish embryos with a mutation in uhrf1 have a defect in lens formation (Tittle et al., 2011), fail to expand their liver bud, and die during the larval period (Sadler et al., 2007). uhrf1 heterozygous adults do not regenerate their liver following surgical resection (Sadler et al., 2007). These defects suggest that uhrf1 is required for hepatocyte proliferation. Additional studies in tissue culture cells demonstrate that UHRF1 depletion causes cell cycle arrest (Bonapace et al., 2002; Jenkins et al., 2005; Tien et al., 2011), hypersensitivity to DNA damage and chemotherapeutic agents (Muto et al., 2002; Arima et al., 2004), or apoptosis (Abbady et al., 2003; Tien et al., 2011). Conversely, high levels of UHRF1 are found in a number of human cancer cell lines (Mousli et al., 2003; Jenkins et al., 2005) and primary tumors (Hopfner et al., 2000; Crnogorac-Jurcevic et al., 2005) and, in some cell types, overexpression of UHRF1 increases cell proliferation (Hopfner et al., 2002; Jenkins et al., 2005). Therefore UHRF1 may be a prime target for therapies aimed at sensitizing cancer cells to genotoxic chemotherapeutics.
While the mechanism by which UHRF1 contributes to cell division and apoptosis is not well understood, the role of UHRF1 as an important regulator of the epigenome is better characterized. It is likely that the function of UHRF1 is highly conserved, as there is a 67% overall amino acid identity between zebrafish and human UHRF1 and an even stronger homology between the individual domains, each of which has been associated with important epigenetic processes. The SRA domain, which is responsible for recognizing hemimethylated DNA, recruiting the major DNA methyltransferases DNMT1 (Bostick et al., 2007; Sharif et al., 2007), DNMT3a, and DNMT3b (Meilinger et al., 2009) and the histone deacetylase HDAC1 (Unoki et al., 2004), is nearly 90% identical to that of the zebrafish and is also remarkably well conserved in plants (Johnson et al., 2007; Woo et al., 2007; Hashimoto et al., 2008). UHRF1 binds preferentially to methylated H3K9 marks, in part through its tandem Tudor (60% homology) domains (Rottach et al., 2010), and recruits the histone methyltransferase G9a via its SRA and RING (72% homology) domains (Kim et al., 2009). The RING domain also possesses E3 ligase activity and is known to ubiquitinate histone H3 (Jenkins et al., 2005; Citterio et al., 2004). Finally, the PHD domain (72% homology) has been shown to function in the reorganization of pericentric heterochromatin (Papait et al., 2008) and to preferentially bind unmethylated histone H3R2 (Rajakumara et al., 2011; Wang et al., 2011). Whether UHRF1 reads and writes all of these epigenetic marks simultaneously is not known, nor is it clear whether UHRF1 serves the same function in different cell types. However, a role for UHRF1 in methylation is clearly conserved, as both mammalian cells and zebrafish embryos that lack Uhrf1 have genome-wide hypomethylation (Bostick et al., 2007; Sharif et al., 2007; Feng et al., 2010). Moreover, the strikingly similar phenotype between the eye (Tittle et al., 2011) and liver (Anderson et al., 2009) phenotypes of dnmt1-deficient zebrafish embryos and uhrf1 mutants (Sadler et al., 2007; Tittle et al., 2011) points to the common function of these two genes. However, epistasis experiments suggest that the loss of DNA methylation may account for some, but not all, of the embryonic phenotypes associated with uhrf1 loss (Tittle et al., 2011). Given the important role that epigenetics plays in regulating both the activation of the zygotic genome that occurs early in development as well as in the lineage-specific regulation of developmentally important genes (Mudbhary and Sadler, 2011), it is predicted that Uhrf1 may be required for coordinating the multiple and complex epigenetic marks that rapidly change during embryonic development.
Despite the emerging understanding of how and where UHRF1 functions, little is known about how UHRF1 itself is regulated. The dynamic expression pattern of uhrf1 mRNA during embryonic development (Sadler et al., 2007; Tittle et al., 2011) points to transcriptional control as one mechanism of regulation. Changes in uhrf1 mRNA (Sadler et al., 2007), protein (Bonapace et al., 2002; Arima et al., 2004; Brunet et al., 2008; Kim et al., 2009), and localization (Uemura et al., 2000; Miura et al., 2001; Kim et al., 2009) accompany different cell cycle stages in some cells, but in others, most notably in cancer cells, UHRF1 levels do not change dramatically during the cell cycle (Mousli et al., 2003; Arima et al., 2004; Bronner et al., 2007). UHRF1 expression is controlled by key cell cycle regulators, including the Rb/E2F complex (Abbady et al., 2003), p53/p21Cip1(Arima et al., 2004), and the E1A transcription factor (Bonapace et al., 2002). We hypothesize that other factors that act during the cell cycle may provide an additional level of regulation of UHRF1.
Cyclin-dependent kinases (CDKs) control the major transitions in the mammalian cell cycle. CDK2 activity during G1 and S phase is dependent on either cyclin E (CCNE1) or cyclin A (CCNA2). In vitro, there is significant overlap in substrate specificity between CDK2 partnered with either CCNE1 or CCNA2, whose shared targets include histone H1, pRb, p107, and Cdc6 (Kitagawa et al., 1996; Takeda et al., 2001; Ukomadu and Dutta, 2003a). A notable exception is E2F1, which is only phosphorylated by CCNA2/CDK2 (Krek et al., 1995; Kitagawa et al., 1996). An additional level of CDK2 specificity in vivo is likely achieved through temporal activation, as CDK2 shifts from CCNE1 binding in mid- to late G1 (Ohtsubo et al., 1995; Resnitzky and Reed, 1995) to CCNA2 binding, which drives cells from G1 into S phase (Pagano et al., 1992; Zindy et al., 1992; Resnitzky et al., 1995).
We discovered a putative CDK2 phosphorylation site corresponding to Ser-661 of human UHRF1 (corresponds to Ser-648 in zebrafish; Figure 1A). That UHRF1 is a substrate for CDK2 is supported by the observations that: 1) overexpression of UHRF1 in combination with CDK2 causes quiescent, differentiated cells to reenter the cell cycle (Bonapace et al., 2002); 2) another UHRF family member, UHRF2, interacts with CDK2 (Li et al., 2004); and 3) the expression of uhrf1 and ccna2 are coordinated following partial hepatectomy in both mice and zebrafish (Sadler et al., 2007). Since UHRF1 and CCNA2 are both required in S phase, we asked whether there might be a direct relationship between them. Our data demonstrate that a novel mechanism of UHRF1 regulation via phosphorylation by CCNA2/CDK2 exists and that phosphorylation of UHRF1 on Ser-661 is essential for zebrafish embryogenesis.
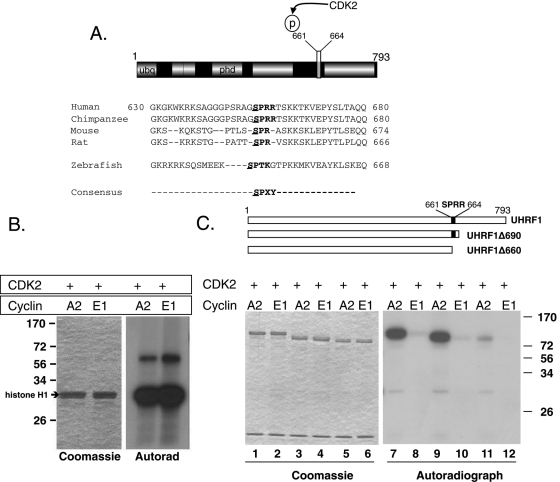
FIGURE 1:
UHRF1 is phosphorylated by CCNA2/CDK2 at Ser-661. (A) Domain structure of UHRF1 and alignment of the conserved CDK2 phosphorylation site in vertebrate UHRF1. Canonical consensus is bolded (SPXY: X = any amino acid; Y = basic amino acid). The putative phosphorylation residue is underlined. (B) Left, Coomassie-stained gel shows equal loading of histone H1. Right, autoradiograph of the phosphorylated histone H1. (C) Top, Schematic representation of UHRF1 truncation mutations used as substrates for CDK2 kinase assays. Black box indicates the CDK2 phosphorylation site (SPRR). Left, Coomassie staining showing equal loading in all lanes. CCNA2/CDK2 phosphorylates a truncated UHRF1 that retains the SPRR motif (lanes 7 and 9) but a truncated mutant lacking SPRR (lane 11) is barely phosphorylated. CCNE1/CDK2 has almost no kinase activity against UHRF1.
RESULTS
UHRF1 is phosphorylated on Ser-661 in vitro by CCNA2/CDK2
Alterations in UHRF1 expression affect cell cycle progression (Hopfner et al., 2002; Muto et al., 2002; Jeanblanc et al., 2005; Jenkins et al., 2005; Tien et al., 2011). UHRF1 mRNA (Arima et al., 2004; Abbady et al., 2005; Sadler et al., 2007; Brunet et al., 2008) and protein (Uemura et al., 2000; Miura et al., 2001; Bonapace et al., 2002; Kim et al., 2009) levels fluctuate during the cell cycle in parallel with CCNA2, raising the possibility that UHRF1 and CCNA2 are functionally linked. The CDK2 consensus sequence is S/TPXY (where X is any amino acid and Y is a basic amino acid). The first and second residues (serine or threonine, followed by proline) are always required, but substituted adducts at the third and fourth amino acid can be phosphorylated (Higashi et al., 1995). Figure 1A illustrates a classic CDK2 phosphorylation site (SPRR) at amino acids 661–664 of UHRF1 and shows the serine-proline to be conserved in all vertebrate homologues of UHRF1, but absent from UHRF2. A canonical Cdk2 site SPTK is present in the zebrafish protein, with Ser-648 as part of the consensus site (Figure 1A). There is one other CDK2 site beginning with Ser-709 (SPFQ); however, this motif is not conserved in mice or zebrafish.
To test whether UHRF1 is a CDK substrate, we assessed in vitro complexes created from activated, recombinant CDK2 paired with either CCNE1 or CCNA2 for their ability to phosphorylate either their common substrate, histone H1, or recombinant His6-UHRF1. While both CCNE1/CDK2 and CCNA2/CDK2 are equally capable of phosphorylating histone H1 (Figure 1B), only CCNA2/CDK2 phosphorylates recombinant His6-UHRF1 (Figure 1C). This phosphorylation is maintained when UHRF1 is truncated at amino acid 690 (UHRF1Δ690), but virtually none occurs when the putative CDK2 site at Ser-661 is removed (UHRF1Δ660; Figure 1C). Therefore phosphorylation by CDK2 must occur at a site between amino acids 660 and 690, thus eliminating Ser-709 as a likely CDK2 substrate. Taken together, our data indicate that UHRF1 is phosphorylated on a CDK2 phosphorylation motif residing between amino acids 660 and 690. Moreover, complexes containing CCNA2 are selective for UHRF1, suggesting that CCNA2/CDK2 may regulate UHRF1 function.
UHRF1 is phosphorylated on Ser-661 in vivo
To determine whether UHRF1 is phosphorylated on Ser-661 in vivo, we generated a polyclonal antibody against a peptide containing a phosphorylated residue on Ser-661 (α-pS661) and then validated its specificity. First, we used this antibody to determine whether UHRF1 in mammalian tissue culture cells can be phosphorylated on this site. Cells were transiently transfected with an empty vector, UHRF1 tagged with the FLAG epitope, or the FLAG-tagged mutant version of UHRF1 with Ser-661 changed to an unphosphorylatable residue (alanine: UHRF1S661A in Figure 2A or glycine: UHRF1S661G in Supplemental Figure S1A). We found that both anti-UHRF1 and anti-FLAG detect a band of the appropriate size in cells transfected with constructs encoding wild-type or mutant forms of UHRF1, whereas α-pS661 recognizes a weak band at the appropriate size in untransfected HCT116 cells (Figure 2A) and a strong band at the same size in cells transfected with wild-type (FLAG-UHRF1) but not in those transfected with UHRF1S661A (Figure 2A) and UHRF1S661G (Figure S1A).
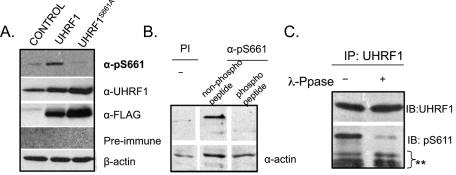
FIGURE 2:
α-pS661 detects phospho-UHRF1 in vivo and phosphorylation is eliminated when Ser-661 is changed to glycine. (A) HCT116 cells transfected with empty plasmid (CONTROL), FLAG epitope–tagged UHRF1 (UHRF1), or UHRF1 in which Ser-661 is changed to an alanine (UHRF1S661A). α-pS661 specifically recognizes a protein of appropriate size in FLAG-UHRF1 cells, but not in FLAG-UHRF1S661A cells, whereas both FLAG-UHRF1 and FLAG-UHRF1S661A are recognized by α-UHRF1 and α-FLAG. (B) Endogenous UHRF1 is phosphorylated on Ser-661. HCT116 cell lysates blotted with Preimmune (PI) or α-pS661 sera that was preincubated with either the phosphorylated or a cognate nonphosphorylated form of the immunizing peptide. Actin used as a loading control. (C) Lysates from HCT116 cells were immunoprecipitated with anti-UHRF1 antibody and then treated with or without λ-phosphatase. Immunoblotting (IB) with α-UHRF1 and with α-pS661 shows that phosphatase treatment reduces recognition of UHRF1 by α-pS661 but not by α-UHRF1.
We next evaluated the ability of α-pS661 to recognize endogenous UHRF1 and to further confirm the specificity of Ser-661 phosphorylation. HCT116 cells immunoblotted with α-pS661 reveal a 95-kDa band (Figure 2, A and B). This immunoreactivity is prevented when α-pS661 is preincubated with the immunizing peptide, but not with the cognate peptide lacking the phosphorylated serine (Figure 2B). A similar result is obtained with mouse livers harvested 48 h after partial hepatectomy (Figure S1B). The band recognized by α-pS661 in HCT116 cells disappears when an immunoprecipitate of the UHRF1 is treated with λ-phosphatase (Figure 2C), confirming that the immunoreactivity of α-pS661 is dependent on the phosphorylation of Ser-661.
The results with colon cancer cells (Figure 2) and mouse hepatocytes (Figure S1B) demonstrate that endogenous UHRF1 is phosphorylated in some cells. Interestingly, we found variability between the amounts of endogenous phosphorylated UHRF1 in different cell types: α-pS661 recognizes a band in the colon cancer line HCT116 but not in the transformed, nonmalignant 293T cell line. However, it is clear that exogenously provided UHRF1 can be phosphorylated in a variety of transformed and malignant mammalian cells (unpublished data). To determine whether UHRF1 phosphorylation is conserved across species and whether it occurs in nontransformed cells, we introduced zebrafish Uhrf1 tagged with a 6X-myc-epitope (6X-myc-Uhrf1) into zebrafish embryos using mRNA injection (Figure S1C) or human UHRF1 tagged with green fluorescent protein (GFP; UHRF1-EGFP) using transgenes (see Figure 3 and Figure 7 later in this paper). Embryos injected at the one-cell stage with mRNA encoding 6X-myc-Uhrf1 were immunoblotted using α-pS661 and α-Myc. Both antibodies detect a single band in pregastrula embryos (Figure S1C) and a faint band is also detected in uninjected embryos (Figure S1, arrow), suggesting that in zebrafish, as in mammalian cells, endogenous Uhrf1 is phosphorylated. Immunofluorescence studies support this: UHRF1-EGFP expressed in zebrafish under either the heat shock promoter (Tg(hsp70:UHRF1-EGFP); see Figure 3B) or the hepatocyte-specific fabp10 promoter (Tg(fabp10:UHRF1-EGFP); see Figure 7 later in this paper) is recognized by α-pS661, as is the endogenous Uhrf1 in some cell types (see Figure 7C later in this paper). Collectively, these findings support the conclusions that both exogenously provided UHRF1 and endogenous UHRF1 are phosphorylated on Ser-661 in vivo in both mammalian and zebrafish cells, that the phosphorylation site on the serine at position 648 in zebrafish corresponds to position 661 in human UHRF1, and that the serine phosphorylated moiety of UHRF1 is enriched in mouse hepatocytes and zebrafish embryonic cells that are actively traversing the cell cycle.
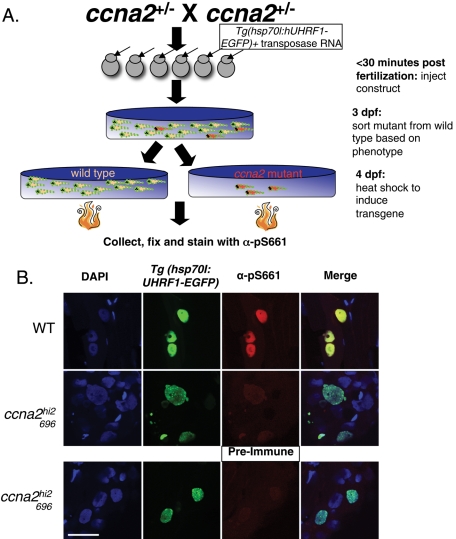
FIGURE 3:
Phosphorylation of UHRF1 requires ccna2. (A) Experimental design. Embryos obtained from a cross of heterozygous ccna2hi2696 mutants were injected (arrow) with a construct containing UHRF1-EGFP driven by the heat shock (hsp70l) promoter. Mutants (red) were separated from wild-type (tan) siblings based on phenotype at 3 dpf, heat-shocked at 4 dpf, and stained for the presence of phosphorylated UHRF1. (B) Immunofluorescence of 4-dpf wild-type (WT) and ccna2hi2696 embryos shows that ccna2hi2696 mutants do not have phosphorylation of UHRF1 despite overexpression of UHRF1, as seen by the GFP, due to mosaic expression from the Tg(hsp70I: UHRF1-EGFP) transgene. Mosaic expression is obtained by this method, so not all nuclei are labeled with GFP. Scale bar: 25 μm.
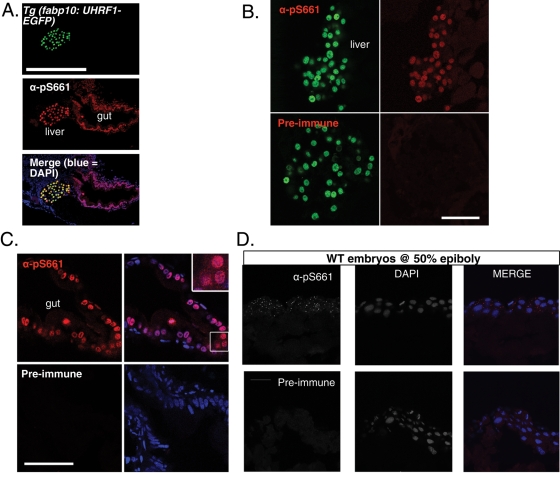
FIGURE 7:
Phosphorylated endogenous Uhrf1 is cytoplasmic. (A) α-pS661 detects both overexpressed and endogenous phosphorylated UHRF1 in zebrafish. Top, GFP signal in hepatocytes of Tg(fabp10:UHRF1-GFP); middle, immunofluorescence with α-pS661; bottom, merge with DAPI. The gut and liver are labeled. Magnification: 20×; scale bar: 250 μm. (B) Sections of 5-dpf zebrafish expressing GFP-tagged UHRF1 under the control of the hepatocyte specific promoter (Tg(fabp10:UHRF1-EGFP)). Left, GFP expression in hepatocytes; right, immunofluorescence with α-pS661 (top) or preimmune (bottom). Confocal images taken with identical parameters. Scale bar: 50 μm. (C) Left, immunofluorescence of enterocytes with α-pS661 or preimmune. Right, merge with DAPI. Inset, 63× magnification of endogenous phosphorylated UHRF1 in the zebrafish gut with predominant nuclear localization. Confocal images taken with identical parameters. Scale bar: 50 μm. (D) Endogenous UHRF1 is phosphorylated in early development as seen with immunofluorescence on cryosections at 50% epiboly. Bottom, left, immunofluorescence on cryosections of embryos at 50% epiboly with preimmune sera; middle, DAPI alone; right, merge. Top, left, immunofluorescence with α-pS661; middle, DAPI alone; right, merge. Magnification: 63×; scale bar: 25 μm.
Phosphorylation of UHRF1 requires ccna2 in zebrafish
Given that CCNA2/CDK2 selectively phosphorylates UHRF1 in vitro (Figure 1), we asked whether this is also the case in vivo. To address this, we used ccna2hi2696 mutant zebrafish, which have a viral insertion in the first coding exon of the ccna2 gene (Figure S2A; Amsterdam et al., 2004). At day 4, these fish have no detectable ccna2 transcript as seen by PCR (Figure S2B), and some features of their phenotype are reminiscent of that seen in uhrf1 mutants (Sadler et al., 2007), including a small liver (Figure S2C). To test whether Ccna2 is required for UHRF1 phosphorylation in vivo, we carried out the experiment design outlined in Figure 3A. UHRF1 tagged with EGFP was introduced to ccna2 mutants by injecting the offspring of a cross between ccna2hi2696 heterozygous adults with a construct expressing EGFP-tagged UHRF1 under a heat shock–inducible promoter (Tg(hsp70I:UHRF1-EGFP)) and containing the Tol2 transposon sites that allow for genome integration (Kawakami, 2007; Kwan et al., 2007). At 3 d postfertilization (dpf), we segregated the ccna2hi2696 mutant embryos from their siblings based on phenotype (see Figure S2C). At 4 dpf, embryos were heat-shocked to induce expression of the transgene in those cells that integrated the construct into the genome. Cryosections from larvae collected 4 h after heat shock were immunostained with α-pS661. While EGFP-expressing cells occur with similar frequency in both wild-type and ccna2hi2696 mutants, α-pS661 recognizes the transgene only in wild-type embryos (Figure 3B). Moreover, there is no staining of endogenous phosphorylated Uhrf1 in any ccna2 mutant cells. While these findings could reflect a defect in the ccna2 mutant cells that precludes UHRF1 phosphorylation, it is also consistent with our in vitro data, suggesting a requirement for CCNA2 in UHRF1 phosphorylation.
uhrf1 is essential for pregastrula development in zebrafish
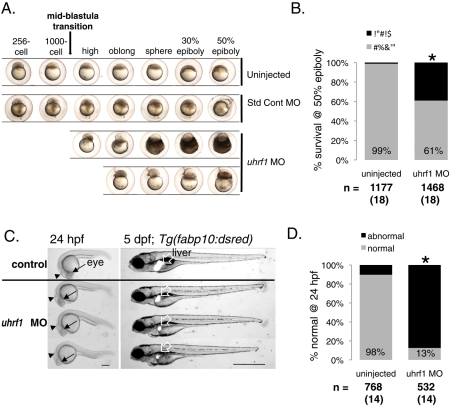
Uhrf1 depletion in mice results in embryonic lethality in early gestation (Muto et al., 2002). In contrast, the uhrf1hi272 zebrafish mutant phenotype is first evident late in development (Sadler et al., 2007), similar to dnmt1 mutant zebrafish (Tittle et al., 2011). Uhrf1 is highly expressed in the early embryo (Sadler et al., 2007; Tittle et al., 2011), and we assume that zebrafish uhrf1hi272 mutants survive early development due to maternally supplied mRNA and/or protein. For most genes, the transition from maternally provided transcripts to those derived from the zygotic genome (i.e., the maternal–zygotic transition) occurs during the midblastula transition. Activation of the zygotic genome during this time is associated with widespread changes in chromatin structure and epigenetic modifications (Newport and Kirschner, 1982b; Kane and Kimmel, 1993; Meehan et al., 2005; Schier, 2007; Tadros and Lipshitz, 2009; Lindeman et al., 2010b; Vastenhouw et al., 2010). We asked whether suppressing translation of both maternal and zygotic uhrf1 mRNA using a morpholino targeting the ATG of the uhrf1 message (Figure S3) would reveal a phenotype earlier in development than is observed in uhrf1hi272 embryos. We found that this was indeed the case: uhrf1 morphant embryos proceed through the blastula period without interruption, but a significant percent (32%) arrest before gastrulation at the high, sphere, or dome stages (Figure 4A). Most of these arrested embryos (80%) die by the time control embryos reach 50% epiboly (Figure 4A). The remaining 20% of arrested embryos do not complete epiboly and die by 24 h postfertilization (hpf). Our observations are based on careful gross inspection of live embryos using a standard dissecting microscope. While the early morphant embryos appear to have the same morphological appearance as the control embryos, our observations cannot exclude the possibility that a subtle cellular defect occurs in morphants prior to the midblastula transition.
FIGURE 4:
uhrf1 is essential for zebrafish development. (A) Early embryonic development of uninjected, standard control morpholino-injected, and uhrf1 morpholino-injected embryos. uhrf1 morphants display a distinct developmental arrest phenotype leading to early embryonic death. Scale bar: 500 μm. (B) By 50% epiboly, uhrf1 morphants exhibit decreased survival to 61% compared with 99% of control. Total number of embryos and experiments are noted under each bar. *, p < 0.0001 by Fisher's exact test. (C) Left, uhrf1 morphants at 24 hpf are characterized by a small head, underdeveloped eye (arrow), and abnormal brain, typified by the depressed midbrain–hindbrain boundary (arrowhead) and dilated ventricle (caudal to arrowhead). Scale bar: 100 μm. Right, uhrf1 morphants at 5 dpf have a small liver as visualized in Tg(fabp10:dsred) zebrafish. All morphants have a small liver in three experiments. Scale bar: 500 μm. (D) uhrf1 morphants have a significant abnormal phenotype at 24 hpf. Total number of embryos and experiments are noted under each bar; *, p < 0.0001 by Fisher's exact test.
Using early embryonic death as the phenotype for binary scoring, we carried out 18 individual experiments involving more than 1400 embryos, demonstrating that only 61% of uhrf1 morphants survive to the 50% epiboly stage (∼6 hpf), compared with 99% in control embryos (Figure 4B; p < 0.0001 by Fisher's exact test). Those that do not arrest or die during gastrulation appear normal, albeit delayed, through bud stage (unpublished data), but are severely affected at 24 hpf (Figure 4C). Nearly all morphants that survive to 24 hpf have a malformed CNS, smaller head size, overall small size (Figure 4, C and D; see also Figure S5B), and significantly smaller eyes (Figure S4), reminiscent of the eye defects observed in uhrf1 mutants (Tittle et al., 2011). By 5 dpf, morphants have a small liver size (Figure 4C, right), similar to uhrf1hi272 mutants (Sadler et al., 2007).
Embryos injected with the same amount of a standard control morpholino or those injected with a higher concentration of another morpholino (targeting p53) do not display early arrest, significant mortality, or phenotypic differences when compared with noninjected embryos (Figures 4A and S5, A and B). Thus we used uninjected embryos as controls throughout. Moreover, reducing nonspecific morpholino toxicity by coinjecting the uhrf1 morpholino with a morpholino targeting p53 (Robu et al., 2007) does not alter the survival at 50% epiboly or 24-hpf phenotype of the uhrf1 morphants (Figure S5, A and B). Therefore the effects of uhrf1 knockdown appear to be specific, causing developmental arrest following the midblastula transition, early embryonic death, and severe morphological abnormalities in the brain, eye, and liver.
Phosphorylation of UHRF1 is required for early zebrafish development
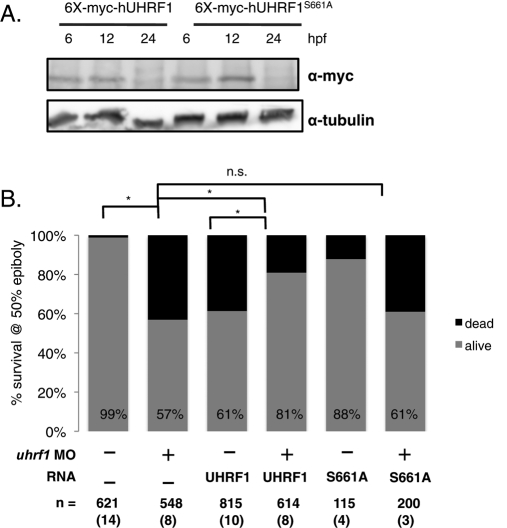
Zebrafish and human UHRF1 share a 67% overall amino acid identity and an even higher homology between the individual domains. If the human UHRF1 gene is the functional orthologue of the zebrafish gene, then it will rescue zebrafish embryos that lack uhrf1. We tested this by injecting zebrafish embryos with RNA encoding UHRF1 to determine whether it could rescue the morphant or mutant phenotypes. Western immunoblots of embryo lysates show that the UHRF1 translated from mRNA injected at the one-cell stage is detectable in embryos at 50% epiboly (Figure 5A; see also Figures S1C and S6A) but not at 24 hpf (Figure 5A). We reasoned that if UHRF1 were sufficient to rescue any consequences of uhrf1 loss, then only the earliest embryonic event, that is, survival through gastrulation at 50% epiboly, requires Uhrf1.
FIGURE 5:
UHRF1 levels are tightly regulated, and phosphorylation of UHRF1 at Ser-661 is essential for zebrafish embryogenesis. (A) Western blot with α-myc of 6, 12, and 24 hpf embryos injected with 400 pg of mRNA encoding full-length WT-UHRF1 and UHRF1S661A. Tubulin is the loading control. (B) Both depletion and overexpression of UHRF1 are deleterious to embryonic survival. Only 57% of morphants and 61% of embryos injected with 6X-myc-UHRF1 survive to 50% epiboly. Survival improves to 81% when the morpholino is coinjected with the 6X-myc-UHRF1 mRNA (compare columns 2–4 and columns 3–4; for both, p < 0.0001 by Fisher's exact test). Coinjection of morpholino and mRNA encoding UHRF1S661A mRNA results in no difference in mortality as compared with morpholino alone; p = 0.9146 by Fisher's exact test. mRNA = 400 pg. The total number of embryos and experiments are noted under each bar.
Embryos were divided into four groups: 1) coinjected with mRNA encoding 6X-myc-UHRF1 plus uhrf1 morpholino, 2) injected with the morpholino alone, 3) injected with the mRNA alone, and 4) uninjected. Each group was scored for early arrest and eventual mortality at the time corresponding to 50% epiboly in their uninjected siblings. Whereas on average only 57% of the morphants survive to 50% epiboly, survival improves to 81% when the morpholino is coinjected with mRNA encoding 6X-myc-UHRF1 (Figure 5B, compare columns 2–4; p < 0.0001 by Fisher's exact test). As expected, since there is no UHRF1 protein remaining in injected embryos at 24 hpf (Figure 5A), 6X-myc-UHRF1 mRNA injection does not rescue the phenotype induced by the uhrf1 morphant at 24 hpf or 5 dpf (unpublished data). Nevertheless, the significant reduction in mortality at 50% epiboly in uhrf1 morphants demonstrates that human and zebrafish UHRF1 are functional orthologues.
Studies in mammalian cells have revealed phenotypes caused by both depletion and enhancement of UHRF1 protein levels. We questioned whether increasing UHRF1 levels in zebrafish embryos induces a phenotype. We found this to be the case, as only 61% of embryos injected with 6X-myc-UHRF1 mRNA survive to 50% epiboly (Figure 5B, column 3; p < 0.0001 by Fisher's exact test compared with uninjected controls). The increased mortality due to overexpression of the myc-tagged wild-type UHRF1 is significantly reduced with coinjection of uhrf1 morpholino, which targets endogenous UHRF1 but not the significantly mismatched human UHRF1 sequence (Figure 5B, column 4; p < 0.0001 by Fisher's exact test). Thus we conclude that both too much and too little UHRF1 are deleterious for the rapid cell cycles and the massive epigenetic modifications that are required for normal progression through the maternal–zygotic transition. Importantly, when the morpholino and the mRNA are coinjected, embryonic survival is restored, likely because the level of Uhrf1 is maintained within the range required for successful early development.
Our finding that Uhrf1 can be phosphorylated in gastrula (Figure S1C) suggests that the machinery required for this phosphorylation is present in early embryos. To test whether phosphorylation is required for Uhrf1 function during the midblastula transition and gastrulation, we coinjected the uhrf1 morpholino with mRNA encoding nonphosphorylatable human and zebrafish UHRF1. Neither UHRF1S661A (Figure 5B, column 6; p = 0.3573 by Fisher's exact test), UHRF1S661G (Figure S6B, column 5; p = 0.2603 by Fisher's exact test), nor Uhrf1S648A (Figure S6C, column 4; n.s. p value = 0.4681) improve morphant survival at 50% epiboly. Mutant versions of the protein are detected at levels equivalent to those in embryos injected with mRNA encoding wild-type UHRF1 (Figures 5A and S6, A and D), suggesting the phenotype is not attributable to differences in UHRF1 levels. Coinjecting the uhrf1 morpholino with zebrafish Uhrf1 mRNA, which has an amino terminus 6X-myc tag and significant mismatch introduced by utilizing the degeneracy of the genetic code in the mRNA clone (Figure S3), improves survival of gastrula from 63% in morphants to 83% in the coinjected embryos (Figure S6C, column 4; p < 0.0001 by Fisher's exact test). In contrast, morphant survival is unchanged (66%) when coinjected with equivalent amounts encoding Uhrf1S648A (Figure S6, C and D; p = 0.4681 compared with morpholino alone). Collectively, these data demonstrate that 1) Uhrf1 can be phosphorylated in pregastrula embryos and 2) phosphorylation is required for Uhrf1 function in early vertebrate development.
Localization of UHRF1 changes with phosphorylation status
All functions ascribed to UHRF1 occur in the nucleus. Accordingly, UHRF1 was initially identified as a nuclear protein (Fujimori et al., 1998). Mutating the phospho-acceptor site does not dramatically alter protein stability (Figures 5 and S6, A and D) or the ability of UHRF1 to interact with DNMT1 (Figure S7), suggesting that there is no gross change in protein structure caused by mutation of this residue. We thus asked whether phosphorylation at Ser-661 alters UHRF1 localization.
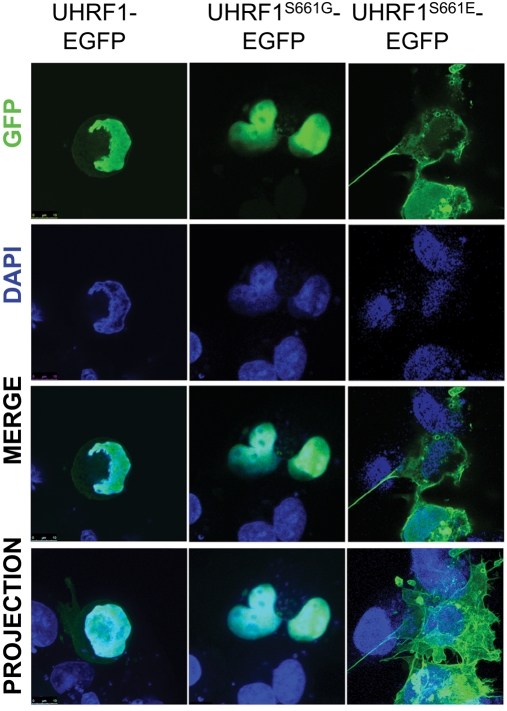
We found that UHRF1-EGFP is exclusively nuclear and not phosphorylated in ccna2 mutant cells UHRF1-EGFP (Figure 3B), which suggests that nonphosphorylatable UHRF1 would also be localized to the nucleus. Indeed, UHRF1S661G is detected only in the nucleus of nontransformed human (Cos7, Figure 6; 293T, Figure S8A) and normal zebrafish cells (Figure S8B). However, in some cell types, such as those in the eye of 2-dpf embryos, UHRF1-EGFP expressed via the heat shock promoter (Tg(hsp70I:UHRF1-EGFP)) is primarily cytoplasmic, whereas nonphosphorylatable (UHRF1S661G) is only detected in the nucleus in all cell types examined (Figure S8C). Therefore we conclude that a lack of phosphorylation does not prevent nuclear localization; however, these data suggest UHRF1 may be cytoplasmic in some scenarios, and this cytoplasmic localization may require phosphorylation on Ser-661. To further address this, we investigated the localization of a phosphomimetic mutation of UHRF1 (UHRF1S661E) and found it to be primarily cytoplasmic in Cos7 (Figure 6) and 293T (Figure S8A) cells. This suggests that phosphorylation at Ser-661 may cause UHRF1 to localize to the cytoplasm.
FIGURE 6:
Localization of UHRF1 changes with its phosphorylation. Cos7 cells transfected with UHRF1-GFP, UHRF1S661G-GFP, or UHRF1S661E-GFP show that phosphorylation of UHRF1 promotes cytoplasmic localization in these cells. Top panel, GFP signal in transfected cells. Second row, DAPI signal only. Third row, merge of top two panels. Bottom panel, maximum projection, merge. Magnification: 63×; scale bar: 10 μm.
To test this, immunofluorescence was performed on mammalian and zebrafish cells using α-pS661 that was affinity-purified by multiple passages through a column containing the cognate peptide before binding to and elution from an affinity column of the immunizing peptide. While this antibody readily recognizes exogenously provided UHRF1 in zebrafish cells, both phosphorylated UHRF1 and total UHRF1 as detected by EGFP fluorescence (see Figures 3B and 7, A and B) and immunofluorescence with anti-GFP (unpublished data) can only be detected in the nucleus,. However, we were unable to reliably detect endogenous phosphorylated UHRF1 in 293T, Cos7, and HepG2 cells (unpublished data). Staining of endogenous phosphorylated Uhrf1 is observed, however, in both the cytoplasm and nucleus of enterocytes in 5-dpf larvae (Figure 7, A and C) and in the cytoplasm of blastula-stage embryos (Figure 7D), corresponding to the stage of development in which Uhrf1 phosphorylation is required. It is curious, however, that there is little to no phosphorylated Uhrf1 identified in any other tissue in 5-dpf zebrafish. While we cannot rule out that this is reflective of the absence of total Uhrf1 expression in other cell types, since we were unable to successfully utilize any UHRF1 antibodies for immunostaining the endogenous zebrafish Uhrf1, our data indicate that both the levels of total UHRF1 and intracellular localization of phosphorylated UHRF1 vary between cells. Whether the differences in localization are dependent on cell type, differentiation status, or cell cycle stage remains to be determined.
DISCUSSION
UHRF1 is a multi-domain protein involved in epigenetic modification of histones and DNA (Unoki et al., 2004; Bostick et al., 2007; Sharif et al., 2007; Karagianni et al., 2008). Both loss- and gain-of-function experiments indicate that UHRF1 contributes to cell cycle progression (Bonapace et al., 2002; Hopfner et al., 2002; Arima et al., 2004; Jeanblanc et al., 2005; Sadler et al., 2007; Brunet et al., 2008; Fang et al., 2009), although it is interesting to note that manipulating UHRF1 most severely affects cell division in nonmalignant cells (Bonapace et al., 2002; Arima et al., 2004; Sadler et al., 2007). In this paper, we show that the conserved serine residue, which lies between the SRA and RING domains (position 661 in humans and 648 in zebrafish), is phosphorylated by CCNA2/CDK2 in vitro and that depleting ccna2 prevents UHRF1 phosphorylation in vivo. We demonstrate the important function of UHRF1 phosphorylation with our finding that UHRF1 is able to rescue pregastrulation mortality caused by uhrf1 depletion. Moreover, we find that either a phospho-mimetic mutant or endogenous phosphorylated UHRF1 is differentially localized compared with total UHRF1 or nonphosphorylatable UHRF1. Together, these data are consistent with a model in which UHRF1 phosphorylation can promote localization to the cytoplasm, where it may carry out an essential, previously unidentified function. An alternative possibility is that changing UHRF1 phosphorylation status does not affect interaction with DNMT1, but it might alter binding affinity for another one of its partners or alter specificity for modified histones and thereby alter the ability of UHRF1 to read the epigenetic code.
Three significant aspects of UHRF1 are revealed by this study. First, very little is known about how UHRF1 is regulated. While levels of UHRF1 mRNA and protein and its localization change during the cell cycle, these effects are not seen in all cell types. Indeed, in several cancer cell lines, UHRF1 levels are constant throughout the cell cycle (Mousli et al., 2003; Bronner et al., 2007). In these cases, it is possible that posttranslational modifications, such as phosphorylation, may provide an additional level of UHRF1 regulation; however, this area has received little attention. One study found that UHRF1 is phosphorylated by protein kinase A at Ser-298 and speculates that this corresponds to increased TOP2A gene expression (Trotzier et al., 2004). The significance of this finding is difficult to interpret, as Ser-298 is not conserved in zebrafish Uhrf1, and we have not found any change in TOP2A expression when UHRF1 levels are manipulated in cancer cells or in zebrafish (Tien et al., 2011; unpublished data). A related protein, UHRF2, does not contain the same Ser-661, but may be phosphorylated at different sites by CDK2 (Li et al., 2004), neither of which is conserved in UHRF1. We speculate that phosphorylation may be a central mechanism for regulating members of the UHRF1 family during specific cell cycle stages.
Second, the finding that UHRF1 is selectively phosphorylated in vitro by CCNA2/CDK2 complexes is unusual, as the CCNA2/CDK2 complex is more discriminating in substrate selection than the less discerning CCNE1/CDK2 pair (Kitagawa et al., 1996; Takeda et al., 2001; Wohlschlegel et al., 2001; Ukomadu and Dutta, 2003a; Su and Stumpff, 2004; Malumbres and Barbacid, 2009). Using the ccna2hi2696 mutant zebrafish, we show that ccna2 is required for in vivo phosphorylation of overexpressed UHRF1, as well as endogenous Uhrf1, in gut epithelial cells (unpublished data). Unfortunately, there is no ccne1 mutant zebrafish to compare the effects of Ccne1 loss on UHRF1 phosphorylation. We predict that Ccne1 knockdown with a morpholino may lead to severe embryonic defects, as seen in invertebrates (Knoblich et al., 1994), or will cause cell cycle anomalies that would perturb Ccna2 synthesis. A similar caveat applies to results with ccna2hi2696 mutants, in which the cell cycle may be perturbed due to the lack of Ccna2, so that the lack of UHRF1 phosphorylation is secondary to a cell cycle defect, not directly caused by loss of Ccna2 per se. Given that UHRF1 and CCNA2 are both expressed and function in S phase (Sadler et al., 2007), the phosphorylation of UHRF1 by CCNA2/CDK2 represents a novel and potentially important means of posttranslational regulation of UHRF1. Although the CDK2 phosphorylation site in UHRF1 is conserved in vertebrates, it does not have the canonical CY motif found in other CDK substrates, raising the possibility that the CDK/cyclin interaction with UHRF1 may occur through noncanonical motifs.
The third important and interesting finding of this study is the requirement for Uhrf1 at a stage of early vertebrate development in which there is massive reorganization of the genome. The uhrf1 mutant appears normal until 4–5 dpf, when defects in the brain, eye, liver, and gut become evident (Sadler et al., 2007; Tittle et al., 2011). This is correlated with global DNA hypomethylation (Feng et al., 2010). The survival of uhrf1 mutants to this late stage of development is likely attributed to maternally provided Uhrf1 protein and message. However, when we deplete both the maternal and zygotic Uhrf1 with a morpholino, we reveal a requirement for this gene in early embryogenesis just following the midblastula transition, when the zygotic genome is activated, (Newport and Kirschner, 1982a; Schier, 2007; Tadros and Lipshitz, 2009), the cell cycles lengthen (Schier and Talbot, 2005), and the embryos prepare for epiboly and gastrulation. At this time, there are global changes in the histone methylation pattern that are associated with both zygotic gene activation and repression (Bernstein et al., 2006; Lindeman et al., 2010a, 2010b; Vastenhouw et al., 2010). Although UHRF1 has been implicated in both DNA methylation and several histone modifications, DNA methylation is the only epigenetic mark that has thus far been reported to depend on Uhrf1 in zebrafish (Feng et al., 2010; Tittle et al., 2011). We were unable to detect major global DNA hypomethylation in pregastrula or 24-hpf morphants (unpublished data). However, this may be attributed to the sensitivity of the assay used. Moreover, alterations in other epigenetic modifications in midblastula uhrf1 morphants could culminate in embryonic death, and these have yet to be examined.
It is interesting that not all morphants display a phenotype early in embryogenesis. However, those that do survive are dramatically affected in the tissues that have been shown to express highest levels of uhrf, that is, the brain, eye, liver, gut, and brachial arches (Sadler et al., 2007; Tittle et al., 2011). We speculate that morphants display defects in proliferation and increased cell death in the affected tissues, which is supported by our preliminary studies revealing a significant increase in acridine orange staining in morphants (J. Chu and K. C. Sadler, unpublished data).
How does phosphorylation at Ser-661 alter UHRF1 function? Changing the phospho-acceptor serine to a glycine or alanine makes UHRF1 exclusively nuclear and does not alter interaction with DNMT1 (Figure S7). However, these nonphosphorylatable forms fail to rescue the morphant phenotype, and they do not induce a phenotype when overexpressed, indicating that UHRF1 that cannot be phosphorylated is not functional in cells of early zebrafish embryos. In contrast, substitution with a phospho-mimetic residue (glutamic acid) leads to cytoplasmic accumulation. Interestingly, although UHRF1 is predominantly nuclear, some wild-type UHRF1 localizes to the cytoplasm in a number of cell types (i.e., Cos7), suggesting that its entrance or exit from the nucleus may be a regulated process. It is possible that cytoplasmic UHRF1 interacts with other proteins, perhaps retaining them in the cytoplasm or even promoting their transport into the nucleus.
In summary, we show that UHRF1 is a substrate for cyclin A/CDK2 in vitro and in vivo, that human UHRF1 is a functional homologue of zebrafish Uhrf1, and that phosphorylation of UHRF1 on Ser-661 is essential for embryonic development. This phosphorylation likely alters the localization of UHRF1 under some conditions. The effect of phosphorylation on epigenetic modifications is the focus of our ongoing studies.
MATERIALS AND METHODS
Zebrafish maintenance, embryo injection, and generation of transgenics
Adult zebrafish were maintained on a 14:10 h light:dark cycle at 28°C. Wild-type Tab 5 and Tab 14 and ccna2hi2696 fish (Amsterdam et al., 2004) were used. Fertilized embryos collected following natural spawning were cultured at 28°C in fish water (0.6 g/l Crystal Sea Marinemix; Marine Enterprises International, Baltimore, MD) containing methylene blue (0.002 g/l). The Mount Sinai School of Medicine Institutional Animal Care and Use Committee approved all protocols.
A morpholino (5′-CACCTGAATCCACATGGCGGCAAAC-3′) targeting the ATG of the uhrf1 transcript, standard control morpholino (CCTCTTACCTCAGTTACAATTTATA), and p53 morpholino (GCGCCATTGCTTTGCAAGAATTG) were obtained from Gene Tools (Philomath, OR). Morpholino (1.68 pg) was injected into embryos at the one- to two-cell stage using a Narishige IM-300 microinjector.
Transgenic fish expressing UHRF1 under the heat shock promoter (Tg(hsp70I:UHRF1-EGFP)) or in hepatocytes (Tg(fabp10:UHRF1-EGFP)) were created using Tol2-mediated transgenesis (Kwan et al., 2007). Two hundred picograms of the Tg construct was coinjected with 200 pg of mRNA encoding the Tol2 transposase into one-cell-stage embryos, creating F0 fish that express the transgene in a mosaic manner. At 5 dpf, embryos expressing the transgene were selected, raised to sexual maturity, and out-crossed. Heat shock protocol consisted of placing embryos in water prewarmed to 38°C and placing them in a 38°C incubator for 45 min. After 45 min, embryos were placed in 28.5°C incubator and monitored for transgene expression via fluorescence microscopy.
In vitro kinase assay
Active cyclin/CDK2 complexes were prepared as previously described (Kaldis et al., 1996). The substrate, recombinant full-length or truncated his6-UHRF1, was prepared by purification of bacterial lysate on a nickel column. Briefly, 2 μg histone H1 or recombinant full-length or truncated UHRF1 was added to 20 ml of kinase reaction buffer (50 mM Tris, pH 7.4, 10 mM MgCl2, 5 mM MnCl2, 5 mM dithiothreitol, 10 mM ATP, and 0.5mCi of [γ-32P]ATP). Ten nanograms of enzyme complex of CDK2 with either CCNA2 or CCNE1 was also added to the reaction. The mixture was incubated at 30°C for 30 min and the reaction was stopped by the addition of an equal volume of 2X sample buffer containing SDS. Samples were analyzed on SDS–PAGE, stained with Coomassie blue, and processed for autoradiography
Antibodies
The polyclonal antibody recognizing phosphorylated Ser-661 in UHRF (α-pS661) was generated by creating a synthetic phosphoserine peptide corresponding to residues 654–659 of human UHRF1 (CGPSRAGS(P04)PRRTSKKT). Following collection of preimmunization serum, rabbits were immunized with the peptide coupled to keyhole limpet hemocyanin. The resulting immune serum was screened for reactivity against UHRF1. A cognate peptide like the phosphorylated serine was also synthesized and used for affinity purification and as a blocking control. The crude antibody was used at 1:1000–1:2000 dilution for Western blots. Antibody for histological analysis was passed multiple times through a column of the cognate peptide before affinity purification with the immunizing peptide. The affinity-purified antibody was used at a dilution of 1:1000. Additionally, the following antibodies were used: anti-UHRF1 (known as ICBP90; 1:2000 except for zebrafish embryos at 1:500; BD Transduction, BD Biosciences, Franklin Lakes, NJ), anti-FLAG (at 1:2000 [Sigma-Aldrich, St. Louis, MO] or 1:1000 [Stratagene, Agilent, Santa Clara, CA]), anti-myc [9E10] (1:1500; Abgent, San Diego, CA), anti-β-actin (1:5000; Sigma), anti-tubulin (1:2000; Developmental Studies Hybridoma Bank, http://dshb.biology.uiowa.edu), and anti-DNMT1 (1:500; Santa Cruz Biotechnology, Santa Cruz, CA).
Cell lines and transfections
HEK293T, HCT116, and Cos7 cells were obtained from ATCC. Cell lines were grown in monolayer in appropriate growth media with 10% fetal bovine serum. Transfections were performed using Invitrogen Lipofectamine 2000 (Carlsbad, CA).
Fluorescence imaging
Tissue culture and zebrafish sections were counterstained with Vectashield containing 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA) and imaged on a Leica TCS SP5 confocal microscope. Images were pseudocolored using Leica LAS AF Lite (Leica Microsystems, Buffalo Grove, IL) or ImageJ software and were further manipulated, if required, using Adobe Photoshop (San Jose, CA).
Cloning
cDNA corresponding to the open reading frame of human UHRF1 (nucleotides 169–2550, accession number NM_001048201) was cloned directionally into the SpeI and EcoRI site of the mammalian plasmid expression vector pEFF-N. cDNA of zebrafish UHRF1 was obtained from Open Biosystems (GenBank Accession BC058055; Thermo Scientific, Lafayette, CO). Ser-661 to glycine and alanine variants (UHRF1S661G, UHRF1S661A, and Uhrf1S648A) were generated using the Stratagene QuikChange site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA). To generate bacterial derived full-length and deletion mutants of UHRF1, we cloned full-length or truncated PCR products of human cDNA into pet28A vector. Invitrogen pCR8/GW/TOPO TA Cloning Kit, in combination with the appropriate vectors for mRNA expression in zebrafish (Villefranc et al., 2007), was used to make the myc-tag UHRF1 constructs for injection into zebrafish embryos (pCS3-6XMT-UHRF1). GFP-tagged UHRF1 used in tissue culture cells was generated using pDest53 from Invitrogen. Degenerate sequencing primers surrounding the zebrafish UHRF1 ATG site were used to ensure no significant overlap with morpholino sequence would occur. Supplemental Table S1 lists primers used for generating constructs. Human Myc-DNMT1 was obtained from X. Li (Mount Sinai School of Medicine).
mRNA rescue
To ensure that the mRNA encoding zebrafish Uhrf1 would not be targeted by the morpholino, we placed six Myc epitopes following the start site of translation, which is the site of action by which morpholinos block translation, and used degeneracy of the genetic code to alter the mRNA sequence of zebrafish uhrf1 to create significant mismatch with morpholino (Figure S3). To rescue uhrf1 morphants, the pCS 3MT DEST vector (Villefranc et al., 2007) containing the coding sequence for human UHRF1 (wild-type or mutants) or zebrafish Uhrf1 was linearized with NsiI, and capped mRNA was synthesized using mMESSAGE mMACHINE (Ambion, Austin, TX). mRNA was purified using lithium chloride, diluted in RNase-free water, and either injected alone or coinjected with 1.68 pg of uhrf1 morpholino into embryos prior to the four-cell stage.
Immunoprecipitation, phosphatase assay, and Western blots
HCT116 cells were harvested and lysed in detergent lysis buffer. The lysate was divided into four equal parts, and each was immunoprecipitated with anti-UHRF1 antibody. The immunoprecipitate was washed three times in sample buffer, which was followed by phosphatase buffer devoid of manganese (50 mM HEPES, 100 mM NaCl, 2 mM EDTA, and 0.01% Brij-35). Each pellet was resuspended in 30 μl of phosphatase buffer containing 1 mM of MnCl2, and 1 μl of λ-phosphatase (New England Biolabs, Ipswich, MA) was added to two of the four samples. Samples were incubated at 30°C for 30 min and reaction was terminated by the addition of 1X Laemmli sample buffer. Immunoprecipitation of FLAG-UHRF1 and MYC-DNMT1 from 293T cells was carried out similar manner, except cells were collected in CO-IP buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 50 mM NaF, and 1% nonidet P-40 with fresh protease inhibitor) and incubated overnight with anti-FLAG–conjugated beads (Sigma) or Negative Control Mouse IgG2a (Dako, Carpinteria, CA) plus Pierce protein A/G agarose beads (Thermo Scientific). Lysates were also washed three times in CO-IP buffer, with a final wash in 50 mM Tris (pH 7.4).
Western blots on cell lysates were performed as previously described (Ukomadu and Dutta, 2003b). For Western blots on zebrafish embryos, 10 embryos at 6 or 24 hpf were homogenized in lysis buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 1% NP-40, 2 mM EDTA, 10% glycerol, and protease inhibitors) and centrifuged, and a 1:5 volume of sample buffer was added to the supernatant to achieve 2% SDS, 5% 2-mercaptoethanol.
Supplementary Material
Acknowledgments
This work was supported by Public Health Service grants 5R01DK080789-02 (C.U. and K.C.S.) and 5T32DK007792-09 (J.C.) from the National Institute of Diabetes and Digestive and Kidney Diseases; grant 5K12HD052890-05 (J.C.) from the Eunice Kennedy Shriver National Institute of Child Health and Human Development; and funding from the Breast Cancer Alliance (K.C.S.), the March of Dimes (K.C.S.), and the Sidney A. Swensrud Foundation (C.U.). Alex Mir and Meghan Walsh provided expert technical support. Confocal laser-scanning microscopy was performed at the Mount Sinai School of Medicine–Microscopy Shared Resource Facility, which is supported with funding from National Institutes of Health–National Cancer Institute shared resources grant 5R24CA095823-04, National Science Foundation Major Research Instrumentation grant DBI-9724504, and National Institutes of Health shared instrumentation grant 1 S10 RR0 9145-01.
Abbreviations used:
- CCNA2
cyclin A2
- CCNE1
cyclin E
- CDK
cyclin-dependent kinase
- dpf
days postfertilization
- GFP
green fluorescent protein
- hpf
hours postfertilization
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-06-0487) on November 9, 2011.
REFERENCES
- Abbady AQ, Bronner C, Bathami K, Muller CD, Jeanblanc M, Mathieu E, Klein JP, Candolfi E, Mousli M. TCR pathway involves ICBP90 gene down-regulation via E2F binding sites. Biochem Pharmacol. 2005;70:570–579. doi: 10.1016/j.bcp.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Abbady AQ, Bronner C, Trotzier MA, Hopfner R, Bathami K, Muller CD, Jeanblanc M, Mousli M. ICBP90 expression is downregulated in apoptosis-induced Jurkat cells. Ann NY Acad Sci. 2003;1010:300–303. doi: 10.1196/annals.1299.052. [DOI] [PubMed] [Google Scholar]
- Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci USA. 2004;101:12792–12797. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RM, et al. Loss of Dnmt1 catalytic activity reveals multiple roles for DNA methylation during pancreas development and regeneration. Dev Biol. 2009;334:213–223. doi: 10.1016/j.ydbio.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arima Y, Hirota T, Bronner C, Mousli M, Fujiwara T, Niwa S, Ishikawa H, Saya H. Down-regulation of nuclear protein ICBP90 by p53/p21Cip1/WAF1-dependent DNA-damage checkpoint signals contributes to cell cycle arrest at G1/S transition. Genes Cells. 2004;9:131–142. doi: 10.1111/j.1356-9597.2004.00710.x. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bonapace IM, Latella L, Papait R, Nicassio F, Sacco A, Muto M, Crescenzi M, Di Fiore PP. Np95 is regulated by E1A during mitotic reactivation of terminally differentiated cells and is essential for S phase entry. J Cell Biol. 2002;157:909–914. doi: 10.1083/jcb.200201025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- Bronner C, Achour M, Arima Y, Chataigneau T, Saya H, Schini-Kerth VB. The UHRF family: oncogenes that are drugable targets for cancer therapy in the near future. Pharmacol Ther. 2007;115:419–434. doi: 10.1016/j.pharmthera.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Brunet J, Pfaff AW, Abidi A, Unoki M, Nakamura Y, Guinard M, Klein JP, Candolfi E, Mousli M. Toxoplasma gondii exploits UHRF1 and induces host cell cycle arrest at G2 to enable its proliferation. Cell Microbiol. 2008;10:908–920. doi: 10.1111/j.1462-5822.2007.01093.x. [DOI] [PubMed] [Google Scholar]
- Citterio E, Papait R, Nicassio F, Vecchi M, Gomiero P, Mantovani R, Di Fiore PP, Bonapace IM. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol Cell Biol. 2004;24:2526–2535. doi: 10.1128/MCB.24.6.2526-2535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crnogorac-Jurcevic T, et al. Proteomic analysis of chronic pancreatitis and pancreatic adenocarcinoma. Gastroenterology. 2005;129:1454–1463. doi: 10.1053/j.gastro.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Fang Z, Xing F, Bronner C, Teng Z, Guo Z. ICBP90 mediates the ERK1/2 signaling to regulate the proliferation of Jurkat T cells. Cell Immunol. 2009;257:80–87. doi: 10.1016/j.cellimm.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Feng S, et al. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA. 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori A, Matsuda Y, Takemoto Y, Hashimoto Y, Kubo E, Araki R, Fukumura R, Mita K, Tatsumi K, Muto M. Cloning and mapping of Np95 gene which encodes a novel nuclear protein associated with cell proliferation. Mamm Genome. 1998;9:1032–1035. doi: 10.1007/s003359900920. [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–829. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi H, Suzuki-Takahashi I, Taya Y, Segawa K, Nishimura S, Kitagawa M. Differences in substrate specificity between Cdk2-cyclin A and Cdk2-cyclin E in vitro. Biochem Biophys Res Commun. 1995;216:520–525. doi: 10.1006/bbrc.1995.2653. [DOI] [PubMed] [Google Scholar]
- Hopfner R, Mousli M, Jeltsch JM, Voulgaris A, Lutz Y, Marin C, Bellocq JP, Oudet P, Bronner C. ICBP90, a novel human CCAAT binding protein, involved in the regulation of topoisomerase IIα expression. Cancer Res. 2000;60:121–128. [PubMed] [Google Scholar]
- Hopfner R, Mousli M, Oudet P, Bronner C. Overexpression of ICBP90, a novel CCAAT-binding protein, overcomes cell contact inhibition by forcing topoisomerase II alpha expression. Anticancer Res. 2002;22:3165–3170. [PubMed] [Google Scholar]
- Jeanblanc M, Mousli M, Hopfner R, Bathami K, Martinet N, Abbady AQ, Siffert JC, Mathieu E, Muller CD, Bronner C. The retinoblastoma gene and its product are targeted by ICBP90: a key mechanism in the G1/S transition during the cell cycle. Oncogene. 2005;24:7337–7345. doi: 10.1038/sj.onc.1208878. [DOI] [PubMed] [Google Scholar]
- Jenkins Y, et al. Critical role of the ubiquitin ligase activity of UHRF1, a nuclear RING finger protein, in tumor cell growth. Mol Biol Cell. 2005;16:5621–5629. doi: 10.1091/mbc.E05-03-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, Jacobsen SE. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol. 2007;17:379–384. doi: 10.1016/j.cub.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldis P, Sutton A, Solomon MJ. The Cdk-activating kinase (CAK) from budding yeast. Cell. 1996;86:553–564. doi: 10.1016/s0092-8674(00)80129-4. [DOI] [PubMed] [Google Scholar]
- Kane DA, Kimmel CB. The zebrafish midblastula transition. Development. 1993;119:447–456. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- Karagianni P, Amazit L, Qin J, Wong J. ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol Cell Biol. 2008;28:705–717. doi: 10.1128/MCB.01598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K. Tol2: a versatile gene transfer vector in vertebrates. Genome Biol. 2007;8(suppl 1):S7. doi: 10.1186/gb-2007-8-s1-s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Esteve PO, Jacobsen SE, Pradhan S. UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res. 2009;37:493–505. doi: 10.1093/nar/gkn961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, et al. The consensus motif for phosphorylation by cyclin D1-Cdk4 is different from that for phosphorylation by cyclin A/E-Cdk2. EMBO J. 1996;15:7060–7069. [PMC free article] [PubMed] [Google Scholar]
- Knoblich JA, Sauer K, Jones L, Richardson H, Saint R, Lehner CF. Cyclin E controls S phase progression and its down-regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell. 1994;77:107–120. doi: 10.1016/0092-8674(94)90239-9. [DOI] [PubMed] [Google Scholar]
- Krek W, Xu G, Livingston DM. Cyclin A-kinase regulation of E2F-1 DNA binding function underlies suppression of an S phase checkpoint. Cell. 1995;83:1149–1158. doi: 10.1016/0092-8674(95)90141-8. [DOI] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Li Y, Mori T, Hata H, Homma Y, Kochi H. NIRF induces G1 arrest and associates with Cdk2. Biochem Biophys Res Commun. 2004;319:464–468. doi: 10.1016/j.bbrc.2004.04.190. [DOI] [PubMed] [Google Scholar]
- Lindeman LC, Reiner AH, Mathavan S, Alestrom P, Collas P. Tiling histone H3 lysine 4 and 27 methylation in zebrafish using high-density microarrays. PLoS One. 2010a;5:e15651. doi: 10.1371/journal.pone.0015651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeman LC, Winata CL, Aanes H, Mathavan S, Alestrom P, Collas P. Chromatin states of developmentally-regulated genes revealed by DNA and histone methylation patterns in zebrafish embryos. Int J Dev Biol. 2010b;54:803–813. doi: 10.1387/ijdb.103081ll. [DOI] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- Meehan RR, Dunican DS, Ruzov A, Pennings S. Epigenetic silencing in embryogenesis. Exp Cell Res. 2005;309:241–249. doi: 10.1016/j.yexcr.2005.06.023. [DOI] [PubMed] [Google Scholar]
- Meilinger D, Fellinger K, Bultmann S, Rothbauer U, Bonapace IM, Klinkert WE, Spada F, Leonhardt H. Np95 interacts with de novo DNA methyltransferases, Dnmt3a and Dnmt3b, and mediates epigenetic silencing of the viral CMV promoter in embryonic stem cells. EMBO Rep. 2009;10:1259–1264. doi: 10.1038/embor.2009.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura M, Watanabe H, Sasaki T, Tatsumi K, Muto M. Dynamic changes in subnuclear NP95 location during the cell cycle and its spatial relationship with DNA replication foci. Exp Cell Res. 2001;263:202–208. doi: 10.1006/excr.2000.5115. [DOI] [PubMed] [Google Scholar]
- Mousli M, Hopfner R, Abbady AQ, Monte D, Jeanblanc M, Oudet P, Louis B, Bronner C. ICBP90 belongs to a new family of proteins with an expression that is deregulated in cancer cells. Br J Cancer. 2003;89:120–127. doi: 10.1038/sj.bjc.6601068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudbhary R, Sadler KC. Epigenetics, development, and cancer: zebrafish make their ARK. Birth Defects Res C Embryo Today. 2011;93:194–203. doi: 10.1002/bdrc.20207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto M, Kanari Y, Kubo E, Takabe T, Kurihara T, Fujimori A, Tatsumi K. Targeted disruption of Np95 gene renders murine embryonic stem cells hypersensitive to DNA damaging agents and DNA replication blocks. J Biol Chem. 2002;277:34549–34555. doi: 10.1074/jbc.M205189200. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982a;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: II. control of the onset of transcription. Cell. 1982b;30:687–696. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- Ohtsubo M, Theodoras AM, Schumacher J, Roberts JM, Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol Cell Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. EMBO J. 1992;11:961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papait R, et al. The PHD domain of Np95 (mUHRF1) is involved in large-scale reorganization of pericentromeric heterochromatin. Mol Biol Cell. 2008;19:3554–3563. doi: 10.1091/mbc.E07-10-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumara E, et al. PHD finger recognition of unmodified histone H3R2 links UHRF1 to regulation of euchromatic gene expression. Mol Cell. 2011;43:275–284. doi: 10.1016/j.molcel.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnitzky D, Hengst L, Reed SI. Cyclin A-associated kinase activity is rate limiting for entrance into S phase and is negatively regulated in G1 by p27Kip1. Mol Cell Biol. 1995;15:4347–4352. doi: 10.1128/mcb.15.8.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnitzky D, Reed SI. Different roles for cyclins D1 and E in regulation of the G1-to-S transition. Mol Cell Biol. 1995;15:3463–3469. doi: 10.1128/mcb.15.7.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottach A, Frauer C, Pichler G, Bonapace IM, Spada F, Leonhardt H. The multi-domain protein Np95 connects DNA methylation and histone modification. Nucleic Acids Res. 2010;38:1796–1804. doi: 10.1093/nar/gkp1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler KC, Krahn KN, Gaur NA, Ukomadu C. Liver growth in the embryo and during liver regeneration in zebrafish requires the cell cycle regulator, uhrf1. Proc Natl Acad Sci USA. 2007;104:1570–1575. doi: 10.1073/pnas.0610774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier AF. The maternal-zygotic transition: death and birth of RNAs. Science. 2007;316:406–407. doi: 10.1126/science.1140693. [DOI] [PubMed] [Google Scholar]
- Schier AF, Talbot WS. Molecular genetics of axis formation in zebrafish. Annu Rev Genet. 2005;39:561–613. doi: 10.1146/annurev.genet.37.110801.143752. [DOI] [PubMed] [Google Scholar]
- Sharif J, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- Su TT, Stumpff J. Promiscuity rules? The dispensability of cyclin E and Cdk2. Sci STKE. 2004;2004:pe11. doi: 10.1126/stke.2242004pe11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136:3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- Takeda DY, Wohlschlegel JA, Dutta A. A bipartite substrate recognition motif for cyclin-dependent kinases. J Biol Chem. 2001;276:1993–1997. doi: 10.1074/jbc.M005719200. [DOI] [PubMed] [Google Scholar]
- Tien AL, Senbanerjee S, Kulkarni A, Mudbhary R, Goudreau B, Ganesan S, Sadler KC, Ukomadu C. UHRF1 depletion causes a G2/M arrest, activation of DNA damage response and apoptosis. Biochem J. 2011;435:175–185. doi: 10.1042/BJ20100840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tittle RK, Sze R, Ng A, Nuckels RJ, Swartz ME, Anderson RM, Bosch J, Stainier DY, Eberhart JK, Gross JM. Uhrf1 and Dnmt1 are required for development and maintenance of the zebrafish lens. Dev Biol. 2011;350:50–63. doi: 10.1016/j.ydbio.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotzier MA, Bronner C, Bathami K, Mathieu E, Abbady AQ, Jeanblanc M, Muller CD, Rochette-Egly C, Mousli M. Phosphorylation of ICBP90 by protein kinase A enhances topoisomerase IIα expression. Biochem Biophys Res Commun. 2004;319:590–595. doi: 10.1016/j.bbrc.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Uemura T, Kubo E, Kanari Y, Ikemura T, Tatsumi K, Muto M. Temporal and spatial localization of novel nuclear protein NP95 in mitotic and meiotic cells. Cell Struct Funct. 2000;25:149–159. doi: 10.1247/csf.25.149. [DOI] [PubMed] [Google Scholar]
- Ukomadu C, Dutta A. Inhibition of cdk2 activating phosphorylation by mevastatin. J Biol Chem. 2003a;278:4840–4846. doi: 10.1074/jbc.M208658200. [DOI] [PubMed] [Google Scholar]
- Ukomadu C, Dutta A. p21-dependent inhibition of colon cancer cell growth by mevastatin is independent of inhibition of G1 cyclin-dependent kinases. J Biol Chem. 2003b;278:43586–43594. doi: 10.1074/jbc.M307194200. [DOI] [PubMed] [Google Scholar]
- Unoki M, Bronner C, Mousli M. A concern regarding the current confusion with the human homolog of mouse Np95, ICBP90/UHRF1. Radiat Res. 2008;169:240–244. doi: 10.1667/RR1209.1. [DOI] [PubMed] [Google Scholar]
- Unoki M, Nishidate T, Nakamura Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene. 2004;23:7601–7610. doi: 10.1038/sj.onc.1208053. [DOI] [PubMed] [Google Scholar]
- Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, Rinn J, Schier AF. Chromatin signature of embryonic pluripotency is established during genome activation. Nature. 2010;464:922–926. doi: 10.1038/nature08866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villefranc JA, Amigo J, Lawson ND. Gateway compatible vectors for analysis of gene function in the zebrafish. Dev Dyn. 2007;236:3077–3087. doi: 10.1002/dvdy.21354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, et al. Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger. Cell Res. 2011;21:1379–1382. doi: 10.1038/cr.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel JA, Dwyer BT, Takeda DY, Dutta A. Mutational analysis of the Cy motif from p21 reveals sequence degeneracy and specificity for different cyclin-dependent kinases. Mol Cell Biol. 2001;21:4868–4874. doi: 10.1128/MCB.21.15.4868-4874.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Pontes O, Pikaard CS, Richards EJ. VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev. 2007;21:267–277. doi: 10.1101/gad.1512007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy F, Lamas E, Chenivesse X, Sobczak J, Wang J, Fesquet D, Henglein B, Brechot C. Cyclin A is required in S phase in normal epithelial cells. Biochem Biophys Res Commun. 1992;182:1144–1154. doi: 10.1016/0006-291x(92)91851-g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.