Background: Protein-tyrosine kinase 6 (PTK6) is a non-receptor tyrosine kinase that is aberrantly expressed in several types of human cancer.
Results: PTK6 directly phosphorylates p130 CRK-associated substrate (p130CAS).
Conclusion: PTK6 promotes peripheral adhesion complex formation and prostate cancer cell migration by phosphorylating p130CAS and activating ERK5.
Significance: These studies define a novel PTK6-p130CAS-ERK5 signaling cascade in cancer cells.
Keywords: Adhesion, Migration, Scaffold Proteins, Signal Transduction, Tyrosine Protein Kinase (Tyrosine Kinase), BRK, ERK5, FAK, SRC, Sik
Abstract
Protein-tyrosine kinase 6 (PTK6) is a non-myristoylated intracellular tyrosine kinase evolutionarily related to Src kinases. Aberrant PTK6 expression and intracellular localization have been detected in human prostate tumors. In the PC3 prostate cancer cell line, the pool of endogenous activated PTK6, which is phosphorylated on tyrosine residue 342, is localized at the membrane. Expression of ectopic membrane-targeted PTK6 led to dramatic morphology changes and formation of peripheral adhesion complexes in PC3 cells. Peripheral adhesion complex formation was dependent upon PTK6 kinase activity. We demonstrated that p130 CRK-associated substrate (p130CAS) is a novel direct substrate of PTK6, and it works as a crucial adapter protein in inducing peripheral adhesion complexes. Activation of ERK5 downstream of p130CAS was indispensable for this process. Knockdown of endogenous PTK6 led to reduced cell migration and p130CAS phosphorylation, whereas knockdown of p130CAS attenuated oncogenic signaling induced by membrane-targeted PTK6, including ERK5 and AKT activation. Expression of membrane-targeted PTK6 promoted cell migration, which could be impaired by knockdown of p130CAS or ERK5. Our study reveals a novel function for PTK6 at the plasma membrane and suggests that the PTK6-p130CAS-ERK5 signaling cascade plays an important role in cancer cell migration and invasion.
Introduction
Protein-tyrosine kinase 6 (PTK6),2 also called breast tumor kinase (BRK) and Src-related intestinal kinase (Sik), was first identified in cultured human melanocytes (1) and later cloned from human breast tumor cells (2) and normal mouse epithelial cells (3, 4). PTK6 is structurally similar to Src family tyrosine kinases but lacks an N-terminal myristoylation consensus sequence, allowing it to localize to different cellular compartments, including the nucleus (5). PTK6 and Src share a similar optimal substrate sequence that includes the residues X−2(I/E)−1Y0(D/E)1(D/E)2, although Src prefers acidic amino acids at −3 and −4 positions, whereas PTK6 shows strong acidic preference at +1 and +2 positions (6–8). Several substrates are phosphorylated by both PTK6 and Src, including AKT (9), STAT3 (10), STAT5b (11), β-catenin (12), paxillin (13), and p190RhoGAP (14).
Up-regulated PTK6 expression has been detected in different types of cancers, including breast carcinomas (15–17), colon cancer (18), ovarian cancer (19), head and neck cancers (20), and metastatic melanoma cells (21). Aberrant translocation of PTK6 from the nucleus to the cytoplasm was observed in prostate tumors (22), and cytoplasmic retention of PTK6 promotes growth of prostate tumor cells (23). PTK6 is involved in different oncogenic signaling pathways that promote cancer cell proliferation and migration (9–11, 13, 14, 17, 24, 25).
p130 CRK-associated substrate (p130CAS) was first identified as a hyperphosphorylated protein in v-CRK- and v-Src-transformed cells (26, 27). The human gene encoding p130CAS, BCAR1 (breast cancer resistance 1), was identified in a retroviral insertion screen for genes that promote resistance to the antiestrogen tamoxifen (28). p130CAS is concentrated at focal adhesions (29). Following activation of integrin signaling, focal adhesion kinase (FAK) and Src phosphorylate p130CAS at several tyrosine residues, which provide binding sites for the adaptor protein CRK. This leads to activation of the small GTPase RAC, inducing membrane ruffling and cytoskeleton remodeling and promoting cell migration (30–32). p130CAS is essential for Src-induced transformation of primary fibroblasts (33). Overexpression of p130CAS in murine mammary tumor virus-HER2/Neu mice results in multifocal mammary tumors with significantly reduced latency (34). A human breast cancer study showed that tumor levels of p130CAS were inversely correlated with relapse-free survival and overall survival time (35). Higher p130CAS expression was also detected in metastatic prostate cancer compared with localized prostate lesions that correlated with EGF receptor expression (36).
Ectopic expression of active Src in KM12C cells induces the formation of peripheral adhesion complexes, which are focal adhesion-like structures. Vinculin, paxillin, FAK, and integrins were enriched in these discrete structures at the tips of membrane protrusions. Src-induced formation of peripheral adhesions relies on integrin αv and β1, FAK, and ERK1/2 signaling cascades (37, 38). Src is also a central mediator in forming podosome/invadopodium structures in a wide range of cells; these structures play critical roles in cell migration and invasion (39). Podosomes/invadopodia are dynamic, actin-rich adhesion structures that also share common molecular components with focal adhesions, including FAK, vinculin, and integrins (40).
We show here that p130CAS is a novel substrate of PTK6 that serves as an important adapter protein in the formation of peripheral adhesion complexes induced by membrane-targeted PTK6. Unlike Src, PTK6-induced peripheral adhesion complexes appeared to be FAK- and ERK1/2-independent. Instead, activation of the ERK5 signaling pathway downstream of p130CAS was crucial for the formation of peripheral adhesion complexes. Membrane-targeted PTK6 promoted cell migration through p130CAS and ERK5, whereas knockdown of endogenous PTK6 in PC3 cells impaired cell migration.
EXPERIMENTAL PROCEDURES
Antibodies
Anti-human PTK6 (C-18 and G-6), anti-mouse PTK6 (C-17), anti-FAK (C-20), anti-phosphotyrosine (PY20), anti-SP1 (PEP2), and anti-HER2/Neu (C-18) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-phosphotyrosine clone 4G10 and anti-P-PTK6 (Tyr-342) antibodies were purchased from Millipore (Bedford, MA). Antibodies directed against AKT, P-AKT (Thr-308), P-AKT (Ser-473), ERK1/2, P-ERK1/2 (Thr-202/Tyr-204), ERK5, P-ERK5 (Thr-218/220), P-p130CAS (Tyr-165), and Myc tag (9B11) were purchased from Cell Signaling Technology (Danvers, MA). Antibodies directed against paxillin, p130CAS, and β-catenin were purchased from BD Pharmingen. Antibodies directed against α-tubulin (T-9026), β-actin (AC-15), and vinculin were purchased from Sigma-Aldrich. Anti-rat P-130Cas (Tyr-762) antibody, which recognizes human P-p130CAS Tyr-664, was purchased from Abcam (Cambridge, MA), and anti-glutathione S-transferase (GST) tag antibody was purchased from Covance (Cumberland, VA). Donkey anti-rabbit or sheep anti-mouse antibodies conjugated to horseradish peroxidase were used as secondary antibodies (Amersham Biosciences) and detected by chemiluminescence with SuperSignal West Dura extended duration substrate from Pierce.
Plasmids and siRNAs
Myc-tagged full-length human wild type (WT), active (YF), and kinase-defective (kinase-dead mutant (KM)) PTK6 in the pcDNA3 vector as well as Myc-tagged NLS-PTK6 and Palm-PTK6 constructs in the pcDNA4-TO vector have been described previously (9, 12). Coding sequences from the pcDNA4-TO constructs were subcloned into the pBABE-puro vector (Cell Biolabs, Inc., San Diego, CA). The GST-PTK6 (mouse) constructs were described previously (9). The pEBG-p130CAS plasmid (plasmid 15001; Ref. 41) was obtained from Addgene (Cambridge, MA). The siRNA against p130CAS was purchased from Dharmacon (Lafayette, CO). The sequence has been reported previously (42): 5′-GGTCGACAGTGGTGTGTAT-3′. Dicer-substrate siRNAs against ERK5, FAK, and PTK6 were purchased from the Integrated DNA Technologies predesigned DsiRNA library (Coralville, IA). The sequence for Dsi-PTK6 is 5′-AGGTTCACAAATGTGGAGTGTCTGC-3′. The sequences for Dsi-ERK5 are 5′-GCAGCTATCTAAGTCACAGGTGGAG-3′and 5′-ACTAGTGCTCAGTGACAATGACAGA-3′. The sequence for Dsi-FAK is 5′-GCAATGGAGCGAGTATTAAAGGACT3′.
Cell Culture and Transfections
The human embryonic kidney cell line 293 (HEK-293) (ATCC CRL-1573), the mouse embryonic fibroblast cell line SYF (ATCC CRL-2459), the human prostate cancer cell lines PC3 (ATCC CRL-1435) and LNCaP (ATCC CRL-1740), and the human breast cancer cell lines T47D (ATCC HTB-133) and MDA-MB-231 (ATCC HTB-26) were cultured according to ATCC guidelines in the recommended medium. The benign prostatic hyperplasia epithelial cell line BPH1 (kindly provided by Dr. Simon Hayward, Vanderbilt University, Nashville, TN) was cultured in RPMI 1640 medium containing 5% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin (43). Transfections were performed using the Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer's instructions.
Protein Lysates and Fractionation
Transfected cells were rinsed twice with cold PBS and lysed in 1% Triton X-100 lysis buffer (1% Triton X-100, 20 mm HEPES, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 10 mm sodium pyrophosphate, 100 mm NaF, 5 mm iodoacetic acid, 0.2 mm phenylmethylsulfonyl fluoride (PMSF), protease inhibitor mixture (Roche Applied Science)) 18–24 h after transfection. Cell fractionation of prostate and breast cancer cell lines was performed using the ProteoExtract subcellular proteome extraction kit from Calbiochem according to the manufacturer's instructions. One-tenth volume of each fraction was subjected to SDS-PAGE and transferred onto Immobilon-P membranes (Millipore) for immunoblotting.
Immunoprecipitation and GST Pulldown Assay
Immunoprecipitations were performed with 500 μg of total cell lysates and 0.5 μg of specific antibodies under overnight incubation at 4 °C. 30 μl of protein A-Sepharose CL-4B beads (GE Healthcare) was added and incubated for 1 h after which the beads were washed four times in wash buffer (1% Triton X-100, 20 mm HEPES, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 10 mm sodium pyrophosphate). After removing the supernatant in the final wash, samples were resuspended in 30 μl of 2× reducing Laemmli sample buffer and boiled for 5 min. The proteins retained on the protein A beads were subjected to SDS-PAGE on 8% gels and transferred onto Immobilon-P membranes for immunoblotting. For the GST pulldown assay, GST fusion proteins were expressed in BL21 cells (Stratagene, La Jolla, CA). GST protein purification and the subsequent processes were performed as described previously (9).
Immunostaining
PC3 cells were seeded in 4- or 8-well chamber slides that were uncoated or coated with collagen I (Sigma-Aldrich). Cells were washed with PBS, fixed in Carnoy's solution (6:3:1 ethanol:chloroform:acetic acid), then blocked with 3% BSA for 1 h, and incubated with primary antibodies overnight. After washing, samples were incubated with biotinylated anti-rabbit or anti-mouse secondary antibodies and then incubated with fluorescein isothiocyanate (FITC)-conjugated avidin (Vector Laboratories, Burlingame, CA). For double staining, FITC-conjugated anti-mouse secondary antibodies (Sigma-Aldrich) were used to detect primary antibodies made in mouse (green), and biotinylated anti-rabbit secondary antibodies (Vector Laboratories) were used and then incubated with rhodamine-conjugated avidin to detect primary antibodies made in rabbit (red). Slides were mounted in Vectashield fluorescent mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories). Cells were then observed using standard UV, rhodamine, or FITC filters under 40× and 63× differential interference contrast oil immersion objectives using a Zeiss LSM 5 PASCAL confocal microscope. Images were obtained with an Axiocam HRc color digital camera and LSM 5 PASCAL software (Zeiss, Jena, Germany).
In Vitro Kinase Assays
Kinase assays were performed with 50 ng of recombinant human PTK6 (Invitrogen) and 1 μg of recombinant human p130CAS (Novus Biologicals, Littleton, CO) in 30 μl of kinase buffer (10 mm HEPES, pH 7.5, 150 mm NaCl, 2.5 mm dithiothreitol (DTT), 0.01% Triton X-100, 10 mm MnCl2) with or without 200 μm ATP for 10 min at 30 °C. The reaction was stopped by adding 30 μl of 2× reducing Laemmli sample buffer and boiling. Samples were subjected to SDS-PAGE and transferred onto Immobilon-P membranes for immunoblotting.
Reversed-phase Microcapillary/Tandem Mass Spectrometry (LC/MS/MS)
For all mass spectrometry (MS) experiments, p130CAS immunoprecipitates were separated using SDS-PAGE, the gel was stained with Coomassie Brilliant Blue R-250 (Bio-Rad) and destained, and the protein band was excised. Samples were subjected to reduction with 10 mm DTT for 30 min, alkylation with 55 mm iodoacetamide for 45 min, and in-gel digestion with modified trypsin or chymotrypsin overnight at pH 8.3 followed by LC/MS/MS. LC/MS/MS was performed using an EASY-nLC splitless nanoflow HPLC system (Proxeon Biosciences, Denmark) with a self-packed 75-μm-inner diameter × 15-cm C18 Picofrit column (New Objective, Woburn, MA) coupled to an LTQ-Orbitrap XL mass spectrometer (Thermo Scientific, San Jose, CA) in the data-dependent acquisition and positive ion mode at 300 nl/min with one full MS Fourier transform scan followed by six MS/MS spectra. MS/MS spectra collected via collision-induced dissociation in the ion trap were searched against the concatenated target and decoy (reversed) single entry Na+K+-ATPase and full Swiss-Prot protein databases using Sequest (Proteomics Browser Software, Thermo Scientific) with differential modifications for Ser/Thr/Tyr phosphorylation (+79.97) and the sample processing artifacts Met oxidation (+15.99), deamidation of Asn and Gln (+0.984), and Cys alkylation (+57.02). Phosphorylated and unphosphorylated peptide sequences were identified if they initially passed the following Sequest scoring thresholds against the target database: 1+ ions, Xcorr ≥ 2.0, Sf ≥ 0.4, p ≥ 5; 2+ ions, Xcorr ≥ 2.0, Sf ≥ 0.4, p ≥ 5; and 3+ ions, Xcorr ≥ 2.60, Sf ≥ 0.4, p ≥ 5 against the target protein database. Passing MS/MS spectra were manually inspected to be sure that all b- and y-fragment ions aligned with the assigned sequence and modification sites. Determination of the exact sites of phosphorylation was aided using FuzzyIons and GraphMod, and phosphorylation site maps were created using ProteinReport software (Proteomics Browser Software suite, Thermo Scientific). False discovery rates of peptide hits (phosphorylated and unphosphorylated) were estimated below 1.50% based on reversed database hits.
Titanium Dioxide (TiO2) Phosphopeptide Enrichment
Half of the digested peptide pool was reserved for enrichment with the Phos-trap phosphopeptide enrichment kit containing titanium dioxide (TiO2)-coated magnetic beads (PerkinElmer Life Sciences) according to the manufacturer's protocol. Briefly, peptide mixtures containing phosphopeptides were acidified with binding buffer and incubated with 20 μl of 20× TiO2 magnetic beads diluted in 180 μl of HPLC grade water with continuous shaking for 1 h at room temperature followed by washing three times with binding buffer and one time with washing buffer. Phosphopeptides were then incubated with 35 μl of basic elution buffer with continuous shaking. Elution buffer was then transferred to a 12 × 32-mm autosampler vial with 50 μl of HPLC A buffer, and the final solution was concentrated to 5 μl using a SpeedVac prior to injection via LC/MS/MS.
Retrovirus Production and Transduction
pBABE-puro plasmids were transfected into Phoenix Ampho cells using Lipofectamine 2000 (Invitrogen). Retrovirus was collected 48 and 72 h later. PC3 cells were infected with retrovirus at a multiplicity of infection of 500 for 24 h. Stable cell pools were selected in F-12K medium containing 10% fetal bovine serum (FBS) and 2 μg/ml puromycin for a week.
Migration Chamber Assays and Wound Healing Assays
PC3 cells were transfected with PTK6 siRNA or scrambled control siRNA for 24 h and then serum-starved for another 24 h. 5 × 104 cells were plated in the top chamber of a Transwell (24-well insert; pore size, 8 μm; Corning) and incubated with 1% FBS-containing F-12K medium. 20% FBS-containing F-12K medium was added to the lower chamber as a chemoattractant. After 18 h, cells that did not migrate through the pores were removed by a cotton swab, and the cells on the lower surface of the membrane were stained by crystal violet. For the wound healing assay, cells expressing vector or Palm-PTK6-YF were grown to confluence in 6-well plates and serum-starved for 24 h. Wounds were carefully made across the cell monolayer, and the medium was replaced by fresh complete F-12K medium. Cell migration was monitored for 48 h. Images were taken under the phase-contrast microscope using 10× magnification.
Statistics
For the analysis of the NCBI human genome microarray data set GDS2545, which contains 171 human prostate samples, results are shown as the mean ± S.E. For all the other cell studies, data represent the mean of at least three independent experiments ±S.D. p values were determined using the one-tailed Student's t test (Microsoft Excel 2010). A difference was considered statistically significant if the p value was equal to or less than 0.05. Quantitative analyses of immunoblots were performed using the NIH ImageJ program (44).
RESULTS
Membrane-targeted PTK6 Induces Formation of Peripheral Adhesion Complexes
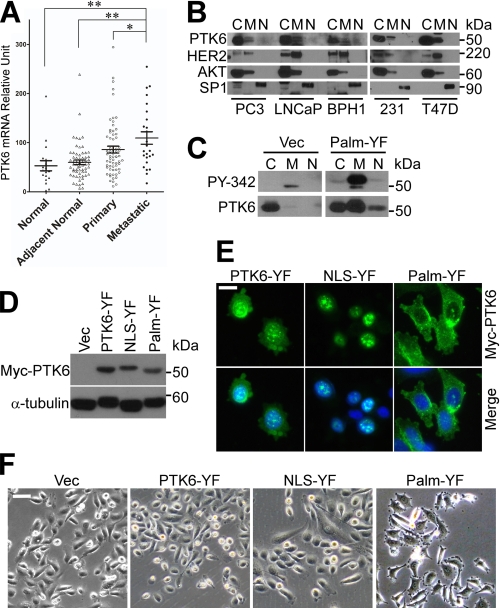
To investigate the role of PTK6 in prostate cancer, we compared PTK6 expression in normal human prostate tissue, normal tissue adjacent to the tumor, primary and metastatic prostate tumors using the NCBI human genome microarray data set GDS2545 (45). PTK6 mRNA levels were increased in metastatic prostate tumor samples (Fig. 1A), suggesting an oncogenic role for PTK6 in prostate cancer metastasis. Next, we examined the localization of PTK6 in two prostate cancer cell lines, PC3 (androgen receptor-negative) and LNCaP (androgen receptor-positive), and a benign prostatic hyperplasia cell line, BPH1 (43). Cells were fractionated into cytoplasmic, membrane/organelle, and nuclear compartments. In all three prostate cell lines, PTK6 was primarily localized in the cytoplasm and membrane compartments (Fig. 1B). Similar results were obtained in the MDA-MB-231 and T47D breast cancer cell lines (Fig. 1B).
FIGURE 1.
Membrane-targeted PTK6 induces formation of peripheral adhesion complexes. A, increased PTK6 mRNA expression was detected in human metastatic prostate cancer samples by analyzing the NCBI human genome microarray data set GDS2545 (*, p < 0.05; **, p < 0.01). Error bars represent standard error of the mean. B, intracellular localization of PTK6 was examined in prostate cells (PC3, LNCaP, and BPH1) and breast cancer cells (MDA-MB-231 and T47D). Cells were fractionated into cytoplasmic (C), membrane/organelle (M), and nuclear (N) compartments. AKT, HER2, and SP1 were used as loading controls for cytoplasmic, membrane/organelle, nuclear compartments, respectively. C, the membrane pool of PTK6 is the active pool. Total cytoplasmic, membrane/organelle, and nuclear PC3 cell lysates of PC3 cells were subjected to immunoblotting with PTK6 and phospho-PTK6 (PY-342) antibodies. D, immunoblot analysis of total cell lysates of PC3 cells stably expressing vector (Vec), PTK6-YF, NLS-PTK-YF, or Palm-PTK6-YF was performed using anti-Myc tag (PTK6) and -α-tubulin antibodies. E, intracellular localization of exogenous PTK6 was examined by indirect immunofluorescence with anti-Myc tag antibody. Myc-tagged PTK6 immunoreactivity was visualized with FITC (green). Cells were counterstained with DAPI (blue). The size bar denotes 20 μm. F, peripheral adhesion complexes were induced in Palm-PTK6-YF-expressing cells. Phase-contrast images of PC3 cells stably expressing vector, PTK6-YF, NLS-PTK-YF, or Palm-PTK6-YF are shown. The size bar denotes 50 μm.
A variety of data indicate that PTK6 may activate oncogenic signaling when localized at the membrane or within the cytoplasm, whereas it has a growth inhibitory effect when targeted to the nucleus (12, 23, 46, 47). This is probably due to its access to specific substrates in different intracellular compartments, but the detailed mechanisms are still unclear. Interestingly, although the majority of detectable PTK6 was cytoplasmic in PC3 cells, the activated pool of the endogenous kinase that is phosphorylated on tyrosine residue 342 was associated with the membrane (Fig. 1C, Vec). Phosphorylation of PTK6 at tyrosine 342 (Tyr(P)-342) is associated with its activation (48) and is detected with a PTK6 phosphospecific Tyr(P)-342 antibody.
To address the significance of PTK6 activation at the membrane, we targeted expression of different PTK6 isoforms to different cellular compartments in PC3 cells. A myristoylation/palmitoylation sequence (Palm) from the Src family kinase Lyn was added to the N terminus of PTK6 for membrane targeting. Mutation of the inhibitory tyrosine residue 447 to phenylalanine (YF) results in a constitutively active form of PTK6 (48). As determined by fractionation assays, exogenous Palm-PTK6-YF protein was at the membrane, but significant levels could also be detected in the cytoplasm perhaps due to its retention there by an as of yet unidentified factor in PC3 cells (23). Only the membrane pool of exogenous Palm-PTK6-YF was highly phosphorylated on tyrosine residue 342 (Fig. 1C). This is similar to the endogenous PTK6 expressed in the vector control cells (Fig. 1C). These data suggest that membrane localization of PTK6 is critical for its activation. In addition, an SV40 nuclear localization signal (NLS) was added to the N terminus of PTK6 for nuclear targeting.
Equivalent levels of ectopic Myc-tagged PTK6 were expressed in PC3 stable cell lines (Fig. 1D). Immunofluorescence using anti-Myc tag antibody showed that Palm-PTK6-YF was localized in membrane and cytoplasmic compartments, whereas NLS-PTK6-YF was in the nucleus. Non-targeted PTK6-YF was localized both in the cytoplasm and nucleus (Fig. 1E). Overexpression of Palm-PTK6-YF induced striking formation of focal adhesion-like structures around cell membrane (Fig. 1F), which are similar to the peripheral adhesion complexes induced by Src in KM12C cells (37). This was not observed in vector-, PTK6-YF-, or NLS-PTK6-YF-expressing cells, indicating that this phenotype is dependent on PTK6 membrane localization. In addition to the dramatic cell morphology change, these cells also showed a scattering phenotype with fewer cell-cell contacts (Fig. 1F).
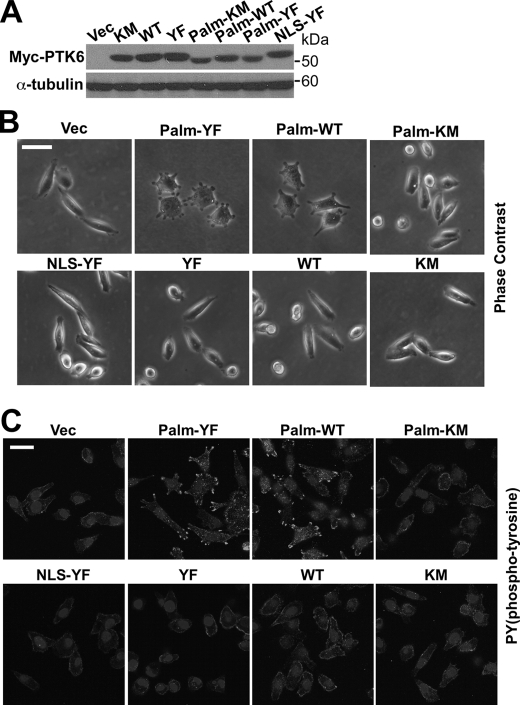
To determine whether formation of peripheral adhesion complexes is dependent on PTK6 kinase activity, cell lines stably expressing Palm-PTK6-WT, PTK6-WT, Palm-PTK6-KM, and PTK6-KM were constructed. All cell lines expressed comparable levels of ectopic PTK6 (Fig. 2A). Peripheral adhesion structures formed in PC3 cells expressing Palm-PTK6-YF and Palm-PTK6-WT but not kinase-dead Palm-PTK6-KM, indicating that formation of these adhesion structures depends on both PTK6 kinase activity and membrane localization (Fig. 2B). Because PTK6 is a tyrosine kinase, phosphotyrosine signaling was checked by immunofluorescence. Only Palm-PTK6-YF- and Palm-PTK6-WT-expressing PC3 cells showed highly induced phosphotyrosine signaling in peripheral adhesion complexes around the plasma membrane (Fig. 2C).
FIGURE 2.
Formation of peripheral adhesion complexes depends on PTK6 kinase activity and membrane localization. A, immunoblot analysis of total cell lysates of PC3 cells stably expressing vector (Vec), Myc-tagged PTK6-KM, PTK6-WT, PTK6-YF, Palm-PTK6-KM, Palm-PTK6-WT, Palm-PTK6-YF, or NLS-PTK-YF was performed with anti-Myc tag and α-tubulin antibodies. B, peripheral adhesion complexes only form in Palm-PTK6-YF- or Palm-PTK6-WT-expressing cells. Phase-contrast images of PC3 cells described in A are shown. The size bar denotes 50 μm. C, induced phosphotyrosine signaling in peripheral adhesion complexes was detected in Palm-PTK6-YF- or Palm-PTK6-WT-expressing cells. PC3 cells were stained with anti-phosphotyrosine antibodies and visualized with FITC, which labels the peripheral adhesion complexes. Cells were counterstained with DAPI (nuclei). The size bar denotes 50 μm.
Integrin and Growth Factor Receptor Signaling Promotes Peripheral Adhesion Complex Formation
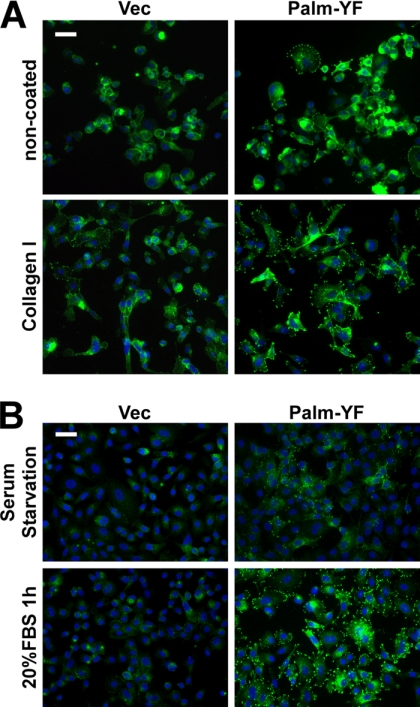
Integrins αv and β1 were reported to be essential for the formation of peripheral adhesion complexes induced by active Src in KM12C cells (37, 38). We next investigated the role of integrin signaling in PTK6-induced peripheral adhesion formation. Collagen I is a ligand of the integrin receptor and is often used to activate integrin signaling. Palm-PTK6-YF-expressing PC3 cells were able to form more peripheral adhesions on collagen I-coated chamber slides compared with non-coated chamber slides. Peripheral adhesions were visualized by anti-phosphotyrosine immunostaining (Fig. 3A). These data indicate that integrin receptors work upstream of PTK6, and activated integrin signaling promotes PTK6-induced peripheral adhesion formation.
FIGURE 3.
Integrin and growth factor signaling promote formation of peripheral adhesion complexes. A, Palm-PTK6-YF-expressing PC3 cells form more peripheral adhesions on collagen I-coated plates than on non-coated plates. PC3 cells stably expressing Palm-PTK6-YF or vector (Vec) were seeded in collagen I-coated or non-coated chamber slides for 24 h. Cells were stained with anti-phosphotyrosine antibodies (green) and counterstained with DAPI (blue). The size bar denotes 50 μm. B, 1-h 20% FBS stimulation after 24-h serum starvation promotes the formation of peripheral adhesion complexes in Palm-PTK6-YF-expressing PC3 cells. Peripheral adhesion complexes were visualized by anti-phosphotyrosine immunostaining (green). Cells were counterstained with DAPI (blue). The size bar denotes 50 μm.
PTK6 is involved with several growth factor receptors, including EGF receptor, HER2, IGF-R1, and MET (17, 24, 25, 49). Therefore, we checked whether growth factor stimulation is able to promote the formation of peripheral adhesions. Palm-PTK6-YF-expressing cells showed diminished phosphotyrosine signaling in peripheral adhesion complexes after 24-h serum starvation that could be reactivated following 1-h 20% FBS stimulation (Fig. 3B), indicating that growth factor receptors work upstream of PTK6 in regulating peripheral adhesion formation.
Phospho-p130CAS Is Enriched in Peripheral Adhesion Complexes
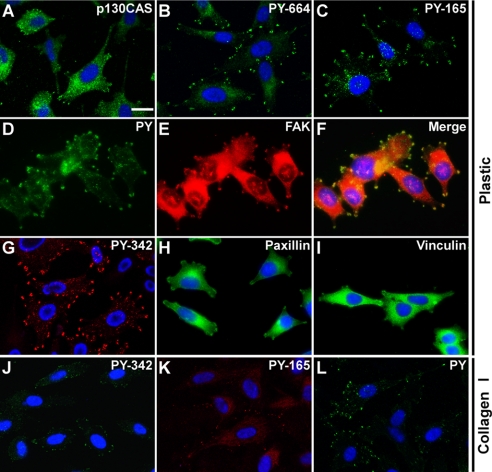
To examine components of Palm-PTK6-YF-induced peripheral adhesion complexes, PC3 cells stably expressing Palm-PTK6-YF were stained with antibodies against proteins found in focal adhesion complexes, including p130CAS, vinculin, paxillin, and FAK (29, 37). As evident by immunofluorescence, p130CAS, vinculin, paxillin, and FAK were detected in adhesion complexes induced by Palm-PTK6-YF (Fig. 4, A, E, H, and I). Tyrosine phosphorylated p130CAS was detected by specific antibodies against tyrosine residues 165 in the substrate domain and 664 in the C-terminal domain of p130CAS. Although p130CAS was mainly localized in the cytoplasm, phospho-p130CAS Tyr(P)-165 and Tyr(P)-664 were highly enriched in the peripheral adhesion complexes (Fig. 4, A–C). Palm-PTK6-YF-induced phosphotyrosine signaling colocalized with FAK in peripheral adhesion complexes (Fig. 4, D–F). Although exogenous Palm-PTK6-YF was in both the membrane and cytoplasmic compartments (Fig. 1E), phospho-PTK6 (Tyr(P)-342) was concentrated in peripheral adhesion complexes (Fig. 4G), suggesting that some components of peripheral adhesion complexes are important for PTK6 activation. Although it has been reported that Src-induced formation of peripheral adhesion complexes depends on Src-induced tyrosine phosphorylation of FAK (37), knockdown of FAK in PC3 cells did not affect formation of peripheral adhesion complexes induced by Palm-PTK6-YF (supplemental Fig. S1).
FIGURE 4.
Phospho-p130CAS, vinculin, paxillin, and FAK are enriched in peripheral adhesion complexes. A–I, indirect immunofluorescence of PC3 cells stably expressing Palm-PTK6-YF was performed using anti-p130CAS (A), phospho-p130CAS (PY-664) (B), phospho-p130CAS (PY-165) (C), phosphotyrosine (PY) (D), FAK (E), phospho-PTK6 (PY-342) (G), paxillin (H), and vinculin (I) antibodies. F is a merge of D and E. J–L, PC3 cells expressing vector controls were grown at collagen I-coated chamber slides, and indirect immunofluorescence was performed using anti-phospho-PTK6 (PY-342) (J), phospho-p130CAS (PY-165) (K), phosphotyrosine (PY) (L) antibodies. Cells were counterstained with DAPI (blue). The size bar denotes 20 μm.
To test whether endogenous PTK6 is involved in focal adhesion formation, collagen I-coated plates were utilized to activate integrin signaling because the fluorescence intensity on non-coated chamber slides is too weak to distinguish focal adhesions. Phosphotyrosine signaling, endogenous activated PTK6 (Tyr(P)-342), and phospho-p130CAS (Tyr(P)-165) were detected in focal adhesions induced by collagen I (Fig. 4, J–L).
p130CAS Is a Direct Substrate of PTK6
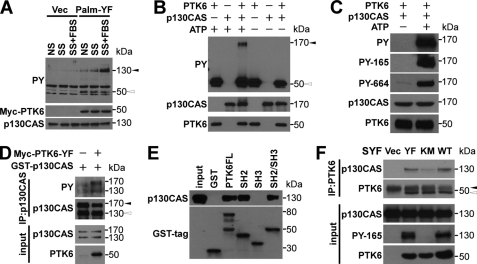
To identify the potential targets of PTK6 that are involved in peripheral adhesion complex formation, anti-phosphotyrosine immunoblotting was performed using vector- or Palm-PTK6-YF-expressing PC3 cell lysates. Tyrosine phosphorylation of a protein around 130 kDa was higher under normal serum, serum starvation, or FBS stimulation in Palm-PTK6-YF-expressing cells compared with vector control cells (Fig. 5A), suggesting that the 130-kDa protein might be a preferred substrate of PTK6. Because p130CAS was reported as a highly tyrosine phosphorylated protein (27) and phospho-p130CAS was induced in peripheral adhesion complexes induced by Palm-PTK6-YF (Fig. 4, B and C), we examined the possibility that p130CAS is a direct substrate of PTK6. An in vitro kinase assay was performed with recombinant human PTK6 and p130CAS protein. Immunoblotting using anti-phosphotyrosine antibody showed that p130CAS could be directly phosphorylated by PTK6 in the presence of ATP (Fig. 5B). Tyrosine residues 165 and 664 of p130CAS were both phosphorylated by PTK6 in vitro (Fig. 5C).
FIGURE 5.
PTK6 directly phosphorylates p130CAS. A, increased tyrosine phosphorylation of a 130-kDa protein was observed in the presence of Palm-PTK6-YF. PC3 cells stably expressing Palm-PTK6-YF or vector (Vec) were grown in normal serum (NS), serum-starved for 48 h (SS), or serum-starved and then stimulated with 20% FBS for an hour (SS+FBS). Lysates were analyzed by immunoblotting with anti-phosphotyrosine (PY), Myc tag, and p130CAS antibodies. The black arrowhead points at a 130-kDa band, and the white arrowhead points at a 50-kDa band (phospho-Palm-PTK6). B, PTK6 directly phosphorylates p130CAS in an in vitro kinase assay. Recombinant human PTK6 and human p130CAS were incubated in kinase buffer with or without ATP for 10 min at 30 °C. Immunoblot analysis was performed using anti-phosphotyrosine, p130CAS, and PTK6 antibodies. The black arrowhead points at phospho-p130CAS, and the white arrowhead points at phospho-PTK6. C, immunoblot analysis of an in vitro kinase assay as described in B was performed using anti-phosphotyrosine (PY), phospho-p130CAS (PY-165), phospho-p130CAS (PY-664), p130CAS, and PTK6 antibodies. D, tyrosine phosphorylation of both endogenous p130CAS and exogenous GST-p130CAS was induced in the presence of active PTK6 in HEK-293 cells. p130CAS was immunoprecipitated from lysates of HEK-293 cells co-expressing GST-p130CAS and PTK6-YF. p130CAS tyrosine phosphorylation was analyzed by immunoblotting with anti-phosphotyrosine and p130CAS antibodies. The black arrowhead points at exogenous GST-p130CAS, and the white arrowhead points at endogenous p130CAS. E, PTK6 interacts with p130CAS through its SH2 domain. Glutathione-Sepharose CL-4B beads binding with GST fusion proteins were used to pull down p130CAS from HEK-293 cell lysates. Bound p130CAS was analyzed by immunoblotting with anti-p130CAS antibody. 10% of the lysates added to the pulldown reaction served as input. F, interaction between p130CAS and PTK6 depends on tyrosine phosphorylation of p130CAS. PTK6 was immunoprecipitated from lysates of SYF cells expressing vector, PTK6-YF, PTK6-KM, or PTK6-WT. p130CAS association was analyzed by immunoblotting with anti-p130CAS and PTK6 antibodies. The black arrowhead points at the band of IgG heavy chain, and the white arrowhead points at immunoprecipitated PTK6. SYF cell lysates were analyzed by immunoblotting with anti-p130CAS, phospho-p130CAS (PY-165), and PTK6 antibodies as input. IP, immunoprecipitation.
We transfected a GST-p130CAS plasmid with and without the PTK6-YF expression construct to examine the ability of PTK6 to phosphorylate p130CAS in vivo in HEK-293 cells. Immunoprecipitation of p130CAS protein followed by anti-phosphotyrosine immunoblotting showed that tyrosine phosphorylation of both endogenous (bottom band) and exogenous (top band) p130CAS was induced by PTK6-YF (Fig. 5D). A mobility shift was observed for the phospho-p130CAS band in the presence of PTK6, suggesting that PTK6 phosphorylates p130CAS at multiple residues. This is supported by LC/MS/MS of phospho-p130CAS from an in vitro kinase assay performed with purified proteins that showed that 11 tyrosine residues in the substrate domain are phosphorylated by PTK6 (supplemental Fig. S2).
We performed pulldown assays with purified GST-PTK6 fusion proteins to identify the domains of PTK6 involved in its interactions with p130CAS (50). All PTK6 SH2 domain-containing fusion proteins (GST-PTK6FL, GST-SH2, and GST-SH2+SH3), but not the SH3 domain alone, were able to pull down p130CAS from HEK-293 cell lysates, suggesting that the PTK6 SH2 domain interacts with p130CAS (Fig. 5E). We next investigated the interaction of PTK6 and p130CAS in SYF cells (Src−/−, Yes−/−, Fyn−/− mouse embryonic fibroblasts). SYF cells were utilized to avoid interference of Src family kinases, which also phosphorylate p130CAS. Overexpression of PTK6-WT or PTK6-YF induced the phosphorylation of p130CAS on tyrosine residue 165 (Fig. 5F). Immunoprecipitation of PTK6 followed by anti-p130CAS immunoblotting showed that p130CAS interacted with wild type PTK6 and constitutively active PTK6-YF, whereas the interaction between p130CAS and kinase-dead PTK6-KM was markedly attenuated (Fig. 5F). Although PTK6-KM is kinase-defective, p130CAS might be phosphorylated by other tyrosine kinases, including FAK and PYK2 (30, 51), facilitating PTK6-KM/p130CAS interactions. These data suggest that PTK6 kinase activity is important for the interaction between p130CAS and PTK6.
p130CAS Mediates Oncogenic Signaling Induced by Membrane-targeted PTK6
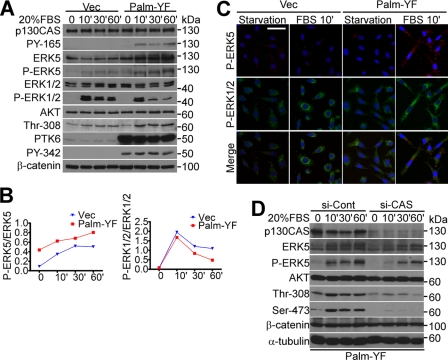
ERK1/2 signaling is critical for the assembly of Src-induced peripheral adhesion complexes in colon cells (38). PTK6 has been coupled with ERK5 in regulating breast cancer cell migration following MET receptor activation (49, 52). Therefore, we examined whether MAPK/ERK signaling is induced by Palm-PTK6-YF in PC3 cells. Following 20% FBS stimulation, increased phosphorylation of p130CAS (Tyr-165) was detected in Palm-PTK6-YF-expressing PC3 cells as well as increased ERK5 activation (Fig. 6, A and B). Total ERK5 levels were also up-regulated. However, ERK1/2 activation upon FBS stimulation was lower in the Palm-PTK6-YF-expressing cells (Fig. 6, A and B) perhaps due to a compensatory effect among MAPK family members (49). Immunofluorescence staining showed induction of phospho-ERK5 but not phospho-ERK1/2 in Palm-PTK6-YF-induced peripheral adhesion complexes after FBS stimulation (Fig. 6C). Immunoblotting also showed higher AKT activation upon FBS stimulation in Palm-PTK6-YF-expressing cells (Fig. 6A). Based on these data, we hypothesized that membrane-targeted PTK6 phosphorylates the scaffold protein p130CAS, creating multiple docking sites for interacting proteins and thereby promoting MAPK/ERK5 and AKT signaling in peripheral adhesion complexes. To test this, p130CAS was knocked down by siRNAs in Palm-PTK6-YF-expressing PC3 cells (Fig. 6D). Both AKT and ERK5 signaling cascades induced by Palm-PTK6-YF were attenuated after p130CAS knockdown (Fig. 6D). These data supported our hypothesis that p130CAS works as a scaffold protein to convey the downstream signaling induced by membrane-targeted PTK6.
FIGURE 6.
p130CAS mediates oncogenic signaling induced by Palm-PTK6-YF. A, increased p130CAS tyrosine phosphorylation and ERK5 and AKT activation were detected in Palm-PTK6-YF-expressing PC3 cells following FBS stimulation. PC3 cells stably expressing PTK6-Palm-YF or vector (Vec) were serum-starved for 48 h and stimulated with 20% FBS for 10, 30, or 60 min. Immunoblotting was performed with anti-p130CAS, phospho-p130CAS (PY-165), ERK5, phospho-ERK5, ERK1/2, phospho-ERK1/2, AKT, phospho-AKT (Thr-308), PTK6, phospho-PTK6 (PY-342), and β-catenin antibodies. B, increased ERK5 activation correlates with decreased ERK1/2 activation upon FBS stimulation in Palm-PTK6-YF-expressing cells. Relative band densities from A were quantified with NIH ImageJ software (44). The density of phospho-ERK5 and phospho-ERK1/2 was normalized by the density of total ERK5 and total ERK1/2, respectively. C, increased ERK5 activation in Palm-PTK6-YF-expressing PC3 cells upon FBS stimulation was detected by indirect immunofluorescence. PC3 cells expressing vector or Palm-PTK6-YF were serum-starved for 48 h and stimulated with 20% FBS for 10 min. Phospho-ERK1/2 immunoreactivity was visualized with FITC (green), whereas phospho-ERK5 was detected with rhodamine (red). Cells were counterstained with DAPI (blue). The size bar denotes 50 μm. D, ERK5 and AKT activation was diminished upon knockdown of p130CAS. PTK6-Palm-YF-expressing PC3 cells were transfected with p130CAS siRNAs (si-CAS) or control siRNAs (si-Cont) for 24 h, then-serum starved for 48 h, and stimulated by 20% FBS for 10, 30, or 60 min. Total cell lysates were analyzed by immunoblotting with anti-p130CAS, ERK5, phospho-ERK5, AKT, phospho-AKT (Thr-308 and Ser-473), β-catenin, and α-tubulin antibodies.
p130CAS and ERK5 Are Essential for Formation of Peripheral Adhesion Complexes
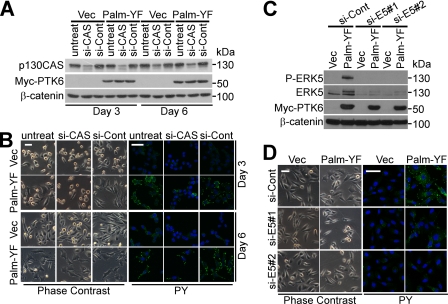
To investigate roles for p130CAS in the formation of peripheral adhesion complexes, p130CAS-targeted siRNAs were used to knock down p130CAS expression in PC3 cells. p130CAS protein decreased to 20% of the level of control cells 3 days after transfection (Fig. 7A). Formation of peripheral adhesion complexes induced by Palm-PTK6-YF was disrupted upon p130CAS knockdown, and the cells become round shaped compared with fully spread control cells with protrusions (Fig. 7B). Induced phosphotyrosine signaling in peripheral adhesion complexes also dramatically decreased upon p130CAS knockdown as evident by anti-phosphotyrosine immunofluorescence (Fig. 7B). Interestingly, 6 days after siRNA transfection, p130CAS protein was re-expressed to 50% of the level of control cells (Fig. 7A), which restored the ability of Palm-PTK6-YF-expressing cells to form peripheral adhesion complexes (Fig. 7B); phosphotyrosine signaling was again induced (Fig. 7B). These data indicate that formation of peripheral adhesion complexes is reversible, relying on the scaffold protein p130CAS.
FIGURE 7.
p130CAS and ERK5 are crucial for forming peripheral adhesion complexes. A, p130CAS protein is knocked down by siRNAs at day 3 and re-expressed at day 6. PC3 cells were transfected with p130CAS siRNAs or control siRNAs and harvested at 3 or 6 days. Total cell lysates were analyzed by immunoblotting with anti-p130CAS, Myc tag (PTK6), and β-catenin antibodies. B, Palm-PTK6-YF-induced peripheral adhesion complexes are disrupted upon p130CAS knockdown (si-CAS) and reassembled once p130CAS is re-expressed at day 6. Phase-contrast images and indirect immunofluorescence using anti-phosphotyrosine (PY) antibodies are shown. Size bar denotes 50 μm. C, ERK5 protein is knocked down by two ERK5 siRNAs. PC3 cells were transfected with ERK5 siRNAs (si-E5) or control siRNAs (si-Cont) and harvested at 3 days. Total cell lysates were analyzed by immunoblotting with anti-ERK5, phospho-ERK5, Myc tag, and β-catenin antibodies. D, peripheral adhesion complexes are disrupted upon ERK5 knockdown. Phase-contrast images and indirect immunofluorescence using anti-phosphotyrosine antibodies are shown. Size bars denote 50 μm. Vec, vector.
Because Palm-PTK6-YF expression led to activation of ERK5 in peripheral adhesion complexes (Fig. 6), we examined whether ERK5 signaling is required for the formation of peripheral adhesion complexes. ERK5 was knocked down by two different siRNAs (Fig. 7C). Two ERK5 bands were observed in the control siRNA-treated Palm-PTK6-YF-expressing cells of which the top band co-migrated with the phospho-ERK5 band shown in the anti-P-ERK5 immunoblotting (Fig. 7C). Knockdown of ERK5 with either siRNA abolished peripheral adhesion complex formation as well as phosphotyrosine signaling induced by Palm-PTK6-YF (Fig. 7D), suggesting that ERK5 is also crucial for peripheral adhesion complex formation.
PTK6 Promotes Cell Migration through p130CAS and ERK5 Signaling
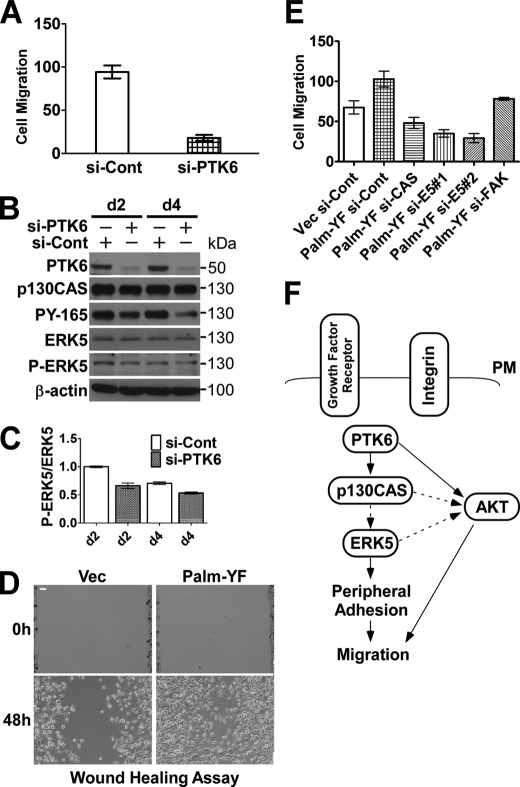
The p130CAS-CRK signaling complex plays important roles in activating RAC and promoting RAC-driven cell migration and invasion (32). PTK6 has been shown to be coupled with ERK5 in regulating MET-mediated cell migration (49, 53). We examined whether PTK6 promotes cell migration through phosphorylation of p130CAS and activation of ERK5. Endogenous PTK6 was knocked down in PC3 cells using siRNA against PTK6, and the effect of knockdown lasted up to 4 days after transfection (Fig. 8B). In Transwell migration assays, the ability of PC3 cells to migrate to the bottom side of the membrane was largely impaired after PTK6 knockdown (Fig. 8A). This was associated with a substantial reduction of p130CAS phosphorylation and a modest decrease in ERK5 phosphorylation in PTK6 knockdown cells (Fig. 8B). There was an ∼30% decrease in the phospho-ERK5/ERK5 ratio in PTK6 knockdown cells at days 2 and 4, respectively (Fig. 8C).
FIGURE 8.
PTK6 promotes cell migration through p130CAS and ERK5 signaling. A, PC3 cell migration is diminished upon PTK6 knockdown in Transwell chamber assays using 20% FBS as a chemoattractant. B, knockdown of PTK6 by siRNA in PC3 cells lasts 4 days. Cells were transfected with control siRNA (si-Cont) or PTK6 siRNA for 24 h and harvested at 2 (d2) or 4 (d4) days after transfection. Total cell lysates were analyzed by immunoblotting with anti-PTK6, p130CAS, phospho-p130CAS (PY-165), ERK5, P-ERK5, and β-actin antibodies. C, knockdown of PTK6 results in decreased ERK5 phosphorylation in PC3 cells. Relative band densities from B were quantified with NIH ImageJ software (44). Data were collected from two individual experiments, and S.D. is shown. D, PC3 cells stably expressing Palm-PTK6-YF show increased migration in wound healing assays. Representative images at 0 and 48 h are shown. Size bars denote 50 μm. E, membrane-targeted PTK6 promotes cell migration through phosphorylation of p130CAS and activation of ERK5. PC3 cells stably expressing Palm-PTK6-YF show greater cell migratory ability than control cells in Transwell chamber assays using 20% FBS as a chemoattractant. Knockdown of FAK causes a modest decrease, whereas knockdown of p130CAS or ERK5 (E5) results in a substantial reduction of cell migration induced by Palm-PTK6-YF. Error bars in A, C, and E represent standard deviation. F, a proposed model shows how PTK6 regulates peripheral adhesion complex formation and promotes migration. Integrin and growth factor receptor receptors are upstream of PTK6. PTK6 phosphorylates p130CAS near the plasma membrane (PM) and then activates ERK5 signaling, inducing formation of peripheral adhesion complexes and promoting migration. PTK6 has been shown to directly target AKT and promote AKT activation (9). Palm-PTK6-YF-expressing PC3 cells show increased AKT activation upon FBS stimulation that can be impaired by knockdown of p130CAS. AKT was reported to promote migratory and invasive ability of squamous carcinoma cells (62). Solid line arrows indicate direct regulation, whereas dotted line arrows represent proposed or indirect regulation. Vec, vector.
We examined the migratory ability of Palm-PTK6-YF-expressing PC3 cells, which formed peripheral adhesion complexes where phosphorylated p130CAS and ERK5 were induced (Figs. 4 and 6). In wound healing assays, Palm-PTK6-YF-expressing cells were able to close the wound at a much faster rate than vector control-expressing cells (Fig. 8D). In Transwell chamber assays, these cells also showed a 60% increase in the number of migrating cells when 20% FBS-containing medium was used as a chemoattractant (Fig. 8E). In Palm-PTK6-YF-expressing PC3 cells, knockdown of FAK caused a small but significant reduction of cell migration, whereas knockdown of p130CAS or ERK5 largely impaired cell migratory ability (Fig. 8E), indicating the essential roles of p130CAS and ERK5 in PTK6-mediated cell migration. The oncogenic signaling induced by PTK6, which is critical for cell migration, is more dependent on p130CAS and ERK5 than on FAK.
DISCUSSION
We identified p130CAS as a novel substrate of PTK6. Using phosphorylation-specific antibodies, we showed that PTK6 phosphorylated p130CAS within its substrate domain, which contains several YXXP motifs as well at the C-terminal Y664DYVHL motif (Fig. 5C). Using mass spectrometry, we identified 11 tyrosine residues within the p130CAS substrate domain that can be phosphorylated by PTK6 in vitro. Although no phosphorylation of the YDYVHL motif was detected using LC/MS/MS probably due to low abundance of YDYVHL-containing peptides, we did detect phosphorylation of this motif with a phosphospecific antibody (Fig. 5C and supplemental Fig. S2). In the canonical model, p130CAS is phosphorylated by FAK at the YDYVHL motif upon integrin activation. Then Src kinase binds to the pYDpYVHL motif (where pY is phosphotyrosine) through its SH2 domain and further phosphorylates the p130CAS substrate domain, creating binding sites for CRK (30). Knockdown of FAK expression did not affect formation of peripheral adhesion complexes induced by Palm-PTK6-YF (supplemental Fig. S1), and we hypothesize that PTK6 does not rely on FAK to initiate the phosphorylation at the p130CAS C-terminal YDYVHL motif. PTK6 might directly target the YDYVHL motif and then further phosphorylate the substrate domain, activating signaling downstream of p130CAS.
We showed that expression of membrane-targeted PTK6 induced the formation of peripheral adhesion complexes at the cell periphery, and this depended on PTK6 kinase activity and membrane localization (Figs. 1 and 2). We did not observe the rosette ring structure of F-actin bundles in those peripheral structures by rhodamine-conjugated phalloidin staining that is commonly observed in Src-induced podosomes/invadopodia (54), suggesting that this is a different adhesion structure.
PTK6 shares only 44% amino acid identity with Src (55). The structures of the PTK6 SH2 and SH3 domains have unique features that distinguish it from Src family kinases and may modulate its recognition of interacting proteins and substrates (56, 57). Although the Src SH3 domain is important for its interaction with and phosphorylation of p130CAS (58), PTK6 interacts with p130CAS through its SH2 domain (Fig. 5E). Differential phosphorylation of p130CAS tyrosine residues and distinct domain/domain interactions may lead to the formation of distinct scaffolding complexes at the plasma membrane.
Activating integrin signaling by collagen I was able to promote the formation of peripheral adhesion complexes induced by Palm-PTK6-YF, suggesting that integrin receptors are upstream of PTK6 (Fig. 3). In addition, growth factor receptors are also involved, probably by activating PTK6 activity upon ligand binding (Fig. 3). PTK6 participates in different growth factor receptor signaling pathways, including EGF receptor, HER2, IGF-R1, and MET (17, 24, 25, 49). It will be of great interest to further identify specific integrin receptors and growth factor receptors that are involved in PTK6-induced peripheral adhesion formation.
Like Src-induced peripheral adhesions in KM12C cells (37), we have shown that peripheral adhesion complexes induced by Palm-PTK6-YF contain paxillin, vinculin, and FAK (Fig. 4). FAK and ERK1/2 signaling cascades are important mediators downstream of Src (37, 38). However, here we did not detect roles for FAK or p42/p44 ERK1/2 (supplemental Fig. S1 and Fig. 6, A and C). Instead, we found that p130CAS and ERK5 serve as important regulators of peripheral adhesion complex formation induced by Palm-PTK6-YF. Phospho-p130CAS and phospho-ERK5 were enriched in peripheral adhesions, and knockdown of either protein disrupted the formation of peripheral adhesion complexes (Figs. 6 and 7). Knockdown of p130CAS in PC3 cells attenuated ERK5 activation stimulated by FBS, indicating that ERK5 works downstream of p130CAS (Fig. 6D).
We previously reported that PTK6 directly phosphorylates AKT at two tyrosine residues, 315 and 326, promoting AKT activation in response to epidermal growth factor (9). Consistent with this, overexpression of Palm-PTK6-YF in PC3 cells promoted serum-induced AKT activation (Fig. 6A). Knockdown of p130CAS protein largely impaired serum-induced AKT activation (Fig. 6D), suggesting that intact p130CAS scaffold complexes are important for activation of AKT signaling. It has been reported that ERK5 is required for AKT activation and VEGF-mediated survival of microvascular endothelial cells (59). Activated ERK5 signaling was also found to promote survival of fibroblasts via AKT-dependent inhibition of FoxO3a (60).
Prostate cancer is the second most common cancer and second leading cause of cancer-related deaths in American men (61). Most prostate cancer-related deaths are due to advanced disease resulting from lymphatic, blood, or contiguous local spread. Here we have shown that overexpression of membrane-targeted PTK6 induced the formation of peripheral adhesion complexes and increased migration in prostate cancer cells. This requires p130CAS and ERK5 signaling because knockdown of p130CAS or ERK5 impaired cell migration induced by membrane-targeted PTK6 (Fig. 8E). Increased AKT signaling might also contribute to cell migration (Fig. 6A) because AKT has been reported to regulate the epithelial to mesenchymal transition in squamous cell carcinoma lines and promote migration and invasion (62). In contrast, knockdown of PTK6 reduced PC3 cell migration, and this was accompanied by decreased p130CAS phosphorylation and ERK5 activation (Fig. 8, A and B). Our studies suggest that aberrant expression and relocalization of PTK6 in prostate cancer cells may stimulate multiple oncogenic signaling pathways, including p130CAS, ERK5, and AKT, and promotes peripheral adhesion formation and cell migration (Fig. 8F). Therefore, PTK6 might be a beneficial target as part of a therapeutic regimen to treat prostate cancer.
Supplementary Material
Acknowledgments
We thank Dr. Jessica J. Gierut for sharing unpublished data and helpful discussions and Priya Mathur for helpful comments.
This work was supported, in whole or in part, by National Institutes of Health Grants DK044525 and DK068503 (to A. L. T.).

This article contains supplemental Figs. S1 and S2.
- PTK6
- protein-tyrosine kinase 6
- CAS
- CRK-associated substrate
- FAK
- focal adhesion kinase
- NLS
- nuclear localization signal
- SH
- Src homology.
REFERENCES
- 1. Lee S. T., Strunk K. M., Spritz R. A. (1993) Oncogene 8, 3403–3410 [PubMed] [Google Scholar]
- 2. Mitchell P. J., Barker K. T., Martindale J. E., Kamalati T., Lowe P. N., Page M. J., Gusterson B. A., Crompton M. R. (1994) Oncogene 9, 2383–2390 [PubMed] [Google Scholar]
- 3. Siyanova E. Y., Serfas M. S., Mazo I. A., Tyner A. L. (1994) Oncogene 9, 2053–2057 [PubMed] [Google Scholar]
- 4. Vasioukhin V., Serfas M. S., Siyanova E. Y., Polonskaia M., Costigan V. J., Liu B., Thomason A., Tyner A. L. (1995) Oncogene 10, 349–357 [PubMed] [Google Scholar]
- 5. Derry J. J., Richard S., Valderrama Carvajal H., Ye X., Vasioukhin V., Cochrane A. W., Chen T., Tyner A. L. (2000) Mol. Cell. Biol. 20, 6114–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shin D. S., Kim Y. G., Kim E. M., Kim M., Park H. Y., Kim J. H., Lee B. S., Kim B. G., Lee Y. S. (2008) J. Comb. Chem. 10, 20–23 [DOI] [PubMed] [Google Scholar]
- 7. Kreegipuu A., Blom N., Brunak S., Järv J. (1998) FEBS Lett. 430, 45–50 [DOI] [PubMed] [Google Scholar]
- 8. Songyang Z., Cantley L. C. (1995) Trends Biochem. Sci. 20, 470–475 [DOI] [PubMed] [Google Scholar]
- 9. Zheng Y., Peng M., Wang Z., Asara J. M., Tyner A. L. (2010) Mol. Cell. Biol. 30, 4280–4292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu L., Gao Y., Qiu H., Miller W. T., Poli V., Reich N. C. (2006) Oncogene 25, 4904–4912 [DOI] [PubMed] [Google Scholar]
- 11. Weaver A. M., Silva C. M. (2007) Breast Cancer Res. 9, R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Palka-Hamblin H. L., Gierut J. J., Bie W., Brauer P. M., Zheng Y., Asara J. M., Tyner A. L. (2010) J. Cell Sci. 123, 236–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen H. Y., Shen C. H., Tsai Y. T., Lin F. C., Huang Y. P., Chen R. H. (2004) Mol. Cell. Biol. 24, 10558–10572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shen C. H., Chen H. Y., Lin M. S., Li F. Y., Chang C. C., Kuo M. L., Settleman J., Chen R. H. (2008) Cancer Res. 68, 7779–7787 [DOI] [PubMed] [Google Scholar]
- 15. Barker K. T., Jackson L. E., Crompton M. R. (1997) Oncogene 15, 799–805 [DOI] [PubMed] [Google Scholar]
- 16. Mitchell P. J., Barker K. T., Shipley J., Crompton M. R. (1997) Oncogene 15, 1497–1502 [DOI] [PubMed] [Google Scholar]
- 17. Xiang B., Chatti K., Qiu H., Lakshmi B., Krasnitz A., Hicks J., Yu M., Miller W. T., Muthuswamy S. K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12463–12468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Llor X., Serfas M. S., Bie W., Vasioukhin V., Polonskaia M., Derry J., Abbott C. M., Tyner A. L. (1999) Clin. Cancer Res. 5, 1767–1777 [PubMed] [Google Scholar]
- 19. Schmandt R. E., Bennett M., Clifford S., Thornton A., Jiang F., Broaddus R. R., Sun C. C., Lu K. H., Sood A. K., Gershenson D. M. (2006) Cancer Biol. Ther. 5, 1136–1141 [DOI] [PubMed] [Google Scholar]
- 20. Lin H. S., Berry G. J., Fee W. E., Jr., Terris D. J., Sun Z. (2004) Arch. Otolaryngol. Head Neck Surg. 130, 311–316 [DOI] [PubMed] [Google Scholar]
- 21. Easty D. J., Mitchell P. J., Patel K., Flørenes V. A., Spritz R. A., Bennett D. C. (1997) Int. J. Cancer 71, 1061–1065 [DOI] [PubMed] [Google Scholar]
- 22. Derry J. J., Prins G. S., Ray V., Tyner A. L. (2003) Oncogene 22, 4212–4220 [DOI] [PubMed] [Google Scholar]
- 23. Brauer P. M., Zheng Y., Wang L., Tyner A. L. (2010) Cell Cycle 9, 4190–4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kamalati T., Jolin H. E., Fry M. J., Crompton M. R. (2000) Oncogene 19, 5471–5476 [DOI] [PubMed] [Google Scholar]
- 25. Irie H. Y., Shrestha Y., Selfors L. M., Frye F., Iida N., Wang Z., Zou L., Yao J., Lu Y., Epstein C. B., Natesan S., Richardson A. L., Polyak K., Mills G. B., Hahn W. C., Brugge J. S. (2010) PLoS One 5, e11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kanner S. B., Reynolds A. B., Parsons J. T. (1991) Mol. Cell. Biol. 11, 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reynolds A. B., Kanner S. B., Wang H. C., Parsons J. T. (1989) Mol. Cell. Biol. 9, 3951–3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brinkman A., van der Flier S., Kok E. M., Dorssers L. C. (2000) J. Natl. Cancer Inst. 92, 112–120 [DOI] [PubMed] [Google Scholar]
- 29. Petch L. A., Bockholt S. M., Bouton A., Parsons J. T., Burridge K. (1995) J. Cell Sci. 108, 1371–1379 [DOI] [PubMed] [Google Scholar]
- 30. Tachibana K., Urano T., Fujita H., Ohashi Y., Kamiguchi K., Iwata S., Hirai H., Morimoto C. (1997) J. Biol. Chem. 272, 29083–29090 [DOI] [PubMed] [Google Scholar]
- 31. Sakai R., Iwamatsu A., Hirano N., Ogawa S., Tanaka T., Mano H., Yazaki Y., Hirai H. (1994) EMBO J. 13, 3748–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smith H. W., Marra P., Marshall C. J. (2008) J. Cell Biol. 182, 777–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Honda H., Oda H., Nakamoto T., Honda Z., Sakai R., Suzuki T., Saito T., Nakamura K., Nakao K., Ishikawa T., Katsuki M., Yazaki Y., Hirai H. (1998) Nat. Genet. 19, 361–365 [DOI] [PubMed] [Google Scholar]
- 34. Cabodi S., Tinnirello A., Di Stefano P., Bisarò B., Ambrosino E., Castellano I., Sapino A., Arisio R., Cavallo F., Forni G., Glukhova M., Silengo L., Altruda F., Turco E., Tarone G., Defilippi P. (2006) Cancer Res. 66, 4672–4680 [DOI] [PubMed] [Google Scholar]
- 35. Dorssers L. C., Grebenchtchikov N., Brinkman A., Look M. P., van Broekhoven S. P., de Jong D., Peters H. A., Portengen H., Meijer-van Gelder M. E., Klijn J. G., van Tienoven D. T., Geurts-Moespot A., Span P. N., Foekens J. A., Sweep F. C. (2004) Clin. Cancer Res. 10, 6194–6202 [DOI] [PubMed] [Google Scholar]
- 36. Fromont G., Vallancien G., Validire P., Levillain P., Cussenot O. (2007) Prostate 67, 268–273 [DOI] [PubMed] [Google Scholar]
- 37. Avizienyte E., Wyke A. W., Jones R. J., McLean G. W., Westhoff M. A., Brunton V. G., Frame M. C. (2002) Nat. Cell Biol. 4, 632–638 [DOI] [PubMed] [Google Scholar]
- 38. Avizienyte E., Fincham V. J., Brunton V. G., Frame M. C. (2004) Mol. Biol. Cell 15, 2794–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ayala I., Baldassarre M., Giacchetti G., Caldieri G., Tetè S., Luini A., Buccione R. (2008) J. Cell Sci. 121, 369–378 [DOI] [PubMed] [Google Scholar]
- 40. Weaver A. M. (2006) Clin. Exp. Metastasis 23, 97–105 [DOI] [PubMed] [Google Scholar]
- 41. Zvara A., Fajardo J. E., Escalante M., Cotton G., Muir T., Kirsch K. H., Birge R. B. (2001) Oncogene 20, 951–961 [DOI] [PubMed] [Google Scholar]
- 42. Sanders M. A., Basson M. D. (2005) J. Biol. Chem. 280, 23516–23522 [DOI] [PubMed] [Google Scholar]
- 43. Hayward S. W., Dahiya R., Cunha G. R., Bartek J., Deshpande N., Narayan P. (1995) In Vitro Cell Dev. Biol. Anim. 31, 14–24 [DOI] [PubMed] [Google Scholar]
- 44. Rasband W. S. (1997–2011) ImageJ, United States National Institutes of Health, Bethesda, MD [Google Scholar]
- 45. Yu Y. P., Landsittel D., Jing L., Nelson J., Ren B., Liu L., McDonald C., Thomas R., Dhir R., Finkelstein S., Michalopoulos G., Becich M., Luo J. H. (2004) J. Clin. Oncol. 22, 2790–2799 [DOI] [PubMed] [Google Scholar]
- 46. Ie Kim H., Lee S. T. (2009) J. Biochem. 146, 133–139 [DOI] [PubMed] [Google Scholar]
- 47. Brauer P. M., Zheng Y., Evans M. D., Dominguez-Brauer C., Peehl D. M., Tyner A. L. (2011) PLoS One 6, e14789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Qiu H., Miller W. T. (2002) J. Biol. Chem. 277, 34634–34641 [DOI] [PubMed] [Google Scholar]
- 49. Castro N. E., Lange C. A. (2010) Breast Cancer Res. 12, R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vasioukhin V., Tyner A. L. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 14477–14482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Astier A., Manié S. N., Avraham H., Hirai H., Law S. F., Zhang Y., Golemis E. A., Fu Y., Druker B. J., Haghayeghi N., Freedman A. S., Avraham S. (1997) J. Biol. Chem. 272, 19719–19724 [DOI] [PubMed] [Google Scholar]
- 52. Ostrander J. H., Daniel A. R., Lofgren K., Kleer C. G., Lange C. A. (2007) Cancer Res. 67, 4199–4209 [DOI] [PubMed] [Google Scholar]
- 53. Locatelli A., Lange C. A. (2011) J. Biol. Chem. 286, 21062–21072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Seals D. F., Azucena E. F., Jr., Pass I., Tesfay L., Gordon R., Woodrow M., Resau J. H., Courtneidge S. A. (2005) Cancer Cell 7, 155–165 [DOI] [PubMed] [Google Scholar]
- 55. Serfas M. S., Tyner A. L. (2003) Oncol. Res. 13, 409–419 [DOI] [PubMed] [Google Scholar]
- 56. Qiu H., Miller W. T. (2004) Oncogene 23, 2216–2223 [DOI] [PubMed] [Google Scholar]
- 57. Ko S., Ahn K. E., Lee Y. M., Ahn H. C., Lee W. (2009) Biochem. Biophys. Res. Commun. 384, 236–242 [DOI] [PubMed] [Google Scholar]
- 58. Pellicena P., Miller W. T. (2001) J. Biol. Chem. 276, 28190–28196 [DOI] [PubMed] [Google Scholar]
- 59. Roberts O. L., Holmes K., Müller J., Cross D. A., Cross M. J. (2010) J. Cell Sci. 123, 3189–3200 [DOI] [PubMed] [Google Scholar]
- 60. Wang X., Finegan K. G., Robinson A. C., Knowles L., Khosravi-Far R., Hinchliffe K. A., Boot-Handford R. P., Tournier C. (2006) Cell Death Differ. 13, 2099–2108 [DOI] [PubMed] [Google Scholar]
- 61. American Cancer Society (2010) Cancer Facts & Figures 2010, American Cancer Society, Atlanta, GA [Google Scholar]
- 62. Grille S. J., Bellacosa A., Upson J., Klein-Szanto A. J., van Roy F., Lee-Kwon W., Donowitz M., Tsichlis P. N., Larue L. (2003) Cancer Res. 63, 2172–2178 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.