Background: Tryptophan side chains can influence the binding of amphipathic peptides to biological membranes.
Results: The cytotoxic activity of model helical amphipathic peptides was markedly influenced by the positions of tryptophan residues in the sequence.
Conclusion: Tryptophan residues located adjacent to a hydrophobic helical portion created the most potent cytotoxic peptides.
Significance: More potent anticancer helical peptides can now be designed.
Keywords: Antimicrobial Peptides, Cancer Therapy, Liposomes, Nuclear Magnetic Resonance, Peptide Conformation, Anticancer Peptides, Cationic Antimicrobial Peptides, Peptide Structure
Abstract
The cytotoxic activity of 10 analogs of the idealized amphipathic helical 21-mer peptide (KAAKKAA)3, where three of the Ala residues at different positions have been replaced with Trp residues, has been investigated. The peptide's cytotoxic activity was found to be markedly dependent upon the position of the Trp residues within the hydrophobic sector of an idealized α-helix. The peptides with Trp residues located opposite the cationic sector displayed no antitumor activity, whereas those peptides with two or three Trp residues located adjacent to the cationic sector exhibited high cytotoxic activity when tested against three different cancer cell lines. Dye release experiments revealed that in contrast to the peptides with Trp residues located opposite the cationic sector, the peptides with Trp residues located adjacent to the cationic sector induced a strong permeabilizing activity from liposomes composed of a mixture of zwitterionic phosphatidylcholine and negatively charged phosphatidylserine (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC)/1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine (POPS)) (2:1) but not from liposomes composed of zwitterionic phosphatidylcholine, POPC. Fluorescence blue shift and quenching experiments revealed that Trp residues inserted deeper into the hydrophobic environment of POPC/POPS liposomes for peptides with high cytotoxic activity. Through circular dichroism studies, a correlation between the cytotoxic activity and the α-helical propensity was established. Structural studies of one inactive and two active peptides in the presence of micelles using NMR spectroscopy showed that only the active peptides adopted highly coiled to helical structures when bound to a membrane surface.
Introduction
Membrane-active peptides can broadly be divided into two classes: antibacterial peptides like magainins (1), cecropins (2), cathelicidins (3), and defensins (4) and cytotoxic peptides like melittin (5) and mastoperans (6). In addition, some peptides, such as the cyclotides, display a range of different biological activities, in which an action on the cell membrane most likely is involved (7, 8). Both antibacterial and cytotoxic peptides may perturb the barrier function of cell membranes, which may eventually lead to cell lysis and cell death. Antibacterial peptides selectively kill prokaryotic cells, whereas cytotoxic peptides have a lytic activity against both prokaryotic and eukaryotic cells. Whereas cytotoxic peptides are generally considered to be highly toxic against normal eukaryotic cells, several antibacterial peptides have been found that are able to kill certain types of tumor cells at concentrations that are not toxic toward non-malignant cells (9–11). Peptides that are able to kill cancer cells have been found to depolarize the transmembrane potential of cancer cells at the same rate and concentration as for the biological activity, and this has led to the suggestion of a killing mechanism that involves perturbations of the plasma membrane. Because these peptides are highly cationic, anionic membrane constituents, such as phosphatidylserine, a phospholipid with a negatively charged head group, that is found on the outer leaflet of cancer cells, may function as a target for cationic peptides (12, 13). It is noteworthy that an elevated phosphatidylserine expression of 3–9% has been found in the outer leaflet of cancer cells (14). Furthermore, several of these cationic peptides exert a toxic effect against multidrug-resistant cell lines and potentiate the toxic effect of doxorubicin against such multidrug-resistant tumor cells (15). In addition, intratumoral administration of a cationic anticancer peptide in animal models has resulted in a long term, specific cellular immunity against the treated tumor (16). Papo et al. (17, 18) have demonstrated that enzymatically stable cationic peptides with a high specificity for cancer cells versus normal cells inhibit tumor growth in animal models after intravenous injections. Thus, these peptides confer a promising novel strategy for developing new anticancer agents that may overcome some of the obstacles in cancer therapy (for reviews, see Refs. 14 and 19). In the literature, it has been common to loosely refer to peptides that are able to kill cancer cells as possessing anticancer or antitumor activities; however, in practice, one is often only measuring the cytotoxic activity. In the present study, we have applied an in vitro assay to evaluate the ability of peptides to kill cancer cells, and we are thus referring to the observed biological activity as cytotoxic activity.
Many membrane-disturbing peptides share two common features: amphipathicity and a net positive charge. The cationic residues seem to be important for an initial electrostatic interaction with negatively charged membrane components, whereas hydrophobic moieties appear to be important for membrane disruption (20). The linear peptides usually are flexible and unfolded in aqueous solutions, but they can organize into amphipathic structures upon binding to negatively charged membranes (20, 21). Whereas a lot is known regarding the structural parameters that are critical for antimicrobial activity of cationic membrane-active peptides (22), much less is known regarding the structural basis for tumor cell specificity. Earlier studies have, however, revealed that a stable secondary structure is more important for activity against eukaryotic cells than for antimicrobial activity (23). We have also demonstrated that the angle subtended by the cationic residues in helical peptides is an important factor that modulates cytotoxic activity and tumor cell specificity (23). Additionally, it has been found that the number of aromatic residues in the lipophilic part of membrane-active peptides is critical for cytotoxic activity (24). The aromatic amino acid Trp has been singled out as being of special importance among the proteinogenic aromatic amino acids due to its large size, shape, and aromatic character (25–29).
In the present study, the influence of the position of Trp residues on the anticancer activity of membrane-active peptides has been explored. Based on the idealized amphipathic helical 21-mer peptide (KAAKKAA)3, 10 synthetic peptides were produced where three of the Ala residues in different positions have been replaced with Trp. These were investigated for their in vitro cytotoxicity and their ability to interact with and disrupt model membrane systems. The propensity to form α-helices under different conditions has been investigated for all peptides, and in addition, the three-dimensional structures for selected peptides in the presence of model membrane systems have been elucidated through nuclear magnetic resonance (NMR) experiments.
EXPERIMENTAL PROCEDURES
Materials
All Fmoc-amino acids, resins, and chemicals used during peptide synthesis, cleavage, and precipitation were purchased from PerSeptive Biosystems (Warrington, UK), Fluka (Buchs, Switzerland), Advanced ChemTech (Louisville, KY), Applied Biosystems (Warrington, UK), and Sigma-Aldrich. Fetal bovine serum (FBS) was obtained from Biochrom KG (Berlin, Germany), and l-glutamine was from Invitrogen. All culture media were prepared at the University of Tromsø (Tromsø, Norway). 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)3 was obtained from Sigma. 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) was obtained from Genzyme Pharmaceuticals (Cambridge, MA). 1-Palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine (POPS) was purchased from Avanti Polar Lipids (Alabaster, AL). Sodium dodecyl-d25 sulfate (SDS-d25), dodecylphosphocholine-d38 (DPC-d38), sodium 2,2-dimethyl-2-silapentane-5-sulfonate (DSS), and D2O were purchased from Cambridge Isotopes laboratories, Inc. (Andover, MA). Calcein and Triton X-100 were obtained from Sigma, whereas Sephadex® G-50 medium was obtained from Amersham Biosciences.
Peptide Synthesis
Synthesis and purification of the peptides were performed as described previously (25). Briefly, the peptides were synthesized in an automated fashion on a Milligen 9050 Plus PepSynthesizer (Milligen, Milford, MA) using standard solid-phase Fmoc chemistry and preloaded PAC-PEG-PSTM resins. The peptides were purified by reversed phase high pressure liquid chromatography (RP-HPLC) (Delta-PakTM C18, 100 Å, 15 μm, 25 × 100 mm; Waters Corp., Milford, MA). The purified peptides were shown to be homogeneous (>95%) by analytical RP-HPLC (Delta-PakTM C18, 100 Å, 15 μm, 3.9 × 150 mm; Waters Corp.), and the integrity of the peptides was checked by positive ion electrospray ionization mass spectrometry on a VG Quattro quadropole mass spectrometer (VG Instruments Inc., Altringham, UK).
Cell Cultures
The murine fibrosarcoma cell line MethA (30), the human colorectal adenocarcinoma cell line HT-29 (ATCC; HTB-38), and the human mammary carcinoma cells MT-1 (kindly provided by Dr. Fodstad, Norwegian Radium Hospital, Oslo, Norway) were all grown in RPMI 1640 medium. The human embryonic fibroblast cell line MRC-5 was grown in minimum essential medium Eagle. Media for MethA, HT-29, MT-1, and MRC-5 were supplemented with 10% heat-inactivated FBS and 1% l-glutamine. The MethA cell line was grown in suspension, whereas the other cell lines were cultured as monolayers. All cells were incubated in a humidified atmosphere of 5% CO2 at 37 °C.
In Vitro Cytotoxicity
To assess the cytotoxicity of the peptides for the different cell lines, the MTT (Sigma) reduction assay was employed (31). The assays were performed in 96-well microtiter plates (Falcon, BD Biosciences). The seeding densities of the different cell lines were predetermined to ensure that all of the cell lines were at similar subconfluence levels before use. HT-29 cells (4 × 104 cells/well), MT-1 cells (1.5 × 104 cells/well), and MRC-5 cells (1 × 104 cells/well) were seeded 16 h prior to use. After the incubation period, the growth medium was removed, and the cells were washed with serum-free RPMI 1640 medium prior to use. MethA cells (4 × 104 cells/well) were seeded the same day as the experiment was performed. The seeded cells were stimulated with peptide concentrations ranging from 1 to 500 μg/ml, dissolved in serum-free RPMI 1640 medium, and allowed to incubate for 4 h at 37 °C. Cells in serum-free RPMI 1640 medium functioned as a negative control, whereas cells treated with 1% Triton X-100 in RPMI 1640 medium functioned as a positive control. Following incubation, 10 μl (adherent cells) or 20 μl (cells in suspension) of MTT solution (5 mg of MTT/ml of phosphate-buffered saline) was added to each well, and the incubation was continued for 2 h. A volume of 80 μl/well or 130 μl was removed from the non-adherent cells and the adherent cells, respectively. 100 μl of 0.04 m HCl in isopropyl alcohol was added to dissolve the formazan crystals, and the plates were gently agitated for 1 h. Optical density was measured spectrophotometrically at 590 nm on a microtiter plate reader (Thermomax Molecular Devices, Sunnyvale, CA). Cell viability was determined relative to the control, and the final results were recorded. The EC50 for each peptide was obtained from the curve of cell viability versus peptide concentration and taken from the concentration at which cell viability was 50%.
Vesicle Preparation
Large unilamellar vesicles (LUVs) of POPC and a mixture of POPC/POPS (2:1) were prepared as described previously (32). Briefly, a defined amount of lipid was dissolved in a chloroform/methanol mixture (2:1, v/v). The solvents were evaporated at 40 °C using a rotary evaporator, forming a thin lipid film. The lipid film was hydrated with Hepes buffer (10 mm Hepes and 150 mm NaCl, 1 mm EDTA at pH 7.4) to produce multilamellar vesicles. The liposome dispersion was subjected to three freeze-thaw cycles and then passed 11 times each through Isopore® membrane filters of decreasing pore size (0.8, 0.4, 0.2, and 0.1 μm) using a mini extruder (Avestin, Ottawa, Canada) to produce LUVs. Small unilamellar vesicles (SUVs) for CD measurements were prepared by hydrating the lipid film (POPC and POPC/POPS (2:1)) in 10 mm potassium phosphate buffer (pH 7.4). The lipid suspensions were sonicated for 10 min using the ultrasonic homogenizer LABSONIC U (B. Braun Biotech International GmbH) with a probe tip of type 5 T, for 10 min with intervals of a few s (31). The size of the liposomes was determined by dynamic laser light scattering using a Nicomp Submicron Particle Sizer (Particle Sizing Systems, Inc., Santa Barbara, CA). Monomodal size distributions were fitted in all cases with mean diameter between 100 and 130 nm for LUVs, between 20 and 25 nm for SUVs, and ∼400 nm for multilamellar vesicles.
Dye Leakage Experiments
Dye leakage experiments were performed using a PerkinElmer Life Sciences Luminescence Fluorimeter LS-50 B (Beaconsfield, UK). Calcein buffer (25 mm calcein and 10 mm Hepes, 150 mm NaCl, 1 mm EDTA (pH 7.4)) was entrapped in vesicles composed of POPC and POPC/POPS (2:1). Remaining free calcein was removed from the vesicles by gel filtration on a Sephadex G-50 column. Lipid concentration was determined by quantitative phosphorus analysis (33). Aliquots of the liposome suspensions were diluted using Hepes buffer to a final concentration of 50 μm lipid and incubated for 5 min with concentrations of peptide solution ranging from 5 to 200 μg/ml. Calcein release from LUVs was assessed fluorometrically by measuring the increase in the fluorescence intensity resulting from the decrease in the level of self-quenching (excitation at 489 nm and emission at 520 nm). The fluorescence intensity corresponding to 100% calcein release was determined by the addition of 10% (w/v) Triton X-100 solution. Relative leakage was calculated according to the equation, ((Fx − F0) × 100)/(Ft − F0) where Fx is the intensity measured at a given concentration of peptide, F0 is the intensity of the liposomes (background), and Ft is the intensity after lysis by Triton X-100.
Fluorescence Spectroscopy
Peptide-phospholipid interactions were studied by monitoring the changes in the maximum wavelength of the Trp fluorescence emission spectra of the peptides upon the addition of LUVs (POPC and POPC/POPS (2:1)) using a PerkinElmer Luminescence Fluorimeter LS-50 B. Five individual emission spectra for each peptide, with and without lipid vesicles, were recorded between 310 and 450 nm, using an excitation of 280 nm (bandwidths were 10 nm with a scan speed of 60 nm/s). The concentrations of the peptides and lipids in these experiments were 2.1 and 50 μm, respectively.
Circular Dichroism (CD) Analyses
CD spectra were recorded on a Jasco J-810 spectropolarimeter (Jasco International Co., Ltd., Tokyo Japan) calibrated with ammonium d-camphor-10-sulfonate (Icatayama Chemicals, Tokyo Japan). Measurements were performed with 1.4 mm POPC and POPC/POPS liposomes or various concentrations of 1,1,1,3,3,3-hexafluoropropan-2-ol (hexafluoroisopropanol; HFIP) or in a phosphate buffer (0 and 50% (v/v)) at 23 °C by using a quartz cuvette (Starna, Essex, UK) with a path length of 0.1 cm. All of the measurements were performed with a peptide concentration of 0.10 mg/ml in 10 mm potassium phosphate buffer (pH 7.4). Samples were scanned five times at 20 nm/min with a bandwidth of 1 nm and a response time of 1 s, over the wavelength range 190–260 nm. The data were averaged, and the spectrum of a peptide-free control sample was subtracted. The α-helical content of the various peptides was calculated after smoothing (means-movement, convolution width 5) from the mean residue ellipticity at 222 nm ([θ]222) using the formula, ƒH = [θ]222/(40,000(1 − 2.5/n)), where ƒH and n represent the α-helical content and the number of peptide bonds, respectively (34). All measurements were conducted at least twice.
NMR Spectroscopy
All NMR samples were prepared by dissolving 2–3 mg of the peptide in 0.5 ml of 90:10 H2O/D2O and adjusted to pH 3.5–4.5. Initial NMR data were acquired on each peptide before adding 9.6 mg of SDS-d25 and readjusting the pH. To duplicate peptide samples 22.2 mg of DPC-d38 was added instead. All NMR spectra were acquired at 25 °C on a Bruker Avance 700-MHz NMR spectrometer equipped with a 5-mm TBI 3-axis gradient probe. Two-dimensional TOCSY and NOESY spectra were collected for the micelles using mixing times of 80 ms for the two-dimensional TOCSY and 100 ms for the two-dimensional NOESY. A two-dimensional NOESY spectrum for the peptide in solution was acquired with a mixing time of 500 ms. Spectra were acquired with 2048 × 512 or 2048 × 600 data points in the F2 and F1 dimensions using spectral widths of 8400 Hz. Water suppression for the two-dimensional spectra was performed using the excitation sculpting technique (35). The two-dimensional TOCSY and NOESY NMR spectra were processed with NMRPipe and analyzed with the NMRView 5.0.4 (36) software package on workstations operating with the Redhat 8.0 version of the Linux operating system. The two-dimensional data were zero-filled once in each dimension and Fourier-transformed with a shifted sine-bell function. Samples were externally referenced to DSS.
Structural Calculations
The assignment of the peptide chemical shifts was determined using standard methods (37). Upon completion of the proton assignments, NOE-based distance restraints were collected from NOESY spectra and automatically allocated to short, medium, and long range distance interactions based upon peak volume. Broad dihedral restraints were used to confine the bond angles to the allowed Ramanchandran space. The peptide structures were determined using the programs CNS 1.0 and ARIA 1.2 (38, 39). ARIA calculations were initiated using default parameters except in the final ARIA run, where the number of structures generated in the seventh and eighth iterations was increased to 40 and 100. Also, in the eighth iteration, the 20 lowest energy structures were used for statistical analysis. Molecular structures were viewed using MOLMOL 2K.2 (40) and analyzed using PROCHECK 3.4 (41, 42). Residues recognized by MOLMOL as having potential hydrogen bonds in at least 60% of the structures were noted, and hydrogen bonds were introduced in a final ARIA calculation.
RESULTS
Peptide Design
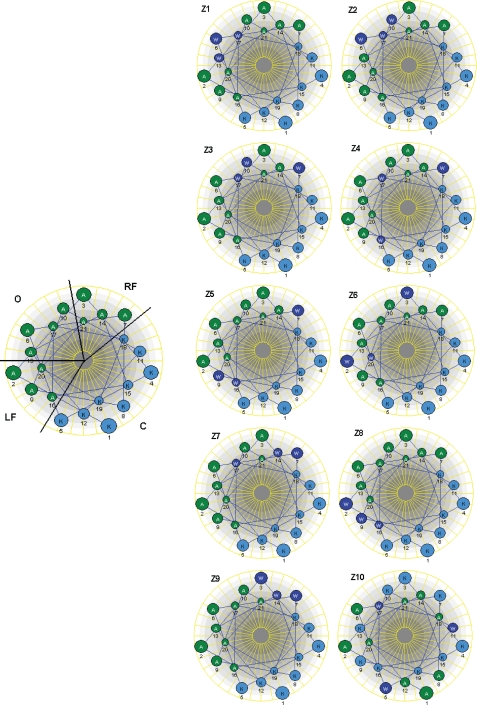
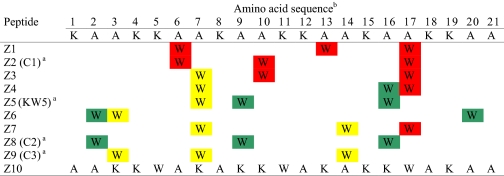
In order to investigate how the positioning of Trp residues in an idealized α-helical, model amphipathic peptide (KAAKKAA)3 (43) affected the cytotoxic activity, specific Ala residues at different positions in the lipophilic sector of the peptide were replaced by Trp residues. Preliminary experiments showed that three Trp residues appear to be a minimum for obtaining significant cytotoxic activity against the tumor cell lines tested; for example, we found that a peptide based on the (KAAKKAA)3 sequence encompassing Trp residues in positions 9 and 16 did not display any activity within the concentration range tested (up to 1000 μg/ml; data not shown). We have divided the hydrophobic sector of the Z peptides into three distinct sectors: a left flanking sector encompassing residues 2, 9, 16, and 20; a right flanking sector encompassing residues 3, 7, 14, and 21; and a third sector encompassing residues 6, 10, 13, and 17, which is located opposite to the cationic region in the Edmundson helical wheel projection (Fig. 1). By subsequently shuffling the Trp residues from the sector opposite to cationic sector to the sectors adjacent to cationic sector by exchanging their positions with different Ala residues, nine distinct Z peptide analogs were constructed. In addition, for comparison, a scrambled peptide (Z10), with the same net amino acid content but without any predicted amphipathic helical or β-strand conformation, was also included in the study (Table 1).
FIGURE 1.
Edmundson helical wheel projection of the model peptide. The lipophilic sector is divided into three sectors (right flank (RF), left flank (LF), and opposite (O)). The peptide also has a cationic sector (C). Different peptide analogues were constructed by placing Trp in different positions within the lipophilic sector.
TABLE 1.
Amino acid sequence of the analogs of model peptide (KAAKKAA)3
b Red, Trp residues are located in the sector opposite the cationic sector (O); green, Trp residues are located in the sector flanking the left side of the cationic sector (LF); yellow, Trp residues are located in the sector flanking the right side of the cationic sector (RF). See the legend to Fig. 1 for sector abbreviations.
Cytotoxic Activity
All of the peptides were tested against murine fibrosarcoma (MethA) and human mammary carcinoma (MT-1) cells and colorectal adenocarsinoma (HT-29) cells. The cytotoxic activity of the peptides against the cell lines is compiled in Table 2. Active peptides were also tested against normal fibroblasts (MRC-5 cells), and the activity against these cells followed the same trend as observed for the cancer cell lines (Table 2). The scrambled peptide Z10 demonstrated no activity against any of the cell lines tested. Except for peptide Z9, the HT-29 cells were less sensitive to the active peptides relative to the MethA and MT-1 cells. The peptides Z1 and Z2 with all three tryptophans located opposite to the cationic sector (Fig. 1) displayed no activity against the MethA and HT-29 cells and only had low activity against the MT-1 cells. A similar pattern in relative activities was obtained with peptide Z3, with two tryptophans located opposite the cationic sector and one Trp located adjacent to the cationic sector. The Z4 peptide with one Trp located in each sector (i.e. one Trp in each sector flanking the cationic sector and one Trp in the opposite sector) displayed low activity against all three tumor cell lines. It is noteworthy that the Z7 peptide with one Trp located in the opposite sector and two tryptophans located in the sector flanking the right side of the cationic sector were highly active against all three tumor cell lines tested. The peptides Z8 and Z9 with all three tryptophans located in one of the sectors flanking the cationic sector were more active against all three cancer cell lines than the peptides Z5 and Z6 with two tryptophans in one flanking sector and one Trp in the other flanking sector. Hence, the most active peptides were the peptides with all three Trp residues located simultaneously in one of the sectors flanking the cationic sector. It is also noteworthy that the peptide Z9 with three tryptophans located in the right flanking sector was almost 2-fold more active against MethA and HT-29 than peptide Z8 with three tryptophans located in the left flanking sector.
TABLE 2.
In vitro cytotoxicity of the model peptide analogs
| Peptide | Cytotoxic activity (EC50)a,b |
|||
|---|---|---|---|---|
| MethA | MT-1 | HT-29 | MRC-5 | |
| μm | ||||
| Z1 | >211 | 186 | >211 | NDc |
| Z2 | >211 | 173 | >211 | ND |
| Z3 | >211 | 128 | >211 | ND |
| Z4 | 155 | 68 | 173 | 78 |
| Z5 | 42 | 28 | 67 | 27 |
| Z6 | 17 | 16 | 31 | 23 |
| Z7 | 15 | 15 | 21 | 20 |
| Z8 | 19 | 14 | 23 | 17 |
| Z9 | 9 | 16 | 13 | 14 |
| Z10 | >211 | >211 | >211 | ND |
a The maximum concentration of the peptide used was 211 μm, whereas the minimum concentration tested was 4.2 μm. The results are an average value of three independent experiments performed in duplicate.
b EC50 corresponds to the peptide concentration where 50% of the cells are killed.
c ND, not determined.
Dye Leakage
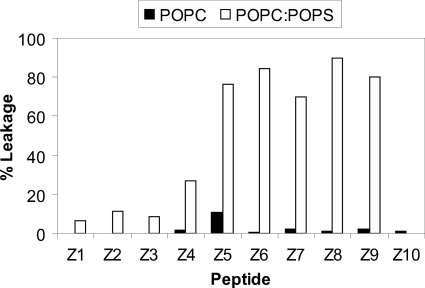
The ability of the peptides to disrupt model membranes was investigated by studying dye release from phospholipid vesicles upon the addition of peptide using a standard calcein fluorescence assay (44). A general trend for all the peptides investigated was a low ability to cause leakage in POPC vesicles (see Fig. 2). The leakage was 11% or lower relative to control for all of the peptides. As expected, the ability to cause leakage in the negative POPC/POPS membranes was much higher. Here all of the relatively active peptides, with two or more Trp residues in one of the flank sectors (Z4, Z5, Z6, Z7, Z8, and Z9), induced a leakage above 50% even at the lowest concentration tested (5 μg/ml). The inactive peptides, those with only one or no Trp residues in the flanking sectors (Z1, Z2, Z3, Z4, and Z10), induced no or low leakage even at the highest concentration tested (200 μg/ml).
FIGURE 2.
Peptide-induced calcein leakage from neutral POPC and negative POPC/POPS liposomes. The results are presented as a percentage of released calcein in POPC and POPC/POPS liposome dispersions with 200 μg/ml of the peptides Z1–Z10. The error in each individual leakage experiment is estimated to be ±5%.
Fluorescence Spectroscopy
The fluorescence spectrum for all peptides either in buffer or in the presence of POPC or POPC/POPS LUVs was measured. Z1, with all Trp residues located opposite to the cationic sector, exhibited an emission maximum at 356 nm in aqueous solution (blank). Interaction with neutral membranes (POPC) did not give any blue shift, whereas the interaction with the negatively charged membranes POPC/POPS gave rise to a blue shift of 13.3 nm. The results for all of the peptides tested are given in Table 3. All of the peptides showed similar results as Z1, but those with strong anticancer activity displayed a slightly larger blue shift than the inactive peptides in the presence of POPC/POPS (2:1). The blue shift that was measured was in general between 13 and 20 nm, with Z9 (the most active peptide) as an exception with 29 nm. Z10 had the lowest blue shift at 8.7 nm.
TABLE 3.
Fluorescence spectroscopy analysis in model membrane systems
| Peptide | Positions of Trp residuesa | Fluorescence blue shiftb |
|
|---|---|---|---|
| POPC | POPC/POPS | ||
| nm | |||
| Z1 | 3 O | −1.7 | 13.3 |
| Z2 | 3 O | 0 | 14 |
| Z3 | 1 RF/2 O | 0 | 13.4 |
| Z4 | LF/O/ RF | 0 | 16 |
| Z5 | 1 RF/2 LF | 0 | 18.5 |
| Z6 | 1 RF/2 LF | −1 | 16.9 |
| Z7 | 2 RF/1 O | 1.6 | 12.8 |
| Z8 | 3 LF | 0 | 17.7 |
| Z9 | 3 RF | 2.5 | 29 |
| Z10 | Scramble | 0 | 8.7 |
a O, opposite; RF, right flank; LF, left flank.
b Blue shift was determined by subtracting emission wavelength of peptide in contact with liposome from wavelength of peptide in buffer (blank).
Circular Dichroism
Far-UV CD spectroscopic investigations of the Z peptides in 10 mm potassium phosphate buffer confirmed that all peptides were unfolded under these conditions with less than 7% α-helicity (Table 4). The CD spectra of the Z peptides in 50% HFIP, a solvent known to induce and stabilize secondary structures in peptides (45), were characteristic of helical conformation. The helicity of the peptides ranges from 24 to 66%, with the highest helical content in the Z6 peptide with two Trp residues in the left flanking sector and one Trp in the right flanking sector (Table 4). The helicities of Z8 with three Trp residues in the left flanking sector and Z9 with three Trp residues in the right flanking sector were calculated to be 35 and 38%, respectively. Considerably larger differences in helicity were found in the presence of negatively charged (POPC/POPS) SUV suspensions ranging from 6 to 27%. In general, the helicity of the peptides with a relative high cytotoxic activity (Z5, Z6, Z7, Z8, and Z9) was higher than the helicity of the peptides with low or no cytotoxic activity (Z1, Z2, Z3, and Z4). The helicity of the Z peptides with three or two Trp residues in the sector opposite the cationic sector was only 6–7%. The highest helicity was found in the peptides Z8 and Z6 with three tryptophans in the right flanking sector or two and one Trp in the right flanking and in the left flanking sectors, respectively. The helicity in (POPC) SUV suspensions was very low for all the peptides, ranging from 0–8%. Thus, the degree of helicity estimated by CD for the peptides in the presence of negatively charged (POPC/POPS) liposomes seems to correlate with their cytotoxic activities.
TABLE 4.
Helical content of the Z peptides under different conditions
| Peptide | Positions of Trp residuesa | Helicity |
|||
|---|---|---|---|---|---|
| Buffer | HFIP | POPC | POPC/POPS | ||
| % | % | % | % | ||
| Z1 | 3 O | 0 | 24 | 0 | 7 |
| Z2 | 3 O | 2 | 24 | 2 | 7 |
| Z3 | 1 RF/2 O | 2 | 31 | 2 | 6 |
| Z4 | LF/O/ RF | 6 | 28 | 7 | 11 |
| Z5 | 1 RF/2 LF | 2 | 26 | 2 | 19 |
| Z6 | 2 RF/1 LF | 7 | 66 | 8 | 26 |
| Z7 | 2 RF/1 O | 2 | 27 | 2 | 17 |
| Z8 | 3 LF | 2 | 35 | 3 | 27 |
| Z9 | 3 RF | 3 | 38 | 3 | 21 |
| Z10 | Scramble | - | - | - | - |
a O, opposite; RF, right flank; LF, left flank.
NMR Experiments
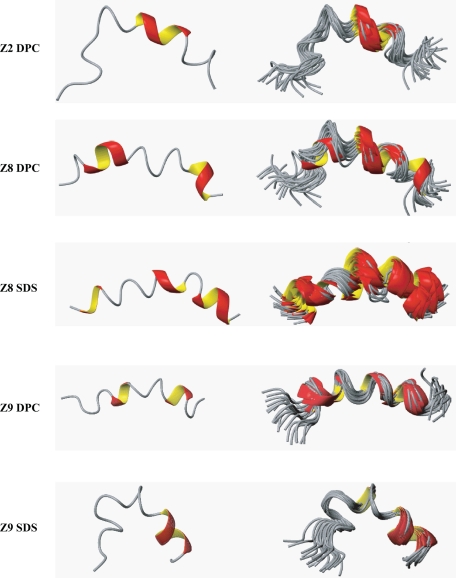
Initial studies began by determining the structure of three peptides, one inactive and two active, in the presence of micelles. Micelles usually provide the most convenient method for determining high resolution NMR structures because vesicles are much larger and NMR signals broaden to the point where the standard two-dimensional experiments cannot be used (46, 47). The two-dimensional NOESY spectra of all of the individual peptides in aqueous solution did not indicate the presence of any regular secondary structure. However, in the presence of SDS-d25 or DPC-d38 micelles, the active Z8 and Z9 peptides both produced spectra with numerous NOE interactions, indicating that a preferred conformation had been adopted. The calculated backbone ribbon structures (see below) of the 20 lowest energy conformations are shown in Fig. 3. The Z2 peptide, however, only adopted a structure with DPC-d38 and not in the presence of SDS-d25 micelles. Upon closer inspection, the two-dimensional NOESY spectra of each of the three peptides with SDS-d25 micelles displayed few (if any) amide proton interactions for the first few N-terminal residues, indicating a lack of well defined structure in this portion of each peptide. In general, the spectra showed surprisingly good dispersion in the fingerprint region although there were nine lysines and nine alanine residues in each peptide. The NOE summaries of the peptides in the presence of micelles are shown in Fig. 4. From all of these summaries, there are numerous medium range NOE interactions that are consistent with mostly helical structures. These interactions are distributed over most of the peptide length except for Z2 with DPC-d38 micelles, which only has a large number of medium range NOE interactions from Lys-8 to Lys-18.
FIGURE 3.
Ribbon structures for the lowest energy (left) and 20 lowest energy conformations (right) of each peptide in the presence of micelles. N termini are at the left of each structure. The structures were generated with MOLMOL (40).
FIGURE 4.
NOE summary and chemical shift index values for peptide micelle samples of Z2-DPC-d38 (A) Z8-DPC-d38 (B), Z8-SDS-d25 (C), Z9-DPC-d38 (D), and Z9-SDS-d25 (E).
The chemical shift index is another method of identifying regular secondary structure in peptides and proteins (48). For the conditions examined, the Z2 peptide shows incomplete helical structure near the peptide termini. Because there are inevitably going to be correlations in the two-dimensional NOESY that have more than one possible proton-proton assignment, a method such as the NOE summary is required to assess the structures calculated. Overall, the combination of all interactions in the NOE summaries along with the chemical shift index information provides a good visual indication of the basis for the peptide structures.
Structure Calculations
The statistics from the lowest 20 conformations produced by ARIA are shown in supplemental Table 1. The average number of NOE interactions per residue ranges from 15 for Z2 in DPC-d38 to 24 for Z8 in DPC-d38; however, these values are only a general indication of the number of constraints used because the termini of some of the peptides were lacking in NOE interactions and therefore not well structured. Root mean square deviation values obtained for the backbone overlap of the 20 lowest structures of each peptide are taken from the central portion of the peptide because root mean square deviation values get increasingly large as residues from the termini are incorporated into the calculation. Visually, the agreement of structural overlap is shown in Fig. 3, which shows the backbone ribbon structures of the lowest energy conformation along with all 20 overlapping conformations. All structures show various degrees of curvature over the peptide length. This has been seen in many other helical antimicrobial peptides, and the curvature may arise from the use of micelles, which do not provide a flat surface, unlike membrane bilayers (22).
The structures of the peptide backbones along with their lysine and aromatic side chains are shown in Fig. 5. The aromatic side chains tend to locate on one side of the peptide for the Z8 and Z9 peptides with DPC-d38, whereas the Z2 peptide has aromatic side chains on both sides of the backbone. Furthermore, the Z8 and Z9 peptides with SDS-d25 display the most amphipathic character of all of the conditions studied. The Z8 peptide determined bound to DPC-d38 also shows amphipathic character, albeit with a more obscured distinction between charged and aromatic side chains.
FIGURE 5.
Ribbon structures for the lowest energy conformations of each peptide in the presence of micelles. N termini are at the left of each structure. Lysine side chains are indicated in blue, and aromatic side chains are shown in red. The structures were generated with MOLMOL (40).
DISCUSSION
(KAAKKAA)3 is a model antimicrobial peptide that is designed to be perfectly amphipathic when it adopts a helical conformation. The parent peptide itself did not display any cytotoxic activity, and three Trp residues had to be introduced in place of Ala to obtain significant activity against malignant cell lines. The role of the aromatic amino acids in membrane-active peptides with cytotoxic and/or antimicrobial effect has been emphasized in several recent studies (23–28, 49–58); thus, it is perhaps not surprising that high cytotoxic activity can be obtained by the introduction of Trp residues. In this work, we were specifically interested in elucidating how the positioning of the Trp residues along an α-helical amphipathic peptide influenced the activity against cancer cell lines as well as the ability of the peptide to interact with model membrane systems and to adopt an α-helical structure under these conditions.
The basis for this study was formed by the design of 10 peptides, where three Ala residues at different positions within the hydrophobic sector of the idealized amphipathic α-helical 21-mer peptide (KAAKKAA)3 were replaced by Trp residues (Table 1). As highlighted in Fig. 1, in the design process, the hydrophobic sector could be further divided into three parts denoted the left and right flanking region and the “opposite” sector. Interestingly, the two peptides with all three Trp residues located in the opposite sector (peptides Z1 and Z2) showed no or very low cytotoxic activity. The shifting of the Trp residues from the opposite sector to either one of the two flanking regions, led to a significant difference in cytotoxic activity (Table 2). The two peptides with all three Trp residues located either on the left flank (peptide Z8) or on the right flank (peptide Z9) displayed the highest activity. The active peptides were also tested for their cytotoxic activity against normal fibroblasts (MRC-5 cells), and it was found that peptides with high activity against the cancer cell lines also displayed high activity against this normal cell line (Table 2). Thus, simply by repositioning the Trp residues within the hydrophobic part of the idealized helix, significant changes in the cytotoxic activity of the peptides from inactive to active and highly active peptides were obtained. Our data clearly illustrate that the cytotoxic activity of the peptides was dependent on Trp positioning and that the most efficient peptides could be obtained when two or more Trp residues were positioned close to the cationic sector.
In an attempt to elucidate why the position of Trp is so critical for the cytotoxic activity, the interactions of the different peptides with both zwitterionic (POPC or DPC) and anionic (POPC/POPS or SDS) membrane systems were investigated. Zwitterionic neutral lipid headgroups are usually found on the outer leaflet of the membranes of healthy eukaryotic cells, whereas negatively charged phosphatidylserine headgroups become exposed on the surface of cancer cells (14, 59). The fluorescence emission maximum of the indole side chain of Trp can be used to estimate the positioning of Trp residues in biological membranes. An intermediate fluorescence emission maximum of 330–345 nm is compatible with a polar/non-polar interface location, whereas a larger blue shift indicates a deeper penetration of the Trp indole side chain into a more hydrophobic environment (27). All peptides in this study showed a change in the emission maximum in this area in the presence of the POPC/POPS vesicles, suggesting that all of the peptides are localized in the membrane interface (60). The peptide's favored interfacial location of membranes appears to stem in part from its content of Trp residues. It has been found that aromatic side chains, particularly the indole ring of Trp, strongly prefer partitioning into the interfacial region of membranes (61–63). In our case, no clear cut trends can be found; however, the most active peptide, Z9, displayed by far the highest blue shift in POPC/POPS.
The active Z peptides were also able to lyse membranes, depending on the phospholipid composition of the vesicles investigated. Calcein leakage from vesicles containing negatively charged lipid headgroups was found to be much higher than the leakage from vesicles containing neutral zwitterionic lipids (Fig. 2). The active Z peptides prefer to bind to vesicles containing phospholipids with negatively charged headgroups, and in doing so, they induce efficient dye leakage from the vesicles, thus supporting the idea that electrostatic interactions play a dominant role in the binding process (64).
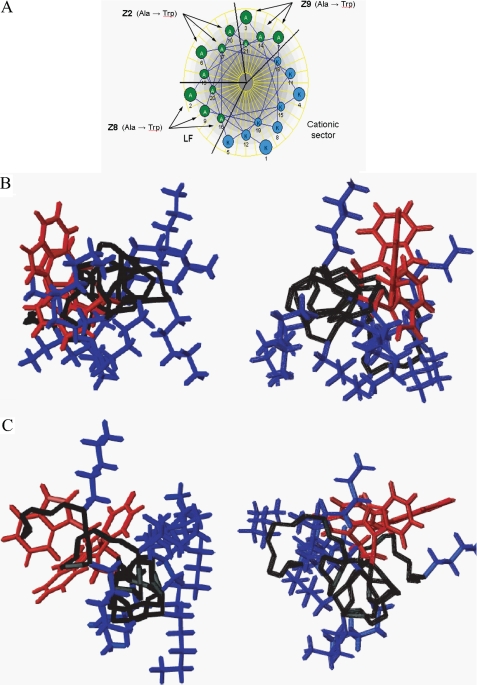
Most membrane protein structures elucidated have regular α-helical or β-sheet secondary structure (65). As such, the formation of backbone hydrogen bonds is energetically favorable in membrane interfaces. Also, a cationic amphipathic structure would be best suited for maximizing both electrostatic and hydrophobic interactions with the membrane (66). Structural studies using CD spectroscopy revealed that all the peptides have an unordered structure in aqueous solution, which is common with antimicrobial peptides, such as magainin and cecropin A, and a number of de novo designed peptides (43). However, the presence of an anionic membrane surface allowed the peptides to adopt a predominantly distorted/dynamic helical structure, which is sufficient to destabilize membranes. It was implicit in the design strategy that these peptides would have a higher propensity to form a helical conformation due to a high content of helix-stabilizing residues (67, 68). The structure of the peptides is generally helical in the presence of HFIP. This is consistent with the hypothesis that electrostatic interactions between the cationic amino acid residues and the anionic systems are important for structuring and functionality of the peptides. A factor such as ionic strength is expected to influence the aqueous conformation of the membrane lipids. In other words, the initial interaction and selective recognition are electrostatically driven, but after binding, hydrophobic interactions play an important role, due to the partial amphipathic structure of the peptide. Woody and Dunker have noted that the presence of two Trp residues per 20 amino acids produces a measurable effect on the [θ]222 (69). This contribution has to be considered when analyzing the CD data because this is the critical wavelength for the calculation of the helical content (69). Peptides investigated by NMR were selected based on Trp positioning and cytotoxic activity, and the structural results provide an interesting evaluation of the actual conformation of the designed peptides. Fig. 6 illustrates the placement of the aromatic residues in the Edmundson helical wheel projection of the (KAAKKAA)3 peptide. The Z2 peptide was designed with all three Trp residues located in the sector opposite to the cationic region, whereas for Z8 and Z9, the Trp residues reside in the left flank sector and the right flank sector, respectively. In the resulting micelle systems, the inactive Z2 peptide failed to form a complete helical structure in DPC-d38, although in HFIP, a structure with helical character was induced (see CD data above). The active Z8 and Z9 peptides in both types of micelles adopt highly coiled, mostly helical structures, similar to those found in HFIP solution. However, in both DPC-d38 and SDS-d25 micelles, the location of the aromatic side chains of Z8 and Z9 are opposite to the bulky lysine side chains, which occupy approximately an 180° sector in a helical wheel representation (Fig. 6). It would seem that the size of the bulky aromatic side chains blur the distinction between right and left flanking and opposite sectors, making it too small of a classification to be visualized in this system.
FIGURE 6.
Comparison of the Edmundson helical wheel projection (A) for the (KAAKKAA)3 peptide along with illustrations of the Z8 (left) and Z9 (right) peptides in DPC-d38 (B) and SDS-d25 (C) as viewed along the backbone from the N terminus. Lysine side chains are indicated in blue, and aromatic side chains are shown in red. The structures were generated with MOLMOL (40).
In contrast to many other cytotoxic and anticancer peptides, the active Z peptides displayed a low tumor cell versus normal cells specificity (Table 2). We have earlier investigated and identified structural parameters that were critical for tumor cell specificity versus normal cells (23, 24). By combining the information regarding the critical role of Trp residues obtained in the present study with structural parameters that affect toxicity against normal cells, it should be possible to prepare peptides with low toxicity toward normal cells while maintaining high activity against cancer cells.
In conclusion, we have found that the position of the three Trp residues in the 21-mer model amphipathic peptides is absolutely critical for generating linear peptides with cytotoxic activity. The most active peptides with the Trp residues located adjacent to the cationic sector seemed to be more able to adopt amphipathic structures in the presence of anionic membranes than the peptides with the Trp residues located opposite to the cationic sector. Furthermore, our results indicate that an embedding of the Trp residues into the lipid membrane may be more easily facilitated after an initially driven electrostatic interaction when located close to the cationic sector.
Supplementary Material
Acknowledgments
We thank Mari Wikman and Cristiane de A. Cavalcanti Jacobsen for technical assistance.
This work was supported by operating grants from the Research Council of Norway and the Norwegian Cancer Society (to Ø. R.) and from the Alberta Cancer Foundation (to H. J. V.).

This article contains supplemental Table 1.
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- DPC-d38
- dodecylphosphocholine-d38
- DSS
- sodium 2,2-dimethyl-2-silapentane-5-sulfonate
- HFIP
- hexafluoroisopropanol (1,1,1,3,3,3-hexafluoropropan-2-ol)
- LUV
- large unilamellar vesicle
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPS
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine
- SDS-d25
- sodium dodecyl-d25 sulfate
- SUV
- small unilamellar vesicle
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- NOE
- nuclear Overhauser effect
- NOESY
- NOE spectroscopy
- TOCSY
- total correlation spectroscopy.
REFERENCES
- 1. Zasloff M. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 5449–5453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Steiner H., Hultmark D., Engström A., Bennich H., Boman H. G. (1981) Nature 292, 246–248 [PubMed] [Google Scholar]
- 3. Zanetti M. (2004) J. Leukocyte Biol. 75, 39–48 [DOI] [PubMed] [Google Scholar]
- 4. Ganz T., Selsted M. E., Szklarek D., Harwig S. S., Daher K., Bainton D. F., Lehrer R. I. (1985) J. Clin. Invest. 76, 1427–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Habermann E., Jentsch J. (1967) Hoppe-Seyler's Z. Physiol. Chem. 348, 37–50 [PubMed] [Google Scholar]
- 6. Hirai Y., Ueno Y., Yasuhara T., Yoshida H., Nakajima T. (1980) Biomed. Res. 1, 185–187 [Google Scholar]
- 7. Huang Y. H., Colgrave M. L., Daly N. L., Keleshian A., Martinac B., Craik D. J. (2009) J. Biol. Chem. 284, 20699–20707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Henriques S. T., Huang Y. H., Rosengren K. J., Franquelim H. G., Carvalho F. A., Johnson A., Sonza S., Tachedjian G., Castanho M. A., Daly N. L., Craik D. J. (2011) J. Biol. Chem. 286, 24231–24241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cruciani R. A., Barker J. L., Zasloff M., Chen H. C., Colamonici O. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 3792–3796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baker M. A., Maloy W. L., Zasloff M., Jacob L. S. (1993) Cancer Res. 53, 3052–3057 [PubMed] [Google Scholar]
- 11. Ohsaki Y., Gazdar A. F., Chen H. C., Johnson B. E. (1992) Cancer Res. 52, 3534–3538 [PubMed] [Google Scholar]
- 12. Schröder-Borm H., Bakalova R., Andrä J. (2005) FEBS Lett. 579, 6128–6134 [DOI] [PubMed] [Google Scholar]
- 13. Iwasaki T., Ishibashi J., Tanaka H., Sato M., Asaoka A., Taylor D., Yamakawa M. (2009) Peptides 30, 660–668 [DOI] [PubMed] [Google Scholar]
- 14. Papo N., Shai Y. (2005) Cell. Mol. Life Sci. 62, 784–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Johnstone S. A., Gelmon K., Mayer L. D., Hancock R. E., Bally M. B. (2000) Anticancer Drug Des. 15, 151–160 [PubMed] [Google Scholar]
- 16. Berge G., Eliassen L. T., Camilio K. A., Bartnes K., Sveinbjørnsson B., Rekdal O. (2010) Cancer Immunol. Immunother. 59, 1285–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Papo N., Braunstein A., Eshhar Z., Shai Y. (2004) Cancer Res. 64, 5779–5786 [DOI] [PubMed] [Google Scholar]
- 18. Papo N., Seger D., Makovitzki A., Kalchenko V., Eshhar Z., Degani H., Shai Y. (2006) Cancer Res. 66, 5371–5378 [DOI] [PubMed] [Google Scholar]
- 19. Hoskin D. W., Ramamoorthy A. (2008) Biochim. Biophys. Acta 1778, 357–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wieprecht T., Beyermann M., Seelig J. (1999) Biochemistry 38, 10377–10387 [DOI] [PubMed] [Google Scholar]
- 21. Wieprecht T., Apostolov O., Beyermann M., Seelig J. (1999) J. Mol. Biol. 294, 785–794 [DOI] [PubMed] [Google Scholar]
- 22. Haney E. F., Vogel H. J. (2009) Annu. Rep. NMR Spectrosc. 65, 1–51 [Google Scholar]
- 23. Yang N., Stensen W., Svendsen J. S., Rekdal Ø. (2002) J. Pept. Res. 60, 187–197 [DOI] [PubMed] [Google Scholar]
- 24. Yang N., Lejon T., Rekdal O. (2003) J. Pept. Sci. 9, 300–311 [DOI] [PubMed] [Google Scholar]
- 25. Haug B. E., Svendsen J. S. (2001) J. Pept. Sci. 7, 190–196 [DOI] [PubMed] [Google Scholar]
- 26. Haug B. E., Andersen J., Rekdal O., Svendsen J. S. (2002) J. Pept. Sci. 8, 307–313 [DOI] [PubMed] [Google Scholar]
- 27. Vogel H. J., Schibli D. J., Jing W., Lohmeier-Vogel E. M., Epand R. F., Epand R. M. (2002) Biochem. Cell Biol. 80, 49–63 [DOI] [PubMed] [Google Scholar]
- 28. Chan D. I., Prenner E. J., Vogel H. J. (2006) Biochim. Biophys. Acta 1758, 1184–1202 [DOI] [PubMed] [Google Scholar]
- 29. Eliassen L. T., Haug B. E., Berge G., Rekdal O. (2003) J. Pept. Sci. 9, 510–517 [DOI] [PubMed] [Google Scholar]
- 30. Seljelid R. (1986) Biosci. Rep. 6, 845–851 [DOI] [PubMed] [Google Scholar]
- 31. Mosmann T. (1983) J. Immunol. Methods 65, 55–63 [DOI] [PubMed] [Google Scholar]
- 32. Mayer L. D., Hope M. J., Cullis P. R. (1986) Biochim. Biophys. Acta 858, 161–168 [DOI] [PubMed] [Google Scholar]
- 33. Eibl H., Lands W. E. (1969) Anal. Biochem. 30, 51–57 [DOI] [PubMed] [Google Scholar]
- 34. Scholtz J. M., Qian H., York E. J., Stewart J. M., Baldwin R. L. (1991) Biopolymers 31, 1463–1470 [DOI] [PubMed] [Google Scholar]
- 35. Hwang T. L., Shaka A. J. (1995) J. Magn. Reson., Ser. A 112, 275–279 [Google Scholar]
- 36. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 37. Wüthrich K. (1986) in NMR of Proteins and Nucleic Acids, pp. 117–175, John Wiley & Sons, Inc., New York [Google Scholar]
- 38. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 39. Nilges M., O'Donoghue S. I. (1998) Prog. Nucl. Magn. Reson. Spectrosc. 32, 107–139 [Google Scholar]
- 40. Koradi R., Billeter M., Wüthrich K. (1996) J. Mol. Graph. 14, 51–55 [DOI] [PubMed] [Google Scholar]
- 41. Morris A. L., MacArthur M. W., Hutchinson E. G., Thornton J. M. (1992) Proteins 12, 345–364 [DOI] [PubMed] [Google Scholar]
- 42. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 43. Javadpour M. M., Juban M. M., Lo W. C., Bishop S. M., Alberty J. B., Cowell S. M., Becker C. L., McLaughlin M. L. (1996) J. Med. Chem. 39, 3107–3113 [DOI] [PubMed] [Google Scholar]
- 44. Matsuzaki K., Murase O., Fujii N., Miyajima K. (1996) Biochemistry 35, 11361–11368 [DOI] [PubMed] [Google Scholar]
- 45. Wei J., Fasman G. D. (1995) Biochemistry 34, 6408–6415 [DOI] [PubMed] [Google Scholar]
- 46. Henry G. D., Sykes B. D. (1994) Methods Enzymol. 239, 515–535 [DOI] [PubMed] [Google Scholar]
- 47. Opella S. J. (1997) Nat. Struct. Biol. 4, 845–848 [PubMed] [Google Scholar]
- 48. Wishart D. S., Sykes B. D., Richards F. M. (1992) Biochemistry 31, 1647–1651 [DOI] [PubMed] [Google Scholar]
- 49. Strøm M. B., Rekdal O., Svendsen J. S. (2000) J. Pept. Res. 56, 265–274 [DOI] [PubMed] [Google Scholar]
- 50. Haug B. E., Skar M. L., Svendsen J. S. (2001) J. Pept. Sci. 7, 425–432 [DOI] [PubMed] [Google Scholar]
- 51. Strøm M. B., Rekdal O., Svendsen J. S. (2002) J. Pept. Sci. 8, 431–437 [DOI] [PubMed] [Google Scholar]
- 52. Strøm M. B., Rekdal O., Svendsen J. S. (2002) J. Pept. Sci. 8, 36–43 [DOI] [PubMed] [Google Scholar]
- 53. Haug B. E., Stensen W., Stiberg T., Svendsen J. S. (2004) J. Med. Chem. 47, 4159–4162 [DOI] [PubMed] [Google Scholar]
- 54. Haug B. E., Stensen W., Svendsen J. S. (2007) Bioorg. Med. Chem. Lett. 17, 2361–2364 [DOI] [PubMed] [Google Scholar]
- 55. Haug B. E., Stensen W., Kalaaji M., Rekdal Ø., Svendsen J. S. (2008) J. Med. Chem. 51, 4306–4314 [DOI] [PubMed] [Google Scholar]
- 56. Hansen T., Alst T., Havelkova M., Strøm M. B. (2010) J. Med. Chem. 53, 595–606 [DOI] [PubMed] [Google Scholar]
- 57. Schibli D. J., Nguyen L. T., Kernaghan S. D., Rekdal Ø., Vogel H. J. (2006) Biophys. J. 91, 4413–4426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Haug B. E., Strøm M. B., Svendsen J. S. (2007) Curr. Med. Chem. 14, 1–18 [DOI] [PubMed] [Google Scholar]
- 59. Utsugi T., Schroit A. J., Connor J., Bucana C. D., Fidler I. J. (1991) Cancer Res. 51, 3062–3066 [PubMed] [Google Scholar]
- 60. Jing W., Hunter H. N., Hagel J., Vogel H. J. (2003) J. Pept. Res. 61, 219–229 [DOI] [PubMed] [Google Scholar]
- 61. Hu W., Lee K. C., Cross T. A. (1993) Biochemistry 32, 7035–7047 [DOI] [PubMed] [Google Scholar]
- 62. Kachel K., Asuncion-Punzalan E., London E. (1995) Biochemistry 34, 15475–15479 [DOI] [PubMed] [Google Scholar]
- 63. Yau W. M., Wimley W. C., Gawrisch K., White S. H. (1998) Biochemistry 37, 14713–14718 [DOI] [PubMed] [Google Scholar]
- 64. Matsuzaki K., Mitani Y., Akada K. Y., Murase O., Yoneyama S., Zasloff M., Miyajima K. (1998) Biochemistry 37, 15144–15153 [DOI] [PubMed] [Google Scholar]
- 65. Fiedler S., Broecker J., Keller S. (2010) Cell. Mol. Life Sci. 67, 1779–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schibli D. J., Hwang P. M., Vogel H. J. (1999) Biochemistry 38, 16749–16755 [DOI] [PubMed] [Google Scholar]
- 67. Padmanabhan S., Marqusee S., Ridgeway T., Laue T. M., Baldwin R. L. (1990) Nature 344, 268–270 [DOI] [PubMed] [Google Scholar]
- 68. Chakrabartty A., Schellman J. A., Baldwin R. L. (1991) Nature 351, 586–588 [DOI] [PubMed] [Google Scholar]
- 69. Woody R. W., Dunker A. K. (1996) in Circular Dichroism and the Conformational Analysis of Biomolecules (Fasman G. D., ed) pp. 109–157, Plenum Press, New York [Google Scholar]
- 70. Yang N. (2003) Structural Features of Amphipathic Membrane Active Peptides and Their Potential to Modulate Antitumor Activity and Tumor Cell Specificity Versus Antibacterial Activity, Ph.D. thesis, University of Tromsø, Tromsø, Norway [Google Scholar]
- 71. Fadnes B., Rekdal O., Uhlin-Hansen L. (2009) BMC Cancer 9, 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.