Abstract
Bioassay-guided fractionation of two cyanobacterial extracts from Papua New Guinea has yielded hoiamide D in both its carboxylic acid and conjugate base forms. Hoiamide D is a polyketide synthase (PKS)/non-ribosomal peptide synthetase (NRPS)-derived natural product that features two consecutive thiazolines and a thiazole, as well as a modified isoleucine residue. Hoiamide D displayed inhibitory activity against p53/MDM2 interaction (EC50= 4.5 μM), an attractive target for anticancer drug development.
Keywords: marine cyanobacteria, natural products, lipopeptide, anticancer, spectroscopy
The protein p53 is a well characterized tumor suppressor that acts as a transcription factor to regulate cell cycle dynamics, apoptosis, and DNA repair.1 It is encoded by the gene TP53 that is inactivated in nearly 60% of all human malignancies.2 Structurally, p53 harbors an NH2 terminal transactivation domain, a DNA binding domain in the center, and a C-terminal locus for regulatory function and tetramerization.3 Its function is regulated either by mutation or deletion of the parent gene or through regulatory feedback by MDM2. MDM2, a murine ubiquitin ligase, negatively regulates p53 function by a variety of mechanisms, including degradation through proteosomal association and ubiquitylation. MDM2 also facilitates nuclear export with its export signal and binds to p53’s transactivation domain, thereby repressing transcriptional activity.3,4 Recent studies have also shown MDM2 to interact, albeit more weakly, with the C-terminal and core domains of p53.5 In theory, disruption of any of these regulatory functions by MDM2 is a viable strategy to reactivate p53, especially through inhibition of the p53/MDM2 binding interaction.
Based on site directed mutagenesis, the essential amino acids of the p53 N-terminal binding domain that constitute the majority of the p53/MDM2 interaction and binding energy are Phe19-Trp23-Leu26, with the three terminal atoms of the phenyl ring, the indole nitrogen of tryptophan (Trp), and the isopropyl group of leucine (Leu) serving as the pertinent pharmacophore.3,6 Thus, the p53/MDM2 interaction is largely hydrophobic, and the binding interface of the two proteins is also quite small, making small, peptidic or non-peptidic molecular mimics of the p53 binding site good candidates for inhibitors of p53/MDM2 interaction.3,6 This hydrophobic binding domain has been exploited to rationally design synthetic p53/MDM2 inhibitors with excellent affinity. Three major classes of synthetic p53/MDM2 inhibitors, the nutlins, spiro-oxindoles, and benzodiazepinediones, have taken advantage of the Phe19-Trp23-Leu26 binding core to displace p53 and bind MDM2 with much greater affinity and in vitro potency. However, maintaining in vivo potency has been problematic for many of these inhibitors due to their poor pharmacokinetic profiles.7,8
Marine cyanobacteria are one of Nature’s most ancient organisms and offer a plethora of bioactive secondary metabolites, many of which are active against validated targets of human disease. Of the 678 marine cyanobacterial natural products reported in the literature, the majority is polyketide synthase-non-ribosomal peptide synthetase derived.9 There is a predominance of nonpolar amino acids in cyanobacterial peptide-derived compounds, and leucine and phenylalanine, two essential amino acids of the p53 binding pocket, are relatively common constituents in these peptidic compounds. Approximately half of all amino acids utilized in cyanobacterial natural products are modified with the most frequent modifications including N-methylation, N’-N’-dimethylation, ketide extension, and halogenation to a lesser extent.9 Thus, many features of marine cyanobacterial compounds, including their molecular weight distribution (median 604 Da), lipophilicity, and chemical diversity, lend themselves to favorable bioactivity and medicinal chemistry profiles, particularly as it pertains to p53/MDM2 inhibition. Herein, we describe hoiamide D (1), a peptide-derived p53/MDM2 inhibitor, isolated in both its acid and carboxylate forms, from two separate collections of the Papua New Guinea cyanobacterium Symploca sp. Hoiamide D displays promising inhibitory activity towards p53/MDM2 interaction (EC50= 4.5 μM).
Samples of the spongy, purple cyanobacterium Symploca sp. were collected by SCUBA near Kape Point, Papua New Guinea in 2006. The collection was extracted with CH2Cl2:MeOH (2:1) to yield a crude extract (1.37 g) that was further processed using normal phase silica vacuum liquid chromatography to generate nine subfractions (A-I).10 The ninth and most polar fraction (I) displayed inhibitory activity in the p53/MDM2 assay and was further purified using RP C18 solid phase extraction to yield an additional four relatively pure subfractions. The most polar subfraction (>95% pure) contained the new natural product, hoiamide D (23.6 mg), and harbored the greatest inhibitory activity (EC50= 4.5 μM).
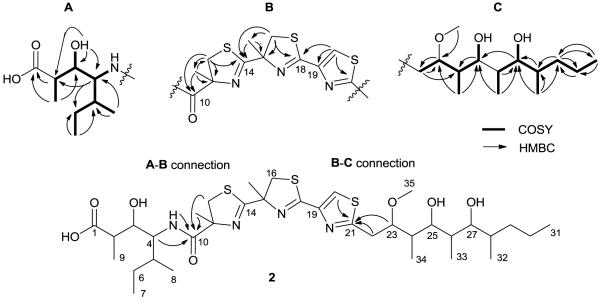
The HRESIMS measurement of hoiamide D (1), [M+H]+ m/z 743.3535, established its formula as C35H58N4O7S3 with an isotopic pattern consistent with the presence of three sulfur atoms.11 The IR spectrum of 1 displayed an absorption band at 3381 cm−1, indicating the presence of a hydroxy group while the 1H NMR spectrum revealed two distinctive low-field shifts, a sharp singlet at δH 8.22 (δC 122.6) and a doublet at δH 6.85 that were indicative of a substituted alkene and an amide, respectively. A singlet at δH 3.23 (δC 56.2) revealed the presence of an O-Me substituent, and two sharp high-field singlets between 1.4-1.6 ppm were suggestive of deshielded methyl groups. 2D COSY, HSQC, TOCSY, and HMBC experiments were subsequently used to elucidate three partial structures (A-C) with key HMBC correlations used to provide attachment points and complete the planar structure, as described below.
TOCSY data afforded a two-spin system for substructure (A) with one spin system consisting of H-2/H-3/H-9 and the other spin system consisting of H4-H8. A COSY correlation between H-4 and the amide proton at δH 6.85, as well as HMBC correlations between H-4/C-5, H3-8/C-5, H-6a/C-3, and H3-7/C-6, supported construction of a modified isoleucine with a hydroxy at C-3 (δC 71.8, δH 3.51). The C-3 methine was coupled to the C-2 methine via a COSY correlation and through to the C-1 carbonyl with a 3-bond HMBC correlation. A methyl (δC 13.8, δH 0.85) was then assigned to C-2 via H-2/C-9 and H3-9/C-1 HMBC correlations, thus completing the planar substructure (A) or 4-amino-3-hydroxy-2,5-dimethylheptanoic acid (Ahdhe), a structural feature characteristic of the hoiamide class (Figure 2).
Figure 2.
Partial structures of 1 and 2 with key COSY and HMBC correlations.
Substructure (B) featured two consecutively coupled α-methyl thiazoline rings and a thiazole. Preliminary data analysis suggested that two methylenes with chemical shifts δH 3.59/3.13 (δC 40.9); δH 3.74/3.76 (δC 42.2) were adjacent to heteroatoms, and a low-field singlet at δH 8.21 (δC 122.6) implied the presence an olefinic proton of a heterocycle. Atom counting and the presence of three deshielded quaternary carbons C-14/18/21 (δC 177.7, 162.5, 169.9) also suggested that these sp2 carbons were proximate to a nitrogen and a sulfur atom in each ring. HMBC correlations were then used to construct the rings of the triheterocyclic system, as follows. A correlation between the C-12 methylene (δC 40.9) and the deshielded sp2 C-14 (δC 177.7) permitted assignment of C-12 adjacent to the sulfur atom with C-14 placed between the sulfur and nitrogen atoms. An additional HMBC correlation between H2-12 and the relatively shielded quaternary carbon at C-11 (δC 84.4) necessitated placement of C-11 adjacent to a nitrogen atom and between the C-12 methylene and the carbonyl at C-10. Further HMBC correlations between H3-13/C-10/11/12 permitted assignment of an α-methyl group on C-11 of the thiazoline ring. In a similar fashion, the second thiazoline ring was constructed with HMBC correlations between the C-16 methylene, the deshielded sp2 C-18 (δC 162.5), and the relatively shielded quaternary carbon at C-15 (δC15 83.6). Additional correlations between H3-17/C-15/16 permitted assignment of an α-methyl group on C-15 of the second thiazoline ring as well. The third ring was constructed with HMBC correlations from a low-field olefinic proton at δH 8.21 (δC 122.6) to two sp2 quaternary carbons at C-19/21which implied the presence of a thiazole ring and satisfied the remaining degrees of unsaturation for the molecule. Further correlations between H-16a/16b/C-14 and H-20/C-18 demonstrated the consecutive nature of the triheterocyclic ring system to complete substructure (B).
Substructure (C) consisted of an extended 10-carbon chain with two hydroxy and three methyl groups, and a methoxy substituent. COSY correlations were used to consecutively assign the H-22 methylene with three methines H-23/24/25. An additional COSY correlation was used to assign a methyl group at C-24 (δH24 2.08), and an HMBC correlation between C-23 and the H-35 O-methyl allowed placement of the O-methyl at this position. COSY correlations were also used to assign the H-26/27/28 methines and two methyl groups at C-26 (δH261.52) and C-28 (δH281.54), respectively. A 3-bond HMBC correlation between H3-33 and C-25 allowed connection of C25/C26. Further COSY correlations were then used to place the H-29 and H-30 methylenes adjacent to the terminal C-31 methyl group. HMBC correlations between the C-28 methine and both H-29a/H-29b protons then permitted connection of C-28/29. Accounting of the remaining atoms and chemical shift reasoning allowed hydroxy groups to be placed at C-25 (δC 69.4) and C-27 (δC 72.7) to complete substructure (C).
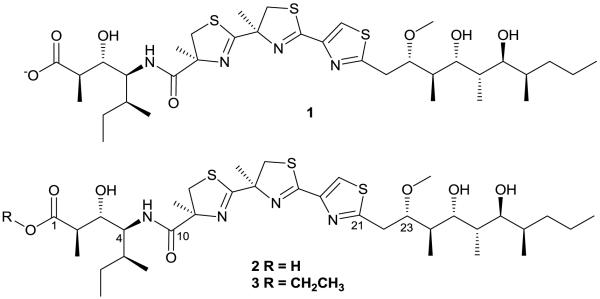
Finally, a key HMBC correlation between the C-10 carbonyl and the H-13 α-methyl connected substructure (A) with substructure (B), and HMBC correlations between C21 and H22/H23 permitted connection of substructure (B) to (C), thereby completing the proposed planar structure of 1 (Figure 1).
Figure 1.
Structures of the conjugate base of hoiamide D (1), the acid form of hoiamide D (2), and hoiamide C (3).
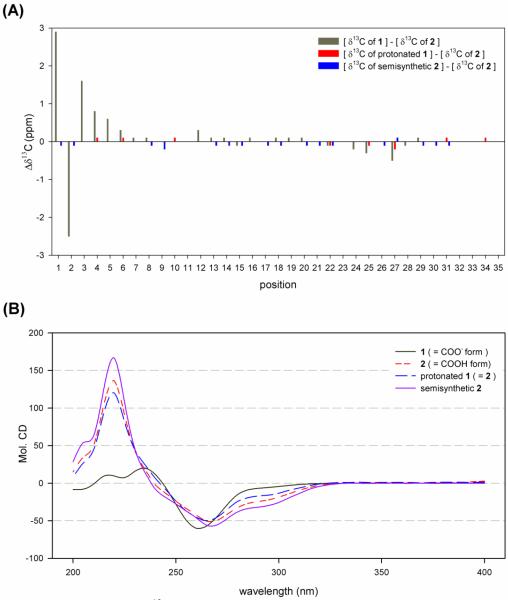
Colon cancer selectivity-guided fractionation of a second collection of mat-forming Symploca sp. obtained near Kolaio island, Papua New Guinea in 2003 yielded 2 with a HRESI [M+H]+ m/z 743.3535 mass identical to 1.12,13 Independent structure elucidation of 2 led to the same planar structure of 1. However, subsequent analysis of spectroscopic and analytical data revealed subtle differences between 1 and 2 that suggested the possibility of either different stereoisomers, different ionic states, or some other small difference. First, ΔδC values of C-1-C-4 and C-6 and ΔδH values of H-2 and H-3 are significant between 1 and 2 with the greatest ΔδC nearly 3 ppm at C-1 (Figure 3a). One hypothesis accounting for the carbon shift difference was the possibility of different stereoisomers. However, coupling constant analysis did not reveal significant differences between 1 and 2. Another possibility to account for the carbon shift disparity was a difference in ionic states. The more deshielded carbon of 1 (δC 179.5) suggests 1 may be the conjugate base of 2. Second, 1 and 2 were obtained by different chromatographic conditions; 1 was isolated under neutral conditions while 2 was purified with an acidified buffer. A LC-ESI-MS co-injection experiment, with simultaneous elution of 1 and 2 in various chromatographic conditions, also provided supporting evidence of constitutional equivalence but different ionic states. Third, the IR spectrum of 1 showed absorption of carbonyls at 1661 and 1580 cm−1, indicating the presence of an amide and carboxylate anion while the spectrum of 2 showed strong absorption of a carbonyl at 1673 cm−1, indicating the presence of an amide and carboxylic acid. The working hypothesis of different ionic states was finally proven by protonation of 1 with 0.1N trifluoroacetic acid, followed by comparison of spectroscopic data (supplemental info).14 The 1H and 13C data of protonated 1 are spectroscopically identical to those of 2, and optical rotation values are similar. In addition, protonation of 1 changed chiroptical property of 1 and the CD absorption curve of protonated 1 is almost identical to that of 2 and semisynthetic 2. Therefore, the proposed planar structure of 1 was amended to reflect its negatively charged state (Figure 1), and the HRESI [M+H]+ m/z 743.3535 mass of 1 is likely the result of reduction during electrospray ionization.
Figure 3.
Comparison of 13C NMR chemical shifts and CD spectra of 1 (-COO− form), 2 (-COOH form), protonated 1 and semisynthetic 2 (A) 13C NMR chemical shifts comparison; (B) CD absorption curve of 1 and 2 with protonated 1 and semisynthetic 2.
To ascertain the absolute configuration of the Me-Cys residues, compound 1 was subjected to ozonolysis and oxidative workup prior to acid hydrolysis. The hydrolysate was analyzed by chiral HPLC and only d(S)-2-methylcysteic acid was detected.15
To determine the configuration of the remaining 10 stereocenters, semisynthetic hoiamide D, prepared by base hydrolysis of hoiamide A, was used for comparative analysis.16 Semisynthetic hoiamide D was identical to 2 by LC-ESI-MS analyses (retention time, UV spectrum, and m/z value) and 1H and 13C NMR data analyses, had a similar optical rotation value and an identical CD spectrum to protonated 1 and 2 which indicated that natural hoiamide D (1 and 2) and semisynthetic hoiamide D were stereochemically identical (Figure 3b). The absolute configuration of natural hoiamide D (1 and 2) was thus determined as 2R, 3S, 4S, 5S, 11S, 15R, 23S, 24R, 25R, 26S, 27S and 28R.
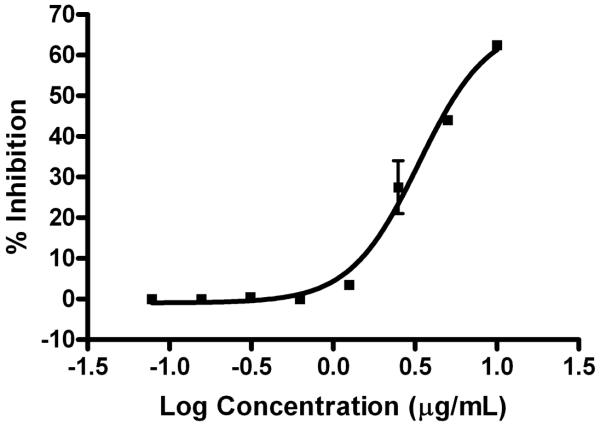
The carboxylate anion (1) was screened in a panel of assays to determine its biological activity, and its greatest bioactivity was inhibition of p53/HDM2 (human homologue) interaction. A HTR-FRET-based competition assay was used to characterize antagonists of the p53/HDM2 binding interaction based on the ability of antagonist molecules to compete with biotin-labeled p53 protein (bio-QETFSDLWKLLP-Ac) binding to GST-tagged HDM2 (GST-MPRFMDYWEGLN).17 FRET was detected (ex 320 nm, em 615, 665 nm) when donor labeled, Europium (Mab GST-Eu cryptate, Cisbio 61GSTKLB), and acceptor labeled, XLent (streptavidin-XLent, Cisbio 611SAXLB), complexes were in close proximity due to the anti-GST/GST-HDM2 interaction and the streptavidin/biotin-p53 interaction. The carboxylate anion (1) inhibited the p53/HDM2 protein binding (EC50= 4.5 μM; 95% confidence interval: 2.8-7.3 μM) with minimal cytotoxicity to the mammalian H460 cell line at 40 μM (16% ± 1), compared to control.18 Compound 2 was not tested in the p53/HDM2 assay, although the carboxyl group would be deprotonated at the pH (7.4) of the media utilized in the assay.
Chlorofusin (IC50= 4.6 μM), isolated from a marine fungus Fusarium sp., was the first natural product reported to inhibit p53/MDM2 interactions.19 A second natural product inhibitor, the non-peptidic hexylitaconic acid (IC50= 230 μM), was isolated from a marine fungus Arthrinium sp. in 2006, followed by sempervirine (IC50= 29.4 μM).20,21 Hoiamide D is the second peptide-based natural product inhibitor of p53/MDM2 interaction reported, and is one of the most potent. Of note, however, there are rationally designed synthetic inhibitors, such as the spiro-oxindoles and benzodiazepinediones, with nanomolar efficacy.
Hoiamide D joins three other hoiamides isolated to date, comprising a recently described class of cyanobacterial compounds featuring a triheterocylic system. Hoiamide A and B are cyclic whereas hoiamide C and D are linear.22,23 In fact, hoiamide C may be an extraction artifact of hoiamide D given the utilization of ethanol in the storage conditions of the biological material. Hoiamide A and B exhibited potent inhibition of calcium oscillation in rat cortical neurons and micromolar activation of voltage-gated sodium channels (VGSC) with hoiamide A reported to be a partial agonist at site 2 of the mammalian VGSC.23 Both hoiamide C and D were inactive in these assays, suggesting that a cyclic architecture is required for this potent neuromodulatory activity.23 However, the activity of hoiamide D (1) in the p53/HDM2 assay suggests that this molecular class has the capacity to interact with a variety of cellular targets, and thus, members of this group should be attractive targets for total synthesis24 and evaluation in a diversity of biological assays.
Supplementary Material
Figure 4.
Inhibition of p53/MDM2 binding by hoiamide D (1). EC50= 4.5 μM; 95% confidence interval: 2.8-7.3 μM (n=2).
Acknowledgments
We thank the government of Papua New Guinea for permission to make these cyanobacterial collections, A. Pereira for the synthetic standard and technical expertise, T. Suyama for helpful advice, J. Wingerd and T. Byrum for the H-460 cytotoxicity and Neuro-2A screening, and the TSRI and UCSD mass spectrometry facilities for their analytical services. This work was supported by NIH grants CA 52955, NS 053398, and NIH/NCI CA100851.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Data
Supplementary data associated with this article can be found, in the online version, at.
References and Notes
- 1.Mandinova A, Lee SW. Sci. Trans. Med. 2011;3:64rv1. doi: 10.1126/scitranslmed.3001366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nigro JM, Baker SJ, Preisinger AC, Jessup JM, Hostetter R, Cleary K, Bigner SH, Davidson N, Baylin S, Devilee P, Glover T, Collins FS, Weslon A, Modali R, Harris CC, Vogelstein B. Nature. 1989;342:705. doi: 10.1038/342705a0. [DOI] [PubMed] [Google Scholar]
- 3.Chene P. Mol. Cancer Res. 2004;2:20. [PubMed] [Google Scholar]
- 4.Kulikov R, Letienne J, Kaur M, Grossman SR, Arts J, Blattner C. Proc. Natl. Acad. Sci. U.S.A. 2010;107:10038. doi: 10.1073/pnas.0911716107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poyurovsky MV, Katz C, Laptenko O, Beckerman R, Lokshin M, Ahn J, Byeon IJ, Gabizon R, Mattia M, Zupnick A, Brown LM, Friedler A, Prives C. Nat. Struct. Mol. Biol. 2010;17:982. doi: 10.1038/nsmb.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chene P. Nat. Rev. Cancer. 2003;3:102. doi: 10.1038/nrc991. [DOI] [PubMed] [Google Scholar]
- 7.Dickens MP, Fitzgerald R, Fischer PM. Semin. Cancer Biol. 2010;20:10. doi: 10.1016/j.semcancer.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Patel S, Player MR. Expert Opin. Invest. Drugs. 2008;17:1865. doi: 10.1517/13543780802493366. [DOI] [PubMed] [Google Scholar]
- 9.Tidgewell K, Clark BR, Gerwick WH. In: Comprehensive Natural Products Chemistry. Mander L, Lui H-W, editors. Vol. 2. Elsevier; Oxford: 2010. pp. 141–188. [Google Scholar]
- 10.Compound 1: pale green solid; [α]D −30 (c 0.05, MeCN); UV (MeCN) λmax (log ε) 252 (3.54); IR (neat) vmax 3381, 2963, 2929, 1661, 1580, 1518, 1458, 1404, 1154, 1084, 1034, 1018 cm−1; 1H NMR (DMSO, 500 MHz) and 13C NMR (DMSO, 75 MHz) data, see Table 1; (+)-HRESIMS [M+H]+ m/z 743.3535 (calcd for C35H59N4O7S3, 488.3546).
- 11.The collection (PNG-04-26-06-3) was extracted six times with heat using CH2Cl2:MeOH (2:1) and was further processed on silica gel using vacuum liquid chromatography with a 100% hexanes-EtOAc-MeOH gradient to generate nine subfractions A-I. Fraction I eluted with 100% MeOH and harbored the greatest p53/MDM2 inhibitory activity. Fraction I (259 mg) was subsequently filtered using a Waters RP C18 SPE cartridge with a MeOH gradient to yield four relatively pure subfractions. The most polar subfraction exhibited the greatest activity and was further examined.
- 12.Compound (2): pale yellow oil; [α]D 19.6 (c 0.05, MeCN); UV (MeOH) λmax (log ε) 250 (3.49); IR (neat) vmax 3378, 2965, 2930, 1674, 1521, 1458, 1382, 1299, 1203, 1139, 1088, 1026 cm−1; 1H NMR (DMSO, 500 MHz) and 13C NMR (DMSO, 75 MHz) data, see Table 1; (+)-HRESIMS [M+H]+ m/z 743.3535 (calcd for C35H59N4O7S3, 488.3546).
- 13.The collection (PNG-12-16-03-4) was extracted repeatedly with CH2Cl2-MeOH (2:1) and further fractionated by silica gel vacuum column chromatography (VLC) to produce nine fractions (A-I). Fraction I was found to possess slight colon cancer selective inhibitory activity. This fraction was subjected to RP Sep-pak followed by RP HPLC [Phenomenex Hydro RP C18, 300 Å, 10 × 250 mm, 68% CH3CN/H2O (0.001% trifluoroacetic acid) at 3 mL/min] to afford hoiamide D (2.4 mg).
- 14.Compound 1 (3.0 mg) was treated with 0.1N trifluoroacetic acid for 1 hr and dried under N2.
- 15.A sample of 1 (2.1 mg) was dissolved in 3 mL of CH2Cl2 and treated with ozone at 25° C for 15 min. The sample was dried and further treated with 1 mL of H2O2-HCOOH (1:2) at 70° C for 30 min. l(R)-2-Methylcysteic acid (2.0 mg) and d(S)-2-methylcysteic acid (2.5 mg) standards were prepared and treated as above ( Pattenden G, Thom SM, Jones MF. Tetrahedron. 1993;49:2131. ). The dry sample 1 was then hydrolyzed using 6N HCl (0.5 mL) for 2 hr at 110° C and analyzed by chiral HPLC [Phenomenex Chirex 3126 (D), 4.6 × 50 mm; 2 mM CuSO4 in H2O at 0.3 mL/min; detection at 230 nm] to yield d(S)-2-methylcysteic acid with a retention time of 9.7 min (d-MeCysA tR= 10.0 min; l-MeCysA tR = 12.0 min).23
- 16.A semi-synthetic standard of hoiamide D was prepared according to Choi H, Pereira AR, Cao Z, Shuman CF, Engene N, Byrum T, Matainaho T, Murray TF, Mangoni A, Gerwick WH. J Nat Prod. 2010;73:1411. doi: 10.1021/np100468n. [α]D +25.2 (c 0.05, MeCN); UV (MeOH) λmax (log ε) 250 (3.52)
- 17.The FRET buffer used for all experiments was phosphate buffered saline (pH 7.4), 100 mM KF, 0.1% bovine serum albumin, and 5 mM ß-mercaptoethanol. Biotin-P53 (500 nM, final) was preincubated for 20 min with test compound (10-0.0781 μg/mL; 2-fold serial dilution), and a master mix was then added containing GST-HDM2 (2.5 nM, final), Mab GST-Eu cryptate (17 nM, final), and streptavidin XLent (9 nM, final); the solution was incubated for 1 hr. Data were collected in duplicate using the PerkinElmer EnVision 2103 Multilabel Reader using excitation filter 320nm, emission filters 665nM (to detect XLent) and 615 nM (to detect Europium), respectively, and the optical module Lance Eu/APC Dual 452. Percent inhibition compared to DMSO control was calculated for each concentration of test compound. Dose-response graph was generated using Graphpad Prism software (Graphpad Software Inc., San Diego, CA). The EC50 values were determined by non-linear regression analysis using a logistic equation.
- 18.Alley MC, Scudiero DA, Monks A, Hursey ML, Czerwinski MJ, Fine DL, Abbott BJ, Mayo JG, Shoemaker RH, Boyd MR. Cancer Res. 1988;48:589. [PubMed] [Google Scholar]
- 19.Duncan SJ, Gruschow S, Williams DH, McNicholas C, Purewal R, Hajek M, Gerlitz M, Martin S, Wrigley SK, Moore M. J. Am. Chem. Soc. 2001;123:554. doi: 10.1021/ja002940p. [DOI] [PubMed] [Google Scholar]
- 20.Tsukamoto S, Yoshida T, Hosono H, Ohta T, Yokosawa H. Biorg. Med. Chem. Lett. 2006;16:69. doi: 10.1016/j.bmcl.2005.09.052. [DOI] [PubMed] [Google Scholar]
- 21.Sasiela CA, Stewart DH, Kitagaki J, Safiran YJ, Yang Y, Weissman AM, Oberoi P, Davydov IV, Goncharova E, Beutler JA, Mcmahon JB, O’Keefe BR. J. Biomol. Screening. 2008;13:229. doi: 10.1177/1087057108315038. [DOI] [PubMed] [Google Scholar]
- 22.Pereira A, Cao Z, Murray TF, Gerwick WH. Chem. Biol. 2009;16:893. doi: 10.1016/j.chembiol.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi H, Pereira AR, Cao Z, Shuman CF, Engene N, Byrum T, Matainaho T, Murray TF, Mangoni A, Gerwick WH. J. Nat. Prod. 2010;73:1411. doi: 10.1021/np100468n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Xu Z, Ye T. Organic Lett. 2011;13:2506. doi: 10.1021/ol2007567. [DOI] [PubMed] [Google Scholar]
Table 1.
1H and 13C NMR data for the conjugate base (1) and acid (2) forms of hoiamide D (DMSO-d6).
| Compound (1) |
Compound (2) |
|||||
|---|---|---|---|---|---|---|
| Position | δH (J in Hz)a | δ Cb | HMBCa | δH (J in Hz)a | dCb | HMBCa |
| 1 | 179.5 | 176.5 | ||||
| 2 | 1.70, m | 41.6 | 9 | 2.17, dt (6.8, 10.9) | 44.1 | 1, 3, 9 |
| 3 | 3.51, d (10.8) | 71.8 | 1 | 3.81, m | 70.2 | 2 |
| 4 | 3.52, t (10.4) | 53.5 | 1, 5 | 3.53, d (9.6) | 52.7 | 2, 5, 6, 8, 10 |
| 5 | 1.61, m | 36.1 | 1.58, m | 35.5 | 7 | |
| 6a | 1.49, m | 25.4 | 3, 7 | 1.44, m | 25.1 | 7 |
| 6b | 1.04, m | 1.07, m | 5, 7, 8 | |||
| 7 | 0.85, t (7.1) | 11.1 | 6 | 0.84, t (7.2) | 11.0 | 5, 6 |
| 8 | 0.88 d (6.8) | 15.7 | 4, 5 | 0.89, d (6.3) | 15.6 | 4, 5, 6 |
| 9 | 0.85, d (4.1) | 13.8 | 1, 2, 3 | 0.92, d (7.2) | 13.6 | 1, 2, 3 |
| NH | 6.85, d (9.7) | 4, 10 | 6.93, d (9.8) | 10 | ||
| 10 | 173.3 | 173.3 | ||||
| 11 | 84.4 | 84.4 | ||||
| 12a | 3.59, d (11.6) | 40.9 | 10, 11, 13, 14 | 3.61, d (11.3) | 40.6 | 10, 11, 13, 14 |
| 12b | 3.13, d (11.6) | 10, 11, 13, 14 | 3.15, d (11.3) | 10, 11, 13, 14 | ||
| 13 | 1.45, s | 25.7 | 10, 11, 12 | 1.44, s | 25.6 | 10, 11, 12 |
| 14 | 177.7 | 177.6 | ||||
| 15 | 83.6 | 83.7 | ||||
| 16a | 3.74, d (11.4) | 42.2 | 14, 15, 17, 18 | 3.78, d (11.1) | 42.1 | 14, 15, 17, 18 |
| 16b | 3.46, d (11.4) | 14, 15, 17, 18 | 3.45, d (11.1) | 14, 15, 17, 18 | ||
| 17 | 1.57, s | 26.0 | 14, 15, 16 | 1.62, s | 26.0 | 14, 15, 16 |
| 18 | 162.5 | 162.4 | ||||
| 19 | 146.4 | 146.5 | ||||
| 20 | 8.21, d (1.0) | 122.6 | 18, 19, 21 | 8.19, s | 122.5 | 18, 19, 21 |
| 21 | 169.9 | 169.9 | ||||
| 22a | 3.17, m | 33.0 | 21 | 3.16, dd (3.7, 15.3) | 33.1 | 21, 23, 24 |
| 22b | 2.88, dd (10.5, 15.1) | 21, 23 | 2.90, dd (11.0, 15.3) | 21, 23, 24 | ||
| 23 | 3.88, d (10.5) | 80.6 | 21 | 3.86, d (11.0) | 80.5 | 21, 22, 34, 35 |
| 24 | 2.08, m | 35.8 | 34 | 2.09, m | 35.9 | 34 |
| 25 | 3.75, d (10.4) | 69.4 | 24 | 3.74, d (10.0) | 69.7 | 24, 33 |
| 26 | 1.52, m | 37.3 | 1.54, m | 37.3 | 27, 33 | |
| 27 | 3.33, d (7.6) | 72.7 | 26 | 3.33, m | 73.1 | 25, 26, 32 |
| 28 | 1.54, m | 33.7 | 33 | 1.53, m | 33.8 | 27, 32 |
| 29a | 1.32, m | 36.7 | 28 | 1.31, m | 36.6 | 27, 28, 30, 31, 32 |
| 29b | 1.17, m | 28, 30 | 1.18, m | 27, 28, 30, 31, 32 | ||
| 30 | 1.30, m | 19.9 | 1.30, m | 19.9 | 29, 31 | |
| 31 | 0.85, t (7.1) | 14.3 | 30, 32 | 0.86, t (7.3) | 14.3 | 29, 30 |
| 32 | 0.75, d(6.4) | 12.8 | 27, 28 | 0.76, d (6.6) | 12.8 | 28, 29 |
| 33 | 0.71, d (7.1) | 9.0 | 25, 26, 27 | 0.71, d (6.7) | 9.0 | 25, 26, 27 |
| 34 | 0.72, d (7.5) | 9.6 | 23, 24, 25 | 0.73, d (6.6) | 9.6 | 23, 25 |
| 35 | 3.22, s | 56.2 | 23 | 3.21, s | 56.2 | 23 |
| 1-OH | 12.0, s | |||||
| 3-OH | 5.32 brs | 5.29, d (4.7) | 2, 3, 4 | |||
| 26-OH | 4.50, s | 4.27, s | ||||
| 27-OH | 4.23, s | |||||
500 and 600 MHz for 1H NMR, HMBC
75 MHz for 13C NMR
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.