Background: Cyclotides are a family of plant-derived defense peptides.
Results: Parigidin-br1, a novel cyclotide, shows insecticidal activity in vivo and in vitro. Mechanistic insights into the activity were provided by theoretical and electron microscopic studies.
Conclusion: The cyclotide disrupts insect cell membranes and has potential applications as a biotechnological insecticide.
Significance: The study provides an enhanced understanding of cyclotide activity against a sugarcane insect pest.
Keywords: Electron Microscopy (EM), Insect, Molecular Docking, Peptides, Plant Defense, Diatraea saccharalis, Palicourea rigida, Cyclotide, Insecticidal, Plant Defense
Abstract
Cyclotides are a family of plant-derived cyclic peptides comprising six conserved cysteine residues connected by three intermolecular disulfide bonds that form a knotted structure known as a cyclic cystine knot (CCK). This structural motif is responsible for the pronounced stability of cyclotides against chemical, thermal, or proteolytic degradation and has sparked growing interest in this family of peptides. Here, we isolated and characterized a novel cyclotide from Palicourea rigida (Rubiaceae), which was named parigidin-br1. The sequence indicated that this peptide is a member of the bracelet subfamily of cyclotides. Parigidin-br1 showed potent insecticidal activity against neonate larvae of Lepidoptera (Diatraea saccharalis), causing 60% mortality at a concentration of 1 μm but had no detectable antibacterial effects. A decrease in the in vitro viability of the insect cell line from Spodoptera frugiperda (SF-9) was observed in the presence of parigidin-br1, consistent with in vivo insecticidal activity. Transmission electron microscopy and fluorescence microscopy of SF-9 cells after incubation with parigidin-br1 or parigidin-br1-fluorescein isothiocyanate, respectively, revealed extensive cell lysis and swelling of cells, consistent with an insecticidal mechanism involving membrane disruption. This hypothesis was supported by in silico analyses, which suggested that parigidin-br1 is able to complex with cell lipids. Overall, the results suggest promise for the development of parigidin-br1 as a novel biopesticide.
Introduction
The challenge of improving food production to maintain an estimated 9.5 billion people worldwide by 2050 is very significant. Moreover, the growing demand for biofuel and cattle feed crops has driven increasing interest in agricultural productivity, given that arable land is finite (1). Allied to these factors, the ability to reduce damage to plants caused by pests and pathogens is also important because insect predation currently still leads to substantial crop losses despite the use of conventional pesticides (2). Although these pesticides can control insects, they also cause significant environmental damage due to high toxicity and low selectivity among species affected, which include key pollinators such as bees (3). Given these problems, there is a great interest in developing molecules with potentially lower off-target effects than conventional insecticides. An alternative to topical pesticides is plant bioengineering. Plants can be genetically modified to express insecticidal biomolecules in specific tissues in order to be more selective to target pests (4). This technology is used commonly in many countries, and in 2009, 134 million hectares of genetically modified plants were grown, representing 9% of the 1.5 billion hectares of crops in the world (5). After resistance was first reported (6), researchers started to develop crops with more than one defensive protein, each with a different mechanism of action, to retard insect resistance, but there is a need for further development of novel insecticidal agents.
Among compounds with insecticidal activity extracted from plants, cyclotides have been explored because of their stability and diverse range of functions (7). Cyclotides comprise a peptidic circular backbone that is cross-linked by a cystine knot formed by three conserved disulfide bonds (8). Their biosynthesis involves excision from precursor proteins, and the cyclization process is thought to be mediated by an asparaginyl endopeptidase, which acts both to excise the peptide from the precursor at Asn (or Asp) residues, and catalyze the ligation reaction (9). Cyclotides are extremely stable owing to their cystine knot motif (10) and are resistant to thermal, chemical, and enzymatic degradation (11). To date, two major subclasses of cyclotides have been described, Möbius and bracelet, with the classification based on the presence or absence of a cis-proline in loop 5 of the cyclotide backbone, respectively (12).
Cyclotides are distributed widely in the plant kingdom, found among the Violaceae, Rubiaceae, Fabaceae, and Cucurbitaceae families (7, 13) where a single plant can express a suite of these peptides with tissue-specific expression in the roots, leaves, stems, and other tissues (7). In addition to their stability, cyclotides are well known for their various bioactivities, which include uterotonic (14), anti-HIV (15), antibacterial (16), hemolytic (17), neurotensin antagonism (18), anti-tumor (19), antifungal (20), nematocidal (21), molluscicidal (22), and insecticidal activities (23). The last activity presents great potential for the development of cyclotides as agribusiness tools (10). Unlike other small insecticidal disulfide-rich peptides, the action of cyclotides seems not be due to inhibition of digestive enzymes (24, 25). Rather, recent studies on Helicoverpa punctigera larvae showed that cyclotides cause larval midgut membrane disruption (26). Based on studies that show the tendency of cyclotides to form tetramers and octamers (27), it is probable that these peptides accumulate on the membrane surface where they form multimeric structures, leading to pores, resulting in membrane disruption and alteration of the osmotic balance. These effects lead to lower food absorption, impaired larval development, and higher insect mortality (26, 28, 29).
Here, we report the identification and characterization of parigidin-br1, a bracelet cyclotide from Palicourea rigida, a plant of the Rubiaceae family from the Brazilian Cerrado. The expression of this cyclotide was evaluated across different seasons and in different plant tissues. Significantly, parigidin-br1 shows potent in vivo activity against Diatraea saccharalis, an insect responsible for great economic losses in sugarcane plantations. Parigidin-br1 was also active in vitro against an ovarian cell line of Spodoptera frugiperda SF-9. The relationship between structure and function was explored to determine the specificity of parigidin-br1, which appears to be a promising insecticidal agent.
EXPERIMENTAL PROCEDURES
Purification of Parigidin-br1
Different samples (leaves, inflorescences, and peduncles) of Palicourea rigida, popularly known as “bate-caixa” in Brazil, were collected from 10 individual plants at Ermida Dom Bosco, located in Brasilia DF, Brazil (15° 47′ 51″ S, 47° 48′ 38″ W), and pooled. Sampling occurred in two periods: in August during the dry season and in December during the rainy season when leaves, inflorescences, and peduncles were collected. Samples were ground in liquid nitrogen and extracted with dichloromethane and methanol (1:1, v/v) for 12 h, followed by filtration through cotton fibers. Subsequently, each sample was subjected to a liquid-liquid extraction, and the aqueous phase was collected and lyophilized. The lyophilized material was acidified with 2% acetic acid and applied onto an open column with polyamide resin (Fluka), followed by size exclusion chromatography, using a SuperdexMT Peptide 10/300 GL column (GE Healthcare) equilibrated with 30% acetonitrile/0.1% TFA at a flow rate of 0.5 ml·min−1. The peptide fraction was lyophilized and resuspended in 0.1% TFA and further purified using a reverse-phase analytical HPLC Vydac C18 column (2.6 × 150 mm), with a nonlinear acetonitrile gradient (5–95%) at a flow rate of 1.0 ml·min−1. Peptide elution was monitored at 216 and 280 nm. HPLC fractions were applied onto an ultra-fast liquid chromatographer and eluted with a linear gradient (5–95% acetonitrile), at a flow rate of 400 μl·min−1 at 40 °C, using a Shimpak C18 analytical column (3.0 × 5 mm). The yield of paragidin-br1 was determined by weighing the HPLC-purified peptide.
Molecular Mass Analyses by Mass Spectrometry
Leaves and stems were collected in the dry and rainy seasons. Extracts were concentrated through a Ziptip (PerfectPure C18 Tip, Eppendorf) following the manufacturer's instructions. An aliquot of 1 μl of the eluted sample was mixed in a saturated solution of α-cyano-4-hydroxycinnamic acid (1:3, v/v), deposited on an AnchorChip 600-mm MALDI plate, and dried at room temperature. Molecular masses of compounds were determined using a MALDI-TOF Ultraflex III mass spectrometer (Bruker Daltonics).
Purified parigidin-br1 was reduced, alkylated, and digested with endoproteases to linearize any cyclic peptides present in the extract. One hundred μg of purified, lyophilized peptide was dissolved in 100 μl of 100 mm ammonium bicarbonate (pH 8.5). To this solution, 10 μl of 50 mm dithiothreitol was added, and the sample was incubated for 30 min at 60 °C under N2. Ten μl of 100 mm iodoacetamide was added, and the sample was incubated at room temperature for 15 min. Aliquots (20 μl) were taken and digested with trypsin, chymotrypsin, endoproteinase Glu-C, or a combination of these (1 μl of 1 μg/μl). Peptides were left to digest overnight at 37 °C and then quenched by acidification with 1% formic acid. MALDI-TOF analyses were conducted using an UltrafleXtreme TOF-TOF instrument (Bruker Daltonics) as described previously (13). Linearized cyclotide-containing crude plant extracts were analyzed on a QStar® Elite hybrid LC-MS/MS system (Applied Biosystems/MDS SCIEX) equipped with a nano-electrospray ionization source. MALDI-TOF/TOF MS/MS data were interpreted manually, and the peptide sequence was determined. LC-MS/MS data were searched against a custom cyclotide database using ProteinPilot (version 4.0, AB/Sciex).
Bactericidal Assays
Escherichia coli sp. ATCC 8739 and Staphylococcus aureus ps. ATCC 25923 were used to evaluate antibacterial activity. Bacterial cells were replicated in liquid Mueller-Hinton medium, under stirring for 2–3 h, at 37 °C. Antimicrobial activity was determined using the broth micro-dilution method described previously (30). The growth curve of the original culture was established by monitoring bacterial growth, observing the optical density at 595 nm every half-hour. The optimal time for the inoculum to reach the middle of its exponential phase was determined. To determine the relationship between cfu and optical density, we counted the viable bacterial cells in a suspension, where an aliquot was collected every 30 min, diluted 1:10, and micro-droplets of the suspensions were plated on Mueller-Hinton medium plus agar (15 g·liter−1). The tests were conducted on bacteria in multi-well plates from the original culture. For this, inoculum was prepared, first being diluted to ∼5 × 104 cfu·ml−1 per well. The volume did not exceed 10% of the volume of the well containing Mueller-Hinton liquid. Sterile distilled water was used as a negative control and chloramphenicol (40 μg·ml−1) was used as a positive control. Multi-well plates (TPP - 96 well) were inoculated and then incubated with slight agitation at 37 °C for 2–3 h. Bacterial growth was monitored at 595 nm every 30 min during the exponential phase of bacterial growth. The amount of bacterial growth in the positive control was compared with the amount of growth in the negative control, to determine the end points of growth. The minimum inhibitory concentration was determined as the lowest concentration that inhibits 100% of bacterial growth.
Evaluation of Hemolytic Activity
Fresh human blood was collected from a healthy donor in heparin vacuum blood collection tubes and used to determine the hemolytic activity of peptides as described previously (31). Red blood cells were washed with PBS and centrifuged at 500 × g, after which the supernatant was removed. This procedure was repeated five times. Test peptides, at a range of concentrations (39 μm, 19.5 μm, 9.75 μm, 4.8 μm, 2.4 μm, 1.2 μm, and 0.6 μm), were mixed with 50 μl of the erythrocyte solution (1% of erythrocytes in PBS). A positive control, 0.1% Triton X-100, and negative control, PBS, were included. After 1 h, the tubes were centrifuged at 3500 × g for 15 min, and 100 μl of each supernatant was applied to a 96-well microplate. Hemolysis was measured as absorbance at 540 nm, using a microplate reader (Powerwave HT, Biotek). The percentage of hemolysis was calculated relative to the positive and negative controls: Triton X-100 was taken as 100% hemolysis and PBS as 0%.
Bioassays against Insects
In vivo toxicity toward insects was measured using a method described previously (23), with minor modifications. Neonate larvae of D. saccharalis, popularly known as the sugarcane borer, were treated with an artificial diet consisting of soybean meal (4.2%), wheat germ (3.2%), sugar (5.5%), agar (1.9%), including methylparaben (0.2%), vitamin solution (1.2%), sorbic acid, and ascorbic acid (0.2%). Diets were supplemented with 1 μm parigidin-br1. Cry1aC protein at a concentration of 200 μg·ml−1 was used as a positive control, and the negative control consisted of water plus a non-supplemented diet. For each diet (negative control, positive control, and sample), 12 larvae were placed in multi-well plates (TPP); each well contained one larva and 1 ml of diet. This volume of diet was sufficient for larval feeding for the complete assay period, rendering a change of diet unnecessary. To determine that peptide was not degraded during the bioassay, MALDI-TOF analyses of the diet were performed and demonstrated the continuous presence of parigidin-br1 (data not shown). The growth of the larvae was monitored for 15 days, in a 30 °C environment with a cycle of 16 h of light and 8 h of dark. From day 6 to day 15, on every second day, the larvae were observed using a magnifying glass to evaluate mortality. Fifteen days after the beginning of the experiment, the larvae were weighed and measured.
Cell Viability Tests using Insect Cell Lines
Cell viability tests were performed using insect cell line SF-9 from lepidopteran S. frugiperda, obtained from the envelope of the ovary pupae (32). The cells were maintained at 27 °C in TNM-FH medium supplemented with 10% (v/v) FCS, and 1% penicillin-streptomycin-amphotericin. Cultures in early stationary phase were split every 5 days (typical cell density, 1 × 106 cell·ml−1) with a starting density of 1 × 105 cells·ml−1. Tests were performed at three different concentrations of the peptide (1, 5, and 10 μm), and as negative control cells were left without treatment. Subsequently, the insect cells were seeded in microtiter six well plates at the early stationary phase (with density of cells in 1 × 106 cells·ml−1). The plates were incubated at 27 °C and analyzed at 24, 48, and 72 h. The cell suspension was added to the solution of 0.4% trypan blue dye (1:1, v/v) and observed in a Neubauer chamber using light microscopy. Cells were stained to perform cell counting. All tests were evaluated in triplicate. For the statistical analysis, the 50% cytotoxic concentration (CC50) and their lower and upper fiducial limits (95% confidence limits) were calculated by Probit (33). Data were compared by one-way analysis of variance followed by Tukey's test when significant differences were found at p = 0.05 using SigmaStat (version 2.03) for Windows (SPSS, Inc., Chicago, IL).
TEM
SF-9 ovary cells were treated with the peptide concentrations of 1, 5, and 10 μm for 72 h and analyzed with transmission electron microscopy (TEM).3 Cells were fixed using 2.5% glutaraldehyde in 0.1 m sodium cacodylate buffer (pH 6.8), and kept at 4 °C overnight. The fixed material was centrifuged at 4500 × g for 5 min, and the supernatant was discarded. The pellet was resuspended in cacodylate buffer and centrifuged three times before being post-fixed in 2% osmium tetroxide for 2 h. Post-fixed cells were centrifuged and washed with Milli-Q water five times. The pellet was resuspended in 0.5% uranyl acetate and stored overnight at 4 °C in the dark. For resin infiltration, a progressive ethanol dehydration series (10, 30, 50, 70, 90, and 100%) was conducted, with a 10-min immersion in each solution, followed by centrifugation. Using a tissue rotator (EMS, Hatfield, PA), ethanol was gradually substituted with Spurr resin, following the manufacturer's protocol. Samples were placed in rubber molds and polymerized for 48 h in an oven at 70 °C. They were cut into semi-thin sections of 500 nm for identification of areas of interest with light microscopy and then into 50-nm ultrathin sections using an ultramicrotome (LKB Ultrotome III) for TEM. The ultrathin sections were stained with uranyl acetate 2% for 1 h in the dark. The analysis was performed in a transmission electron microscope (JEOL 1011) at 80 Kv.
Localization of Parigidin-br1-FITC Binding
FITC was coupled covalently to parigidin-br1 by a method described previously (34) with minor modifications. FITC solution (20 mg in 1 ml of anhydrous dimethyl sulfoxide) was added in 0.5 m sodium carbonate buffer at pH 9.5 before use, followed by the addition of parigidin-br1 (20 μm peptide per 1 mg of FITC). The mixture was incubated in a dark glass tube and rotated for 1 h at room temperature. The FITC-labeled parigidin-br1 was recovered by overnight dialysis under slow shaking against distilled water. Parigidin-br1-FITC complexes were evaluated by MALDI-TOF mass spectrometry to confirm FITC binding. Cell viability assays with SF-9 cells were performed as described previously (33). Three treatments were assessed: treatment with parigidin-br1; parigidin-br1 coupled with FITC; and SF-9 cells in the absence of FITC and parigidin-br1. Cells were incubated with the different solutions and evaluated after 3 and 24 h at room temperature in a fluorescence microscope AXIOPHOT (Zeiss), utilizing the FITC filter (Chroma Technology Corp.). The images were captured using AxionVision software (Zeiss).
Phylogenetic Relations
Phylogenetic analysis was carried out using the maximum parsimony method (35). Branches corresponding to partitions reproduced in <50% of trees were collapsed. The initial trees were obtained with the random addition of sequences (10 replicates). The maximum parsimony tree was obtained using the Close-Neighbor-Interchange algorithm with search level three in initial trees. All alignment gaps were treated as missing data. Phylogenetic analyses were conducted in MEGA4 (36). The bootstrap statistical test (37) was used to confirm the reliability of the inferred tree.
Homology Modeling
Initially, PSI-BLAST was used to find the best templates for homology modeling. The sequence of circulin B showed 77% identity, and the Protein Data Bank structure 2ERI was used as template (38). Fifty theoretical three-dimensional peptide structures were constructed using templates by Modeler (version 9.8) (39). The final model was evaluated by measuring geometry, stereochemistry, and energy distributions. This was performed using PROSA II to analyze packing and solvent exposure characteristics and PROCHECK for additional analysis of stereochemical quality (40). In addition, the root mean square deviation was calculated by overlap of Cα traces and backbones onto the template structure through the 3DSS program (41). Protein structures were visualized and analyzed on Swiss protein data bank viewer (version 3.7) (42) and PyMOL (43).
Parigidin-br1-Dodecylphocholine (DPC) Micelle Docking Analysis
The HEX molecular docking (44) program (version 6.1) was used to elucidate possible modes of interaction of parigidin-br1with DPC micelles (neutral lipid), consisting of 65 DPC lipids, after 1100 ps of relaxation. Briefly, this procedure performed global rotational and translational space scan by using Fourier transformations, which rank the output according to surface complementarity and electrostatic characteristics. A list of 100 complexes of cyclotide-micelle was ranked and evaluated according to spatial restraint, salt bridge formation, hydrogen bond, and hydrophobic interaction for both peptides. Validation was carried out according to biochemical data, stereochemical limitation, and formation length, utilizing PyMOL for visualization (43). The interaction area was calculated through the number of contact points (interactions) of amino acid residues with lipid compounds.
RESULTS
Cyclotide Analyses from P. rigida in Different Seasons and Tissues
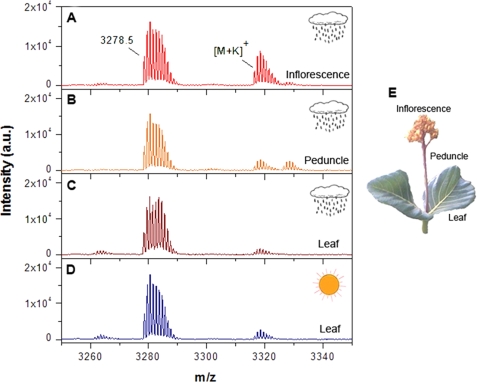
To analyze possible differences in the expression of cyclotides in P. rigida in different seasons, samples were collected from the same location at different times of the year. Before starting the purification procedure, the leaves in dry season and wet season, inflorescence, and peduncle were analyzed by MALDI-TOF mass spectrometry to test for the presence of cyclotides. The extracts showed a wide diversity of peptides with masses in the range 2900–3500 Da, typically the range of masses expected for cyclotides. In general, the expression of the peptides appeared to vary with the season (dry versus wet) as well as with the plant tissue. However, one peptide, identified below as parigidin-br1, was found to present in different plant tissues (leaves, inflorescences, and peduncles) as well under different climatic conditions (Fig. 1).
FIGURE 1.
Mass spectral comparison of different P. rigida tissues: inflorescence (A), peduncle (B), and leaves from wet (C) and dry (D) seasons, showing the parigidin-br1 and respective potassium adducts. The tissues are shown in E.
Cyclotide Purification
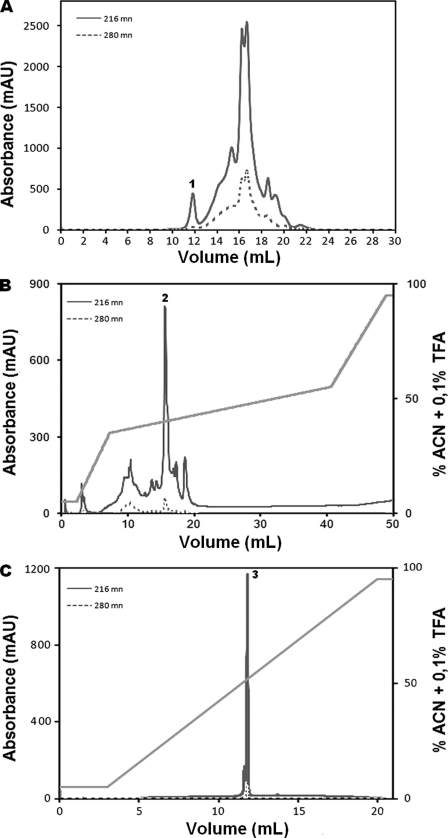
The peptide with m/z 3278.1 was selected for characterization based on its presence in all analyzed extracts (both seasons and all plant parts) and also owing to its relatively higher amount in chromatography runs of leaf extracts from the wet season. The extract was subjected to various purification steps, and the fractions were analyzed by MALDI-TOF MS, confirming the presence of a cyclotide with an m/z of 3278.1 in fraction 1, along with 20 other peptides (Fig. 2A). Fraction 1 was further purified by reversed phase HPLC, and 10 fractions were collected in total (Fig. 2B). Analysis by MALDI-TOF MS showed a peptide with a mass of 3278.1 Da in RP-HPLC fraction 2. Several other minor components were observed in this HPLC fraction (Fig. 2B). A second round of reversed phase purification was undertaken using ultrafast liquid chromatography. The chromatographic profiles showed only two fractions, as observed by MALDI-TOF MS. Fraction 3 corresponded to the pure peptide (Fig. 2C).
FIGURE 2.
Purification profile of P. rigida cyclotide. A, size exclusion chromatography profile of the leaf extract from the wet season. The cyclotide fraction is marked 1. B, reverse-phase chromatographic separation of fraction 1 from size exclusion chromatography using a non-linear gradient of acetonitrile (ACN; 13–50%), represented by the diagonal line. The partially purified fraction is marked 2 on the chromatogram. C, ultra-fast liquid chromatographer chromatographic purification of fraction 2, where fraction 3 corresponds to the purified paragidin-1. mAU, milliabsorbance units.
De Novo Sequencing
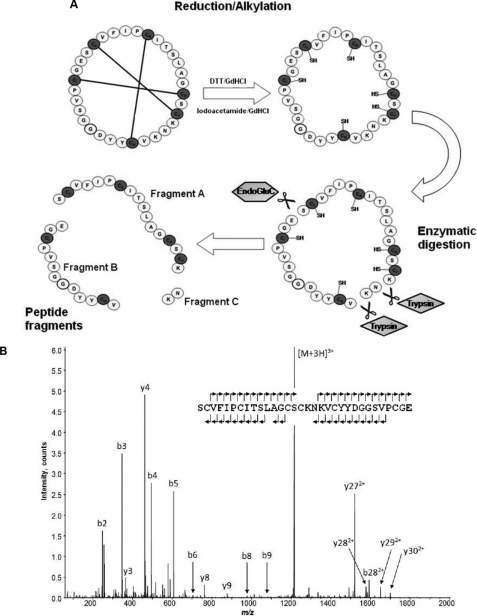
Reduction and alkylation of the peptide of mass 3278.1 Da resulted in a modified mass of 3644.4 Da as observed by MALDI-TOF MS. This corresponded to an increase of 348 Da, consistent with the presence of six cysteine residues. Nanospray-MS of the reduced and alkylated form of the peptide was performed. Peaks at m/z 1823.22+, 1215.83+, and 912.14+ corresponded to a peptide mass of 3644.4 Da. Enzymatic digestion (combined trypsin and endoproteinase Glu-C) was performed and two fragments named A (mass, 1958.8 Da) and B (mass, 1461.5 Da) were identified. MS/MS analysis elucidated the sequence of the major peak as SC*VFIPC*ITSLAGCSC*K (1958.9 Da), where the asterisk indicates that the cysteines are alkylated. The same procedure was performed with fragment B, with sequence VC*YYDGGSVPC*GE (1461.6 Da; Fig. 3).
FIGURE 3.
A, schematic representation of the reduction, alkylation, and enzymatic digestion protocol applied to a cyclotide fraction from HPLC purification. Combined digestion using trypsin and endoproteinase Glu-C resulted in three fragments: A, B, and C. B, MS/MS spectra were collected for fragments A and B allowing assignment of their peptide sequence. Fragment C was too small to allow MS/MS sequencing. A single digestion strategy using only endoproteinase Glu-C revealed the full-length peptide sequence.
MS/MS sequencing cannot distinguish between the isobaric residues Ile and Leu. Amino acid analysis of the peptide was performed to determine the number of each of these residues in the sequence and revealed two Ile residues and one Leu. To determine the location of these residues, chymotrypsin digestion was employed, as chymotrypsin cleaves amide bonds C-terminal to Leu (as long as they are not followed by Pro). The proteolytic fragment IPCITSL was detected, allowing unambiguous identification of the Ile/Leu locations (supplemental Fig. 2).
In addition to the proteolytic peptide fragments described above, an ion at m/z 1215.91 (mass, 3644.73 Da) was identified, corresponding to the linearized peptide after reduction, alkylation, and digestion with endoproteinase Glu-C (supplemental Fig. 1). A combination of HPLC, MS, and amino acid analyses allowed the characterization of a cyclotide containing 32 amino acids, with a mass of 3278.1 Da, which was named parigidin-br1. It contained six cysteine residues that typically form the three disulfide bridges responsible for stabilizing the CCK motif of cyclotides (45). From the residues present in loop 5, it was classified as belonging to the bracelet subfamily of cyclotides. It was present in the plant extract at a concentration of 0.1 mg/kg plant dry weight.
Biological Activities
To determine whether parigidin-br1 had antimicrobial activity, an assay was performed using the micro-dilution method in accordance with M7-A6 National Committee for Clinical Laboratory Standards, against a Gram-positive S. aureus ATCC 25923 and a Gram-negative E. coli ATCC 8739 (30). This assay showed no inhibitory activity (data not shown) against bacteria. Parigidin-br1 showed hemolytic activity of 41% at a concentration of 40 μm and 28% at a concentration of 20 μm (data not shown).
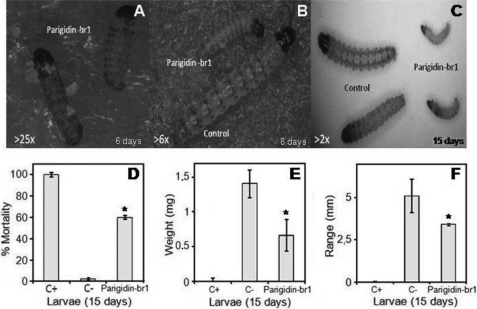
To assess the potential of paradigin-br-1 as an insecticidal agent, its activity against an important pest of sugarcane and against an insect cell line was examined. In vivo evaluation was conducted with administration of 1 μm parigidin-br1 in the diet of neonate larvae of D. saccharalis, and in vitro tests were performed with cultured SF-9 cells from S. frugiperda. Parigidin-br1 showed insecticidal activity in both assays. In the in vivo studies, with administration of parigidin-br1 in amounts comparable with endogenous levels in plants, parigidin-br1 caused reduction in growth and a significant mortality rate compared with insects fed a control diet (Figs. 4, A–C). The experiment was performed over 15 days, with measurements starting from the sixth day and repeated every 2 days. At the end of the experiment, the average mortality observed in the larval diet containing the peptide was 60% (Fig. 4D). Larval weight in the presence of parigidin-br1 was ∼0.6 mg, whereas control larvae had an average weight of 1.4 mg, a reduction of 43% (Fig. 4E). Larvae treated with the parigidin-br1 diet and the control diet were ∼3.4 and 5.1 mm in size, respectively (Fig. 4F). The cyclotide-fed diet reduced the larval size by a statistically significant 33% (p < 0.01).
FIGURE 4.
Insect bioassays. Larvae of D. saccharalis were treated on an artificial diet with 1 μm parigidin-br1. A, 25× magnified image of neonate larvae on the sixth day of the experiment. B, 6× magnified image of the eighth day of the experiment. It was possible to observe a difference in development between the treated larvae on diets containing parigidin-br1 (smaller larvae) and those fed on the negative control diet (larger larvae). C, by the 15th day of the experiment, the difference in development of the larvae treated with parigidin-br1 compared with the larvae of the control is very distinct (image magnified 2×). D, larval mortality after 15 days of the feeding trial. E, larval weight variation. F, graphical representation of developing larvae on the 15th day of the experiment. The positive control in these experiments was a Cry1aC protein diet (200 μg·ml−1), and the negative control was a water diet. The asterisk represents a statistical evaluation carried out using a Welch two Sample t test, giving a confidence interval of 95%, p value = 3.172e-07, p < 0.01. Error bars represent S.D. Each assay was performed in triplicate.
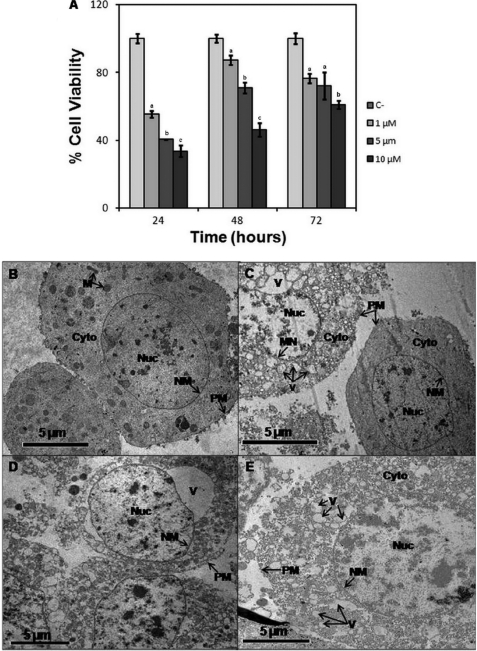
In vitro tests were performed at three different concentrations (1, 5, and 10 μm) of parigidin-br1, showing cytotoxic effects on SF-9 cells, showing significant statistical differences for all assessments compared with the control. Cell viability decreased with the increase of peptide concentration at 24, 48, and 72 h. The viability at 72 h was ∼76% at 1 μm, 72% at 5 μm, and 61% at 10 μm (Fig. 5A). Using these data with the Probit statistical analysis program we determined the CC50 values for parigidin-br1 to be 1.7 μm at 24 h, 10.3 μm at 48 h and 73.1 μm at 72 h. It was observed that a minimum concentration of 1 μm paragidin-br1 was required to induce membrane rupture and activity correlating with previous studies of cyclotide-lytic activity (29, 46). At a paragidin-br1 concentration of 10 μm, 61% mortality against SF-9 cells was observed in 24 h. The presence of resistant cells within the cell culture increased the cell count at 48 and 72 h; and for this reason, the activity was reduced when compared with that observed at 24 h, also suggesting a dose-response activity. To analyze the stability of parigidin-br1 at the end of the cell viability assay, aliquots of containing parigidin-br1 were collected and compared with medium without cyclic peptides. These aliquots were analyzed by MS to assess whether peptide degradation had occurred. MS analysis reveals the presence of parigidin-br1 with a molecular mass of 3278.10 Da and none degraded form (data not shown).
FIGURE 5.
Analysis of the in vitro effects and images of transmission electronic microscopy of SF-9 ovary cells treated with different concentrations of parigidin-br1 performed over 72 h. A, cytotoxic activity of parigidin-br1 over SF-9 cells analyzed at 24, 48, and 72 h. B, negative control SF-9 cells. C, cells treated with 1 μm parigidin-br1. D, cells treated with 5 μm parigidin-br1. E, cells treated with 10 μm parigidin-br1. Labels and arrows indicate the mitochondria (M), cytoplasm (Cyto), nuclei (Nuc), nuclear membrane (NM), plasma membrane (PM), and vacuole (V). Error bars represent S.D. Each assay was performed in triplicate. At each time of evaluation, significant differences are indicated by different letters Tukey (p ≤ 0.05).
Transmission Electron Microscopy
After 72 h of the insect cytotoxicity experiment, SF-9 cells were separated, and a transmission electron microscopy scan was performed. The negative control (Fig. 5B) showed a relatively healthy cell, allowing visualization the plasma membrane, nuclear membrane and the cytoplasm of mitochondria and some organelles. In contrast, in cells treated with 1 μm parigidin-br1 (Fig. 5C), formation of vacuoles in the cytoplasm was observed, with bleaching and breakage of the core at various points of the plasma membrane. Moreover, in cells treated with 5 μm of peptide (Fig. 5D), clear damage was observed, including the presence of large vacuoles and leakage of cytoplasmic contents into the extracellular environment due to rupture in the plasma membrane. Similar effects were observed in cells treated with 10 μm parigidin-br1 (Fig. 5E), and a dead cell was seen, where the plasma membrane no longer existed due to degradation such as the presence of several vacuoles and absence of organelles in the cytoplasm. These data suggest that the insecticidal activity of parigidin-br1 occurs through its interaction with the plasma membrane of cells.
Fluorescence Microscopy of Parigidin-br1-FITC Binding
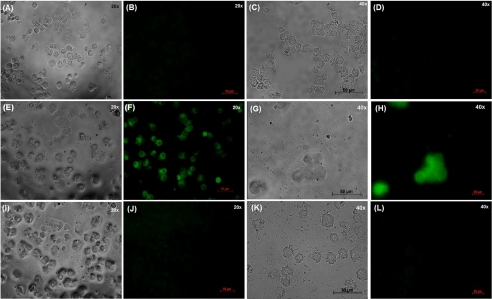
The activity of parigidin-br1-FITC was determined using an in vitro cell viability assay as described previously (33). The FITC-labeled peptide was observed with a molecular mass of 4056.76 Da (the mass difference was due to the addition of two FITC molecules (389.38 Da) at Lys21 and Lys24 of loop 5). Parigidin-br1 was FITC-labeled, and SF-9 cells were incubated in the presence and absence of parigidin-br1-FITC (10 μm) and parigidin-br1 (10 μm) without labeling for 24 h. Control cells observed under white light after 3 h (Fig. 6A) showed normal cellular structures with well defined internal organelles. After 24 h, control cells were observed once more under white light (Fig. 6C), also showing normal cellular development. The control cells were also analyzed at both times under fluorescent light (Fig. 6, B and D), and no fluorescence was observed, confirming that there was no unspecific labeling. Cells incubated with parigidin-br1-FITC were analyzed after 3 and 24 h under white light (Fig. 6, E and G). After 3 h, a regular dimension structure of cells (data also observed by TEM, Fig. 5), with a clear presentation of vacuoles and undefined organelles was observed. Furthermore, when cells treated with parigidin-br1-FITC were evaluated by fluorescent light after 3 h (Fig. 6F), it was possible to visualize a thin layer of fluorescence throughout the cell surface, showing a deposition of parigidin-br1-FITC onto the cell membrane. Moreover, after 24 h, few cells remained (Fig. 6G), with those present appearing swollen. Fluorescent analyses (Fig. 6H) of these cells showed a strong fluorescence signal, which suggests an internalization of parigidin-br1. Finally, the action of parigidin-br1 unlabeled with FITC (Fig. 6, I and K) presented the same pattern previously detected by TEM analyses (Fig. 5), showing lysed cells with leakage of the cellular contents apparent. Under fluorescence analyses (Fig. 6, J and L), no fluorescence was detected.
FIGURE 6.
Fluorescence micrographs showing parigidin-br1-FITC (10 μm) binding in SF9 cells membranes. A and C, shown is a light microscopy of SF-9 cells at 3 h and 24 h, respectively. B and D, shown is a fluorescence microscopy of SF-9 cells at 3 h and 24 h, respectively. E and G, shown is a light microscopy of SF-9 cells incubated with parigidin-br1 labeled with FITC at 3 h and 24 h, respectively. F and H, shown is a fluorescence microscopy of SF-9 cells incubated with parigidin-br1 labeled with FITC at 3 h and 24 h, respectively. I and K, shown is a light microscopy of SF-9 cells incubated with parigidin-br1 at 3 h and 24 h, respectively. J and L, shown is a fluorescence microscopy of SF-9 cells incubated with parigidin-br1 at 3 h and 24 h, respectively.
Molecular Phylogeny and Homology Modeling of Parigidin-br1
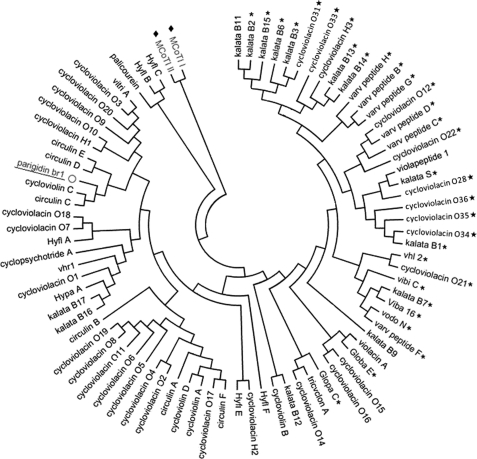
An alignment of parigidin-br1 with other members of the bracelet subfamily was performed (Fig. 7), and a phylogenetic tree was constructed to assess the sequence similarity between parigidin-br1 and other cyclotides of the bracelet, Möbius, and trypsin inhibitor subfamilies (Fig. 8). In general, parigidin-br1 and other bracelet cyclotides are grouped on the left side of the tree, Möbius members on the right, and trypsin inhibitory members on the upper left (MCoT-I and MCoT-II). At the bottom of the tree, a mixture of the two cyclotide subfamilies was found, including kalata B9, violacin A, cycloviolacin O15 and O16, belonging to the bracelet subfamily, and Globa E and Glop C belonging to the Möbius subfamily. The primary sequence of parigidin-br1 was compared with other sequences in CyBase (17), and showed most similarity with circulin B (PDB 2ERI), found in Chassalia parvifolia, and belonging to the Rubiaceae family. On the basis of 75% identity, circulin B was chosen as a template for modeling studies (38). A parigidin-br1 three-dimensional model was built and validated using Procheck. The Ramachandran map indicated that 84% of residues were located in most favorable regions, 12% in allowed regions, and 4% in generously allowed regions. No residue was in the disallowed regions. Additional validation was obtained by the sum of several parameters such as dihedral angles and main-chain covalent forces denominated by the g-factor, which was −0.17. This g-factor indicated that the three-dimensional structure was within an acceptable quality range. The root mean square deviation between the circulin B structure solved by NMR and the model of parigidin-br1 was 0.47 Å, suggesting that the parigidin-br1 model is a reliable structure.
FIGURE 7.
ClustalW alignment of selected cyclotides. Paragidin-br1 was aligned with bracelet cyclotides obtained from Cybase (17). Secondary structural elements are represented above the alignment. The spiral represents the helix 310, and arrows represent β-sheet portions. The conserved Cys are labeled with Roman numerals I–VI, and the black lines represent the connectivity between the disulfide bonds.
FIGURE 8.
Phylogenetic tree assembled by neighbor joining. Subfamilies of Möbius (star), bracelet (no symbol), and trypsin inhibitor cyclotides (filled diamonds) are represented graphically. Parigidin-br1, a bracelet cyclotide, is underlined.
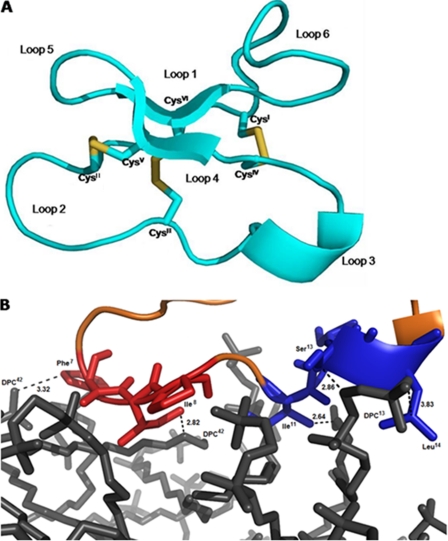
The modeled structure had a total hydrophobic content of 43% neutral net charge, one anti-parallel β-sheet, one 310 helix, and three disulfide bonds (Cys1-Cys17, Cys5-Cys19, and Cys10-Cys24) that stabilize structure. Using the Antimicrobial Peptide Predictor tool from the antimicrobial peptide database, structural similarity with β-structure-like defensins (∼30–50 amino acids) and a saposin-like helical bundle fold (∼80 amino acids; Fig. 9A) were identified (47–49).
FIGURE 9.
A, three-dimensional theoretical model of parigidin-br1 built by Modeler (version 9v8). The drawing was generated with PyMOL. Disulfide bonds are represented by sticks (yellow). B, modeled interaction between parigidin-br1 and a DPC micelle. Highlighted in red, loop 2 of paradigin-br1 is comprised of hydrophobic residues (83%) and likewise in blue, loop 3 has a hydrophobic content of 71%. The dodecylphosphocholine is shown in gray. The figure was generated using PyMOL.
Parigidin-br1:DPC Docking Analysis
To help elucidate the mechanism of interaction of parigidin-br1 with insect membranes, a search of the composition of insect cell membranes was undertaken. They are composed mainly (35%) of phosphatidylcholine (DPC), a neutral lipid (50), and for this reason, a theoretical micelle-membrane composed of DPC was constructed to study cyclotide-SF-9 membrane interactions. This study indicated that parigidin-br1 can bind with DPC micelles via a hydrophobic region in loop 2 (Phe7 and Ile8) and loop 3 (Ile11, Ser13, and Leu14). These loops have a hydrophobic ratio of 83% (loop 2) and 71% (loop 3). Two hydrophobic interactions were observed in loop 2, among the carbons (CE1) of Phe7 and Ile8 with the carbons in DPC42 (C3 and C21) with distances of 3.32 and 2.82 Å, respectively. In addition, three hydrophobic interactions were observed in loop 3 among the carbons atoms of Ile11, Ser13, and Leu14 with DPC13 (C21, C5, and C1) at a distance of 2.64, 2.86, and 3.83 Å (Fig. 9B). Similar interactions were described in experimental studies of cyclotide:DPC micelle interactions (51), confirming that bracelet cyclotides use hydrophobic regions in loops 2 and 3 to form a strong interaction with membranes (51).
DISCUSSION
Cyclotides are part of the defense system of plants against pathogens and insects (23). To understand more about the expression of cyclotides, we investigated their distribution in different parts of P. rigida as well as the influence of climatic variations. We observed that some cyclotides are expressed in specific plant tissues, but others, like parigidin-br1, are found in multiple parts of the plant, independent of climatic variations.
The first study to imply that seasonal or geographical differences might influence expression of cyclotides was reported by Gran et al. (14). During studies in the Congo, he noted that women gathered parts of the plant Oldenlandia affinis during the rainy season and dried them for later use (14) to assist childbirth/labor via drinking a tea made with the leaves. The first systematic study to monitor cyclotide expression in different climatic regions was performed for cyclotides expressed in Violaceae species cultivated in Australia and Sweden. Plants were monitored over 14 months and it was found that Violaceae species examined in Sweden in April showed higher cyclotide levels; this period corresponds to warmer Spring temperatures. In contrast, for the Violaceae species in Australia, the expression remained more constant (52, 53). In another study on the distribution of cyclotides in nine species of Hybanthus (Violaceae) over the Australian continent, it was observed that plants located in southern and central Australia expressed larger amounts of cyclotides than those in hotter northern regions (54). Such seasonal and geographical variations might reflect the induction or inhibition of cyclotide expression, e.g. a plant might increase the production of certain cyclotides to repel pests of a particular season, while maintaining basal expression of other cyclotides such as kalata B1 and B2 in O. affinis, which appear to be expressed all year round (55). In the current study, parigidin-br1 was detected in several plant tissues both in the dry and rainy season, suggesting that it is probably part of the constitutive defense of P. rigida. Although in some plants, individual cyclotides are expressed in just one tissue, there is precedent for broader expression, such as seen here for parigidin-br1. For example, studies with Viola hederacea showed that kalata B1 and Varv peptide A were present in extracts of leaf, flower, and root (45, 52), and similarly, vhr1 and cycloviolacin H4 were observed in roots and bulbs (56). Overall, the data presented here support the pattern of some cyclotides being broadly expressed, whereas others are more tissue-specific. The yield of parigidin-br1 of ∼0.1 mg per gram of dry plant material (0.01%) is lower than that reported for kalata B1 of ∼0.1% (54), consistent with previous observations that not all cyclotides are expressed at similar amounts.
Potential activity of parigidin-br1 against Gram-negative (E. coli) and Gram-positive (S. aureus) bacteria was tested for but gave negative results. Antimicrobial activity has been reported for kalata B1 and B2, circulin A and B, cyclopsychotride A, and cycloviolacin O2 (16, 20), although the activity is generally weak under high salt conditions and conflicting data have been reported (57). Although cyclotides have been demonstrated to interact with membranes and membrane models (51, 57, 58), it is not clear that they should be regarded generally as antimicrobial peptides, and the studies here on parigidin-br1 support this view.
By contrast, with its lack of antimicrobial activity, parigidin-br1 has potent insecticidal activity, suggesting the potential for bioinsecticide development, as has been proposed for kalata B1. That cyclotide showed a mortality rate of 50% against neonate larvae of H. punctigera at a concentration of 0.8 μmol·g−1 and 20% against H. armigera larvae (23, 59). Similar data have been reported for kalata B2 against H. armigera (59). The mechanism of action of cyclotides toward insect pests is believed to involve membrane disruption (29). In support of this proposal, electron microscopy studies with parigidin-br1 reported here show a clear deleterious effect on an insect cell line, consistent with membrane disruption causing cell death. This proposal was confirmed with fluorescence microscopy studies, where parigidin-br1-FITC was observed on the surface of SF-9 cells after 3 h, and after 24 h, a clear cellular disruption was observed. Our results are similar to those reported for the incubation of kalata B1 (60), labeled with fluorescein-conjugate avidin, with adult female nematodes that had been chemically ligated to inhibit feeding. In that report, transverse sectional analyses showed the presence of fluorescently labeled peptide in all internal organs and a thin layer of fluorescence on the worm surface, suggesting that cyclotides increase the permeability of the external nematode membranes (60).
NMR studies on kalata B1 have shown that it binds to DPC micelles through hydrophobic residues, including Trp19, Pro20, and Val21 in loop 5, Leu27, Pro28, and Val29 in loop 6, and Val6 in loop 2. Additionally, Arg24 and Glu3 are involved in electrostatic interactions (51). In a surface plasmon resonance study that assessed the interaction of kalata B1 with lipid membranes, it was observed to bind to phosphatidylethanolamine exposed on the membrane surface. This partnering could then lead to the formation of oligomers and pores within membranes (26, 29). In the morphological study described previously (26), the intake of cyclotides affected the midgut of H. armigera and H. punctigera, causing disruption of the microvilli, blebbing, swelling, and ultimately rupture of the cells of the gut epithelium.
Sequence comparison of parigidin-br1 with other cyclotides shows that it displays many of the common characteristics of cyclotides (61). For example, it contains Phe7 in a region where an aromatic ring is conserved in bracelet cyclotides, a tryptophan being the most frequent residue in this position, but circulin B and C, cyclopsychotride A, and cycloviolacin A, among others, have phenylalanine in this position. It is believed that the conserved aromatic ring at this position might be important for the activity of the bracelet subfamily (61).
A theoretical model of parigidin-br1 was constructed and includes an antiparallel triple-stranded β-sheet, where one strand is composed of residues 14–18 and another strand of residues 21–25, which are linked by a β-turn, forming a β-hairpin. The structure has six conserved cysteine residues that are responsible for the formation of disulfide bonds, CysI-CysIV and CysII-CysV, which (with loops 1 and 4) form a ring that is penetrated by the third disulfide bond, CysIII-CysVI. This arrangement of disulfide bonds is responsible for the formation of a CCK motif (62). Since the discovery and characterization of the first cyclotide, kalata B1, the CCK has been confirmed by NMR analysis in kalata B1 and B2, belonging to the Möbius cyclotides (59, 63, 64); circulin A and B (65), palicourein (66), and cycloviolacin O1 belonging to the bracelet cyclotides (45); and MCoTI-II belonging to the trypsin inhibitors (67, 68). It has also been observed in hybrid cyclotides such as kalata B8 (69). The theoretical parigidin-br1 model developed here suggests the presence of CCK motif and indicates a small 310 helix in loop 3 similar to bracelet cyclotides such as cycloviolacin O1 and O2 and circulin A. The parigidin-br1 310 helix is composed of residues Ser13, Leu14, and Ala15.
A docking analysis between parigidin-br1 and DPC micelles was done based on the expected SF-9 membrane composition (50). The analysis showed that loops 2 and 3, which have high hydrophobicity (83 and 71%, respectively), appeared to be involved in interaction with the headgroup of the membrane. The residues involved are Phe7 and Ile8 in loop 2 and Ile11, Ser13, and Leu14 in loop 3, in agreement with studies on the interaction of membranes with other cyclotides (51), where possible differences in the mechanism of interaction between bracelet and Möbius cyclotides were described. In the Möbius family, interaction occurs through loops 5 and 6, whereas in the bracelet subfamily activity could be related to hydrophobic residues in loops 2 and 3. A recent study of kalata B1 and analogs determined that these cyclotides bind specifically to phosphatidylethanolamine (51, 57). These interactions are favored in regions of the membrane rich in cholesterol and sphingomyelin (57). Parigidin-br1 also acts on the cellular membrane of insects. The parigidin-br1 interaction could be through phosphatidylethanolamine and/or phosphatidylcholine, which are the two most abundant phospholipids in the membranes of SF-9 (50). Membrane binding may facilitate the self-association of cyclotides, leading to pore formation and eventual rupture of the membrane, thereby causing cell death (57, 70). The membrane composition can vary greatly from one organism to another, thus explaining the presence of insecticidal activity and absence of hemolytic activity or antimicrobial against strains of E. coli and S. aureus.
In conclusion, the insecticidal activity of parigidin-br1, demonstrated in an in vitro assay with SF-9 cells and in an in vivo assay against neonate larvae of D. saccharalis, is consistent with the interaction of parigidin-br1 with the lipid membranes of insects. More broadly, the importance of studying the cyclotide family is clear, as cyclotides generally have high structural stability, which is of interest for pharmaceutical, agrochemical, or biotechnological applications.
Supplementary Material
This work was supported by Coordenação de Aperfeiçoamento de Pessoal de Nivel Superior, Conselho Nacional de Pesquisa e Desenvolvimento, Fundação de Amparo a Pesquisa do Distrito Federal, Fundação de Amparo a Pesquisa de Minas Gerais, and Universidade Católica de Brasília. Work at the University of Queensland was supported in part by a grant from the Australian Research Council.

This article contains supplemental Figs. 1 and 2.
- TEM
- transmission electron microscopy
- CCK
- cyclic cystine knot
- DPC
- dodecylphocholine.
REFERENCES
- 1. Biello D. (2008) Biofuels Are Bad for Feeding People and Combating Climate Change in Scientific American, Feb. 7, 2008, 36 [Google Scholar]
- 2. Oerke E. C., Dehne H. W. (2004) Crop Protection 23, 275–285 [Google Scholar]
- 3. Oldroyd B. P. (2007) PLoS Biol. 5, e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Desai P. N., Shrivastava N., Padh H. (2010) Biotechnol. Adv. 28, 427–435 [DOI] [PubMed] [Google Scholar]
- 5. James C. (2009) ISAAA Brief No. 41-2009, Global Status of Commercialized Biotech/GM Crops, pp. 7–11, ISAAA, Ithaca, New York [Google Scholar]
- 6. Bagla P. (2010) Science 327, 1439. [DOI] [PubMed] [Google Scholar]
- 7. Craik D. J. (2010) Toxicon. 56, 1092–1102 [DOI] [PubMed] [Google Scholar]
- 8. Craik D. J. (2009) Trends Plant Sci. 14, 328–335 [DOI] [PubMed] [Google Scholar]
- 9. Saska I., Gillon A. D., Hatsugai N., Dietzgen R. G., Hara-Nishimura I., Anderson M. A., Craik D. J. (2007) J. Biol. Chem. 282, 29721–29728 [DOI] [PubMed] [Google Scholar]
- 10. Daly N. L., Rosengren K. J., Craik D. J. (2009) Adv. Drug Deliv. Rev. 61, 918–930 [DOI] [PubMed] [Google Scholar]
- 11. Benko-Iseppon A. M., Galdino S. L., Calsa T., Jr., Kido E. A., Tossi A., Belarmino L. C., Crovella S. (2010) Curr. Protein Pept. Sci. 11, 181–188 [DOI] [PubMed] [Google Scholar]
- 12. Craik D. J., Anderson M. A., Barry D. G., Clark R. J., Daly N. L., Jennings C. V., Mulvenna J. (2002) Lett. Peptide Sci. 8, 119–128 [Google Scholar]
- 13. Poth A. G., Colgrave M. L., Lyons R. E., Daly N. L., Craik D. J. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 10127–10132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gran L., Sandberg F., Sletten K. (2000) J. Ethnopharmacol. 70, 197–203 [DOI] [PubMed] [Google Scholar]
- 15. Gustafson K. R., McKee T. C., Bokesch H. R. (2004) Curr. Protein Pept. Sci. 5, 331–340 [DOI] [PubMed] [Google Scholar]
- 16. Pränting M., Lööv C., Burman R., Göransson U., Andersson D. I. (2010) J Antimicrob. Chemother. 65, 1964–1971 [DOI] [PubMed] [Google Scholar]
- 17. Wang C. K., Kaas Q., Chiche L., Craik D. J. (2008) Nucleic Acids Res. 36, D206–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Witherup K. M., Bogusky M. J., Anderson P. S., Ramjit H., Ransom R. W., Wood T., Sardana M. (1994) J. Nat. Prod. 57, 1619–1625 [DOI] [PubMed] [Google Scholar]
- 19. Lindholm P., Göransson U., Johansson S., Claeson P., Gullbo J., Larsson R., Bohlin L., Backlund A. (2002) Mol. Cancer Ther. 1, 365–369 [PubMed] [Google Scholar]
- 20. Tam J. P., Lu Y. A., Yang J. L., Chiu K. W. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 8913–8918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Colgrave M. L., Kotze A. C., Huang Y. H., O'Grady J., Simonsen S. M., Craik D. J. (2008) Biochemistry 47, 5581–5589 [DOI] [PubMed] [Google Scholar]
- 22. Plan M. R., Saska I., Cagauan A. G., Craik D. J. (2008) J. Agric. Food Chem. 56, 5237–5241 [DOI] [PubMed] [Google Scholar]
- 23. Jennings C., West J., Waine C., Craik D., Anderson M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10614–10619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gruber C. W., Cemazar M., Anderson M. A., Craik D. J. (2007) Toxicon. 49, 561–575 [DOI] [PubMed] [Google Scholar]
- 25. Chagolla-Lopez A., Blanco-Labra A., Patthy A., Sánchez R., Pongor S. (1994) J. Biol. Chem. 269, 23675–23680 [PubMed] [Google Scholar]
- 26. Barbeta B. L., Marshall A. T., Gillon A. D., Craik D. J., Anderson M. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1221–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nourse A., Trabi M., Daly N. L., Craik D. J. (2004) J. Biol. Chem. 279, 562–570 [DOI] [PubMed] [Google Scholar]
- 28. Wang C. K., Hu S. H., Martin J. L., Sjögren T., Hajdu J., Bohlin L., Claeson P., Göransson U., Rosengren K. J., Tang J., Tan N. H., Craik D. J. (2009) J. Biol. Chem. 284, 10672–10683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang Y. H., Colgrave M. L., Daly N. L., Keleshian A., Martinac B., Craik D. J. (2009) J. Biol. Chem. 284, 20699–20707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Watts J. L., Shryock T. R., Apley M., Bade D. J., Brown S. D., Gray J. T., Heine H., Hunter R. P., Mevius D. J., Papich M. G., Silley P., Zurenko G. E. (2006) Performance Standards for Antimicrobial Disk and Dilution Susceptibility Test for Bacteria Isolated from Animals, 3rd Ed., Vol. 28, pp. 5–99, Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 31. Bignami G. S. (1993) Toxicon 31, 817–820 [DOI] [PubMed] [Google Scholar]
- 32. Vaughn J. L., Goodwin R. H., Tompkins G. J., McCawley P. (1977) In Vitro 13, 213–217 [DOI] [PubMed] [Google Scholar]
- 33. Bliss C. I. (1934) Science 79, 409–410 [DOI] [PubMed] [Google Scholar]
- 34. Jonson J. A., Wofford P. L., Gill R. F. (1995) J. Econ. Entomol. 88, 734–742 [Google Scholar]
- 35. Massatochi N., Kumar S. (2000) Molecular Evolution and Phylogenetics, pp. 73–83, Oxford University Press, New York [Google Scholar]
- 36. Tamura K., Dudley J., Nei M., Kumar S. (2007) Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- 37. Felsenstein J. (1985) Evolution 39, 783–791 [DOI] [PubMed] [Google Scholar]
- 38. Koltay A., Daly N. L., Gustafson K. R., Craik D. J. (2005) Int. J. Pept. Protein Res. Ther. 11, 99–106 [Google Scholar]
- 39. Eswar N., Webb B., Marti-Renom M. A., Madhusudhan M. S., Eramian D., Shen M. Y., Pieper U., Sali A. (2006) Curr. Protoc. Bioinformatics, Chap. 5, Unit 5.6, pp. 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wiederstein M., Sippl M. J. (2007) Nucleic Acids Res. 35, W407–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sumathi K., Ananthalakshmi P., Roshan M. N., Sekar K. (2006) Nucleic Acids Res. 34, W128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guex N., Peitsch M. C. (1997) Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 43. Peters B., Moad C., Youn E., Buffington K., Heiland R., Mooney S. (2006) BMC Struct Biol 6, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ritchie D. W. (2008) Curr. Protein Pept. Sci. 9, 1–15 [DOI] [PubMed] [Google Scholar]
- 45. Craik D. J., Daly N. L., Bond T., Waine C. (1999) J. Mol. Biol. 294, 1327–1336 [DOI] [PubMed] [Google Scholar]
- 46. Burman R., Svedlund E., Felth J., Hassan S., Herrmann A., Clark R. J., Craik D. J., Bohlin L., Claeson P., Göransson U., Gullbo J. (2010) Biopolymers 94, 626–634 [DOI] [PubMed] [Google Scholar]
- 47. Hoover D. M., Chertov O., Lubkowski J. (2001) J. Biol. Chem. 276, 39021–39026 [DOI] [PubMed] [Google Scholar]
- 48. Wang Z., Wang G. (2004) Nucleic Acids Res. 32, D590–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rossmann M., Schultz-Heienbrok R., Behlke J., Remmel N., Alings C., Sandhoff K., Saenger W., Maier T. (2008) Structure 16, 809–817 [DOI] [PubMed] [Google Scholar]
- 50. Marheineke K., Grünewald S., Christie W., Reiländer H. (1998) FEBS Lett. 441, 49–52 [DOI] [PubMed] [Google Scholar]
- 51. Shenkarev Z. O., Nadezhdin K. D., Sobol V. A., Sobol A. G., Skjeldal L., Arseniev A. S. (2006) FEBS J. 273, 2658–2672 [DOI] [PubMed] [Google Scholar]
- 52. Trabi M., Craik D. J. (2004) Plant Cell 16, 2204–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Seydel P., Gruber C. W., Craik D. J., Dörnenburg H. (2007) Appl. Microbiol. Biotechnol. 77, 275–284 [DOI] [PubMed] [Google Scholar]
- 54. Simonsen S. M., Sando L., Ireland D. C., Colgrave M. L., Bharathi R., Göransson U., Craik D. J. (2005) Plant Cell 17, 3176–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Plan M. R., Rosengren K. J., Sando L., Daly N. L., Craik D. J. (2010) Biopolymers 94, 647–658 [DOI] [PubMed] [Google Scholar]
- 56. Chen B., Colgrave M. L., Daly N. L., Rosengren K. J., Gustafson K. R., Craik D. J. (2005) J. Biol. Chem. 280, 22395–22405 [DOI] [PubMed] [Google Scholar]
- 57. Henriques S. T., Huang Y. H., Rosengren K. J., Franquelim H. G., Carvalho F. A., Johnson A., Sonza S., Tachedjian G., Castanho M. A., Daly N. L., Craik D. J. (2011) J. Biol. Chem. 286, 24231–24241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kamimori H., Hall K., Craik D. J., Aguilar M. I. (2005) Anal. Biochem. 337, 149–153 [DOI] [PubMed] [Google Scholar]
- 59. Jennings C. V., Rosengren K. J., Daly N. L., Plan M., Stevens J., Scanlon M. J., Waine C., Norman D. G., Anderson M. A., Craik D. J. (2005) Biochemistry 44, 851–860 [DOI] [PubMed] [Google Scholar]
- 60. Colgrave M. L., Huang Y. H., Craik D. J., Kotze A. C. (2010) Antimicrob. Agents Chemother. 54, 2160–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Craik D. J., Cemazar M., Daly N. L. (2006) Curr. Opin Drug Discov. Devel. 9, 251–260 [PubMed] [Google Scholar]
- 62. Rosengren K. J., Daly N. L., Plan M. R., Waine C., Craik D. J. (2003) J. Biol. Chem. 278, 8606–8616 [DOI] [PubMed] [Google Scholar]
- 63. Saether O., Craik D. J., Campbell I. D., Sletten K., Juul J., Norman D. G. (1995) Biochemistry 34, 4147–4158 [DOI] [PubMed] [Google Scholar]
- 64. Göransson U., Craik D. J. (2003) J. Biol. Chem. 278, 48188–48196 [DOI] [PubMed] [Google Scholar]
- 65. Daly N. L., Love S., Alewood P. F., Craik D. J. (1999) Biochemistry 38, 10606–10614 [DOI] [PubMed] [Google Scholar]
- 66. Bokesch H. R., Pannell L. K., Cochran P. K., Sowder R. C., 2nd, McKee T. C., Boyd M. R. (2001) J. Nat. Prod. 64, 249–250 [DOI] [PubMed] [Google Scholar]
- 67. Felizmenio-Quimio M. E., Daly N. L., Craik D. J. (2001) J. Biol. Chem. 276, 22875–22882 [DOI] [PubMed] [Google Scholar]
- 68. Heitz A., Hernandez J. F., Gagnon J., Hong T. T., Pham T. T., Nguyen T. M., Le-Nguyen D., Chiche L. (2001) Biochemistry 40, 7973–7983 [DOI] [PubMed] [Google Scholar]
- 69. Daly N. L., Clark R. J., Plan M. R., Craik D. J. (2006) Biochem. J. 393, 619–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ireland D. C., Wang C. K., Wilson J. A., Gustafson K. R., Craik D. J. (2008) Biopolymers 90, 51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.