Abstract
Here, we describe a simple and sensitive approach for visual detection of gene mutations based on isothermal strand-displacement polymerase reactions (ISDPR) and lateral flow strip (LFS). The concept was first demonstrated by detecting the R156H-mutant gene of keratin 10 in Epidermolytic hyperkeratosis (EHK). In the presence of biotin-modified hairpin DNA and digoxin-modified primer, the R156H-mutant DNA triggered the ISDPR to produce numerous digoxin- and biotin-attached duplex DNA products. The product was detected on the LFS through dual immunoreactions (anti-digoxin antibody on the gold nanoparticle (Au-NP) and digoxin on the duplex, anti-biotin antibody on the LFS test zone and biotin on the duplex). The accumulation of Au-NPs produces the characteristic red band, enabling visual detection of the mutant gene without instrumentation. After systematic optimization of the ISDPR experimental conditions and the parameters of the assay, the current approach is capable of detecting as low as 1-fM R156H-mutant DNA within 75 min without instrumentation. Differentiation of R156H- and R156C-mutant DNA on the R156 mutation site was realized by using fluorescein- and biotin-modified hairpin probes in the ISDPR process. The approach thus provides a simple, sensitive, and low-cost tool for the detection of gene mutations.
1. Introduction
The detection of gene mutations and specific DNA sequences is of central importance for the diagnosis and treatment of genetic diseases, for the detection of infectious agents, and for reliable forensic analysis (Palecek and Fojta, 2001). Various methods and technologies, such as Northern, Southern and Western blotting; agarose and polyacrylamide gel electrophoresis (Nelson and Cox, 2000); DNA biosensors (Leung et al., 2007); DNA biochips (Brown and Botstein, 1999); polymerase chain reactions (PCR) (Kaltenboeck and Wang, 2005); ligation (Toubanaki et al., 2009); primer extension (Hoogendoorn et al., 1999); and endonuclease digestion (Lyamichev et al., 1999), have been developed for this purpose. Because of high sensitivity and specificity, PCR has become the gold standard in clinical diagnostics and the food industry. However PCR requires precise control of temperature cycling for successful DNA amplification, and the resultant instrumental restraint has been hampering its wider and more versatile applications (point-of-care use in hospitals and miniaturized systems for high-throughput analysis) (Gill and Ghaemi, 2008). Therefore, there is a need for a suitable and cost-effective analytical technology to carry out a rapid, simple, and sensitive analysis of gene mutations and DNA sequences.
Isothermal nucleic acid amplification technologies have been developed to overcome the disadvantages of PCR (Hellyer and Nadeau, 2004). Particularly, isothermal strand-displacement polymerase reaction (ISDPR) in connection with different detection platforms (fluorescence and electrochemistry) has attracted considerable interest (Guo et al., 2009; He et al., 2010a). ISDPR is based on an isothermal amplification process which yields large amounts of labeled DNA products to enhance the signal and the sensitivity of DNA detection (Hellyer and Nadeau, 2004). Although the ISDPR-based fluorescent and electrochemical DNA assays offer high specificity and sensitivity, there are still many challenges. For example, the detection still needs expensive instruments or skilled personnel, preventing its in-field or point-of-care applications, particularly in developing countries. In addition, the false initiation of ISDPRs would result in a high background signal.
Recently, we reported a sensitive, portable nucleic-acid biosensor based on oligonucleotide-modified gold nanoparticle (Au-NP) and a conventional lateral flow device for visual detection of DNA (Mao et al., 2009); the sensitivity and ability to discriminate single base-mismatched DNA and perfect-matched DNA with the device have been explored by using enzyme-Au-NP dual label and hairpin oligonucleotide-modified Au-NPs, respectively (He et al., 2010b; He et al., 2011). However, the sensitivities of such visual detection are still relatively low compared with PCR-based and fluorescence-based isothermal nucleic acid amplification methods. In this article, we report a simple and sensitive approach based on ISDPR and the lateral flow strip (LFS) for the determination of R156 mutations (R156H and R156C) of keratin 10 in Epidermolytic hyperkeratosis (EHK, Mendelian Inheritance in Man no. 113800). EHK is an autosomal dominant skin disease which is caused by mutations in the genes encoding the keratin 1 gene (KRT 1) or the keratin 10 gene (KRT 10) (Rothnagel et al., 1992; Syder et al., 1994). Recent results from the pertinent published work and the Human Gene Mutation Database have shown that the 156 site of the KRT 10 gene is a mutation hot spot. R156H (CGC to CAC, arginine to histidine) and R156C (CGC to TGC, arginine to cysteine) are the most frequent mutations of KRT 10 (Haruna et al., 2007). The concept was first demonstrated by using a biotin-modified hairpin probe and a digoxin-modified primer to determine the R156H-mutant DNA. Visual differentiation of R156H- and R156C-mutant DNA was realized by using fluorescein- and biotin-modified hairpin probes. The attractive characteristics of the new approach are reported in the following sections.
2. Materials and Methods
2.1. Materials
The Airjet AJQ 3000 dispenser, Biojet BJQ 3000 dispenser, Clamshell Laminator, and Guillotine cutting module CM 4000 were from Biodot, LTD (Irvine, CA). A portable strip reader (DT1030) was purchased from Shanghai Goldbio Tech. Co., LTD (Shanghai, China). The polymerase Klenow fragment exo− was purchased from New England Biolabs, Inc. The deoxynucleotide solution mixture (dNTPs), dithiothreitol (DTT), Dimethyl-Sulfoxide (DMSO), anti-biotin antibody, anti-fluorescein antibody, anti-digoxin antibody, human serum albumin (HSA), sucrose, hydroxylamine, Tween 20, Triton X-100, trisodium citrate, bovine serum albumin (BSA), sodium chloride-sodium citrate (SSC) Buffer 20×concentrate (pH 7.0), Tris/Borate/EDTA (TBE) buffer, HAuCl4, and phosphate buffer saline (PBS, pH 7.4, 0.01 M) were purchased from Sigma-Aldrich (St. Louis, MO). Glass fibers (GFCP000800), cellulose fiber sample pads (CFSP001700), laminated cards (HF000MC100), and nitrocellulose membranes (HFB18004) were purchased from Millipore (Billerica, MA). NuSeive® GTG® agarose was purchased from Cambrex Bio Science (Rockland, ME). The DNA oligonucleotide probes and digoxin-modified primers were obtained from Integrated DNA Technologies, Inc. (Coralville, IA). The oligonucleotide sequences were as follows:
R156H-mutant DNA: 5' -CTGAATGACCACCTGGCTGTGTCC - 3'
R156C-mutant DNA: 5' -CTGAATGACTGCCTGGCTGTGTCC - 3'
Wild-type DNA: 5' -CTGAATGACCGCCTGGCTGTGTCC - 3'
Noncomplementay DNA: 5' - TGCAAGGTGTCAGTATAATCCGACGTTTT-3'
Hairpin probe to R156H-mutant DNA: 5' –Bio-TCTTGGACACAGCCAGGTGGTCATTCAGTGTGTCCAAGA-3'
Hairpin probe to R156C-mutant DNA: 5' –Flu-TCTTG GACACAGCCAGGCAGTCATTCAGTGTGTCCAAGA-3'
Primer: 5' - /5DigN/TCTTGGAC-3'
All chemicals used were analytical reagent grade. Other solutions were prepared with ultrapure (>18 MΩ) water from a Millipore Milli-Q water purification system (Billerica, MA).
2.2 Preparation of anti-digoxin-Au-NP conjugates and lateral flow strip (LFS)
Au-NPs with average diameter 15 nm ± 3.5 nm were prepared according to the reported methods with slight modifications (He et al., 2011, also see the details in the supporting information). Conjugation reactions were conducted by adding 5 µL of 10 mg mL−1 anti-digoxin antibody into 1 mL of a tenfold-concentrated Au-NP solution (pH 8.4) followed by incubation at room temperature with periodic gentle mixing for 1 h. Then, a certain volume of 10% BSA was slowly added to the mixture solution to obtain a final concentration of 1%. After gentle stirring for 30 min, the solution was centrifuged at 13,000×g for 15 min. Two phases can be obtained: a clear to pink supernatant of unbound antibodies and a dark red, loosely packed sediment of the anti-digoxin-Au-NP conjugates. The supernatant was discarded, and the soft sediment of anti-digoxin-Au-NP conjugates was rinsed by resuspending in 1 mL of PBS-BSA and collected after a second centrifugation at 13,000×g for 15 min. Finally, the conjugate was resuspended in a 1 mL buffer containing 20 mM of sodium phosphate, 0.25% Tween-20, 10% sucrose, and 5% BSA, and the conjugate was stored at 4°C before further use. LFS was prepared according to the reported methods with slight modifications (He et al., 2011, also see the details in the supporting information).
2.3 Preparation of digoxin- and biotin-attached duplex DNA complexes with ISDPRs
Briefly, ISDPR was performed in a 100-µL, 50-mM Tris-HCl (pH 8.0) buffer consisting of 5.0×10−8 M biotin-modified hairpin probe, 5.0×10−8 M digoxin-modified primer, 3 U polymerase Klenow fragment exo−, 50 µM dNTPs, 6% DMSO, 0.1% BSA, 1 mM DTT, and 5 mM MgCl2. R156H-mutant DNA at different concentrations was then added to every mixture sample solution and incubated at 42°C for 1 h.
2.4 Gel electrophoresis
Gel electrophoresis of the ISDPR product was conducted with a 4% gel (3% NuSeive GTG agarose + 1% Agrose) prepared in 1× TBE (pH = 8.3) at 65 V constant voltage for about 4.5 h. After ethidium bromide staining, gels were scanned using the Chemi Genius Bio Imaging System (Syngene, NJ).
2.5 Visual detection of ISDPR products with LFS
A 100-µL running buffer (10 mM Tris-HCl solution (pH 8.0) containing 5 mM MgCl2, 80×SSC, and 0.5% BSA) was mixed with 10 µL of ISDPR product solution in a 1.5-mL centrifuge tube; then, a LFS was dipped into the mixture. After waiting for 10 min, 60 µL of running buffer were added to wash the LFS. The bands were visualized within 5 min. The intensities of the red bands on the LFS were quantified with a portable strip reader. The optical intensities of the test and the control lines were recorded simultaneously by using “GoldBio strip reader” software, which could search the red bands in a fixed reaction area automatically and then figure out parameters such as peak height and area integral.
3. Results and Discussion
3.1 Principle of detecting mutant gene visually
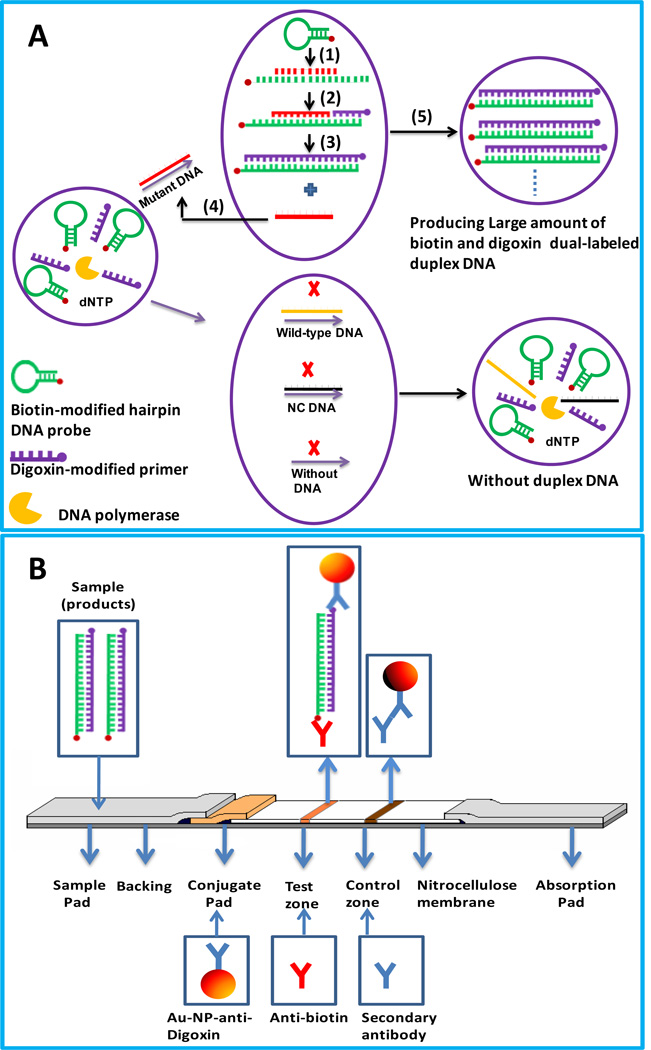
The protocol in this study combines the amplification features of ISDPR with the convenient visual detection of ISDPR products on a LFS. The principle is shown in Scheme 1. ISDPR is used to generate numerous biotin- and digoxin-attached duplex DNAs (Scheme 1A). The ISDPR solution consists of a biotin-modified hairpin probe, a digoxin-modified primer, polymerase, dNTP, and buffer solution. The biotin-modified hairpin probe has a stem-loop structure with biotin linked to the 5-end of the stem. The stem is 11-nt sequences long, and the loop is complementary to the mutant DNA . The digoxin-modified primer is 8-nt sequences long, which is complementary to the stem region of the hairpin probe at the 3-end. In the presence of mutant DNA, the biotin-modified hairpin probe recognizes and hybridizes with it; the probe then undergoes a conformational change, leading to stem separation (Step 1). The digoxin-modified primer thus anneals with the open stem and triggers a polymerization reaction in the presence of dNTPs and polymerase (Step 2). Next in the process of primer extension, the mutant DNA is displaced by the polymerase with strand-displacement activity, after which a complementary DNA is synthesized, forming a biotin- and digoxin-attached duplex DNA (Step 3). Finally, to renew the cycle, the displaced mutant DNA hybridizes with another biotin-modified hairpin probe, triggering another polymerization reaction (Step 4). Throughout this cyclical process, numerous biotin- and digoxin-attached duplex DNAs are produced in the presence of minute amounts of mutant DNAs. In the presence of a wild-type DNA (one-base mismatch) and noncomplementary DNA, and in the absence of mutant DNA (Scheme 1A, bottom), the hairpin probe keeps its original stem-loop structure because of a weak hybridization ability between the wild-type DNA and hairpin probe; lacking a DNA-hybridization reaction in the absence of mutant DNA and in the presence of noncomplementary DNA, the hairpin DNA is unable to anneal with the primer to induce a polymerization reaction. In these cases, there is no duplex DNA complex produced in the reactions.
Scheme 1.
(A) Schematic illustration of the formation of digoxin- and biotin-attached duplex DNA complexes based on isothermal strand-displacement polymerase reactions. (B) Schematic illustration for the visual detection of ISDPR products on a lateral flow strip.
The visual detection of the formed duplex DNA complexes is performed on a LFS, and the principle is shown in Scheme 1B. Anti-digoxin-labeled Au-NPs and anti-biotin antibody were preimmobilized on the conjugate pad and test zone of the LFS, respectively. The secondary antibody, which is against anti-digoxin antibody, was immobilized on the control zone. The LFS was dipped into the mixture of running buffer and ISDPR product. When the solution migrated by capillary action and passed the conjugate pad, it rehydrated the anti-digoxin-Au-NP conjugates. The digoxin on the duplex DNA reacted with the anti-digoxin on the Au-NP surface to form a biotin-duplex DNA-digoxin-anti-digoxin-Au-NP complex and continued to migrate along the strip. The complexes were captured on the test zone by the specific reaction between anti-biotin on the test zone and the biotin of the complexes. The accumulation of Au-NPs in the test zone was visualized as a characteristic red band. Then, the excess anti-digoxin-Au-NP conjugates continued to move and were captured on the control zone by immunoreactions between the secondary antibody and anti-digoxin on the Au-NP surface, thus forming a second red band. In the absence of mutant DNA and in the presence of wild-type DNA and noncomplementary DNA, there was no duplex DNA in the sample solution; therefore, no anti-digoxin-Au-NP conjugate was captured on the test zone of the LFS, and no red band was observed in the test zone. In this case, a red band (control line) on the control zone shows that the LFS was working properly.
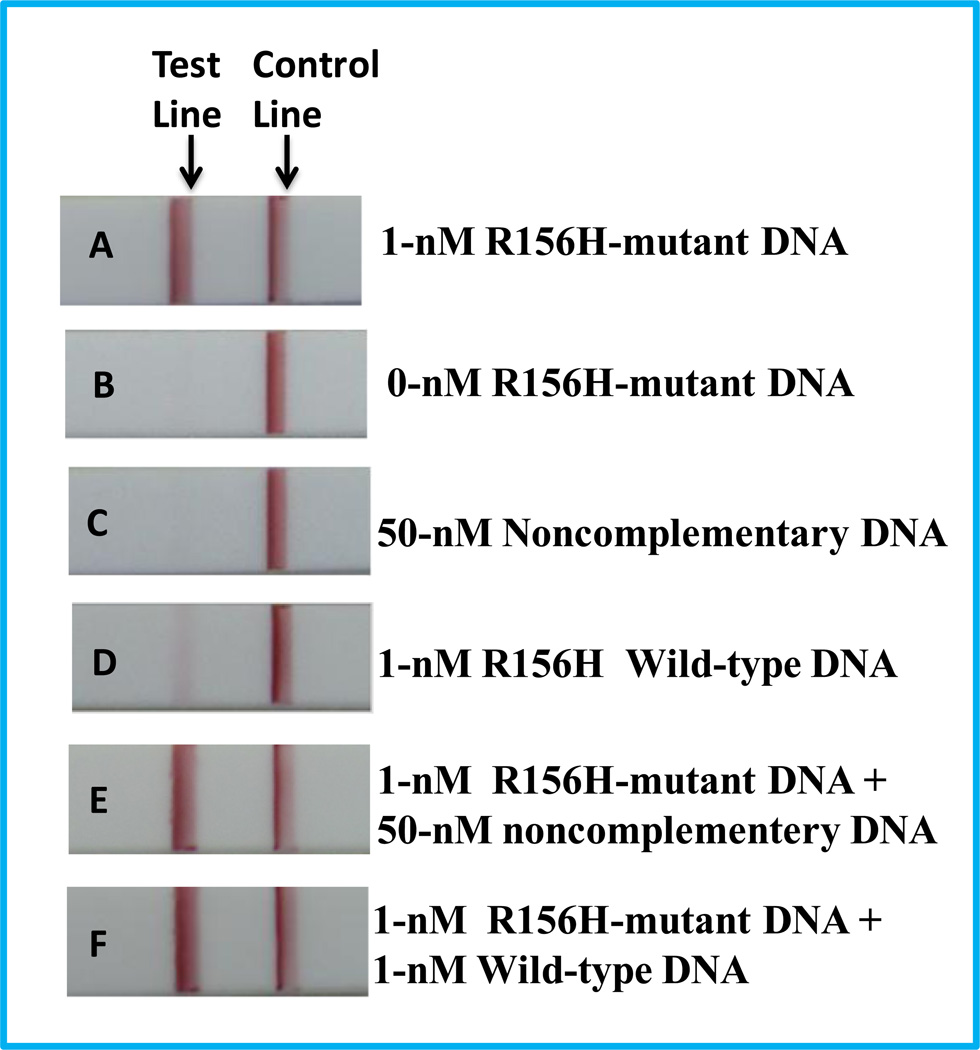
Based on the proposed approach, we first used the R156H-mutant DNA as target DNA and designed a set of biotin-modified hairpin and digoxin-modified primer for ISDPR. Six DNA samples, 1-nM R156H-mutant DNA, 1-nM wild-type DNA, 0-nM mutant DNA, 50-nM noncomplementary DNA, 1-nM R156H-mutant DNA+50-nM noncomplementary DNA, and 1-nM R156H-mutant DNA+1-nM wild-type DNA were used for the ISDPRs, and the products were tested on the LFSs. The typical photo images are shown in Figure 1. As expected, no distinct red band was observed on the test zones of LFSs in the absence of R156H-mutant DNA (Figure 1B) and 50-nM noncomplementary DNA (Figure 1C). A bright red band was observed on the test zone of the LFS with 1-nM R156H-mutant DNA (Figure 1A). The appearance of a red band implied that the Au-NPs were captured on the test zone of the LFS through the duplex DNA via the immunoreactions of digoxin-anti-digoxin and biotin-anti-biotin. A very weak red band was observed on the test zone of LFS with 1-nM wild-type DNA (Figure 1D). The weak band would be caused by the formation of a small amount of biotin- and digoxin-attached duplex DNA from the hybridization reactions between the hairpin DNA and wild-type DNA (one-base mismatched DNA). The intensity difference of the red bands on the test zones in the presence of the mutant DNA and wild-type DNA showed excellent specificity of the assay. The specificity was further demonstrated by detecting the mixture of R156H-mutant DNA and noncomplementary DNA (Figure 1E), and the mixture of R156H-mutant DNA and wild-type DNA (Figure 1F). The responses were similar with those obtained with 1-nM R156H-mutant DNA alone (Figure 1A).
Figure 1.
Typical photo images of LFSs in the presence of 1-nM R156H-mutant DNA(A), 0-nM R156H-mutant DNA(B), 50-nM noncomplementary DNA(C), 1-nM wild-type DNA(D), 1-nM R156H-mutant DNA+50-nM noncomplementary DNA(E), and 1-nM R156H-mutant DNA+1-nM wild-type DNA(F). ISDPR reactions were performed at 42°C for 60 min.
3.2 Verification of the signal amplification of ISDPR for the visual detection of R156H-mutant DNA on LFS
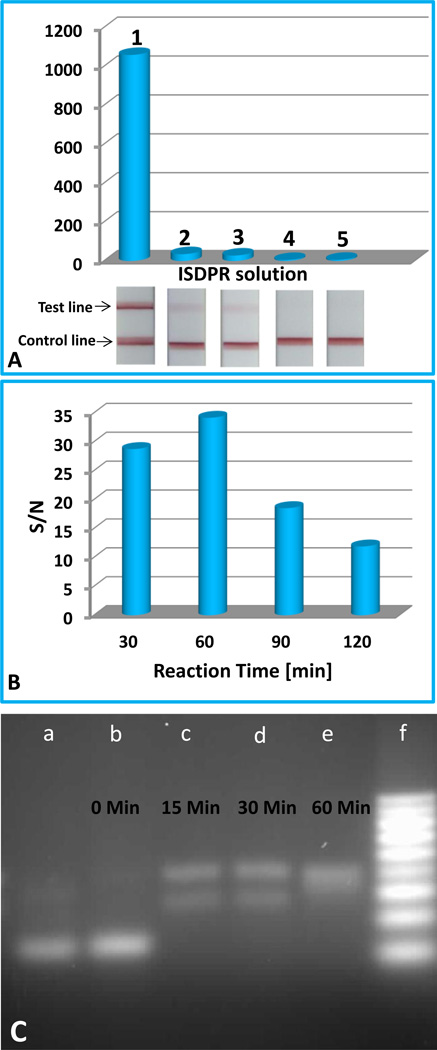
In the current study, ISDPR was used to produce numerous digoxin- and biotin-attached duplex DNA complexes for visual detection of the R156H-mutant DNA. Having proven the suitability of the approach, we investigated the feasibility of this method for amplification of DNA detection. First, we compared the response of 1-nM R156H-mutant DNA on the LFS with and without ISDPR amplification (Figure 2A). In the case without ISDPR (solution 2), polymerase was not added to the sample solution. In order to facilitate the quantitative comparison, intensities of the red bands were recorded using the portable strip reader combined with the “AuBio strip reader” software. Figure 2A presents the typical responses of 1-nM R156H-mutant DNA on the LFSs with (solution 1) and without (solution 2) ISDPR. One can see that the response of the sample with ISDPR was 30 times higher than that without ISDPR. This result indicated that the ISDPR generated numerous duplex DNAs, which increased the number of captured Au-NPs in the LFS test zone, thus enhancing the intensities of test band. We also tested the response of a 1-nM R156H-mutant DNA with ISDPR amplification in the absence of dNTP (solution 3), in the absence of biotin-modified hairpin (solution 4), and/or in the absence of digoxin-modified primer (solution 5). The results are also shown in Figure 2A. In the absence of dNTP, the response of the assay was similar with that in the absence of polymerase (solution 2), indicating that there was no amplification in the absence of dNTP during the ISDPR. In addition, no signal was observed in the absence of biotin-modified hairpin or digoxin-modified primer. The corresponding images of the LFSs are shown in the inset of Figure 2A.
Figure 2.
(A) The optical responses of 1-nM R156H-mutant DNA on the LFSs with different ISDPR solutions. Solution 1: 5.0×10−8 M biotin-modified hairpin probe, 5.0×10−8 M digoxin-modified primer, 3-U polymerase Klenow fragment exo−, 50-µM dNTPs, 6% DMSO, 0.1% BSA, 1-mM DTT, and 5 mM MgCl2 in 50-mM Tris–HCl (pH 8.0) buffer; Solution 2: solution 1 without polymerase Klenow fragment exo−; Solution 3: solution 1 without dNTP; Solution 4: solution 1 without biotin-modified hairpin probe; and Solution 5: solution 1 without digoxin-modified primer. ISDPRs were performed 1 h at 42°C. Corresponding photo images of the LFSs are shown in the inset. (B) The effect of ISDPR time on the S/N ratio of assays. (C) Gel image of ISDPR products in the absence (lane a) and presence of 1-nM R156H-mutant DNA. Lane a: product generated in the absence of R156H-mutant DNA with an ISDPR time of 60 min. Lanes b–d: products generated in the presence of 1-nM R156H-mutant DNA at different ISDPR time intervals: (b) t = 0 min, (c) t = 15 min, (d) t = 30 min, and (e) t = 60 min. Lane e: DNA ladder markers (10, 20, 30, 40, 50, 60, 70, 80, and 90 bp from the bottom). ISDPRs were performed with solution 1 in (A) at 42°C.
To confirm that the amplified signal observed in these experiments was dependent on the ISDPR, we tested the effect of ISDPR time on the response of the assay. The ISDPR time would be proportional to the cycles of the strand displacement; the amount of duplex DNA complexes; and, thus, the response of the assay. Figure 2B presents the effect of the ISDPR time on the signal-to-noise (S/N) ratio of 1-nM R156H-mutant DNA. One can see that the S/N ratio increased when raising the reaction time from 30 to 60 min, indicating that continuous formations of the duplex DNA complexes were the result of circular strand displacement reactions; further increasing the reaction time led to a decreased S/N ratio. The decreased S/N ratio at a longer ISDPR time would be caused by an increased background signal, which would be caused by the confirmation change of the biotin-modified hairpin with a longer reaction time at 42°C, leading to stem separation of the hairpin probe. The digoxin-modified primer thus annealed with the open stem and triggered a polymerization reaction to produce the digoxin- and biotin-attached duplex DNA complex. The effect of the ISDPR time was further verified by the analysis of the ISDPR endpoint product with agarose gel electrophoresis. Figure 2C shows the electrophoresis results of the ISDPR products in the absence and presence of R156H-mutant DNA at various reaction-time intervals. Lane a was the nucleic acid band of ISDPR product in the absence of R156H-mutant DNA; lanes b, c, d, and e were the nucleic acid bands of ISDPR products prepared at 0, 15, 30, and 60 min reaction times, respectively; lane f was loaded with DNA ladder markers (10, 20, 30, 40, 50, 60, 70, 80, and 90 bp from the bottom). The bands on lanes a and b belonged to the hairpin DNA probes, which had 41 bases and 11 bases paired. In the absence of the R156H-mutant (lane a) and 0 min ISDPR time (lane b), no hairpin DNA was consumed, and no duplex DNA was formed. As the reaction time increased to 15 and 30 min, two new bands appeared (lanes c and d). The bottom band was from the formed intermediate product of the hairpin probe-mutant DNA complex that existed in the process of primer extension before the target was displaced (Step 2 in Scheme 1A); the upper band, with a slower migration speed than the intermediate product, was from the formed duplex DNA products. When the ISDPR time reached 60 min (lane e), the bottom band in lane d (30 min) disappeared, and only one band was shown on lane e, indicating that the ISDPR was completed and that the intermediate complexes were converted to the digoxin- and biotin-attached duplex DNA complexes. The results also confirmed the effect of ISDPR time on the response of test band on the LFS (Figure 2B). The signal amplification of ISDPR was also verified by the analysis of the DNA hybridization reaction product without ISDPR with agarose gel electrophoresis (see gel image in Figure S1). In the absence of ISDPR, there was a bright band (lane b in Figure S1) observed at the bottom of the lane, which came from the remaining hairpin DNA probes in the DNA hybridization solution; there was a very weak band observed at the top of the lane, which belonged to the small amount of duplex DNA complexes (mutant DNA-hairpin probe) from the hybridization reactions. These results indicated that the ISDPR did produce a large number of duplex DNA complexes and led to signal amplification.
3.3 Optimization of experimental conditions
To improve sensitivity for the detection of R156H-mutant DNA, the conditions of ISDPR, LFS preparation and detecting conditions were optimized systematically. We studied the effect of the ISDPR temperature and the concentrations of ISDPR components (biotin-modified hairpin probe, digoxin-modified primer, dNTP, and polymerase) on the S/N ratio of the assay (see detailed discussion and Figure S2 in the support information). The best S/N ratio was obtained with 5.0×10−8 M biotin-modified hairpin probe, 5.0×10−8 M digoxin-modified primer, 3-U polymerase Klenow fragment exo−, and 50-µM dNTPs at a temperature of 42°C. The concentration of anti-digoxin antibody used to prepare the anti-digoxin-Au-NP conjugates is one of the most important factors to affect the sensitivity and reproducibility of LFS. The concentrations of anti-digoxin antibody, ranging from 30 µg mL−1 to 60 µg mL−1, were used to prepare the anti-digoxin-Au-NP conjugates, and the conjugate performances were evaluated by detecting 1-nM R156H-mutant DNA (see detailed discussion and Figure S3A in the supporting information). The best S/N ratio was obtained with the concentration of 50 µg mL−1 anti-digoxin antibody. Another factor that affects the sensitivity and reproducibility of the assay is the use of various buffers. In the current study, four types of buffers, 80×SSC+0.5%BSA, 10mM PBS+0.5% BSA, 10 mM Tris-HCl+0.5% BSA, and 80×SSC+10 mM Tris-HCl+0.5% BSA, were used to compare the assay performances (Figure S3B of the support information). The best S/N ratio was given by 80×SSC+10 mM Tris-HCl+0.5% BSA, which may offer an appropriate environment to stabilize the duplex DNA complexes (in the SSC buffer) and the best pH value for the immunoreactions (digoxin and anti-digoxin antibody, biotin and anti-biotin antibody).
3.4 Analytical performances
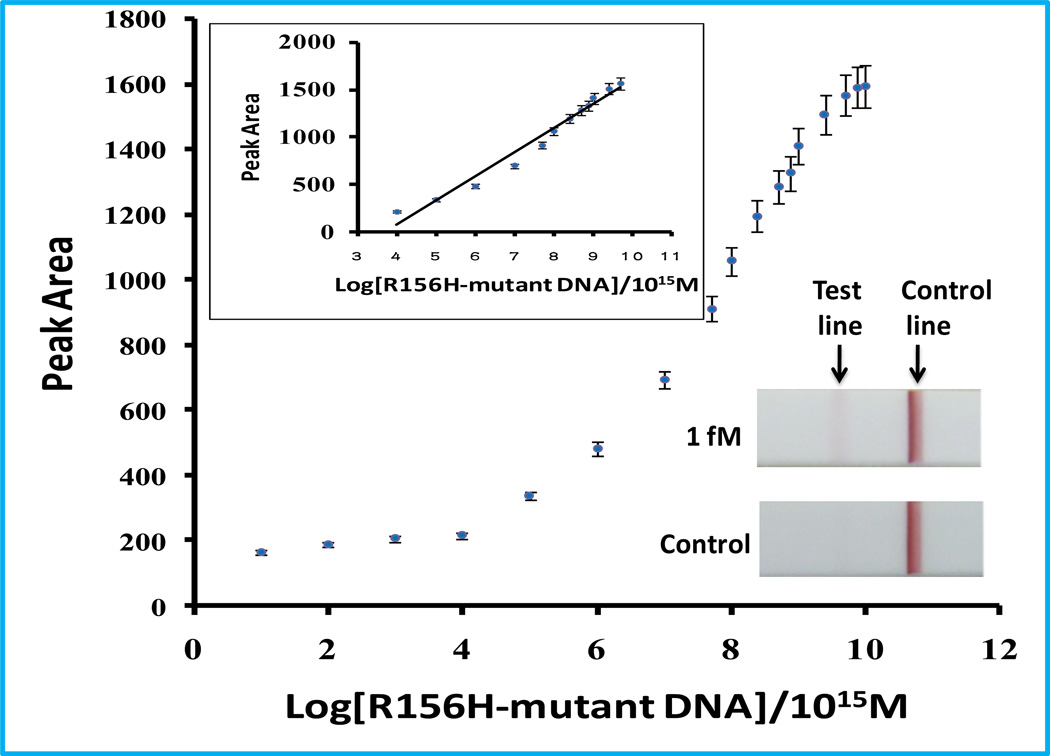
Under the optimal conditions determined above, we examined the analytical performance of the approach with different concentrations of R156H-mutant DNA. For visual detection, the red band in the LFS test zone was observed with as low as 1 fM of R156H-mutant DNA (inset image in Figure 3), which is comparable with that of fluorescence- and electrochemistry-based ISDPR (Guo et al., 2009; He et al., 2010a). Quantitative detections were performed by recording the intensities of the test bands with the portable strip reader. Figure 3 shows the relationship between the intensities of test bands (peak area) and the logarithm of R156H-mutant DNA concentrations. It was found that there was a linear range over the 0.1 pM to 50 nM range, and the detection limit was 0.08 pM (based on S/N=3)
Figure 3.
Calibration curve of the assay with different concentrations of R156H-mutant DNA. The inset is photo images of LFSs after applying the ISDPR product of 0-nM and 1-fM R156H-mutant DNA.
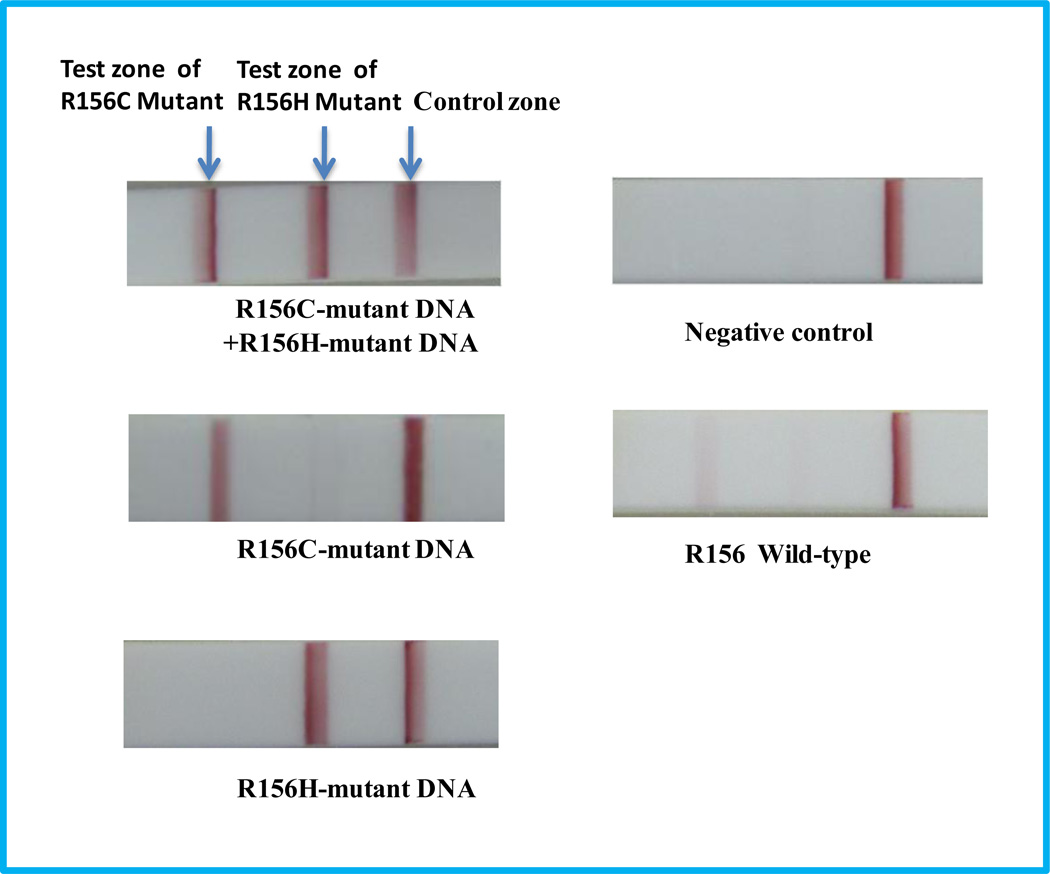
R156H and R156C are the hot mutation spots of KRT 10 (Haruna et al., 2007). In order to differentiate R156H and R156C mutations, we designed a fluorescein-modified hairpin which had a complementary sequence to the R156C-mutant DNA. Two test zones were prepared by dispensing anti-biotin and anti-digoxin antibody solutions on the nitrocellulose membrane of the LFS. The sample solutions, including 1-nM R156H-mutant DNA+1-nM R156C-mutant DNA, 1-nM R156C-mutant DNA , 1-nM R156H-mutant DNA, 0-nM DNA (control), and 1-nM R156 wild-type DNA, were tested after ISDPR in the presence of both biotin-modified hairpin and fluorescein-modified hairpin. The typical photo images of LFSs are shown in Figure 4. Three bright red bands (two test bands and one control band) were observed on the LFS with the sample solution containing both R156H- and R156C-mutant DNAs. The results indicated that two independent ISDPRs were triggered by R156H- and R156C-mutant DNAs, respectively, and two types of duplex DNA complexes, the biotin- and digoxin-attached duplex DNA as well as fluorescein- and digoxin-attached duplex DNA, were produced. The formed duplex complexes were captured on the test zones via the immunoreactions (biotin and anti-biotin antibody, fluorescein and anti-fluorescein antibody). In the case of testing the sample solution containing either R156H- or R156C-mutant DNA, only two red bands (one test band and one control band) were observed. The results indicated that no cross-reaction occurred in the ISDPRs. In the presence of wild-type DNA, very weak bands were observed on the test zones. As expected, there was no test band observed in the LFS test zones in the absence of DNA. These results have shown that the proposed approach is capable to differentiate mutation spots of R156.
Figure 4.
Typical photo images of the LFSs in the presence of 1-nM R156H+1-nM R156C-mutant DNA, 1-nM R156C-mutant DNA, 1-nM R156H-mutant DNA, 0-nM mutant DNA (control), and 1-nM R156 wild-type DNA. Experimental conditions are the same as in Figure 3.
The reproducibility of the assay was assessed by testing various sample solutions. The coefficients of variation of the test (peak area) for 1-nM R156C-mutant DNA, 1-nM R156H-mutant DNA, 1-nM R156 wild-type DNA, 50-nM noncomplementary DNA and 0-nM mutant DNA were 4.9%, 5.4%, 3.0%, 6.5%, and 4.3%, respectively (n=8, see the photo images of the LFSs in Figure S4 of the support information). The overall relative standard deviation was less than 7.0%, which indicates good reproducibility.
4. Conclusions
We have demonstrated a simple and sensitive method for visual detection of gene mutations based on ISDPR and LFS. Compared with conventional PCR and ISDPR, the current approach is much simpler, avoiding the use of specialized instruments and the requirement of highly qualified personnel. The results presented here show that the procedure is a potential platform for the future development of fast and low-cost diagnostic tools to use for point-of-care diagnosis of genetic diseases as well as for detecting infectious agents or warning against biowarfare agents. Further work will test the reliability of the approach with some real samples from local hospitals and clinics, and will compare the results of the test with other standard gene mutation tests, such as PCR.
Supplementary Material
Acknowledgements
This research was supported by Award Number R21CA137703 from the National Cancer Institute. Y. He acknowledges financial support from the Natural Science Foundation of China (NO. 81071286), Guangdong Provincial Natural Science Foundation of China (NO. 10151009503000002), and the Key Research Plan Foundation of the Guangzhou Health Department (NO. 201102A212016).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Brown PO, Botstein D. Nature Genet. 1999;21:33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- Gill P, Ghaemi A. Nucleosides, Nucleotides, and Nucleic Acids. 2008;27:224–243. doi: 10.1080/15257770701845204. [DOI] [PubMed] [Google Scholar]
- Guo Q, Yang X, Wang K, Tan W, Li W, Tang H, Li H. Nucleic Acids Res. 2009;37(3):e20. doi: 10.1093/nar/gkn1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruna K, Suga Y, Mizuno Y, Hasegawa T, Kourou K, Matsuba S, Muramatsu S, Ikeda S. J. Dermatology. 2007;34:545–548. doi: 10.1111/j.1346-8138.2007.00328.x. [DOI] [PubMed] [Google Scholar]
- He Y, Zeng K, Zhang X, Gurung A, Baloda M, Xu H, Liu G. Electrochem. Commun. 2010a;12(7):985–988. [Google Scholar]
- He Y, Zeng K, Gurung A, Baloda M, Xu H, Zhang X, Liu G. Anal. Chem. 2010b;82(17):7169–7177. doi: 10.1021/ac101275s. [DOI] [PubMed] [Google Scholar]
- He Y, Zhang S, Zhang X, Baloda M, Gurung A, Xu H, Zhang X, Liu G. Biosensor and Bioelectronics. 2011;26(5):2018–2024. doi: 10.1016/j.bios.2010.08.079. [DOI] [PubMed] [Google Scholar]
- Hellyer TJ, Nadeau JG. Expert Rev. Mol. Diagn. 2004;4(2):251–261. doi: 10.1586/14737159.4.2.251. [DOI] [PubMed] [Google Scholar]
- Hoogendoorn B, Owen MJ, Oefner PJ, Williams N, Austin J, O’Donovan MC. Hum. Genet. 1999;104:89–93. doi: 10.1007/s004390050915. [DOI] [PubMed] [Google Scholar]
- Kaltenboeck B, Wang C. Adv. Clin. Chem. 2005;40:219–259. doi: 10.1016/S0065-2423(05)40006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A, Shankar PM, Mutharasan R. Sensors and Actuators B. 2007;125:688–703. [Google Scholar]
- Lyamichev V, Mast AL, Hall JG, Prudent JR, Kaiser MW, Takova T, Kwiatkowski RW, Sander TJ, de Arruda M, Arco DA. Nat. Biotechnol. 1999;17:292–296. doi: 10.1038/7044. [DOI] [PubMed] [Google Scholar]
- Mao X, Ma Y, Zhang A, Zhang L, Zeng L, Liu G. Anal. Chem. 2009;81(4):1660–1668. doi: 10.1021/ac8024653. [DOI] [PubMed] [Google Scholar]
- Nelson D, Cox M. Lehninger principles of biochemistry. New York: Worth Publishers; 2000. pp. 1132–1133. [Google Scholar]
- Palecek E, Fojta M. Anal. Chem. 2001;73:75A–83A. doi: 10.1021/ac0123936. [DOI] [PubMed] [Google Scholar]
- Rothnagel JA, Dominey AM, Dempsey LD, et al. Science. 1992;257:1128–1130. doi: 10.1126/science.257.5073.1128. [DOI] [PubMed] [Google Scholar]
- Syder A, Yu QC, Paller AS, Giudice G, Pearson R, Fuchs E. J Clin Invest. 1994;93:1533–1542. doi: 10.1172/JCI117132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toubanaki DK, Christopoulos TK, Ioannou PC, Flordellis CS. Anal. Chem. 2009;81:218–224. doi: 10.1021/ac801870x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.