Background: RECQL4 is a RecQ helicase mutated in Rothmund-Thomson Syndrome (RTS) and has a functional role in DNA replication and repair.
Results: RECQL4-depleted and RTS patient cells show telomere abnormalities and that RECQL4 interacts with telomeric DNA and related proteins.
Conclusion: RECQL4 is involved in telomere maintenance.
Significance: The RecQ helicase RECQL4 is involved in telomere replication and maintenance. This establishes a connection between telomere function and a disease with premature aging phenotype.
Keywords: Aging, DNA Damage, DNA Damage Response, DNA Repair, Telomeres, RecQ Helicase
Abstract
Telomeres are structures at the ends of chromosomes and are composed of long tracks of short tandem repeat DNA sequences bound by a unique set of proteins (shelterin). Telomeric DNA is believed to form G-quadruplex and D-loop structures, which presents a challenge to the DNA replication and repair machinery. Although the RecQ helicases WRN and BLM are implicated in the resolution of telomeric secondary structures, very little is known about RECQL4, the RecQ helicase mutated in Rothmund-Thomson syndrome (RTS). Here, we report that RTS patient cells have elevated levels of fragile telomeric ends and that RECQL4-depleted human cells accumulate fragile sites, sister chromosome exchanges, and double strand breaks at telomeric sites. Further, RECQL4 localizes to telomeres and associates with shelterin proteins TRF1 and TRF2. Using recombinant proteins we showed that RECQL4 resolves telomeric D-loop structures with the help of shelterin proteins TRF1, TRF2, and POT1. We also found a novel functional synergistic interaction of this protein with WRN during D-loop unwinding. These data implicate RECQL4 in telomere maintenance.
Introduction
Telomeres are structures at the ends of linear eukaryotic chromosomes that prevent DNA end-initiated recombination, exonucleolytic DNA degradation, and replication-associated terminal recession. In the absence of an active mechanism to maintain telomere length, the telomere repeat number decreases with each replicative cell cycle. Telomere attrition is associated with genome instability, cell cycle arrest, and senescence or apoptosis. Telomeric DNA is composed of double-stranded tandem repeat sequences (5′-TTAGGG-3′ in human and mouse) followed by a short ssDNA2 3′-overhang (1). In mammals, telomeric DNA is believed to exist as a D-loop, formed by invasion of the 3′-overhang into telomeric dsDNA (2). Telomere-binding proteins, known as shelterin, and associated proteins stabilize the D-loop structure (3). Among the shelterin proteins, POT1 (protection of telomeres 1) binds to single-stranded telomeric DNA (4), and TRF1 and TRF2 (telomere repeat-binding factors 1 and 2) bind to duplex telomeric sequences (5).
The DNA replication machinery is unable to replicate DNA ends, and as cells proliferate telomeric DNA can be lost. This terminal DNA loss is compensated for by telomerase, a reverse transcriptase that adds TTAGGG repeats at the 3′-ends of chromosomal DNA (6). The bulk of telomeric DNA is replicated by the semiconservative DNA replication machinery. The highly repetitive G-rich sequences at the ends of telomeres can adopt unusual DNA secondary structures, such as G-quadruplexes (7) and D-loops (8), which must be resolved prior to replication. Further, telomeres were identified as fragile sites, which are known to form in response to replication stress (9, 10). Additionally, these secondary structures may also prevent the access of repair enzymes to damaged telomeric DNA. Thus, telomeric DNA presents a challenge to DNA replication and repair, and an appropriate coordination between specialized DNA replication/repair proteins and the shelterin proteins is essential for telomere integrity. The RecQ helicases WRN and BLM are known to be involved in telomere maintenance (7, 11–13). The shelterin protein TRF1 interacts with WRN in vitro and modulates its activity at the telomere (11). It has also been suggested that TRF1 may recruit BLM, which can resolve G-quadruplex structures efficiently (10). Likewise, another shelterin protein, TRF2, interacts with both WRN and BLM and stimulates their helicase activities on telomeric D-loops in vitro, suggesting that TRF2 may play a role in the recruitment of these proteins at telomeres (11).
RecQ helicases are a highly conserved family of proteins that play significant roles in DNA metabolic processes including DNA replication, DNA repair, and DNA recombination. Saccharomyces cerevisiae and Schizosaccharomyces pombe each express only a single RecQ helicase (Sgs1 and Rqh1, respectively), whereas five RecQ homologs are expressed in mammalian cells: RECQ1, BLM, WRN, RECQL4, and RECQ5 (14, 15). BLM, WRN, and RECQL4 are linked to autosomal recessive disorders characterized by genomic instability and cancer predisposition. Bloom syndrome and Werner syndrome are associated with defects in BLM and WRN, respectively (16, 17), whereas RECQL4 deficiency is associated with three rare autosomal recessive diseases: Rothmund-Thomson syndrome (RTS), Baller-Gerold syndrome, and RAPADILINO syndrome (18, 19). BLM and WRN play important roles in DNA repair and replication (14, 20–22) and have also been implicated in telomere maintenance.
Although the biological function of RECQL4 is not well established, it has been proposed that it participates in base excision repair, nucleotide excision repair, and homologous recombination (23–26). In addition, some studies suggest that Xenopus laevis RECQL4 is active in the initiation of DNA replication (27, 28). Consistent with this, human RECQL4 interacts with the minichromosome maintenance complex (29) and the origin of replication (30) during replication initiation.
RTS patients who do not die from cancer have a normal life span. However, they show features of “segmental premature aging” such as growth retardation, poikiloderma, hair loss, cataracts, and bony malformations, and thus RTS is considered a premature aging syndrome (31, 32). Interestingly, some RTS patients have phenotypes similar to dyskeratosis congenita, which is caused mainly by telomere abnormalities (33). Further, RTS and Werner syndrome share many clinical features, including developmental abnormalities, premature aging, and a high degree of susceptibility to osteosarcomas (34, 35). WRN interacts with telomeric structures and plays a significant role in telomere replication and repair (11, 36). Recent studies show that RECQL4 has a previously undetected helicase activity with selective DNA substrate specificity (19, 37). RECQL4 also interacts with FEN1 (26), which may also play a role in telomere maintenance (38). Together, these observations suggest that RECQL4, like WRN, may play a role in telomere maintenance.
We set out to look for telomeric abnormalities in RTS patient cells and RECQL4-depleted human cells and characterized interactions between RECQL4, shelterin proteins, telomeric D-loops, and WRN. The results showed that RECQL4 localizes at telomeres in replicative human cells and that the frequency of fragile telomeres is higher in RECQL4-depleted cells than in control cells, especially after DNA replication stress induced by aphidicolin exposure. Although RECQL4 can barely resolve telomeric D-loops, this activity is considerably enhanced in the presence of TRF1 or TRF2. RECQL4 also associates with WRN and stimulates WRN unwinding of telomeric D-loops in vitro. Collectively, our data support the view that RECQL4 participates in telomere maintenance by assisting in D-loop resolution.
EXPERIMENTAL PROCEDURES
Proteins
Wild-type (WT) human RECQL4 and helicase-dead RECQL4 with a C-terminal His9 tag in the pGEX6p1 vector (GE Healthcare) was expressed and purified from Escherichia coli Rosetta2 (DE3) (Novagen) as described previously (19). Recombinant histidine-tagged BLM was overexpressed in S. cerevisiae and purified as described previously (39). Recombinant histidine-tagged wild-type WRN protein, recombinant GST-tagged human POT1 protein, and recombinant histidine-tagged human TRF2 and TRF1 protein were purified using a baculovirus/insect cell expression as described previously (4, 11, 40). Protein concentration was determined by the Bradford assay (Bio-Rad), and purity was determined by SDS-PAGE and Coomassie staining.
Preparation and Characterization of D-loops
All of the unmodified oligonucleotides were from Integrated DNA Technologies (Coralville, IA) and were PAGE-purified by the manufacturer. The modified (8-oxo-2-deoxyguanosine-containing) oligonucleotides were synthesized and purified by The Midland Certified Reagent Co. (Midland, TX). The D-loops were prepared and characterized as described previously (36).
Helicase and Annealing Assays
All assays were performed at least in triplicate. RECQL4 helicase assays were performed as described earlier (19). POT1, TRF1, and TRF2 (amounts indicated in the legends of Figs. 7–9) were added together with RECQL4 when indicated. WRN and BLM helicase reactions were performed and analyzed similarly, only in a different reaction buffer (40 mm Tris, pH 8.0, 4 mm MgCl2, 5 mm DTT, 2 mm ATP, and 100 μg/ml BSA). While investigating the effect of RECQL4 on WRN and BLM helicase activities, the indicated amounts of RECQL4 were added together with WRN or BLM. DNA substrate and protein concentrations were as indicated in the legends of Figs. 7–9.
FIGURE 7.
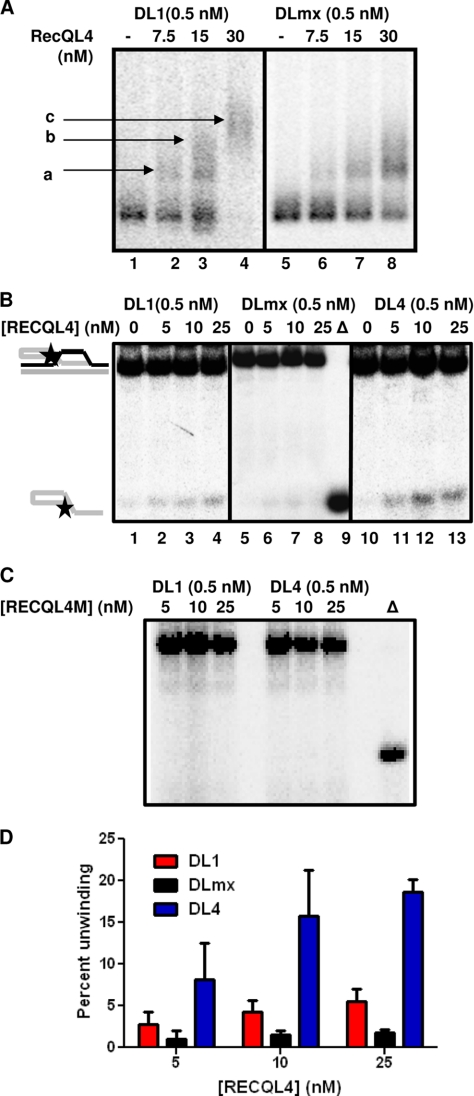
Interaction of RECQL4 with telomeric and non-telomeric D-loops. A, autoradiogram showing binding of 0, 7.5, 15, and 30 nm RECQL4 with 0.5 nm DL1 (lanes 1–4) and 0.5 nm DLmx (lanes 5–8). Bound products are marked as a, b, and c. B, autoradiogram showing the unwinding activity of 0, 5, 10, and 25 nm RECQL4 on telomeric (lanes 1–4, 0.5 nm DL1) and non-telomeric D-loops (lanes 5–8, 0.5 nm DLmx) and oxidatively damaged telomeric D-loops (lanes 10–13, 0.5 nm DL4) in the presence of excess (25×) single-stranded DNA. Δ (lane 9), represents heat-denatured DLmx. C, autoradiogram showing the helicase activity of 5, 10, and 25 nm helicase-dead mutant RECQL4 (RECQL4M) on 0.5 nm DL1 and 0.5 nm DL4 in the presence of excess (25×) single-stranded DNA. D, histogram showing the unwinding activity of 0, 5, 10, and 25 nm RECQL4 on DL1, DLmx, and DL4 in the presence of excess (25×) single-stranded DNA. The error bars represent mean ± S.D., n = 3.
FIGURE 8.
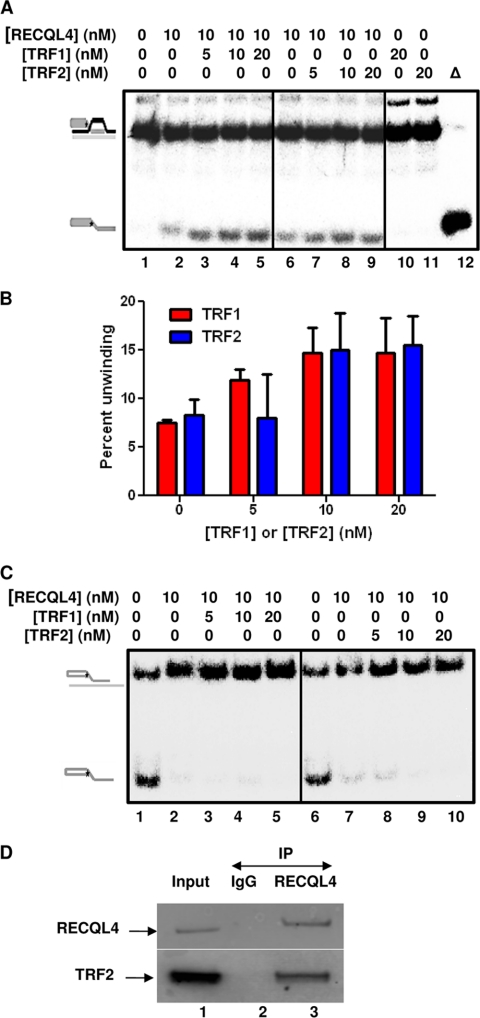
RECQL4 physically and functionally interacts with TRF2. A, autoradiogram showing the unwinding activity of RECQL4 (10 nm) on 0.5 nm DL1 in the presence of 0, 5, 10, and 20 nm TRF1 (lanes 1–5) and TRF2 (lanes 6–9) in the presence of 25× excess single-stranded DNA. Δ (lane 12), represents heat-denatured DL1. 20 nm TRF1 (lane 10) or TRF2 (lane 11) alone does not have any unwinding activity on DL1. B, histogram showing the unwinding activity of RECQL4 (10 nm) on 0.5 nm DL1 in the presence of 0, 5, 10, and 20 nm TRF1 and TRF2 in the presence of 25× excess single-stranded DNA. The error bars represent mean ± S.D., n = 3. C, autoradiogram showing the effect of 5, 10, and 20 nm TRF1 (lanes 1–5) or TRF2 (lanes 6–10) on annealing activity of RECQL4. 0.5 nm radiolabeled oligo SS1 and 2.5× excess oligo BB were used as annealing substrates. D, in vitro pulldown of TRF2 by RECQL4. Lane 1, represents input, showing the bands corresponding to RECQL4 and TRF2. Lane 2, represents co-IP with IgG controls. Lane 3, represents co-IP with antibodies specific to RECQL4. Bands corresponding to anti-RECQL4 and anti-TRF2 antibodies are marked with arrows.
FIGURE 9.
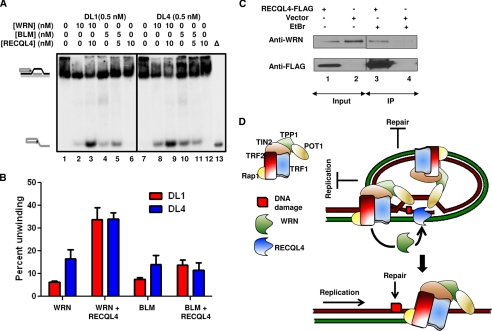
RECQL4 and WRN synergistically unwind telomeric D-loops. A, gel showing the effect of 10 and 5 nm RECQL4 on the unwinding activities of 10 nm WRN and 5 nm BLM, respectively. 0.5 nm DL1 (lanes 1–5) and DL4 (lanes 7–11) were used as substrates. Lanes 6 and 12, represent the unwinding activity of RECQL4 alone on DL1 and DL4, respectively. Δ (lane 13), indicates heat-denatured DL4. B, quantitative analysis of the gel showing the effect of RECQL4 on WRN and BLM unwinding activities on DL1 and DL4 at a 1:1 molar ratio of the corresponding proteins. The error bars represent mean ± S.D., n = 3. C, in vivo co-IP of WRN by FLAG-tagged RECQL4 in U2OS cells. The bands corresponding to anti-WRN and anti-FLAG antibodies are shown. Lanes 1 and 2, the bands in RECQL4-FLAG and empty vector-transduced cells, respectively. The corresponding IPs with FLAG, in the presence of EtBr, are shown in lanes 3 and 4, respectively. D, schematic summarizing the proposed synergistic role of RECQL4 and WRN in telomere maintenance. The DNA replication and repair machinery fails to proceed through the telomeric D-loop, and WRN and RECQL4 are recruited by the shelterin complex (most probably by TRF2, as both of these RecQ helicases interact with it directly) to resolve this structure and thus maintain telomere integrity.
EMSA
RECQL4 (7.5, 15, and 30 nm) was incubated with either 0.5 nm non-telomeric D-loops (DLmx) or 0.5 nm telomeric D-loops (DL1) in a 10-μl reaction buffer containing 30 mm Tris-HCl, pH 7.4, 1 mm DTT, 100 μg/ml BSA, and 50 mm KCl for 15 min on ice. Then 5 μl of stop dye (50% glycerol and 0.05% bromphenol blue) was added. Reactions were kept on ice until loading onto a 1% agarose/20 mm Tris, 10 mm acetic acid, and 0.5 mm EDTA (0.5 × TAE) gel and run in 0.5× TAE at 200 V for 1.5 h at 4 °C. Gel was dried in a gel dryer and exposed to a PhosphorImager screen. The image was scanned and analyzed using ImageQuant TL (GE Healthcare). Assays were performed at least in triplicate, and a representative gel shown.
Cell Culture
U2OS and HeLa cells were maintained in high glucose Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Sigma) and 1% penicillin-streptomycin (Invitrogen) at 37 °C in 5% CO2. Normal (GM00323 and GM01864) and RTS patient cell lines (AG05013 and AG18371) were obtained from Coriell Cell Repository (Camden, NJ). The cells were grown in Amniomax II medium (Invitrogen) and maintained at 37 °C in 5% CO2. Primary BJ cells were grown in Eagle's minimum essential medium (ATCC, Manassas, VA) with 10% fetal bovine serum (Sigma).
shRNA-mediated RECQL4 Knockdown
RECQL4 knockdown (RECQL4 KD) and scrambled control cells (SCR) were prepared in primary BJ, U2OS, and HeLa cells according to the standard protocol using Mission shRNA lentiviral construct TRCN0000051169 from Sigma-Aldrich. Briefly, the shRNA vector was co-transfected with packing plasmid pCMV-dr8.2 DVPR (Addgene, Cambridge, MA) and envelope vector pCMV-VSV-G (Addgene) into human embryonic kidney 293T cells in DMEM with Hyclone FBS (Thermo Fisher Scientific) using FuGENE® 6 transfection reagent (Roche Applied Science) and Opti-MEM (Invitrogen). The medium was collected 48 h after transduction and filtered using a 0.45-μm PVDF membrane filter (Millipore). Lentivirus was applied to the cells, and infections were allowed to proceed for 48 h after which time puromycin was applied to select for those cells that had taken up the lentivirus. Quantitative PCR was performed to confirm that the RECQL4 was knocked down ∼90%, which was later confirmed by Western blotting prior to the cells being used in experiments.
siRNA-mediated RECQL4 Knockdown
100,000 U2OS cells were grown overnight in 6-well plates in 2 ml of antibiotic-free medium (DMEM plus 10% FBS). Then the cells were treated with 100 pmol of either control (Silencer Negative Control #1, Ambion) or RECQL4-targeted siRNA (target sequence CAAUACAGCUUACCGUACA, Dharmacon) in Lipofectamine 2000 reagent (Invitrogen) for 6 h. Cells were then grown overnight and again treated with same siRNAs for 6 h. Cells were then grown in antibiotic (1%)-containing medium and harvested after 72 h, and the RECQL4 level was checked by Western blotting.
Co-immunoprecipitation (co-IP)
For in vitro co-IP, purified WRN (7.5 pmol) and purified RECQL4 (15 pmol) were mixed in a buffer containing 20 mm Tris-HCl, pH 7.4, 20 mm NaCl, 25 mm KCl, 1 mm DTT, 5% glycerol, 2.5 mm MgCl2 and 100 μg/ml BSA and incubated for 90 min at 4 °C. For RECQL4-TRF2 co-IP, the proteins were mixed in a buffer containing 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, and 1× protease inhibitors (Roche Applied Science). Then, precoated antibody-agarose beads (anti-rabbit WRN and normal rabbit IgG, Santa Cruz Biotechnology) were added to the protein mix, and the tubes were incubated for 2 h at 4 °C with end-over-end rotation. After incubation the beads were washed four times in washing buffer containing 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, and 1% Triton X-100, resuspended in 30 μl of 2× SDS loading buffer, and resolved on a 4–12% gradient SDS-PAGE followed by Western analysis. The RECQL4 was detected by polyclonal anti rabbit-RECQL4 (19) and rabbit TrueBlot HRP-conjugated anti-rabbit IgG secondary antibody (eBioscience) using an ECL kit following the manufacturer's protocol (GE Healthcare). For in vivo co-IP of TRF2 by RECQL4, U2OS cells were lysed in cell lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, and 1× protease inhibitors (Roche Applied Science)), and co-IP was performed as described elsewhere using anti-TRF2 (Imgenex, mouse) and anti-RECQL4 (19) (rabbit). For co-IP of WRN with FLAG-RECQL4, U2OS cells were first transduced with FLAG-RECQL4 and empty vector plasmids using Lipofectamine LTX following the manufacturer's protocol (Invitrogen). At 24 h post-transfection, the cells were lysed in 1 ml of cell lysis buffer in the presence of 50 μg/ml ethidium bromide. WRN was detected with polyclonal anti-rabbit WRN antibody (Santa Cruz Biotechnology), and FLAG-RECQL4 was detected with anti-rabbit FLAG antibody (Sigma) using ECL Plus (GE Healthcare).
Immunofluorescence
U2OS or HeLa cells (untreated, RECQL4-specific, or scrambled shRNA-treated, as indicated) were plated on Lab-Tek II chambered glass slides (Thermo-Fisher Scientific) at a density of 20,000 cells/chamber, grown overnight, and then treated with 2 gray IR where indicated. For cell cycle-dependent studies, cells were treated with 2 mm hydroxyurea (Sigma) or 2 mm nocodazole (Sigma) for 16 h as indicated. Hydroxyurea- and nocodazole-containing medium was then replaced by DMEM, and the cells were fixed immediately. Cells were fixed with 4% paraformaldehyde in PBS for 10 min at 37 °C, washed with PBS, and permeabilized with 0.2% Triton X-100 in PBS for 5 min at room temperature. Following a final wash with PBS, cells were blocked overnight with 5% FBS in PBS. Then the cells were treated with primary antibodies for 1 h at 37 °C and secondary antibodies for 30 min at 37 °C after washing in PBS (five times for 3 min). Then the cells were washed again with PBS (five times for 3 min) and treated with Vectashield mounting medium containing DAPI (Vector Laboratories). The following antibodies were used as described under “Results.” Primary antibodies were: 53BP1 (Novus, 1:100, rabbit), TRF1 (Abcam TRF-78, 1:50, mouse), TRF2 (Imgenex, 1:50, mouse), RECQL4 (in-house) (19) (1:100, rabbit or Santa Cruz K-16, 1:100, goat), and WRN (Santa Cruz, 1:100, rabbit). Secondary antibodies were: donkey anti-rabbit Alexa Fluor 488 (1:1000) and donkey anti-mouse Alexa Fluor 647 (1:200 for TRF-78 and 1:1000 for others) (all from Invitrogen). All of the dilutions were prepared in blocking solution (5% FBS). Images were captured on a Nikon Eclipse TE2000 confocal microscope (×40 magnification) with a Hamamatsu C9100-13 camera at −65 °C. Data acquisition and analysis were performed using Volocity 5.5 software (PerkinElmer Life Sciences). Images of individual representative cells were cropped from the original images and are shown in Figs. 2, 3, and 5. To further clarify the coexistence of two signals, images of partially colocalized foci were cropped and visualized as three-dimensional images as shown in Figs. 2, 3, and 5. The Pearson coefficients of these individual foci were calculated using Volocity software. For a better representation (supplemental Fig. 3), colocalization channels (positive products of the differences of the means for the two channels) were generated to highlight the colocalized foci. For quantification purpose, the foci at which two signals are intersected (colocalized foci) are marked and calculated using the same software.
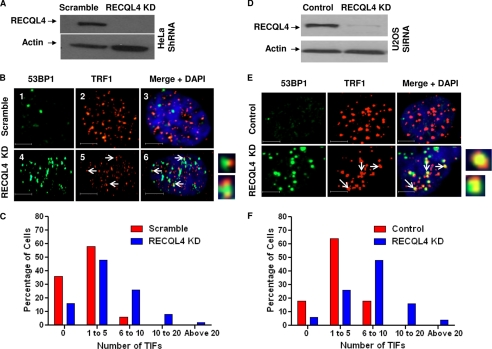
FIGURE 2.
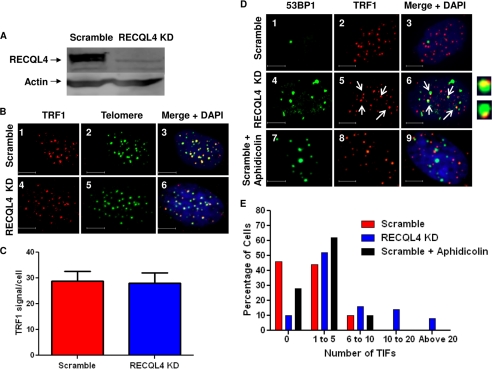
Telomeric 53BP1 foci in RECQL4 knockdown U2OS cells. A, Western blot showing the level of RECQL4 in scrambled shRNA-treated (Scramble) and RECQL4-targeted shRNA-treated (RECQL4 KD) U2OS cells. Bands corresponding to RECQL4 and actin are shown by arrows. B, confocal microscopic images of representative cells showing TRF1 (red) and telomere (green) signals in scrambled shRNA-treated (panels 1 and 2, Scramble) and RECQL4 shRNA-treated (panels 4 and 5, RECQL4 KD) U2OS cells. Colocalized foci are visible as yellow dots in panels 3 and 6. Nuclear staining (with DAPI) is shown in the merged images. Scale bar = 10 μm. C, average numbers of TRF1 signals/cell in scrambled and RECQL4 KD U2OS cells. The error bars represent mean ± S.D., n = 50. D, confocal microscopic images showing colocalization of 53BP1 foci (green) and TRF1 (red) signals in scrambled (panels 1–3) and RECQL4 KD U2OS cells (panels 4–6) and in aphidicolin-treated scrambled U2OS cells (panels 7–9). Nuclear staining with DAPI is shown in the merged image. Some of the colocalized foci are marked with white arrows. Close-up images of some of the colocalized foci are shown next to panel 6. Scale bar = 5 μm. E, histograms showing the frequency distribution of telomeric 53BP1 foci (TIF) in scrambled, RECQL4 KD, and aphidicolin-treated scrambled U2OS cells (n = 70).
FIGURE 3.
Telomeric 53BP1 foci in RECQL4 knockdown HeLa and U2OS cells. A, Western blot showing the level of RECQL4 in scrambled shRNA-treated (Scramble) and RECQL4-targeted shRNA-treated (RECQL4 KD) HeLa cells. Bands corresponding to RECQL4 and actin are shown by arrows. B, confocal microscopic images showing colocalization of 53BP1 foci (green) and TRF1 (red) signals in scrambled (panels 1–3) and RECQL4 KD HeLa cells (panels 4–6). Nuclear staining with DAPI is shown in the merged image. Some of the colocalized foci are marked with white arrows. Close-up images of some of the colocalized foci are shown next to panel 6. Scale bar = 5 μm. C, histograms showing the frequency distribution of telomeric 53BP1 foci (TIF) in scrambled and RECQL4 KD HeLa cells (n = 70). D, Western blot showing the level of RECQL4 in control siRNA-treated (Control) and RECQL4-targeted siRNA-treated (RECQL4 KD) U2OS cells. Bands corresponding to RECQL4 and actin are shown by arrows. E, confocal microscopic images showing colocalization of 53BP1 foci (green) and TRF1 (red) signals in control (panels 1–3) and RECQL4 KD U2OS cells (panels 4–6). Nuclear staining with DAPI is shown in the merged image. Some of the colocalized foci are marked with white arrows. Close-up images of some of the colocalized foci are shown next to panel 6. Scale bar = 5 μm. F, histograms showing the frequency distribution of telomeric 53BP1 foci in control and RECQL4 KD U2OS cells (n = 70).
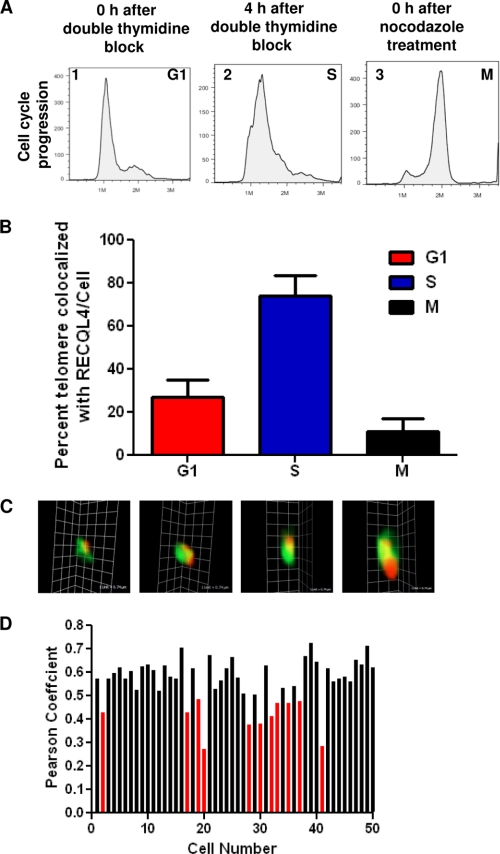
FIGURE 5.
RECQL4 localizes at telomere. A, FACS analysis showing cell cycle progression states of U2OS cells immediately after double thymidine block (panel 1, G1), 4 h after double thymidine block (panel 2, S), and immediately after nocodazole treatment (panel 3, M). B, histograms showing the percentage of telomeric foci colocalized with RECQL4 at G1, S, and M phases. The error bars represent mean ± S.D., n = 50. C, three-dimensional images of four representative colocalized RECQL4 (green) and TRF1 (red) foci. Scale bar = 0.74 μm. D, histogram showing Pearson's correlation coefficients of 50 randomly selected S phase cells.
Fluorescence in Situ Hybridization (FISH)
RECQL4-specific or scrambled shRNA-treated U2OS or HeLa cells were treated with 200 μl of 0.5% colcemid for 3 h. For RTS AG05013 and AG18371 and control GM00323 and GM01864 cells the treatment was for 4 h. Cells were then harvested and immediately incubated in 0.075 m KCl for 20 min in 37 °C followed by fixation in ice-cold (3:1) methanol and glacial acetic acid. Metaphase spreads were then treated with pepsin (1 mg/ml, Sigma) and fixed with 4% formaldehyde. The slides containing the spreads were then dehydrated by ethanol wash (70, 90, and 100%, respectively) and hybridized with a Cy3-labeled p-nitroanilide (CCCTAA)3 C-telomere probe (0.3 mg/ml, Panagene) in hybridization buffer ( 70% formamide, 2.5% blocking protein, 10 mm Tris, pH 7.4, and 2.5 mm MgCl2). Then the slides were washed with washing solution I (70% formamide, 1% BSA, and 10 mm Tris, pH 7.4) and washing solution II (1× TBS and 0.7% Tween 20). After dehydration the spreads were counterstained with DAPI and mounted with ProLong Gold anti-fade reagent (Invitrogen). Images were captured using CytoVisionTM software (Applied Imaging Corp.) on a fluorescence microscope (Axio2, Carl Zeiss, Germany) at ×100 magnification. At least 30 metaphases from three different sets of RECQL4 knockdown and scrambled U2OS and Hela cells or from three different experiments with RTS and control cells were scored manually for telomeric abnormalities (i.e. frequencies of telomere fusion, breaks, and fragile telomeres).
Chromosome Orientation FISH
RECQL4-specific or scrambled shRNA-treated U2OS cells (about 50% confluent) were subcultured in DMEM containing a 3:1 ratio of BrdU/BrdC (Sigma) at a final concentration of 1 × 10−5 m and collected around 16 h for detection of telomere sister chromatid exchange (T-SCE). At 3 h prior to harvest, colcemid (0.1 mg/ml) was added. Metaphase spreads were then prepared as described under “Fluorescence in Situ Hybridization (FISH)” above. Chromosome orientation FISH (CO-FISH) was used to measure the frequency of T-SCE as described previously (41, 42). In brief, the metaphase spreads were treated with RNase A and Hoechst stain followed by UV exposure and exonuclease III (New England Biolabs, 10 units/μl) digestion. Then the spreads were hybridized with Cy3-labeled p-nitroanilide (CCCTAA)3 C-telomere probe as described above. Following washes with wash solution I (70% formamide, 0.1% BSA, 10 mm Tris, pH 7.2) and II (0.1 m Tris, 0.15 m NaCl, 0.08% Tween-20), the spreads were counterstained with DAPI and mounted with ProLong Gold anti-fade reagent. The images were taken as described above under “Fluorescence in Situ Hybridization (FISH).” A chromosome with more than two telomeric DNA signals by Cy3-labeled C-telomere probe was scored as T-SCE-positive. The frequencies of T-SCEs were obtained from at least 50 metaphases from three different RecQ knockdown and scrambled U2OS cells.
Immunofluorescence-FISH
U2OS cells were plated, fixed, and treated with primary and secondary antibodies as mentioned above under “Immunofluorescence.” Then, the cells were fixed by 2% paraformaldehyde, hybridized with a Cy3-conjugated (CCCTAA)3 probe, and stained with DAPI. Images were taken and analyzed as described under “Immunofluorescence.”
Cell Cycle Analysis
Cells were harvested at different time points after hydroxyurea or nocodazole release, washed, and mixed with staining solution containing propidium iodide. The cell cycle progression was detected by flow cytometry using a FACS instrument and analyzed by FlowJo software.
Statistical Analysis
For the frequency distribution of telomeric 53BP1 foci (Figs. 2 and 3), colocalized foci were calculated from at least 70 cells from three independent experiments. For FISH and chromatid orientation FISH experiments, a t test was performed on at least 30 metaphase spreads from three independent experiments using sigma-plot; the p value was obtained. p values between 0.05 and 0.01 are marked with an asterisk and below 0.01 with a double asterisk in Figs. 1 and 4.
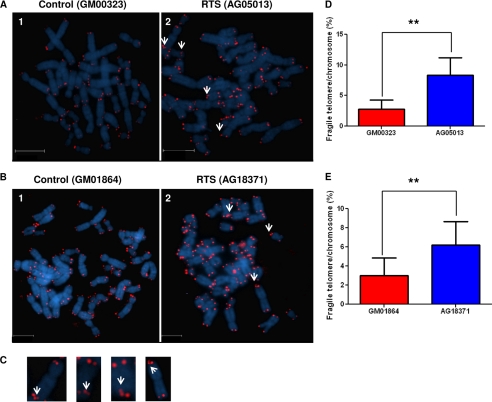
FIGURE 1.
Fragile telomeric ends in RTS patient cells. A and B, microscopic images showing representative metaphase spreads from control cell GM00323 (panel 1) and RTS cell AG05013 (panel 2) (A) and control cell GM01864 (panel 1) and RTS cell AG18371 (panel 2) (B). DAPI (blue) was used for nuclear staining, and the red dots represent the telomeric ends. Some of the fragile telomeres are shown by white arrows. Scale bar represents 95 μm for A and 75 μm for B. C, representative images of chromosomes containing fragile telomeres. Fragile telomeric ends are shown by white arrows. D and E, percent fragile telomeres/chromosome in sex- and age-matched M00323 and AG05013 cells (D) and GM01864 and AG18371 cells (E). The differences in occurrence of fragile telomeres between control and RTS cells are statistically significant (p = 0.006 (D) and p = 0.004 (E); n = 50). The error bars represent mean ± S.D.
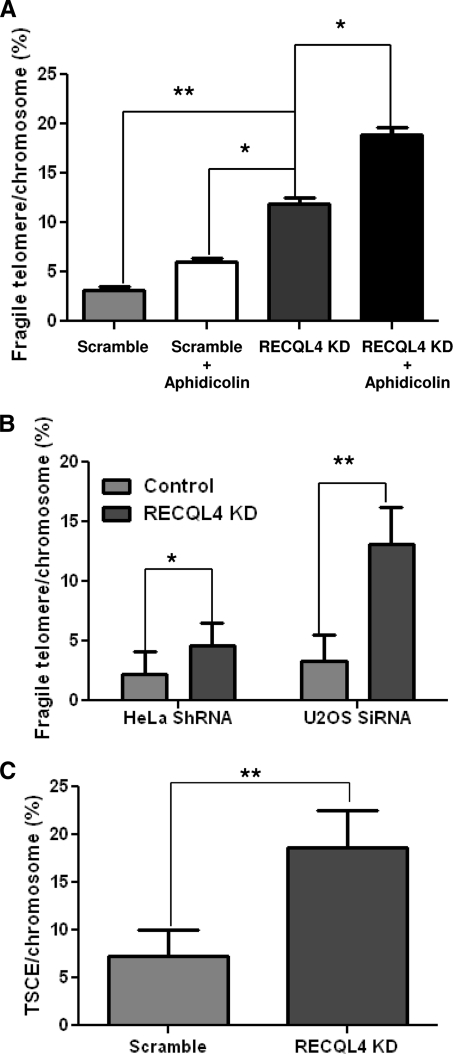
FIGURE 4.
Telomeric abnormalities in RECQL4-depleted U2OS and HeLa cells. A, percent fragile telomeres/chromosome in scrambled, aphidicolin-treated scrambled, RECQL4 KD, and aphidicolin-treated RECQL4 KD U2OS cells. The differences in occurrence of the event between scrambled and RECQL4 KD U2OS cells (**, p = 0.002, n = 50), between aphidicolin-treated scrambled and RECQL4 KD U2OS cells (*, p = 0.01, n = 30), and between RECQL4 KD and aphidicolin-treated RECQL4 KD U2OS cells (*, p = 0.03, n = 50) are statistically significant. Error bars show the standard deviation from the average of three independent experiments. B, percent fragile telomeres/chromosome in scrambled and RECQL4-targeted shRNA HeLa cells and control and RECQL4-targeted siRNA U2OS cells. The differences in occurrence of the event between scrambled and RECQL4 KD HeLa cells (*, p = 0.02, n = 30) and control and RECQL4 KD U2OS cells (**, p = 0.001, n = 30) are statistically significant. Error bars represent the standard deviation from an average of three independent experiments. C, percent T-SCE/chromosome in scrambled and RECQL4-targeted shRNA (RECQL4 KD) U2OS cells. The differences in occurrence of the event between scrambled and RECQL4 KD cells are statistically significant (**, p = 0.002, n = 30). The error bars represent mean ± S.D. of three independent experiments.
RESULTS
RTS Patient Cells Have Telomere Abnormalities
To investigate the possible telomere abnormalities in RTS patient cells, we performed a telomeric FISH assay on two different RTS patient cell lines (AG05013 and AG18371) and compared the results with those from age- and sex-matched human skin fibroblasts (GM00323 and GM01864). The RTS cell line AG05013 possesses compound heterozygous mutations in the RECQL4 gene; one allele contains a 2-base deletion and the other allele has a G→T substitution at the junction of intron 12 and exon 13 that destroys the splicing acceptor sequence. Both mutations are associated with a translational frameshift, whereas the second mutation is predicted to cause deletion in the helicase region of the protein (43). On the other hand, AG18371 is homozygous for a truncating mutation in the RECQL4 gene, an 11-bp deletion at nucleotide g.2746 (g.2746del11) in intron 8, which results in an intron that is too short to be efficiently spliced (44). Both of these RECQL4 mutations co-segregate with RTS (45).
Although the frequency of telomere fusion and breakage was low and comparable in RTS cells and the corresponding control cells, the frequency of fragile telomeres, detected as multiple or diffuse sites of hybridization at a single telomere (Fig. 1, A–C) (10), was almost 3-fold higher (∼ 8%) in AG05013 cells than in control cells GM00323 (∼ 3%) (Fig. 1D). Similarly, the level of this type of aberration was also elevated in RTS cell line AG18371 (6%) compared with the control line GM01864 (∼ 3%) (Fig. 1E). Although the patient cell lines and the corresponding control lines are not isogenic, the similarity of the phenotypes observed in both of the RTS cell lines suggests a potential role of RECQL4 in telomere maintenance. The abnormalities in telomere maintenance could also be an extended effect of general replication abnormalities in the absence of full-length RECQL4.
RECQL4 Depletion Causes Accumulation of Telomeric 53BP1 Foci
To further clarify the role of RECQL4 in protecting telomeric ends, human primary fibroblast (BJ cells) were treated with lentivirus-containing shRNA specific to RECQL4. These RECQL4 shRNA-infected BJ cells ceased to grow almost immediately, and enough cells could not be obtained for analysis. However, U2OS osteoblasts and HeLa cells continued growing after lentiviral infection and shRNA-mediated depletion of RECQL4, and these were used in the following studies. U2OS is a cell type that has been widely used in studies on RecQ helicases, including WRN (11, 46). U2OS cells were infected with lentivirus expressing either scrambled or RECQL4 shRNA, and Western blot analysis showed ∼95% depletion of RECQL4 in RECQL4 shRNA-treated U2OS cells (Fig. 2A). We first looked into the formation of DNA damage response foci at telomeres in these cells.
We used immunofluorescence-based methods to quantify 53BP1 as a marker of DNA damage and TRF1 as a telomeric marker in RECQL4 KD and control U2OS cells. The level of telomeric TRF1 was comparable in RECQL4 KD and control U2OS cells (Fig. 2, B and C). 53BP1 foci were detected in both RECQL4 KD and control cells; however, the average number of 53BP1 foci was significantly higher in RECQL4 KD cells than in control cells. To quantify the 53BP1 foci, the number of foci was determined in at least 50 nuclei using Volocity software. For RECQL4 KD U2OS cells, the average number of 53BP1 foci/nucleus was 12, and about 44% nuclei had more than 10 foci. In control cells, the average number of foci/nucleus was 4, and no nuclei had more than 10 foci (supplemental Fig. 1, A and B).
Interestingly, some of the 53BP1 foci were colocalized with the TRF1 foci (Fig. 2D, panels 1–6). These foci, also known as telomere dysfunction-induced foci (TIFs), are thought to reflect a DNA damage response targeted to telomeric DNA (47). However, some of these TIFs were much larger than the corresponding TRF1-bound region, suggesting that TIFs in RECQL4 KD cells may include a large subtelomeric domain associated with 53BP1. In RECQL4 KD U2OS cells, among the 75 cells studied, ∼ 40% cells had more than five TIFs, whereas only 10% of the scrambled shRNA-treated U2OS cells had more than five TIFs (Fig. 2E). It is well established that RECQL4 plays an important role in replication, and thus the telomeric DNA damage observed in the RECQL4-depleted cells could reflect a general genome replication block rather than a telomere-specific phenomenon. To test this hypothesis, the control U2OS cells were treated with a low dose of aphidicolin (0.1 μm for 24 h), causing replication stress throughout the genome, and the TIFs were quantified. As expected, the number of 53BP1 foci was elevated after aphidicolin treatment. Although there was an increase in the telomeric 53BP1 foci after this replication stress, the effect was much less relative to RECQL4 depletion (Fig. 2, D, panels 7–9, and E). However, the replication stress generated by aphidicolin treatment was dose-dependent, and the level of stress could well be different than the stress produced by RECQL4 depletion. Thus, we compared the amount of 53BP1 foci produced by treatment with alphidicolin with that produced by shRNA-mediated RECQL4 depletion. The frequency distribution shows (supplemental Fig. 1C) that they generate comparable amounts of 53BP1 foci and thus could produce comparable replication stress. Thus, these results indicate that the depletion of RECQ4 induces specific crisis at telomeres, although the role of general replication stress cannot be dismissed.
Additionally, TIFs were analyzed in RECQL4 shRNA- or scrambled shRNA-treated HeLa cells (Fig. 3, A and B), with similar results obtained (Fig. 3C). Further, to exclude any off-target effect caused by the shRNA, U2OS cells were transfected with a control or RECQL4-targeted siRNA (targeting a different sequence than the shRNA approach), and the distribution of telomeric 53BP1 foci was assessed 48 h after the treatment. The RECQL4-targeted siRNA was able to suppress RECQL4 expression considerably (∼90% (Fig. 3D)), and RECQL4-depleted cells generated large numbers of telomeric 53BP1 foci (Fig. 3E). Although almost 80% of the control cells had less than five TIFs, more than 60% of the RECQL4-depleted cells crossed this limit (Fig. 3F). Taken together, these results imply the activation of DNA damage response protein 53BP1 at telomeres as a result of RECQL4 depletion in human cells, signifying a role for RECQL4 in telomere maintenance.
Depletion of RECQL4 Increases Frequency of Fragile Telomeres in Human Cells
The accumulation of 53BP1 foci at telomeres may be caused by replication stress (48) or by inefficient DNA repair machinery. Defective DNA replication and repair machineries at telomeres may also lead to the accumulation of telomere abnormalities such as telomere loss, telomere fusion, telomere breaks, and fragile telomeres (10). As described earlier, we have detected fragile telomeric ends in RTS patient cell lines. Consistent with this observation, the frequency of fragile telomeres was ∼3-fold higher (∼11%) in RECQL4-targeted shRNA-treated U2OS cells than in the control cells (∼4%) (Fig. 4A). The effect of aphidicolin was additive to RECQL4 depletion, resulting in ∼18% fragile telomeres in aphidicolin-treated RECQL4 knockdown U2OS cells. This change was much larger than the ∼1.5-fold increase in fragile telomeres in aphidicolin-treated control cells, Further, RECQL4-depleted HeLa cells and RECQL4-specific siRNA-treated U2OS cells had ∼2- and 4-fold increases in fragile telomeres, respectively, compared with the corresponding control cells (Fig. 4B). Collectively, these results indicate that RECQL4 depletion leads to defects in telomere replication. The increased frequency of fragile telomeres generated in cells lacking RECQL4 may reflect a general role for the protein in DNA replication.
Common fragile sites are prone to undergo sister chromatid exchange (49), and fragile telomere sites caused by conditional deletion of TRF1 also lead to an increase in telomere sister chromatid exchange (10). We tested the frequency of T-SCE in RECQL4-depleted and control U2OS cells using the chromatid orientation FISH technique. The control cells had about 7% T-SCE/chromosome, and the incidence of this event increased more than 2.5-fold (18%) after depletion of RECQL4 (Fig. 4C and supplemental Fig. 2). We also tested the frequency of T-SCE in RECQL4-depleted HeLa cells and could not find any significant increase in this event compared with the control cell (data not shown). It is possible that the presence of a high level of telomerase in HeLa cells suppresses the occurrence of T-SCE.
RECQL4 Associates with Telomeres in Replicating U2OS and HeLa Cells
If RECQL4 plays an important role in telomere maintenance, this protein might associate with the telomere. To test this hypothesis, we investigated the association of RECQL4 with TRF1 by immunofluorescence during different cell cycle phases. Cell cycle progression in U2OS cells was blocked at G1 phase by double thymidine block, and 4 h after thymidine release most of the cells were in S phase. M phase cells were obtained by nocodazole treatment (Fig. 5A). Consistent with the role of RECQL4 in general replication, the level of RECQL4 was considerably higher in S phase than in the G1 and M phases. During S phase on average 75% of TRF1 foci/cell colocalized with the RECQL4 signal (Fig. 5B and supplemental Fig. 3). During G1 and M phases, however, very few TRF1 foci were associated with RECQL4 (Fig. 5B). During S phase, the RECQL4 signal was distributed over the nucleus rather than forming clear foci and thus could possibly be positioned over the TRF1 signal causing false colocalization. To exclude this possibility, we examined the three-dimensional image of each colocalized foci and affirmed that signals from RECQL4 and TRF1 actually coexist (Fig. 5C). Additionally, we analyzed the Pearson's correlation coefficient between the GFP channel (RECQL4) and the Cy5 channel (TRF1) of 50 randomly selected S phase U2OS cells. A Pearson's coefficient value between 0.5 and 1 indicates colocalization between the two channels (50). 80% of the selected cells had Pearson's coefficient value within this range (Fig. 5D) indicating a significant association between RECQL4 and the telomere. We further tested the localization of RECQL4 in HeLa cells during S phase. The confocal microscopic analysis showed that on average 68% of the TRF1 signals/cell were associated with RECQL4 (supplemental Fig. 3B).
To further confirm the association of endogenous TRF1 and TRF2 with RECQL4 in vivo, we performed co-immunoprecipitation experiments using the nuclear extracts from S phase U2OS cells. This experiment was performed in the presence of ethidium bromide, thus eliminating any DNA-mediated association. A rabbit antibody raised against human RECQL4 precipitated endogenous RECQL4 protein, along with endogenous TRF1 and TRF2 (supplemental Fig. 3C, lanes 3 and 6, respectively). Anti-rabbit IgG antibody was not able to precipitate either of these two shelterin proteins, confirming the specificity of this experiment (supplemental Fig. 3C, lanes 3 and 6, respectively). Although, the quantification of this experiment suggests that a very small fraction of TRF2 or TRF1 is associated with REQL4, another RecQ helicase, RECQL5, was not able to immunoprecipitate TRF2 under similar conditions (data not shown). Taken together, these results demonstrate the association of RECQL4 with the telomere and with shelterin proteins TRF1 and TRF2, especially during S phase, further substantiating the involvement of this RecQ helicase in telomere maintenance.
RECQL4 Interacts with Telomeric D-loop Substrates
Telomeric DNA can form different complex structures including G-quadruplex and D-loops in vitro. Although not yet shown in vivo, the resolution of D-loops is thought to be important for telomere maintenance (8). It has been shown that WRN interacts with telomeres in vivo and partially unwinds model telomeric D-loop substrates in vitro (11, 36). Our results suggest that RECQL4 is involved in the preservation of the telomeres and that it localizes to telomeres during S phase when resolution of D-loops is required. Thus, we investigated whether RECQL4 could bind to and unwind DL1 or DLmx in vitro. The oligos used to prepare these substrates and a schematic of these substrates are shown in Fig. 6. The invading strand (boxed in Fig. 6) has four telomere repeats (TTAGGG) in DL1, whereas DLmx has a non-telomeric (but the same G-C content) sequence in the invading strand. RECQL4 bound both substrates efficiently (Fig. 7A). However, with 30 nm RECQL4, all of the DL1 was bound to the protein (Fig. 7A, lane 4), whereas ∼40% DLmx remained unbound (lane 8). Additionally, DL1 was able to bind to multiple RECQL4 proteins and to form more than one slow moving structure (see Fig. 7A, a, b, and c). Although DLmx was able to form structures a and b, the formation of c was not detected (Fig. 7A). This result suggests that RECQL4 may have greater binding affinity toward the telomeric D-loop than toward the non-telomeric D-loop. However, the slightly higher apparent affinity suggested by this experiment could also reflect the binding of multiple copies of RECQL4 to the telomeric D-loop.
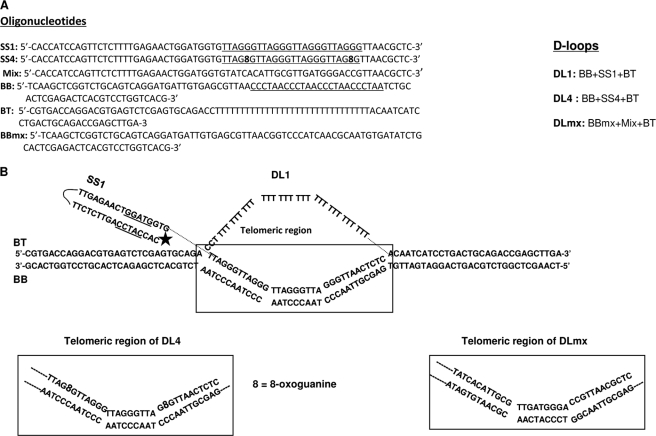
FIGURE 6.
Telomeric D-loop substrates. A, complete list of oligos used to prepare the D-loop structures. B, structures of the D-loop substrates used in this study. The full structure of DL1, composed of three oligos, SS1, BT, and BB, is shown, and the telomeric region is marked. The telomeric regions of DL4 and DLmx are also shown. The rest of the structure remains the same in these three substrates.
Previous studies show that RECQL4 unwinds short DNA duplexes (≤24 bp) (19) and can also unwind longer dsDNA substrates in the presence of excess ssDNA, which counteracts the strong ssDNA annealing activity of RECQL4 (19, 37). RECQL4 (40 nm) was not able to unwind these D-loop substrates in the absence of unlabeled competitor ssDNA (data not shown), likely because of the length of the duplex region. RECQL4 did unwind DL1 with low efficiency in the presence of a 25-fold molar excess of unlabeled competitor ssDNA (Fig. 7, B, lanes 1–4, and D). Interestingly, RECQL4 was not able to unwind the non-telomeric D-loop, DLmx (only 1% resolved strand with 25 nm RECQL4), even in the presence of a 25-fold molar excess of ssDNA (Fig. 7, B, lanes 5–8, and D). To appropriately trap the released unlabeled strand of the D-loops and unmask the helicase activity, ssDNAs SS1 and SSmx were used as competitors for the experiments involving DL1 and DLmx, respectively. Thus, RECQL4, which has very limited substrate specificity (19), is capable of unwinding telomeric DNA substrates with low efficiency. Further, a helicase-dead mutant of RECQL4 (RECQL4M) was not able to unwind DL1 (Fig. 7C) even in the presence of 25× single-stranded DNA SS1, indicating that the unwinding of this D-loop is a result of RECQL4 helicase activity and not of a strand exchange activity.
Telomeric GGG repeats are hot spots for oxidative damage, and both WRN and BLM unwind oxidatively damaged D-loops (37). Here, we demonstrate that RECQL4 unwinds 8-oxoguanine-containing D-loops (Fig. 6B, DL4) with moderate efficiency (Fig. 7, B, lanes 10–13, and D). RECQL4M was not able to unwind this substrate (Fig. 7C). This result shows that RECQL4 resembles WRN and BLM in that it resolves telomeric D-loop substrates more efficiently when they are oxidatively damaged than when they are undamaged (36). Interestingly, bacterial helicase UvrD unwinds undamaged and damaged substrates with equal efficiency (36). Thus, the selectivity toward the oxidatively damaged substrate shown by these RecQ helicases is specific to these proteins and does not signify any differences in stability between these two substrates.
RECQL4 Interacts Physically with Shelterin Protein TRF2
Shelterin proteins POT1, TRF1, and TRF2 stimulate the helicase activity of WRN on telomeric D-loop substrates (11, 36). Because these proteins all are integral to telomere maintenance, we sought to determine whether, individually, they could modulate the helicase activity of RECQL4. The effect of TRF2 and TRF1 on the unwinding ability of RECQL4 on the telomeric D-loop DL1 is shown in Fig. 8, A and B. In the presence of excess ssDNA, 10 nm RECQL4 was able to unwind ∼8% of the 0.5 nm substrate, and this increased to 14% (1.75-fold increase) or 16% (2-fold increase) after the addition of 10 or 20 nm TRF2, respectively. TRF2 (20 nm) alone did not unwind DL1 under these reaction conditions (Fig. 8A, lane 11). Similar results were obtained when RECQL4 helicase assays were carried out in the presence of TRF1. Both TRF1 and TRF2 are double strand-binding proteins and did not interact with the excess single-stranded DNA. To assess whether TRF1 and/or TRF2 directly stimulated the helicase activity of RECQL4 or simply inhibited the strong annealing activity of the protein, we tested the effect of TRF1 and TRF2 on the ability of RECQL4 to anneal oligos SS1 and BB. Our results (Fig. 8C) show that neither TRF1 nor TRF2 has any effect on the annealing activity of RECQL4, suggesting that these two shelterin proteins directly stimulate the helicase activity of RECQL4. Additionally, GST-POT1 (at 5, 10, or 20 nm) stimulated the RECQL4 unwinding of DL1 ∼2-fold in the absence of ssDNA and stimulated the unwinding of DL4 about 6-fold (1.5–10% using 0–20 nm POT1) (supplemental Fig. 4). We understand that the quantitative analysis of catalytic activities of enzymes may have limited meaning when the reaction is assembled with a large excess of proteins. However, RecQ helicases are not very processive in vitro, and it is a common practice in this field to use a large excess of proteins to attain a reasonable activity.
A physical association between WRN and TRF2 has also been reported (51). Here, we have reported that in human cells TRF2 and TRF1 associate with RECQL4. To confirm the direct interaction between full-length RECQL4 and TRF2, we performed co-immunoprecipitation experiments in vitro using purified recombinant proteins. RECQL4 antibody was able to detect TRF2 from a mixture of these two proteins (Fig. 8C, lane 3). However, we were not able to find any physical interaction between RECQL4 and TRF1 in immunoprecipitation experiments, indicating that the association between these proteins may be transient or bridged by TRF2. Collectively, RECQL4 associates physically with TRF2 and shows a functional interaction with TRF1, TRF2, and POT1 because they all stimulate RECQL4 helicase activity on telomeric D-loop substrate.
RECQL4 and WRN Synergistically Resolve Telomeric D-loops
WRN has been implicated in telomere replication in vivo, and both WRN and BLM can resolve telomeric D-loops in vitro. However, their processivity on these structures are poor. In our hands, 5–10 nm WRN or BLM was required to achieve 5–15% unwinding of 0.5 nm undamaged D-loop substrate (DL1), depending on the reaction conditions and activity of the protein preparation (11, 36). The results shown here suggest that RECQL4 also plays an important part in telomere replication. Yet it could only resolve telomeric D-loop substrates in the presence of excess ssDNA or when stimulated by POT1, TRF1, or TRF2. Thus, we investigated whether RECQL4 could work together with WRN or BLM in proficient unwinding of these secondary structures. These experiments were done in the absence of any excess single-stranded DNA, and thus RECQL4 by itself does not have helicase activity on undamaged (DL1) or oxidatively damaged (DL4) telomeric D-loops (Fig. 9A, lanes 6 and 12). Under these reaction conditions, 10 nm WRN and 5 nm BLM, individually, were able to unwind only 5–7% of DL1. Different concentrations of WRN and BLM were used here to get a similar helicase activity at the outset for a better comparison. However, when WRN was preincubated with RECQL4 (1:1 molar ratio) and then added to the substrate, the helicase activity of the former increased almost 6-fold (Fig. 9, A, lanes 2 and 3, and B). RECQL4 was also able to stimulate WRN activity on the 8-oxoguanine-containing D-loop, DL4 (Fig. 9, A, lanes 8 and 9, and B). In contrast, RECQL4 (5 nm) only weakly stimulated BLM helicase (5 nm) on DL1 (Fig. 9, A, lanes 4 and 5, and B) (∼1.5-fold) and had very little effect on the unwinding of DL4 by BLM (Fig. 9A, lanes 10 and 11, and B). 10 nm RECQL4 did not significantly stimulate BLM activity on these substrates either (supplemental Fig. 5). These results suggest that RECQL4 specifically stimulates WRN helicase activity on telomeric D-loops in vitro. The association between WRN and RECQL4 was further examined by in vivo co-immunoprecipitation studies. In this experiment, U2OS cells were transduced with FLAG-tagged RECQL4 or empty vector. Cells were then precipitated with rabbit anti-FLAG antibody, which was able to detect WRN (Fig. 9C, lane 3) from FLAG-RECQL4-expressed cells in the presence of ethidium bromide. These results suggest that RECQL4 and WRN are present in the same complex and, although RECQL4 by itself has very little activity on telomeric D-loops, it can work synergistically with WRN and resolve telomeric D-loops very efficiently.
DISCUSSION
We observed that the depletion of RECQL4 in human cells resulted in increased telomeric fragile telomeres. As for normally fragile sites, replication stress enhanced this phenotype in RECQL4-depleted cells. The accumulation of the DNA damage response factor 53BP1, globally and at the telomere, was also a result of loss of RECQL4. Also, expression of RECQL4 was greatly enhanced in replicative cells, and the amount of RECQL4 at telomeres increased during S phase. These results implicate a malfunction of the telomere replication machinery and of telomere maintenance in the absence of RECQL4.
The predicted sequence loss at telomeres because of the “end replication problem” is about five bases/population doubling (52, 53). Primary human cells lose about 100–200 telomeric sequences/cell division (7, 54). Apart from the end replication problem, some other factors are also involved in the telomere shortening process. One of them is telomeric DNA itself, which is a challenging substrate for the replication machinery. The telomeric single-stranded overhang can invade the double-stranded telomeric region to form a protective loop, known as a telomeric displacement loop or “D-loop” (2, 55). Dissociation of this structure is essential for the proper progression of replication at telomeres, and RecQ helicases WRN and BLM are thought to partially resolve this configuration (56). Here we have shown that RECQL4 can resolve telomeric D-loops in vitro, and like WRN and BLM this activity can be stimulated by shelterin proteins POT1, TRF1, and TRF2. The recently discovered intrinsic helicase activity of RECQL4 (19, 37) is essential for the resolution of complex D-loop structures. Furthermore, the integrity of the SFII helicase domain of RECQL4 is known to be crucial for preventing RTS, Baller-Gerold Syndrome, and RAPADILINO syndromes (25, 57). A RECQL4 mouse model with a defective helicase domain showed most of the phenotypes of the RTS and RAPADILINO human syndromes including defective sister chromatid cohesion, aneuploidy, and cancer predisposition (58). The significance of the SFII domain in cell proliferation and genomic stability was further proven by studies with mouse embryonic fibroblasts derived from the RECQL4 SFII helicase-deficient mice (31, 58). Supporting the importance of the RECQL4 helicase region in telomeric replication, we showed that the frequency of fragile telomeres is elevated in RTS patient cells containing RECQL4 mutated in the helicase region. We have also demonstrated the association between RECQL4 and the shelterin protein TRF2 in vitro and in vivo by immunoprecipitation studies. A shared docking motif, the TRF homology domain, between TRF1 and TRF2 is used for recruitment of different proteins to the telomere, and many of the telomere-associated proteins contain this (F/Y)XLXP motif (59). Interestingly, RECQL4 also contains a putative TRF homology motif, YSLGP, which is encoded by a region of the RECQL4 gene that is deleted in the majority of RAPADILINO patients (57). Additionally, we have demonstrated that RECQL4 interacts with WRN and strongly stimulates its helicase activity on telomeric D-loops. We speculate that RECQL4 is required to unwind a small part of the D-loop, creating a 3′-overhang on which WRN can bind and execute its helicase activity. Thus, we propose that RECQL4 is recruited to telomeres during replication and either resolves D-loops directly, with the help of shelterin proteins, or works synergistically with WRN to dissociate these structures (Fig. 9D). On the other hand, Sfeir et al. (10) have shown recently that BLM knockdown, but not WRN, elevates the fragile telomere phenotype in mice. BLM has been shown to play an important role in G-quadruplex resolution, whereas WRN is important for lagging strand synthesis. As RECQL4 does not interact with G-quadruplexes, it is likely that it interacts with WRN, but not BLM, to resolve the D-loops at telomeres. Additionally, Werner syndrome and RTS individuals have similar phenotypes, and it has been widely speculated that a major cause of Werner syndrome is telomere dysfunction. Our results, implicating a role for RECQL4 in telomere maintenance, further supports the importance of telomere dysfunction in human disease.
Telomeric DNA, being G-rich, is very susceptible to oxidation (60). Although there are several important oxidatively damaged modifications, 8-oxoguanine is the most studied and a biologically significant base lesion. We have previously shown that WRN and BLM are more active on D-loops containing 8-oxoguanine and that POT1 stimulates their activities on damaged D-loops (36). Although, RECQL4 has very poor helicase activity, it is also more proficient in unwinding 8-oxoguanine-containing telomeric D-loops than undamaged D-loops. Therefore, RECQL4 could assist WRN and BLM in promoting DNA repair and replication of telomeres containing oxidatively damaged DNA.
Mammalian cells contain five RecQ helicases, but at present the interactions between these enzymes are poorly understood. Previous studies show that WRN and BLM interact physically and that BLM inhibits the exonuclease activity of WRN (61). This present study demonstrates that RECQL4 stimulates the unwinding of telomeric D-loops by WRN in vitro but does not stimulate BLM helicase activity on the same DNA substrates. These results suggest subtle differences in the enzymatic properties, as well as the selectivity and specialized biological roles of these helicases. BLM may be important in the progression of the replication fork globally, whereas WRN may play a more specific role in telomere replication. Supporting this concept, Bailey and colleagues (62) recently demonstrated that depletion of WRN induces T-SCE, whereas depletion of BLM induces SCE at non-telomeric chromosome regions (i.e. global SCE). In conclusion, we now implicate another RecQ helicase, in addition to WRN, in telomere processing. This presents a new direction for work toward understanding the defects and deficiencies in patients with inactivation of RECQL4.
Supplementary Material
Acknowledgments
We thank Drs. Haritha Vallabhaneni, Jinhu Yin, and Takashi Tadokoro and Mr. Alfred May for technical help and Drs. David Wilson III and Jian Lu for comments.
This work was supported, in whole or in part, by the National Institutes of Health NIA Intramural Program.

This article contains supplemental Figs. 1–5.
- ssDNA
- single-stranded DNA
- dsDNA
- double-stranded DNA
- RTS
- Rothmund-Thomson syndrome
- KD
- knockdown
- SCR
- scrambled
- co-IP
- co-immunoprecipitation
- TIF
- telomere dysfunction-induced foci
- T-SCE
- telomere sister chromatid exchange
- DL1
- telomeric D-loop
- DL4
- 8-oxoguanine-containing D-loop
- DLmx
- non-telomeric D-loop.
REFERENCES
- 1. Smogorzewska A., de Lange T. (2004) Annu. Rev. Biochem. 73, 177–208 [DOI] [PubMed] [Google Scholar]
- 2. Griffith J. D., Comeau L., Rosenfield S., Stansel R. M., Bianchi A., Moss H., de Lange T. (1999) Cell 97, 503–514 [DOI] [PubMed] [Google Scholar]
- 3. de Lange T. (2002) Oncogene 21, 532–540 [DOI] [PubMed] [Google Scholar]
- 4. Lei M., Podell E. R., Cech T. R. (2004) Nat. Struct. Mol. Biol. 11, 1223–1229 [DOI] [PubMed] [Google Scholar]
- 5. Palm W., de Lange T. (2008) Annu. Rev. Genet. 42, 301–334 [DOI] [PubMed] [Google Scholar]
- 6. Greider C. W., Blackburn E. H. (1985) Cell 43, 405–413 [DOI] [PubMed] [Google Scholar]
- 7. Verdun R. E., Karlseder J. (2007) Nature 447, 924–931 [DOI] [PubMed] [Google Scholar]
- 8. Verdun R. E., Karlseder J. (2006) Cell 127, 709–720 [DOI] [PubMed] [Google Scholar]
- 9. Martínez P., Thanasoula M., Muñoz P., Liao C., Tejera A., McNees C., Flores J. M., Fernández-Capetillo O., Tarsounas M., Blasco M. A. (2009) Genes Dev. 23, 2060–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sfeir A., Kosiyatrakul S. T., Hockemeyer D., MacRae S. L., Karlseder J., Schildkraut C. L., de Lange T. (2009) Cell 138, 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Opresko P. L., Otterlei M., Graakjaer J., Bruheim P., Dawut L., Kølvraa S., May A., Seidman M. M., Bohr V. A. (2004) Mol. Cell 14, 763–774 [DOI] [PubMed] [Google Scholar]
- 12. Lillard-Wetherell K., Machwe A., Langland G. T., Combs K. A., Behbehani G. K., Schonberg S. A., German J., Turchi J. J., Orren D. K., Groden J. (2004) Hum. Mol. Genet. 13, 1919–1932 [DOI] [PubMed] [Google Scholar]
- 13. Opresko P. L., Mason P. A., Podell E. R., Lei M., Hickson I. D., Cech T. R., Bohr V. A. (2005) J. Biol. Chem. 280, 32069–32080 [DOI] [PubMed] [Google Scholar]
- 14. Bohr V. A. (2008) Trends Biochem. Sci. 33, 609–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chu W. K., Hickson I. D. (2009) Nat. Rev. Cancer 9, 644–654 [DOI] [PubMed] [Google Scholar]
- 16. Hickson I. D. (2003) Nat. Rev. Cancer 3, 169–178 [DOI] [PubMed] [Google Scholar]
- 17. Yu C. E., Oshima J., Fu Y. H., Wijsman E. M., Hisama F., Alisch R., Matthews S., Nakura J., Miki T., Ouais S., Martin G. M., Mulligan J., Schellenberg G. D. (1996) Science 272, 258–262 [DOI] [PubMed] [Google Scholar]
- 18. Larizza L., Roversi G., Volpi L. (2010) Orphanet J. Rare Dis. 5, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rossi M. L., Ghosh A. K., Kulikowicz T., Croteau D. L., Bohr V. A. (2010) DNA Repair 9, 796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Popuri V., Bachrati C. Z., Muzzolini L., Mosedale G., Costantini S., Giacomini E., Hickson I. D., Vindigni A. (2008) J. Biol. Chem. 283, 17766–17776 [DOI] [PubMed] [Google Scholar]
- 21. Brosh R. M., Jr., Karow J. K., White E. J., Shaw N. D., Hickson I. D., Bohr V. A. (2000) Nucleic Acids Res. 28, 2420–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Walpita D., Plug A. W., Neff N. F., German J., Ashley T. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 5622–5627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Singh D. K., Karmakar P., Aamann M., Schurman S. H., May A., Croteau D. L., Burks L., Plon S. E., Bohr V. A. (2010) Aging Cell 9, 358–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jin W., Liu H., Zhang Y., Otta S. K., Plon S. E., Wang L. L. (2008) Hum. Genet. 123, 643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu Y. (2010) DNA Repair 9, 325–330 [DOI] [PubMed] [Google Scholar]
- 26. Schurman S. H., Hedayati M., Wang Z., Singh D. K., Speina E., Zhang Y., Becker K., Macris M., Sung P., Wilson D. M., 3rd, Croteau D. L., Bohr V. A. (2009) Hum. Mol. Genet. 18, 3470–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matsuno K., Kumano M., Kubota Y., Hashimoto Y., Takisawa H. (2006) Mol. Cell. Biol. 26, 4843–4852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sangrithi M. N., Bernal J. A., Madine M., Philpott A., Lee J., Dunphy W. G., Venkitaraman A. R. (2005) Cell 121, 887–898 [DOI] [PubMed] [Google Scholar]
- 29. Xu X., Rochette P. J., Feyissa E. A., Su T. V., Liu Y. (2009) EMBO J. 28, 3005–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thangavel S., Mendoza-Maldonado R., Tissino E., Sidorova J. M., Yin J., Wang W., Monnat R. J., Jr., Falaschi A., Vindigni A. (2010) Mol. Cell. Biol. 30, 1382–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hoki Y., Araki R., Fujimori A., Ohhata T., Koseki H., Fukumura R., Nakamura M., Takahashi H., Noda Y., Kito S., Abe M. (2003) Hum. Mol. Genet. 12, 2293–2299 [DOI] [PubMed] [Google Scholar]
- 32. Mohaghegh P., Hickson I. D. (2001) Hum. Mol. Genet. 10, 741–746 [DOI] [PubMed] [Google Scholar]
- 33. Walne A. J., Vulliamy T., Beswick R., Kirwan M., Dokal I. (2010) Hum. Mol. Genet. 19, 4453–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stinco G., Governatori G., Mattighello P., Patrone P. (2008) J. Dermatol. 35, 154–161 [DOI] [PubMed] [Google Scholar]
- 35. Vennos E. M., Collins M., James W. D. (1992) J. Am. Acad. Dermatol. 27, 750–762 [DOI] [PubMed] [Google Scholar]
- 36. Ghosh A., Rossi M. L., Aulds J., Croteau D., Bohr V. A. (2009) J. Biol. Chem. 284, 31074–31084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xu X., Liu Y. (2009) EMBO J. 28, 568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Saharia A., Teasley D. C., Duxin J. P., Dao B., Chiappinelli K. B., Stewart S. A. (2010) J. Biol. Chem. 285, 27057–27066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Karow J. K., Newman R. H., Freemont P. S., Hickson I. D. (1999) Curr. Biol. 9, 597–600 [DOI] [PubMed] [Google Scholar]
- 40. Orren D. K., Brosh R. M., Jr., Nehlin J. O., Machwe A., Gray M. D., Bohr V. A. (1999) Nucleic Acids Res. 27, 3557–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bailey S. M., Brenneman M. A., Goodwin E. H. (2004) Nucleic Acids Res. 32, 3743–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zijlmans J. M., Martens U. M., Poon S. S., Raap A. K., Tanke H. J., Ward R. K., Lansdorp P. M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 7423–7428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kitao S., Shimamoto A., Goto M., Miller R. W., Smithson W. A., Lindor N. M., Furuichi Y. (1999) Nat. Genet. 22, 82–84 [DOI] [PubMed] [Google Scholar]
- 44. Cabral R. E., Queille S., Bodemer C., de Prost Y., Neto J. B., Sarasin A., Daya-Grosjean L. (2008) Mutat. Res. 643, 41–47 [DOI] [PubMed] [Google Scholar]
- 45. Wang L. L., Levy M. L., Lewis R. A., Chintagumpala M. M., Lev D., Rogers M., Plon S. E. (2001) Am. J. Med. Genet. 102, 11–17 [DOI] [PubMed] [Google Scholar]
- 46. Liu F. J., Barchowsky A., Opresko P. L. (2010) PLoS One 5, e11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Takai H., Smogorzewska A., de Lange T. (2003) Curr. Biol. 13, 1549–1556 [DOI] [PubMed] [Google Scholar]
- 48. Tripathi V., Nagarjuna T., Sengupta S. (2007) J. Cell Biol. 178, 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Glover T. W., Arlt M. F., Casper A. M., Durkin S. G. (2005) Hum. Mol. Genet. 14, Suppl. 2, R197–R205 [DOI] [PubMed] [Google Scholar]
- 50. Zinchuk V., Zinchuk O. (2008) Curr. Protoc. Cell Biol. 2008, Chapter 4, Unit 4.19 [DOI] [PubMed] [Google Scholar]
- 51. Opresko P. L., von Kobbe C., Laine J. P., Harrigan J., Hickson I. D., Bohr V. A. (2002) J. Biol. Chem. 277, 41110–41119 [DOI] [PubMed] [Google Scholar]
- 52. Lingner J., Hughes T. R., Shevchenko A., Mann M., Lundblad V., Cech T. R. (1997) Science 276, 561–567 [DOI] [PubMed] [Google Scholar]
- 53. Singer M. S., Gottschling D. E. (1994) Science 266, 404–409 [DOI] [PubMed] [Google Scholar]
- 54. Harley C. B., Futcher A. B., Greider C. W. (1990) Nature 345, 458–460 [DOI] [PubMed] [Google Scholar]
- 55. de Lange T. (2004) Nat. Rev. Mol. Cell Biol. 5, 323–329 [DOI] [PubMed] [Google Scholar]
- 56. Gilson E., Géli V. (2007) Nat. Rev. Mol. Cell Biol. 8, 825–838 [DOI] [PubMed] [Google Scholar]
- 57. Siitonen H. A., Sotkasiira J., Biervliet M., Benmansour A., Capri Y., Cormier-Daire V., Crandall B., Hannula-Jouppi K., Hennekam R., Herzog D., Keymolen K., Lipsanen-Nyman M., Miny P., Plon S. E., Riedl S., Sarkar A., Vargas F. R., Verloes A., Wang L. L., Kääriäinen H., Kestilä M. (2009) Eur. J. Hum. Genet. 17, 151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mann M. B., Hodges C. A., Barnes E., Vogel H., Hassold T. J., Luo G. (2005) Hum. Mol. Genet. 14, 813–825 [DOI] [PubMed] [Google Scholar]
- 59. Chen Y., Yang Y., van Overbeek M., Donigian J. R., Baciu P., de Lange T., Lei M. (2008) Science 319, 1092–1096 [DOI] [PubMed] [Google Scholar]
- 60. Wang Z., Rhee D. B., Lu J., Bohr C. T., Zhou F., Vallabhaneni H., de Souza-Pinto N. C., Liu Y. (2010) PLoS Genet. 6, e1000951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. von Kobbe C., Karmakar P., Dawut L., Opresko P., Zeng X., Brosh R. M., Jr., Hickson I. D., Bohr V. A. (2002) J. Biol. Chem. 277, 22035–22044 [DOI] [PubMed] [Google Scholar]
- 62. Hagelstrom R. T., Blagoev K. B., Niedernhofer L. J., Goodwin E. H., Bailey S. M. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 15768–15773 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.