SUMMARY
The β-propeller gene Rv1057 of Mycobacterium tuberculosis is activated by envelope stress and was first characterized as a regulatory target of the TrcRS two-component system (TCS). Rv1057 expression is repressed by TrcRS, and the Rv1057 proximal promoter contains a TrcR binding site. In this study, we determined that Rv1057 is also directly regulated by MprAB, a TCS associated with envelope stress. Multiple potential MprA binding sites (MprA boxes) were identified in the 1 kb intergenic region upstream of Rv1057, and four sites were shown to bind MprA. Although MprA boxes were found in the proximal promoter, analyses suggest that MprA and TrcR do not compete for binding in this region. An MprAB-dependent, detergent-inducible transcriptional start point for Rv1057 was identified downstream of the MprA boxes, and a second TrcR binding site and small ORF of the 13E12 family were discovered in the distal promoter. MprAB was required for activation of Rv1057 during growth in macrophages and under detergent stress, and lacZ promoter constructs suggest the entire intergenic region is utilized during MprAB-dependent activation of Rv1057. These findings indicate that Rv1057 has an extensive and complex promoter, and provide evidence for coordinated regulation of stress response genes by TCSs.
Keywords: mycobacterium, two-component, regulation, envelope stress
INTRODUCTION
Mycobacterium tuberculosis is the causative agent of human tuberculosis and a highly successful pathogen with a global impact on human morbidity and mortality 1. During the course of the disease, the pathogen is exposed to a range of stresses due to environmental changes, nutrient limitation, and immune defense mechanisms, and as in other bacterial pathogens, sigma factors and two-component systems (TCSs) have key roles in adaptation to these stresses.
TCSs consist of a sensory histidine kinase and a response regulator, usually a transcription factor, that is phosphorylated by the cognate kinase in response to a particular stimulus 2, 3. In M. tuberculosis, there are 11 complete TCSs, and several orphan (unpaired) histidine kinases and response regulators 4. Activating stresses and regulatory targets have been identified for a few of these TCSs, and several impact growth of M. tuberculosis in host systems 5, 6. The MtrAB TCS is essential and regulates cell division and cell wall homeostasis 7–10. The DosRST TCS, actually a three-component system that includes the DosS and DosT kinases, activates the dormancy regulon in response to oxygen, nitric oxide, and carbon monoxide levels, and is essential for redox balance and survival during anaerobiosis 11–17. SenX3-RegX enhances survival under phosphate starvation and during growth in mice, and activates genes involved in phosphate transport and the stringent response 18–20. PhoPR regulates genes associated with the ESX-1 secretion system and synthesis of complex surface lipids, and is required for virulence 21–23.
The response regulator MprA and the histidine kinase MprB comprise the MprAB TCS, and mprA mutants exhibit reduced persistence in mice, but have enhanced growth in macrophages 24, 25. MprAB activates SigE, the major sigma factor involved in envelope stress, and is the only mycobacterial TCS known to directly regulate sigma factor genes 25, 26. MprA binding sites (MprA boxes) have been discovered in the promoter of sigE and genes of the SigE regulon, including sigB, pepD, acr2, and mprAB itself 25–29. The MprA box consists of a tandem repeat of the hexamer TCTCAG separated by 5 bases, and whereas the distance between the repeats appears to be critical, there is some variation in the sequence of the hexamers 25, 26, 28, 29.
In this study, we investigated the role of MprAB in regulation of Rv1057, which encodes the only seven-bladed β-propeller protein in M. tuberculosis 30. β-propeller proteins have diverse functions, ranging from cell-to-cell signalling and chromatin condensation in mammalian systems, to photoreception in plants, and antibiotic resistance and envelope maintenance in bacteria 31–35. Rv1057 is activated by detergents, vancomycin, and surfactant proteins 25–27, 30, 36, 37, suggesting it also has a role in maintaining envelope stability. DNA microarray analyses with sigE and mprA mutants indicate that Rv1057 is a member of the SigE and MprAB regulons25–27, consistent with its activation by envelope stresses. However, Haydel and Clark-Curtiss 30 initially characterized Rv1057 as a regulatory target of TrcRS, a TCS of unknown function. An AT-rich sequence in the Rv1057 promoter had similarity to the only known binding site for the response regulator TrcR, which is located in the trcRS promoter 30, 38. Subsequent analyses demonstrated that TrcR bound the 69-bp AT-rich region in the Rv1057 promoter, and that TrcR repressed Rv1057 expression in vitro 30.
As direct coregulation of genes by two TCSs has not been previously described in mycobacteria, we wanted to determine the extent of Rv1057 regulation by MprAB, and specifically, whether it was directly regulated by MprA. Our results indicate that Rv1057 has a large, complex promoter directly regulated by two TCSs.
MATERIALS AND METHODS
Bacterial strains and culture conditions
M. tuberculosis strain Rv-D981 is an mprAB deletion mutant derived from the laboratory strain H37Rv, and Rv-D981Com is the mprAB-complemented strain generated from Rv-D981 25. Strains were grown at 37°C under normal atmospheric conditions, as described 29. Escherichia coli Novablue and Rosetta(DE3)pLysS (Novagen) were used for general cloning purposes and protein expression, respectively.
Construction of promoter-lacZ fusion plasmids
Rv1057 promoter fusions were constructed in the integrating vector pSM128 39 by inserting sections of the Rv1056-Rv1057 intergenic region upstream of lacZ. The entire 1003-bp intergenic region was amplified using primers Rv1057 FuF-1 and Rv1057 FuR-1 (Table S1). The 507-bp proximal and 492-bp distal regions of the Rv1057 promoter were amplified using primer pairs Rv1057 FuF-2/FuR-1 and Rv1057 FuF-1/FuR-3, respectively. Promoter fusion plasmids and the control vector pSM128 were electroporated into H37Rv and Rv-D981. Transformants were selected in the presence of streptomycin (50 µg/ml) and confirmed by PCR analysis. Rv-D981C was not included because mprAB is inserted in the att integration site used by pSM128.
Expression and purification of MprA and TrcR
MprA was expressed in E. coli and purified as described 25. His-tagged TrcR was expressed using plasmid pSH33 and purified essentially as described 40, except after elution from the nickel-nitrilotriacetic acid agarose column, the protein was dialyzed in a Slide-A-lyzer dialysis cassette (Millipore) for 24 hr in each of buffer 1 (20 mM NaH2PO4, 100 mM NaCI, 3 m MDTT), buffer 2 (20 mM NaH2PO4, 50 mM NaCI, 2 mM DTT) and buffer 3 (10 mM Tris, pH8.0, 50 mM NaCI, 1 mM DTT), followed by dialysis in buffer 4 (10 mM Tris, pH8.0, 50 mM NaCI) for 3 days. The protein solution was then centrifuged and concentrated using centrifugal filter devices with a 20 KDa cutoff (Millipore).
Electrophoresis mobility shift assays (EMSAs)
DNA regions were amplified by PCR, end-labeled with [gamma-P32]ATP, incubated with protein, and the reaction mixtures were analyzed on nondenaturing polyacrylamide gels, using described procedures 25, 29. For analyses of MprA boxes, complementary 45-mer oligonucleotides were annealed and end-labeled.
Macrophage infection
THP-1 cells were cultured in RPMI 1640 containing 10% heat-inactivated fetal bovine serum, 0.1 mM non-essential amino acids, 1 mM sodium pyruvate and 9 mg/ml bovine insulin, and were treated with 50nM PMA for 24 hours to stimulate differentiation into macrophages. Bacteria were grown to mid-log phase in 7H9 media, diluted in RPMI 1640 with 10% FCS, and added to each well at an MOI of 1:1. After 2 hours of uptake, cells were washed three times and then incubated with fresh RPMI 1640 with 10% FCS. At each time point, duplicate wells for each infecting strain were processed.
Stress treatment and RNA isolation
Broth cultures were grown to log phase (optical density at 600 nm of 0.3–0.4), and were then exposed to 0.05% SDS or 0.1% deoxycholate for 90 min, or left unexposed (control samples). RNA was extracted and genomic DNA was removed, as described 29. To isolate RNA from intracellular mycobacteria, macrophages were lysed by vortexing in 10 ml extraction buffer (4 M guanidine isothiocyanate, 200 mM Tris-HCI, 300 mM LiCI, 10 mM EDTA, 0.2% Tween-80), and then centrifuged to pellet the bacilli. The pellet was washed and vortexed with the extraction buffer twice before RNA was extracted from the bacteria as outlined above. For the control (reference) sample, RNA was extracted from cells infected with H37Rv for 2 hr.
Reverse-transcription and relative quantification of mRNA by real-time PCR (qRT-PCR)
Experiments were conducted as described 25, 29. Briefly, cDNA was prepared by random priming, and qRT-PCR probes were labeled with 5’-fluorescein phosphoramidite and 3’-6-carboxytetramethylrhodamine. Primer and probe sequences are listed in Supplementary Table S1. Assays were performed using an ABI Prism 7700 thermal cycler, and relative quantities of cDNA were determined from standard curves generated with genomic DNA. Samples were normalized for 16S rRNA.
Primer extension analysis
Three separate primers were used to analyze Rv1057 transcripts (Table S1). Primers were labeled with [gamma-P32] ATP, and annealed to 5 µg of M. tuberculosis RNA, as described 25. Primer extension was performed using the buffers from the Primer extension system-AMV reverse transcriptase kit (Promega), but with the high-temperature-tolerant Thermoscript reverse transcriptase (Invitrogen), using an extension temperature of 55 °C to overcome possible secondary structures.
RESULTS
MprAB is required for activation of Rv1057 under envelope stress
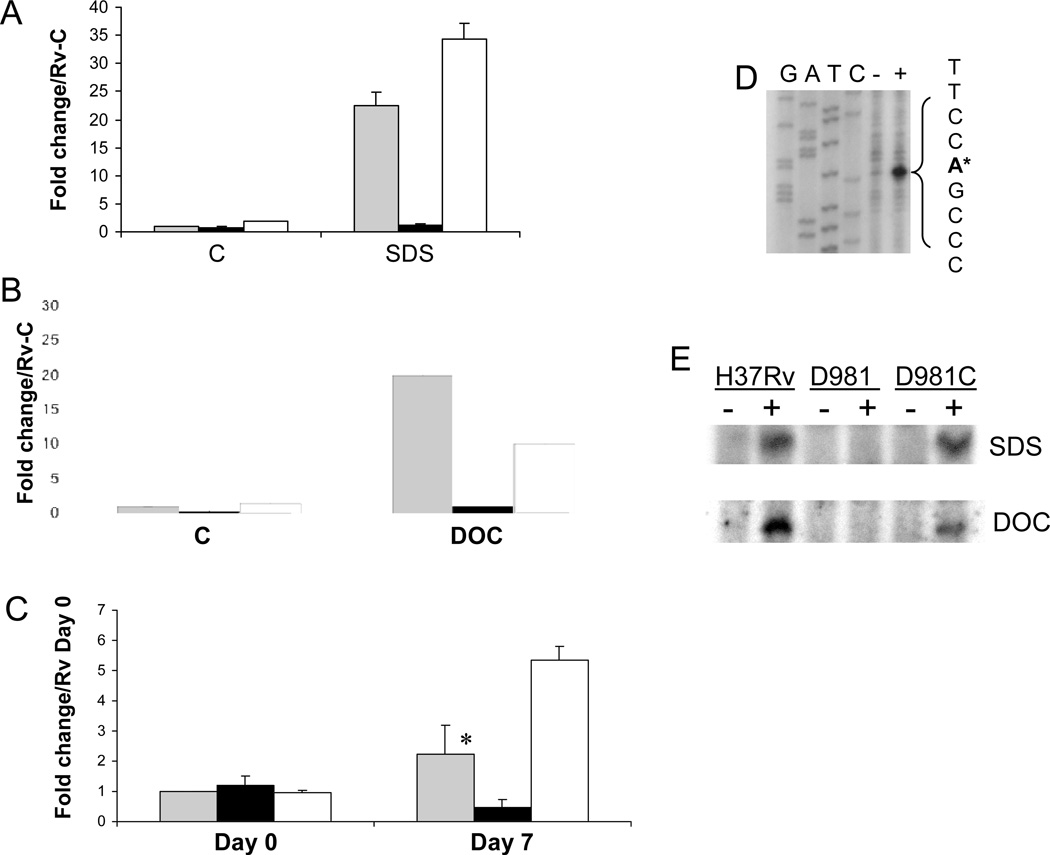
Using qRT-PCR, we determined that Rv1057 was highly induced by SDS exposure in the parental strain H37Rv, but not in our mprAB deletion mutant Rv-D981 (Fig. 1A), confirming the results of our DNA microarray studies 25. Exposure to the bile detergent deoxycholate produced similar results, with Rv1057 induced by 20-fold over control conditions in H37Rv, but by only approximately 2-fold in Rv-D981 (Fig 1B). Induction of Rv1057 by detergent exposure was restored in the mprAB-complemented strain, Rv-D981Com (Fig. 1A, B), indicating that loss of mprAB was responsible for the reduced expression of Rv1057 in Rv-D981.
Figure 1.
MprAB activates Rv1057 under envelope stress. (A–C) Expression of Rv1057 was examined by qRT-PCR using RNA extracted from strains exposed to 0.05% SDS for 90 min (A), 0.1% deoxycholate for 90 min (B), or following infection of THP-1 cells (C). H37Rv (gray bars), Rv-D981 (black bars), and Rv-D981Com (white bars). The “C” on the horizontal axis in (A) and (B) indicates control (untreated) samples. For (C), activated THP-1 cells were infected at a MOI of 1:1, and bacteria were recovered from lysed monolayers at 2 hr (Day 0), or at day 7. Results are shown as fold change over the H37Rv control sample for panels A and B, or over the H37Rv Day 0 sample for panel C. Control and day 0 samples were given values of 1. For all panels, results were normalized for 16S RNA content and show the mean of at least two separate experiments +/− SD. (C) The difference in expression between H37Rv and Rv-D981 at day 7 was analyzed using a Student’s one-tailed T-test, *, P < 0.05.
(D) Rv1057 TSP2 was mapped by primer extension with primer Rv1057 Ext-3, using RNA extracted from H37Rv incubated in the absence (−) or presence (+) of 0.05% SDS. The TSP is marked by an asterisk and was located using a sequence ladder generated with the primer used for primer extension. The GATC labels above the ladder refer to the sequence on the opposite DNA strand. (E) Activation of Rv1057 TSP was compared in H37Rv, Rv-D981 (D981), and Rv-D981Com (D981C) by primer extension. Minus sign, cultures incubated under control conditions. Plus sign, cultures exposed to 0.05 % SDS (upper panel), or 0.1% deoxycholate (lower panel) for 90 min.
Growth in macrophages induces multiple genes associated with envelope stress, including Rv1057 30, 41, so it has been suggested that the intracellular environment causes detergent-like damage to the mycobacterial envelope 41. Therefore, we evaluated the role of MprAB in activation of Rv1057 during intracellular growth. With both H37Rv and Rv-D981Com, Rv1057 transcript levels were higher at day 7 than at day 0 (Fig. 1C). In contrast, Rv1057 transcription decreased in the mprAB deletion mutant, so that by day 7, transcript levels were ~5-fold lower in Rv-D981 than in H37Rv.
We also identified an Rv1057 transcriptional start point (TSP) that was activated by SDS and deoxycholate in H37Rv and Rv-D981Com, but not in Rv-D981 (Fig. 1 D, E). This MprAB-dependent TSP (TSP2) corresponds to an adenosine 181 nucleotides upstream of the predicted start codon of Rv1057 (Fig. 1D), and 95 nucleotides upstream of the previously identified TSP1 30 (Fig. 3A). Transcription from TSP2 was only minimally and occasionally detectable in the absence of detergent stress in H37Rv, as evidenced by the very weak band for the control sample in Fig. 1D.
Figure 3.
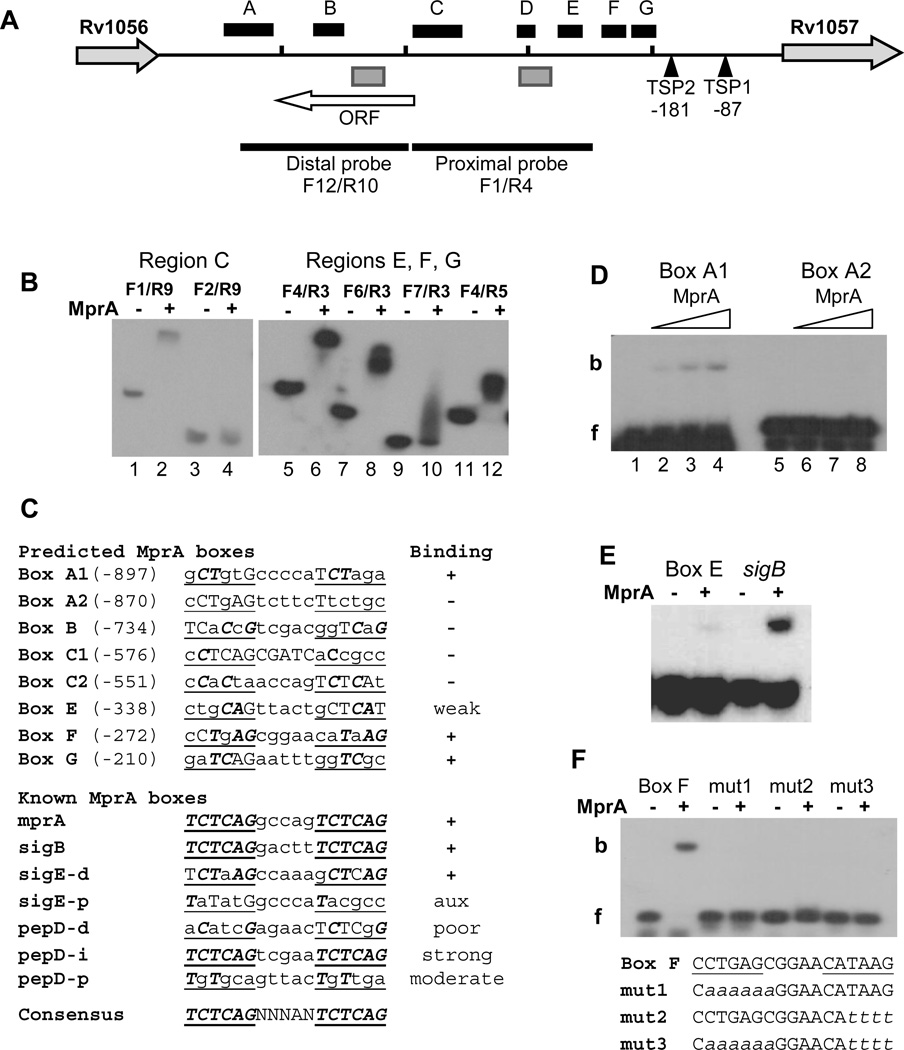
Localization of MprA boxes in the Rv1056-Rv1057 intergenic region. (A) Features of the Rv1056-Rv1057 intergenic region. The horizontal black line represents the chromosome and short vertical bars mark 200-bp sections beginning from the Rv1057 start codon. The location of TSP1 30 and TSP2 (this study) are indicated relative to the Rv1057 start codon. Black boxes labeled A–G designate regions determined to contain at least one MprA box, based on EMSA analyses of overlapping fragments. Gray boxes designate regions containing TrcR binding sites 30 (and this study). The locations of the probes used in Figures 2 A and B are marked by thick black lines. The white horizontal arrow indicates the location and length of a previously unidentified ORF. Rv1057 and Rv1056 are not drawn to scale.
(B) Representative EMSAs used to localize MprA binding in region C (left panel) and regions E, F, and G (right panel). Forward and reverse primers used to generate each probe are noted. (−) and (+) symbols indicate incubation in the absence, or presence, of 2 ug MprA, respectively. Results indicate strong MprA binding to probes F1/R9, F4/R3, F6/R3, and F4/R5.
(C) Comparison of predicted MprA boxes in regions A–G with known MprA boxes. Regions were scanned for sequences with strongest similarity to the published consensus sequence for the MprA box 25, 26, 28. Repeated hexamers of the MprA box half-sites are underlined, bases matching the consensus sequences are capitalized, and capitals in bold italics mark consensus sequence bases present in the same position in each half-site. Names of predicted boxes are derived from the region in which they were found. Two boxes were predicted for each of regions A and C. Numbers in parentheses refer to the position of the first base relative to the Rv1057 start codon. Results of EMSA analysis with 45-bp probes containing the predicted boxes are indicated under “Binding”; (+) and (−) indicate binding, or no binding, by MprA, respectively. Known MprA boxes refer to published sites confirmed by EMSAs, and located in the promoters for the given genes 25, 26, 28. d, p, i refer to distal, proximal, and intermediate promoter regions, respectively 26. Ratings of poor, moderate, and strong were obtained from published data 26, 28. “aux”, auxiliary site (see text for details).
(D–F) MprA was incubated with 45-bp probes containing predicted MprA boxes. (D) Probes containing Box A1 or Box A2 were incubated with the no MprA (Lanes 1 and 5), or 2.5 ug (Lanes 2 and 6), 5 ug (Lanes 3 and 7) or 7.5 ug (Lanes 4 and 8) of MprA. f, free probe; b, bound probe. (E) Comparison of MprA binding to Box E and a sigB promoter control. (−) and (+) symbols indicate incubation in the absence, or presence, of 7.5 ug MprA, respectively. (F) Binding of MprA to probes containing a native or mutagenized Box F. Lowercase italics in the sequences below the gel image indicate mutated bases. (−) and (+) symbols indicate incubation in the absence, or presence, of 2 ug MprA, respectively.
MprA directly regulates Rv1057
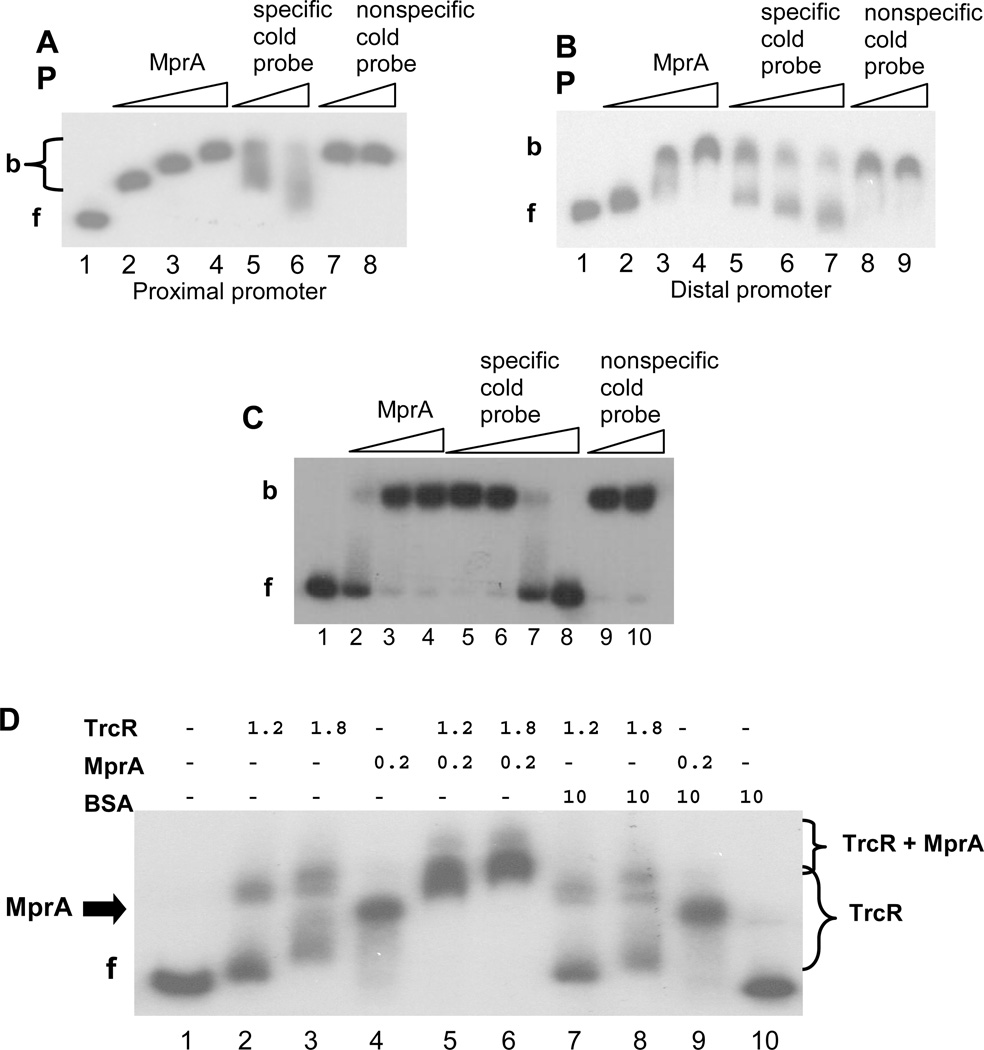
Next, the ability of MprA to bind the Rv1057 promoter was evaluated using EMSAs, and these analyses showed that MprA binds both distal and proximal promoter regions (Fig. 2A and B). Interestingly, at lower concentrations of MprA, there was either a step-ladder pattern of bands (Fig. 2A, Lanes 2–3) or smearing of the shifted band (Fig. 2B, Lanes 2–3). Additionally, although the specific competitor reduced binding to the labeled probe, competition was incomplete and produced a smearing pattern, despite using excess amounts of cold probe (Fig. 2A, Lanes 5–6; Fig. 2B Lanes 5–7). We considered that multiple MprA boxes in each probe might be the basis for these patterns. Indeed, use of a smaller section of the proximal promoter produced a cleaner band shift pattern (Fig. 2C), and high concentrations of specific cold probe effectively competed with labeled probe (Fig. 2C, Lanes 7–8).
Figure 2.
EMSA analyses of MprA binding to the Rv1057 promoter. (A) A 310-bp probe to the proximal region was amplified using primers Rv1057 F1/R4. A fixed amount of probe was incubated with no MprA (Lane 1); 0.25–0.75 µg MprA (Lanes 2–4); 0.75 µg MprA, and 100-fold or 300-fold excess of unlabeled specific probe (Lanes 5 and 6); 0.75 µg MprA, and 100-fold or 200-fold excess of unlabeled nonspecific probe (Lanes 7 and 8). f, free probe; b, bound probe.
(B) A 273-bp probe to the distal region was amplified using primers Rv1057 F12/R10, and a fixed amount was incubated in reaction mixtures containing no MprA (Lane 1); 0.5–1.5 µg MprA (Lanes 2–4); 1.5 µg MprA and 100-fold to 400-fold excess of unlabeled specific probe (Lanes 5–7); 1.5 µg MprA and 50-fold or 200-fold excess of unlabeled nonspecific probe (Lanes 8 and 9).
(C) A 117-bp probe to the proximal region was amplified using primers Rv1057 F4/R5, and a fixed amount was incubated with no MprA (lane 1); 0.5–1.5 µg MprA (Lanes 2–4); 1.5 µg MprA and 100-fold to 400-fold excess of unlabeled specific probe (Lanes 5–8); 1.5 µg MprA and 300-fold or 400-fold of unlabeled nonspecific probe (Lanes 9 and 10).
(D) MprA and TrcR were incubated with the proximal promoter probe F1/R4 separately, or in combination with each other or with BSA. Amounts of each protein are indicated in micrograms. (−) indicates no protein. f, free probe. Brackets indicate range of bands detected in the presence of TrcR alone, or TrcR + MprA, as indicated. The arrow labeled “MprA” marks the position of the band obtained with MprA alone (Lanes 4 and 9).
The proximal region of the Rv1057 promoter contains the binding site for TrcR, the response regulator of the TrcRS TCS 30, so we investigated whether TrcR and MprA compete for binding in this region. When used alone, TrcR produced two different band patterns depending on the amount of protein used (Fig. 2D, Lanes 2–3), consistent with previous observations that the region contains more than one TrcR binding site 30. With both TrcR and MprA present, the lower bands found with TrcR alone were no longer detected, and new more slowly migrating bands appeared (Fig. 2D, Lanes 3, 5, and 6). These data suggest binding is additive and that MprA and TrcR do not occlude each others binding sites. The slower-migrating bands do not appear to be due to nonspecific protein interactions between the two response regulators and the promoter, because incubation of either MprA or TrcR with the control protein BSA yielded patterns similar to those detected for the individual response regulator (Fig. 2C, Lanes 3, 4, 7–9).
Mapping regulatory sites in the Rv1056c-Rv1057 intergenic region
EMSAs with the distal and proximal promoter probes suggested that the Rv1056c-Rv1057 intergenic region contains multiple MprA binding sites (Fig. 2A–C). Further analyses with a series of overlapping DNA fragments revealed seven sections, named A–G, that contained at least one MprA box (Fig. 3A and B, and Supplementary Data Fig. S1). Representative EMSAs for several fragments are shown (Fig. 3B), including the F4/F5 fragment (Fig. 3B, Lanes 11–12) for which an expanded analyses was presented (Fig. 2C). Some fragments produced little or no binding when used separately, but enhanced binding of MprA to adjacent regions (Fig. 3B, Lanes 9–10, and Supplementary Data Fig. S1), suggesting they contain partial or auxiliary binding sites. MprA binding regions were clustered more closely within the proximal promoter (Fig. 3A), but no binding was detected to the region downstream of TSP2 (Fig. 3A, and Fig. S1).
The distal promoter, which we defined as the upstream half of the approximately 1 kb intergenic region, contains at least two sections that bind MprA (Fig. 3A). EMSAs also revealed a TrcR binding site between MprA binding sections B and C (Fig. 3A, and data not shown). This site was contained within a small previously unidentified ORF (Fig. 3A).
Localization of MprA boxes
Sections A–G of the Rv1056-Rv1057 intergenic region were screened for sequences with similarity to the consensus sequence for the MprA box, comprising a tandem repeat of the hexamer TCTCAG, separated by 5 bases 25, 26, 28. Two predicted MprA boxes were found in each of sections A and C, and these and four other predicted MprA boxes (Fig. 3C) were tested for their ability to bind MprA, using 45-mer probes (Table S1). Probes containing boxes A1, E, F, and G were shifted by MprA (Figures 3C–F), but no shift was detected for the remaining four probes (Fig. 3C, D), suggesting the selected sites are not true MprA boxes or that they function only as auxiliary sites, similar to the sigE-p MprA box (Fig. 3C) 26.
Box E bound MprA weakly, as demonstrated by comparison with the strong MprA box from the sigB promoter (Fig. 3C, E). In contrast, of the eight 45-mer probes containing predicted MprA boxes, the probe containing Box F exhibited one of the strongest shifts with MprA (Fig. 3F). To confirm that the selected hexameric repeat in this probe was the actual MprA box, the half-sites were mutated and binding was compared to the native sequence (Fig. 3F). Mutation of either one or both half-sites effectively disrupted MprA binding, supporting the identification of this MprA box.
EMSA data for known MprA boxes suggest that MprA binds most strongly to boxes that are identical to the consensus sequence, such as the sigB promoter box (Fig. 3C), but the exact requirements for MprA binding are unclear as considerable variation in the hexamers appears to be tolerated. We noted, though, that most of the functional sites listed in Fig. 3C contained at least two adjacent bases that matched the consensus sequence and that were in the same position in each half-site, for example, the repeated dinucleotide CT in Box A1. The pepD-d and pepD-p MprA boxes were exceptions, and had poor and moderate MprA binding, respectively 26, 28, although the pepD-p box does have a similar pattern with repetition of the first three bases, two of which match the consensus sequence. However, further studies are required to determine how repeated bases and other features of MprA boxes influence MprA binding.
Identification of the MprAB-dependent Rv1057 promoter
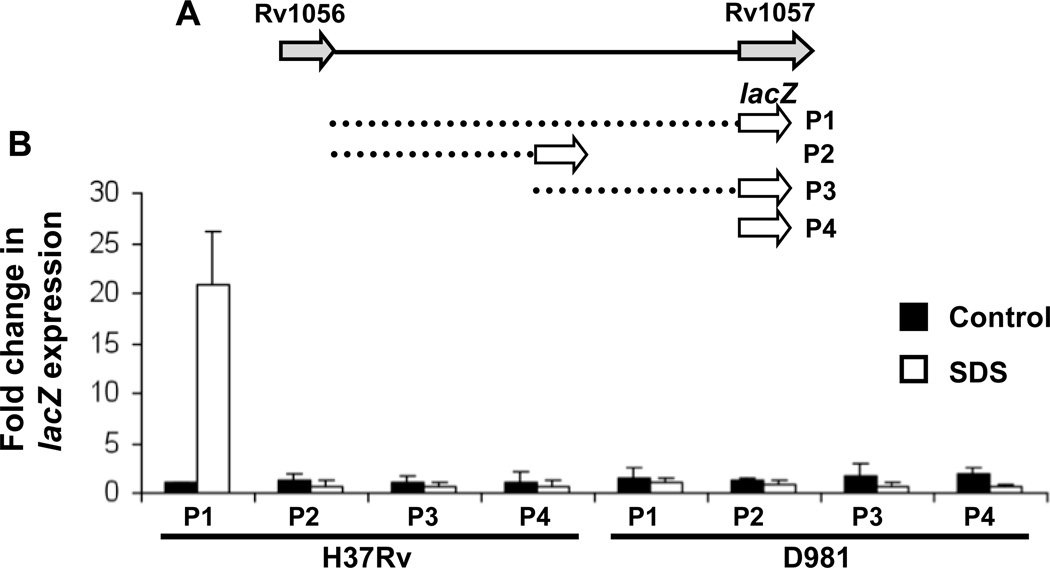
lacZ reporter constructs were generated to identify promoter regions required for activation of Rv1057 under detergent stress. The 1 kb intergenic region, or proximal or distal portions (Fig. 4A) were inserted upstream of lacZ in the integrating vector pSM128 39. Expression of lacZ was evaluated by qRT-PCR due to potential denaturation of β-galactosidase by residual SDS. Induction of lacZ following SDS exposure was detected only with the P1 construct, which contains the entire intergenic region (Fig. 4A and 4B). In addition, induction occurred only in H37Rv and not in Rv-D981 (Fig. 4B). These data are consistent with observations that MprAB activates Rv1057 under envelope stress (Figs. 1 A–E), and suggest that MprA boxes in both the distal and proximal portions of the Rv1057 promoter are required for this activation.
Figure 4.
Delineation of the Rv1057 promoter using lacZ reporter constructs. (A) Dashed lines indicate the sections of the Rv1056-Rv1057 intergenic region inserted upstream of lacZ in constructs P1-P3. P4 represents the promoterless lacZ control pSM128. (B) H37Rv and Rv-D981 were transformed with constructs P1-P4. Recombinants strains were exposed to 0.05% SDS for 90 min or left unexposed (controls). RNA was extracted, and lacZ expression was evaluated by qRT-PCR. Data are the mean +/− SEM from triplicate experiments.
DISCUSSION
Rv1057 encodes the only seven-bladed β-propeller protein in M. tuberculosis 30, and evidence suggests the protein may be a component of the mycobacterial envelope. The β-propeller fold occurs in proteins with diverse functions, including bacterial virulence proteins, but Rv1057 has highest similarity to surface layer proteins of Methanosarcina 30. In DNA microarray studies, Rv1057 was activated by envelope stress induced by exposure to the detergents SDS and Triton-X100 25–27, and the cell wall inhibitor vancomycin 36. Rv1057 is also one of the most highly upregulated genes following exposure to whole lung surfactant 37. In addition, using SCOTS analyses, Haydel and Clark-Curtiss 30 determined that Rv1057 is activated during growth in macrophages, an environment known to activate genes associated with envelope stress, including genes of the SigE regulon 41. Rv1057 may function similarly to TolB, a β-propeller protein that interacts with peptidoglycan-associated proteins and maintains envelope integrity in Gram-negative bacteria 31, 42, 43, and that is one of several Tol proteins essential for resistance to SDS and vancomycin 44, 45.
Our investigations support previous studies indicating a role for MprAB in activation of Rv1057 under detergent stress 25, 26, and further revealed that MprAB activates Rv1057 during growth in macrophages. In contrast, TrcRS suppresses Rv1057, and trcR expression declines during growth in macrophages 30, suggesting that MprAB and TrcRS have opposing effects on Rv1057 expression. MprA and TrcR did not compete for binding in the proximal promoter, and the TrcR binding site discovered in the distal promoter lies between MprA boxes, but further analyses are needed to determine whether the two response regulator compete to regulate Rv1057 activity. Rv1057 was first identified as a regulatory target of TrcRS through the discovery of AT-rich sequences in the promoter with similarity to the TrcR binding site in the trcRS promoter 30, 38. Although the precise binding site for TrcR in the Rv1057 distal promoter has not been determined, it is in the vicinity of the AT-rich sequence GTTTGAA that was previously noted to be highly similar to part of the proximal TrcR binding site 30.
The distal TrcR binding site is located within a small unannotated ORF (Fig. 3A) that is identical to gene MT1086 of strain CDC1551 (www.tbdb.org) 46, and in the same genomic arrangement. MT1086 is a member of the 13E12 repeat family, which may be a type of mycobacterial mobile element 4, 47. No transcripts were detected in this locale by qRT-PCR (X. Pang, unpublished data), suggesting the ORF may be nonfunctional, or that it is repressed by TrcR during normal growth conditions.
We identified at least four new MprA boxes and these supported findings from our laboratory on the acr2 promoter 29, and from the Zahrt laboratory on the pepD promoter 28, showing that MprA can tolerate considerable sequence variation in the hexamers, and MprA box half-sites need not be identical. The Rv1057 promoter is also similar to the acr2 and pepD promoters in having several functional MprA boxes. Surprisingly, the entire Rv1056-Rv1057 intergenic region was required for MprAB-dependent induction of Rv1057 during detergent exposure, suggesting the multiple MprA boxes work cooperatively to activate Rv1057. Although we selected sites strongly resembling known MprA boxes and that were consistent with the EMSA results (Fig. 3B–F, and Fig. S1), four of our predicted MprA boxes did not bind MprA. As noted previously, these sites may be incorrect or they may be auxiliary sites, similar to the sigE-p MprA box, which enhanced binding to the nearby sigE-d MprA box, but alone was unable to bind MprA in EMSAs 26. The MprA box in region D was not localized, but this region has at least two sequences with some similarity to the MprA box consensus sequence, including one at position −421 overlapping the beginning of the proximal TrcR binding site. This position is consistent with EMSA data indicating the presence of an MprA box between positions −429 and −390 (Fig. S1).
Box G was the nearest MprA box to the TSPs, beginning 29 bp upstream of TSP2, the detergent-inducible, MprAB-dependent TSP identified in this study. Based on its position, Box G should overlap a sigma factor binding site, although we did not discern any recognizable sigma factor sites in this region. In particular, no SigE-dependent promoter elements were found. A SigE-like promoter was identified upstream of Rv1057 TSP1 27, 30, but it is not clear whether SigE regulates transcription from this TSP either, as we did not detect induction of TSP1 by detergent (data not shown). Moreover, recent analyses suggest that SigE-dependent promoters have critical residues 48, and these are missing from the putative Rv1057 promoter elements (data not shown). Rv1057 is probably regulated by another sigma factor, possibly SigB, which is a component of the SigE and MprAB regulons 25, 27, 28. However, the sequence GTGG commonly found in SigB-regulated promoters 49, 50 is not present in the −35 regions upstream of the Rv1057 TSPs. It is possible the sigma factor recognition sequences in the Rv1057 promoter are weak, with the multiple MprA boxes in the intergenic region required to stabilize sigma factor binding.
In conclusion, our investigations reveal that the envelope-stress associated gene Rv1057 has an extensive and complex promoter with multiple MprA boxes, that it is activated by MprAB during intracellular growth, and identify it as the first M. tuberculosis gene to be directly regulated by dual TCSs. Additional studies are warranted to define the role of Rv1057 in envelope stress, and to determine whether TrcRS has a more extensive role in coregulation of genes of the MprAB regulon.
Supplementary Material
ACKNOWLEDGMENTS
We thank Buka Samten, Peter Barnes, and Amy Tvinnereim for helpful discussions, and Yuanxin Gu for technical assistance. This research was supported by the Potts Memorial Foundation, by grants from the National Science Foundation of China (NSFC81071328 and NSFC81171540) and from the Shandong Provincal Natural Science Foundation (ZR2010HM007) (to X.P.), and by NIH grant R21 AI063229-01 (to S.T.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Note added in proof: While our manuscript was under review, the Zahrt laboratory reported that MprAB and DosRST coregulate the Rv0081-Rv0088 operon (He H, Bretl DJ et al., 2011, J. Bact. 193:5105–5118), providing further evidence for coordinated regulation of stress response genes by TCSs.
REFERENCES
- 1.Corbett L, Raviglione MC. Global Burden of Tuberculosis: Past, Present and Future. In: Cole ST, Eisenach KD, McMurray DN, Jacobs WR Jr, editors. Tuberculosis and the Tubercle Bacilli. Washington, D.C.: ASM Press; 2005. pp. 3–12. [Google Scholar]
- 2.Hoch JA, Silhavy TJ. Two Component Signal Transduction. Washington, DC.: American Society for Microbiology; 1995. [Google Scholar]
- 3.Hoch JA, Varughese KI. Keeping signals straight in phosphorelay signal transduction. J Bacteriol. 2001;183:4941–4949. doi: 10.1128/JB.183.17.4941-4949.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream M-A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 5.Parish T, Smith DA, Kendall S, Casali N, Bancroft GJ, Stoker NG. Deletion of two-component regulatory systems increases the virulence of Mycobacterium tuberculosis. Infect Immun. 2003;71:1134–1140. doi: 10.1128/IAI.71.3.1134-1140.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rison SCG, Kendall SL, Movahedzadeh F, Stoker NG. The mycobacterial two-component regulatory systems. In: Parish T, editor. Mycobacterium Molecular Microbiology. Wydmondham, Norfolk, UK: Horizon Biosciences; 2005. pp. 29–69. [Google Scholar]
- 7.Zahrt TC, Deretic V. An essential two-component signal transduction system in Mycobacterium tuberculosis. J Bacteriol. 2000;182:3832–3838. doi: 10.1128/jb.182.13.3832-3838.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fol M, Chauhan A, Nair NK, Maloney E, Moomey M, Jagannath C, Madiraju MV, Rajagopalan M. Modulation of Mycobacterium tuberculosis proliferation by MtrA, an essential two-component response regulator. Mol Microbiol. 2006;60:643–657. doi: 10.1111/j.1365-2958.2006.05137.x. doi: PMID: 16629667. [DOI] [PubMed] [Google Scholar]
- 9.Rajagopalan M, Dziedzic R, Al Zayer M, Stankowska D, Ouimet MC, Bastedo DP, Marczynski GT, Madiraju MV. Mycobacterium tuberculosis origin of replication and the promoter for immunodominant secreted antigen 85B are the targets of MtrA, the essential response regulator. J Biol Chem. 2010;285:15816–15827. doi: 10.1074/jbc.M109.040097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen HT, Wolff KA, Cartabuke RH, Ogwang S, Nguyen L. A lipoprotein modulates activity of the MtrAB two-component system to provide intrinsic multidrug resistance, cytokinetic control and cell wall homeostasis in Mycobacterium. Mol Microbiol. 2010;76:348–364. doi: 10.1111/j.1365-2958.2010.07110.x. [DOI] [PubMed] [Google Scholar]
- 11.Sherman DR, Voskuil M, Schnappinger D, Liao R, Harrell MI, Schoolnik GK. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding a-crystallin. Proc Natl Acad Sci USA. 2001;98:7534–7539. doi: 10.1073/pnas.121172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voskuil MI, Schnappinger D, Visconti KC, Harrell MI, Dolganov GM, Sherman DR, Schoolnik GK. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med. 2003;198:705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saini DK, Malhotra V, Dey D, Pant N, Das TK, Tyagi JS. DevR-DevS is a bona fide two-component system of Mycobacterium tuberculosis that is hypoxia-responsive in the absence of the DNA-binding domain of DevR. Microbiology. 2004;150:865–875. doi: 10.1099/mic.0.26218-0. [DOI] [PubMed] [Google Scholar]
- 14.Roberts DM, Liao RP, Wisedchaisri G, Hol WG, Sherman DR. Two sensor kinases contribute to the hypoxic response of Mycobacterium tuberculosis. J Biol Chem. 2004;279:23082–23087. doi: 10.1074/jbc.M401230200. doi: PMID: 15033981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar A, Toledo JC, Patel RP, Lancaster JR, Jr, Steyn AJ. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc Natl Acad Sci USA. 2007;104:11568–11573. doi: 10.1073/pnas.0705054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Honaker RW, Leistikow RL, Bartek IL, Voskuil MI. Unique roles of DosT and DosS in DosR regulon induction and Mycobacterium tuberculosis dormancy. Infect Immun. 2009;77:3258–3263. doi: 10.1128/IAI.01449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leistikow RL, Morton RA, Bartek IL, Frimpong I, Wagner K, Voskuil MI. The Mycobacterium tuberculosis DosR regulon assists in metabolic homeostasis and enables rapid recovery from nonrespiring dormancy. J Bacteriol. 2010;192:1662–1670. doi: 10.1128/JB.00926-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rickman L, Saldanha JW, Hunt DM, Hoar DN, Colston MJ, Millar JB, Buxton RS. A two-component signal transduction system with a PAS domain-containing sensor is required for virulence of Mycobacterium tuberculosis in mice. Biochem Biophys Res Commun. 2004;314:259–267. doi: 10.1016/j.bbrc.2003.12.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sureka K, Dey S, Datta P, Singh AK, Dasgupta A, Rodrigue S, Basu J, Kundu M. Polyphosphate kinase is involved in stress-induced mprAB-sigE-rel signalling in mycobacteria. Mol Microbiol. 2007;65:261–276. doi: 10.1111/j.1365-2958.2007.05814.x. doi: PMID: 17630969. [DOI] [PubMed] [Google Scholar]
- 20.Rifat D, Bishai WR, Karakousis PC. Phosphate depletion: a novel trigger for Mycobacterium tuberculosis persistence. J Infect Dis. 2009;200:1126–1135. doi: 10.1086/605700. [DOI] [PubMed] [Google Scholar]
- 21.Perez E, Samper S, Bordas Y, Guilhot C, Gicquel B, Martin C. An essential role forphoP in Mycobacterium tuberculosis virulence. Mol Microbiol. 2001;41:179–187. doi: 10.1046/j.1365-2958.2001.02500.x. [DOI] [PubMed] [Google Scholar]
- 22.Walters SB, Dubnau E, Kolesnikova I, Laval F, Daffe M, Smith I. The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol Microbiol. 2006;60:312–330. doi: 10.1111/j.1365-2958.2006.05102.x. doi: PMID: 16573683. [DOI] [PubMed] [Google Scholar]
- 23.Frigui W, Bottai D, Majlessi L, Monot M, Josselin E, Brodin P, Garnier T, Gicquel B, Martin C, Leclerc C, Cole ST, Brosch R. Control of M. tuberculosis ESAT-6 secretion and specific T cell recognition by PhoP. PLoS Pathog. 2008;4:e33. doi: 10.1371/journal.ppat.0040033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zahrt TC, Deretic V. Mycobacterium tuberculosis signal transduction system required for persistent infections. Proc Natl Acad Sci USA. 2001;98:12706–12711. doi: 10.1073/pnas.221272198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pang X, Vu P, Byrd TF, Ghanny S, Soteropoulos P, Mukamolova GV, Wu S, Samten B, Howard ST. Evidence for complex interactions of stress-associated regulons in an mprAB deletion mutant of Mycobacterium tuberculosis. Microbiology. 2007;153:1229–1242. doi: 10.1099/mic.0.29281-0. [DOI] [PubMed] [Google Scholar]
- 26.He H, Hovey R, Kane J, Singh V, Zahrt TC. MprAB is a stress-responsive two-component system that directly regulates expression of sigma factors SigB and SigE in Mycobacterium tuberculosis. J Bacteriol. 2006;188:2134–2143. doi: 10.1128/JB.188.6.2134-2143.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manganelli R, Voskuil MI, Schoolnik GK, Smith I. The Mycobacterium tuberculosis ECF sigma factor σE: role in global gene expression and survival in macrophages. Mol Microbiol. 2001;41:423–437. doi: 10.1046/j.1365-2958.2001.02525.x. [DOI] [PubMed] [Google Scholar]
- 28.He H, Zahrt TC. Identification and characterization of a regulatory sequence recognized by Mycobacterium tuberculosis persistence regulator MprA. J Bacteriol. 2005;187:202–212. doi: 10.1128/JB.187.1.202-212.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pang X, Howard ST. Regulation of the α-crystallin gene acr2 by the MprAB two-component system of Mycobacterium tuberculosis. J Bacteriol. 2007;189:6213–6221. doi: 10.1128/JB.00492-07. doi: PMID: 17601788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haydel SE, Clark-Curtiss JE. The Mycobacterium tuberculosis TrcR response regulator represses transcription of the intracellularly expressed Rv1057 gene, encoding a seven-bladed beta-propeller. J Bacteriol. 2006;188:150–159. doi: 10.1128/JB.188.1.150-159.2006. doi: PMID: 16352831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ponting CP, Pallen MJ. A beta-propeller domain within TolB. Mol Microbiol. 1999;31:739–740. doi: 10.1046/j.1365-2958.1999.01168.x. [DOI] [PubMed] [Google Scholar]
- 32.Korczynska M, Mukhtar TA, Wright GD, Berghuis AM. Structural basis for streptogramin B resistance in Staphylococcus aureus by virginiamycin B lyase. Proc Natl Acad Sci U S A. 2007;104:10388–10393. doi: 10.1073/pnas.0701809104. doi: PMID: 17563376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heenan EJ, Vanhooke JL, Temple BR, Betts L, Sondek JE, Dohlman HG. Structure and function of Vps15 in the endosomal G protein signaling pathway. Biochemistry. 2009;48:6390–6401. doi: 10.1021/bi900621w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu M, Grahn E, Eriksson LA, Strid A. Computational Evidence for the Role of Arabidopsis thaliana UVR8 as UV-B Photoreceptor and Identification of Its Chromophore Amino Acids. J Chem Inf Model. 2011;51(6):1287–1295. doi: 10.1021/ci200017f. EpubWu M, Grahn E, Eriksson L: 1287–1295. [DOI] [PubMed] [Google Scholar]
- 35.Xu C, Min J. Structure and function of WD40 domain proteins. Protein Cell. 2011;2:202–214. doi: 10.1007/s13238-011-1018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Provvedi R, Boldrin F, Falciani F, Palù G, Manganelli R. Global transcriptional response to vancomycin in Mycobacterium tuberculosis. Microbiology. 2009;155:1093–1102. doi: 10.1099/mic.0.024802-0. [DOI] [PubMed] [Google Scholar]
- 37.Schwab U, Rohde KH, Wang Z, Chess PR, Notter RH, Russell DG. Transcriptional responses of Mycobacterium tuberculosis to lung surfactant. Microb Pathog. 2009;46:185–193. doi: 10.1016/j.micpath.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haydel SE, Benjamin WH, Jr, Dunlap NE, Clark-Curtiss JE. Expression, autoregulation, and DNA binding properties of the Mycobacterium tuberculosis TrcR response regulator. J Bacteriol. 2002;184:2192–2203. doi: 10.1128/JB.184.8.2192-2203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dussurget O, Timm J, Gomez M, Gold B, Yu S, Sabol SZ, Holmes RK, Jacobs WR, Jr, Smith I. Transcriptional control of the iron-responsive fxbA gene by the mycobacterial regulator IdeR. J Bacteriol. 1999;181:3402–3408. doi: 10.1128/jb.181.11.3402-3408.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haydel SE, Dunlap NE, Benjamin WH., Jr. In vitro evidence of two-component system phosphorylation between the Mycobacterium tuberculosis TrcR/TrcS proteins. Microb Pathog. 1999;26:195–206. doi: 10.1006/mpat.1998.0265. [DOI] [PubMed] [Google Scholar]
- 41.Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C, Schoolnik GK. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bouveret E, Derouiche R, Rigal A, Lloubès R, Lazdunski C, Bénédetti H. Peptidoglycan-associated lipoprotein-TolB interaction. A possible key to explaining the formation of contact sites between the inner and outer membranes of Escherichia coli. J Biol Chem. 1995;270:11071–11077. doi: 10.1074/jbc.270.19.11071. [DOI] [PubMed] [Google Scholar]
- 43.Carr S, Penfold CN, Bamford V, James R, Hemmings AM. The structure of TolB, an essential component of the tol-dependent translocation system, and its protein-protein interaction with the translocation domain of colicin E9. Structure. 2000;8:57–66. doi: 10.1016/s0969-2126(00)00079-4. [DOI] [PubMed] [Google Scholar]
- 44.Bernstein A, Rolfe B, Onodera K. Pleiotropic properties and genetic organization of the tolA,B locus of Escherichia coli K-12. J Bacteriol. 1972;112:74–83. doi: 10.1128/jb.112.1.74-83.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Llamas MA, Ramos JL, Rodríguez-Herva JJ. Mutations in each of the tol genes of Pseudomonas putida reveal that they are critical for maintenance of outer membrane stability. J Bacteriol. 2000;182:4764–4772. doi: 10.1128/jb.182.17.4764-4772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reddy TB, Riley R, Wymore F, Montgomery P, Decaprio D, Engels R, Gellesch M, Hubble J, Jen D, Jin H, Koehrsen M, Larson L, Mao M, Nitzberg M, Sisk P, Stolte C, Weiner B, White J, Zachariah ZK, Sherlock G, Galagan JE, Ball CA, Schoolnik GK. TB database: an integrated platform for tuberculosis research. Nucleic Acids Res. 2009;37:D499–D508. doi: 10.1093/nar/gkn652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davis EO, Dullaghan EM, Rand L. Definition of the mycobacterial SOS box and use to identify LexA-regulated genes in Mycobacterium tuberculosis. J Bacteriol. 2002;184:3287–3295. doi: 10.1128/JB.184.12.3287-3295.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song T, Song SE, Raman S, Anaya M, Husson RN. Critical role of a single position in the-35 element for promoter recognition by Mycobacterium tuberculosis SigE and SigH. J Bacteriol. 2008;190:2227–2230. doi: 10.1128/JB.01642-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee JH, Karakousis PC, Bishai WR. Roles of SigB and SigF in the Mycobacterium tuberculosis sigma factor network. J Bacteriol. 2008;190:699–707. doi: 10.1128/JB.01273-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fontán PA, Voskuil MI, Gomez M, Tan D, Pardini M, Manganelli R, Fattorini L, Schoolnik GK, Smith I. The Mycobacterium tuberculosis sigma factor sigmaB is required for full response to cell envelope stress and hypoxia in vitro, but it is dispensable for in vivo growth. J Bacteriol. 2009;191:5628–5633. doi: 10.1128/JB.00510-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.