Background: IL-27 is a Th17 cell-inhibiting cytokine.
Results: IL-27 activates differential STAT signaling in intestinal epithelial cells resulting in increased epithelial restitution, induction of antibacterial genes (DMBT1 and IDO1), and growth inhibition of intestinal bacteria.
Conclusion: IL-27 mediates epithelial barrier protection and antibacterial responses.
Significance: We identified novel functions of IL-27, including epithelial restitution and a major role in the host response to intestinal bacteria.
Keywords: Bacteria, Cell Proliferation, Cytokine, Cytokine Action, Inflammation, Inflammatory Bowel Disease, Interleukin, Intestine, IL-27, IL-27R
Abstract
The role of the Th17 cell inhibiting cytokine IL-27 in the pathogenesis of inflammatory bowel disease is contradictory. Its effects on the intestinal barrier have so far not been investigated, which was the aim of this study. We show that intestinal epithelial cells (IEC) express both IL-27 receptor subunits IL-27RA and gp130. The IL-27 receptor expression is up-regulated in intestinal inflammation and during bacterial infection. IL-27 activates ERK and p38 MAPKs as well as Akt, STAT1, STAT3, and STAT6 in IEC. IL-27 significantly enhances cell proliferation and IEC restitution. These functions of IL-27 are dependent on the activation of STAT3 and STAT6 signaling pathways. As analyzed by microarray, IL-27 modulates the expression of 428 target genes in IEC (316 up and 112 down; p < 0.05). IL-27 as well as its main target genes are up-regulated in colonic tissue and IEC isolated from mice with dextran sulfate sodium (DSS)-induced colitis. The IL-27-induced expression of the anti-bacterial gene deleted in malignant brain tumor 1 (DMBT1) is mediated by p38 and STAT3 signaling, whereas the activation of the anti-inflammatory and anti-bacterial gene indoleamine 2,3-dioxygenase (IDO1) is dependent on STAT1 signal transduction. IL-27-induced indoleamine 2,3-dioxygenase enzymatic activity leads to growth inhibition of intestinal bacteria by causing local tryptophan depletion. For the first time, we characterize IL-27 as a mediator of intestinal epithelial barrier protection mediated via transcriptional activation of anti-inflammatory and antibacterial target genes.
Introduction
The immunological mechanisms underlying the pathogenesis of inflammatory bowel diseases (IBD),4 which are classified into Crohn's disease (CD) and ulcerative colitis, are so far not completely understood. T helper (Th) 17 cells, a novel subtype of proinflammatory T helper cell, seem to play an important role in the development of IBD (1). Recent evidence suggests that IL-27, a heterodimeric cytokine consisting of EBV-induced gene-3 (EBI3) and p28 (IL-27p28) (2), has a unique role in T cell differentiation. IL-27 inhibits the development of proinflammatory Th17 cells by suppressing the expression of the Th17 master transcription factor RORγt, thereby preventing the production of IL-17A and IL-17F in naive T cells (3). Although IL-27 inhibits the de novo differentiation of Th17 cells, it seems to have little effect on committed Th17 cells (4). Other anti-inflammatory properties of IL-27 include the differentiation of IL-10-secreting Tr1 cells (5) and the up-regulation of IL-10 expression. However, IL-27 induces Th1 immune responses (6) and suppresses Th2 cells (7).
Although IL-27 is primarily produced by activated antigen-presenting cells, the IL-27 receptor is found on T cells, NK cells, mast cells, endothelial cells, monocytes, neutrophils, and epithelial cells such as bronchial epithelial cells. It is composed of two subunits, IL-27RA (also designated WSX-1 or TCCR) and gp130 (8). Whereas IL-27RA is specific for IL-27 signaling, gp130 is also involved in the signal transduction of other cytokines of the IL-6 family such as IL-6, IL-11, oncostatin M, or leukemia inhibitory factor.
The data on the role of IL-27 and its receptors in IBD are so far contradictory, describing protective and proinflammatory effects. For example, it has been shown that deficiency of the IL-27 subunit EBI3 does not influence the Th1-mediated 2,4,6-trinitrobenzene sulfonic acid-induced colitis (9). However, it protects mice from oxazolone-induced colitis, a more Th2-driven model of IBD (9). A very recent study described that knock-out of the IL-27 subunit EBI3 but not IL-27p28 results in increased pathology of spontaneous or T cell transfer-induced colitis (10). In one study, Il27ra knock-out mice were characterized by an earlier onset and increased severity in the experimental DSS colitis model and showed an increase in Th17 cells in gut-associated lymphoid tissue and a decrease in IFN-γ-producing Th1 cells (11). In contrast, another study described a decreased severity of DSS-induced colitis in Il27ra knock-out mice (12). A different study reported that Il27ra knock-out delays the onset of disease in the IL-10 knock-out colitis model (13).
Interestingly, we and others revealed that single nucleotide polymorphisms in the IL27 gene region are associated with the susceptibility to IBD (14, 15). Given the importance of IEC and the disruption of an intact epithelial barrier in the pathogenesis of IBD, we here aimed to analyze the signaling, main target genes, and functions of IL-27 and its receptor complex in IEC that have not been investigated so far.
EXPERIMENTAL PROCEDURES
Reagents and Cell Culture
Human recombinant cytokines, antibodies, and all other reagents used in this study are listed in the supplemental methods. Human colorectal cancer-derived IEC lines HT-29, HCT116, DLD-1, SW480, and T84 were grown in DMEM containing 1% penicillin/streptomycin and 10% FCS in a humidified 5% CO2 atmosphere at 37 °C.
Reverse Transcription and Quantitative PCR (qPCR)
Total RNA was isolated using Qiagen RNeasy kit and was reverse-transcribed with the Transcriptor First Strand cDNA synthesis kit (Roche Applied Science). Real time qPCR (for primer sequences see supplemental Table S1) was performed on a LightCycler480 with SYBR Green PCR Master Mix from Roche Applied Science. Gene expression was normalized to β-actin in the respective samples.
Signal Transduction, Protein Isolation, Gel Electrophoresis, and Immunoblotting
DLD-1 cells were serum-starved overnight and stimulated with 50 ng/ml IL-27 for the indicated time intervals. If applicable, cells were pretreated with specific inhibitors for 1 h. Cells were solubilized in Nonidet P-40 lysis buffer, and the protein concentrations were determined by the Bradford method. Immunoblotting was performed according to standard procedures.
Chromatin Immunoprecipitation (ChIP)
DLD-1 cells were grown in 10-cm dishes until near confluence. Following serum starvation overnight, cells were stimulated with IL-27 or were left unstimulated for 30 min. Formaldehyde was added to a final concentration of 1% for 10 min to cross-link proteins to DNA. After the addition of glycine to quench the formaldehyde, cells were washed with PBS, and chromatin was isolated using the SimpleChIPTM enzymatic chromatin immunoprecipitation kit (Cell Signaling, Boston) according to the manufacturer's instructions. A 2% input control sample was removed from each chromatin preparation before performing immunoprecipitations. ChIP experiments included STAT3 as well as negative and positive control antibody reactions (all antibodies from Cell Signaling). The primers used for DNA quantification by quantitative PCR are listed in supplemental Table S2. PCR data from the immunoprecipitation samples were normalized to the 2% input control chromatin samples. All samples were prepared in triplicate.
Electrophoretic Mobility Shift Assay (EMSA)
Nuclear extracts were prepared according to standard procedures (16) with minor modifications. Details can be found in the supplemental methods. 30 μg of nuclear extract were incubated for 30 min with 100 fmol of a biotin-labeled double-stranded oligonucleotide (TIB MOLBIOL, Berlin, Germany) and 1 μg of poly(dI-dC) in 1× binding buffer (10 mm Tris, pH 7.4, 50 mm KCl, 1 mm DTT). If applicable, 1 μg of phospho-STAT1 antibody or 10 pmol of unlabeled competitor oligonucleotide were added prior to the addition of the labeled oligonucleotide. All oligonucleotide sequences are listed in supplemental Table S3. The samples were separated on a 6% polyacrylamide gel in 0.5× TBE buffer and transferred to a nylon membrane. Gel shift analysis was performed with the LightShift Chemiluminescent EMSA kit from Pierce.
siRNA Transfection
DLD-1 cells were transfected with siRNA using Lipofectamine RNAiMAX (Invitrogen). IL-27 stimulation experiments were performed 48 h post-transfection. For more details see the supplemental methods.
Cell Proliferation and Cell Restitution Assay
Cell proliferation was determined after 48 h using the CyQuant NF cell proliferation assay (Invitrogen). For in vitro cell regeneration assays, a standardized wound was created in confluent cell layers with a sterile pipette tip. Detached cells were removed by several washes with PBS. The cells were stimulated with IL-27 (50 ng/ml) or PBS, and the cell-free area was measured after 24 h. Details are given in the supplemental methods.
Measurement of Indoleamine 2,3-Dioxygenase-1 (IDO1) Enzymatic Activity
IDO1 enzymatic activity was determined by measuring the concentration of its metabolite kynurenine in the cell culture supernatants spectrophotometrically. Briefly, cell culture supernatant was mixed with 30% trichloroacetic acid. Following centrifugation, the supernatant was mixed with an equal volume of Ehrlich's reagent, and the absorbance was measured at 492 nm and compared with a standard curve of defined kynurenine concentrations (0–100 μm).
Assessment of Bacterial Growth Inhibition
Escherichia coli strains WP2 (trp−) and K12 (trp+) were grown on Luria-Bertani (LB) agar plates. A single colony was resuspended in PBS, and aliquots were incubated for 6–8 h in conditioned RPMI medium that was prepared as follows. DLD-1 cells were stimulated with 50 ng/ml IL-27 or were left unstimulated for 48 h. Cells were washed twice with PBS to remove any residual IL-27. Fresh RPMI (containing no antibiotics) was added to all cells and was collected at time intervals between 4 and 72 h as indicated in the respective experiments. Bacterial growth in conditioned RPMI was determined photometrically measuring absorbance at 600 nm. In some experiments, l-tryptophan (10 μg/ml final concentration) was added to the conditioned medium. Additionally, E. coli bacteria in the logarithmic growth phase were serially diluted in sterile PBS and were seeded on LB agar plates to determine the number of colony-forming units (cfu) in the suspension.
Co-culture of E. coli with Intestinal Epithelial Cells
An overnight culture of E. coli grown in DMEM was diluted 1:100 and was incubated until mid-logarithmic phase. DLD-1 cells were grown in 6-well dishes until near confluency, and the cell number was calculated from a separate dish. A 5-, 50-, or 500-fold number of E. coli bacteria was added directly to the DLD-1 cells for 6 h. Alternatively, E. coli bacteria were removed from the overnight culture by centrifugation, and the supernatant was sterile-filtered before it was added to DLD-1 cells in a 1:20 dilution.
Microarray Analysis
IL-27-induced gene expression was analyzed in triplicate using Agilent Whole Human Genome Oligo Microarrays together with a One-color-based hybridization protocol. Differential gene expression was identified by applying appropriate biostatistics to the data set. GeneSpring GX 10 analysis software (Agilent Technologies, Santa Clara, CA) was used to normalize and analyze the raw data. Details can be found in the supplemental methods.
Immunohistochemistry
Immunohistochemistry on paraffin-embedded tissue was performed according to standard procedures. Details are described in the supplemental methods.
Induction of DSS Colitis in Mice and Preparation of Murine IEC
For the induction of acute DSS colitis, 8–10-week-old sex-matched BALB/c mice were given 2.5% DSS in their drinking water for 5 days. DSS was then removed from the drinking water, and mice were euthanized 2 days after cessation of DSS treatment. Assessment of colitis activity and isolation of IEC were performed as described in the supplemental methods.
Colonic Biopsies
All participating patients were recruited at the IBD Center, University of Munich (Germany) and gave written informed consent prior to biopsy sampling. The study was approved by the Ethics Committee of the Medical Faculty of the University of Munich. The diagnosis of CD or ulcerative colitis was determined according to established guidelines. From each patient, two biopsies from macroscopically noninflamed mucosa and two from macroscopically inflamed sites were collected. Detailed patient characteristics are given in supplemental Table S4.
RESULTS
IL-27 Receptor Complex Is Expressed in Intestinal Epithelial Cell Lines and Is Up-regulated under Proinflammatory Conditions
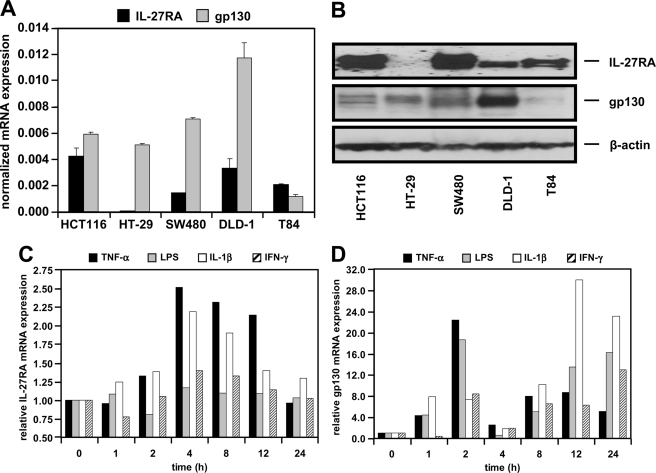
Initial RT-PCR analysis in several human IEC lines (HCT116, HT-29, SW480, DLD-1, and T84) demonstrated expression of IL-27RA and gp130 mRNA in all cell lines tested. However, mRNA expression levels vary between the different cell lines as shown by qPCR (Fig. 1A) and Western blot analysis (Fig. 1B). Based on these data, we chose DLD-1 cells, which express both receptor subunits at high levels, for all following experiments. To analyze IL-27RA and gp130 expression under inflammatory conditions, we treated DLD-1 cells with the proinflammatory cytokines TNF-α, IL-1β, IFN-γ, or with LPS. IL-27RA expression was up-regulated by TNF-α or IL-1β with bell-shaped kinetics, whereas IFN-γ and LPS had no effect (Fig. 1C). All cytokines as well as LPS increased the expression of gp130 in a biphasic manner (Fig. 1D).
FIGURE 1.
Intestinal epithelial cells express the IL-27 receptor complex. A, qPCR analysis reveals differential expression of IL-27RA and gp130 mRNA in IEC. Expression was normalized to β-actin in the respective samples. B, protein of IEC lines as indicated was analyzed by Western blot for IL-27RA and gp130. β-Actin was used as loading control. C, expression of IL-27RA is increased by stimulation with the proinflammatory cytokines TNF-α (50 ng/ml) or IL-1β (10 ng/ml) for the indicated time intervals. D, expression of gp130 mRNA is stimulated biphasically by treatment with TNF-α, LPS (1 μg/ml), IL-1β, or IFN-γ (1000 units/ml). Expression in unstimulated cells (t = 0) was arbitrarily set as 1.
IL-27RA and gp130 Are Expressed in Normal and Inflamed Human Colonic Mucosa
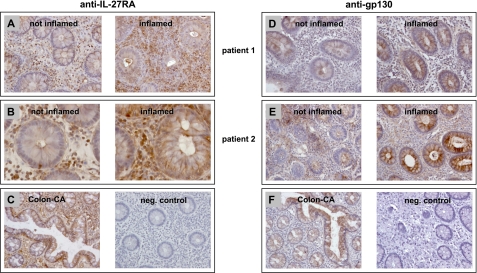
To determine IL-27RA and gp130 expression in primary human colonic tissue, we performed immunohistochemical analyses in inflamed and uninflamed biopsies from CD patients. IL-27RA was expressed at high levels in infiltrating immune cells in uninflamed and even stronger in inflamed intestinal mucosa (Fig. 2, A and B). In normal IEC, IL-27RA was expressed at low levels. However, expression of IL-27RA was up-regulated in IEC from inflamed intestinal biopsies (Fig. 2, A and B). In colon cancer cells, IL-27RA was expressed at higher levels in comparison with normal epithelial cells (Fig. 2C). In the negative control, no staining was observed (Fig. 2C).
FIGURE 2.
IL-27RA and gp130 are expressed in human colonic biopsies. A and B, immunohistochemistry for IL-27RA shows high expression of IL-27RA in infiltrating immune cells and lower expression in IEC in tissue that is not inflamed. In inflamed tissue, IL-27RA expression is increased in IEC as well as in infiltrating cells. C, in colon cancer (CA), IL-27RA is expressed at high levels throughout the tissue (left panel). The negative control shows no unspecific staining. D and E, gp130 is expressed at high levels in IEC and is increased in inflamed tissue. F, staining for gp130 in colon carcinoma cells is similar to that in normal epithelial cells. The negative control confirms specificity of the immunohistochemistry.
gp130 expression was found primarily in epithelial cells of the intestine whereas infiltrating immune cells expressed gp130 at lower levels in uninflamed tissue (Fig. 2, D and E). In inflamed tissue, epithelial cells showed higher expression of gp130 (Fig. 2, D and E). In infiltrating immune cells, gp130 expression was also slightly increased (Fig. 2, D and E). The expression of gp130 in colon carcinoma was comparable with that in normal epithelial tissue, whereas the negative control did not show any staining (Fig. 2F).
IL-27 Is Increased in Inflamed Biopsies from CD Patients
Next, we analyzed expression of IL-27 by qPCR in a total of 40 biopsies from 11 different patients with CD. In 9 out of 11 patients, IL-27 mRNA expression was higher in inflamed regions compared with noninflamed biopsies of the same patients (supplemental Fig. S2). Overall, the average expression levels of IL-27 in inflamed tissue were 46-fold higher than in noninflamed regions (p < 0.05). In patients with ulcerative colitis, no significant difference in IL-27 expression was seen between inflamed and noninflamed tissue (data not shown).
IL-27 Activates the MAPKs ERK and p38 and the Transcription Factors STAT1, STAT3, and STAT6
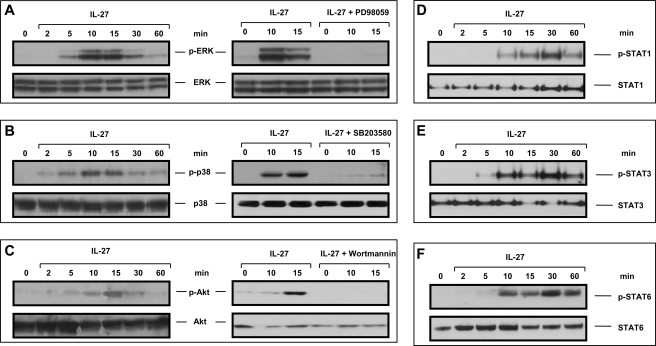
We assessed functionality of the IL-27 receptor complex in IEC by analyzing activation of major signaling pathways. In Western blot experiments using phospho-specific antibodies, we demonstrated activation of the MAPKs ERK (Fig. 3A) and p38 (Fig. 3B) as well as of Akt (Fig. 3C) in DLD-1 cells following IL-27 stimulation. Pretreatment with the MEK1 inhibitor PD98059 completely abolished ERK phosphorylation, whereas the p38 inhibitor SB203580 prevented p38 activation (Fig. 3, A and B). The PI3K inhibitor wortmannin suppressed activation of Akt (Fig. 3C). Following stimulation with 50 ng/ml IL-27 protein, the transcription factors STAT1, STAT3, and STAT6 are strongly phosphorylated (Fig. 3, D–F). The proteins STAT2, STAT4, and STAT5 were not activated by IL-27 in IEC (data not shown).
FIGURE 3.
IL-27 induces MAPK, Akt, STAT1, STAT3, and STAT6 signaling in IEC that is abrogated by specific inhibitors. A, IL-27-induced ERK phosphorylation is completely abolished by the MEK1 inhibitor PD98059. B, p38 is activated by IL-27. The p38 inhibitor SB203580 prevented this activation. C, IL-27 induces phosphorylation of Akt that was suppressed by pretreatment with the PI3K inhibitor wortmannin. D, IL-27 induces phosphorylation of STAT1. E, IL-27 stimulates activation of STAT3. F, STAT6 is phosphorylated following IL-27 stimulation. min, minutes of stimulation; p-, phospho-.
IL-27 Induces Cell Proliferation and Epithelial Barrier Restitution Mediated via STAT3 and STAT6 Activation
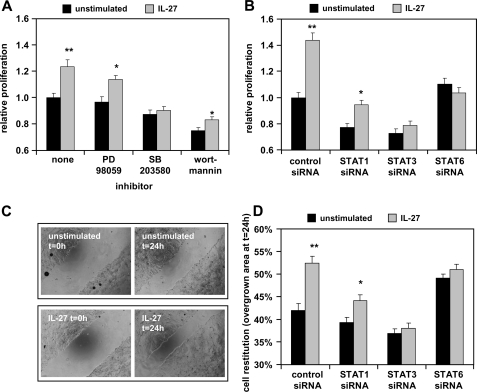
We next determined the influence of IL-27 on the intestinal epithelial barrier by analyzing the effects of IL-27 on IEC proliferation. We show that IL-27 significantly increases cell proliferation of DLD-1 cells (Fig. 4A). Preincubation with specific inhibitors against MEK1, p38, and PI3K revealed that this effect is mediated by p38 but not by the MEK1-ERK or the PI3K-Akt pathway (Fig. 4B). Transfection of DLD-1 cells with specific siRNAs against STAT1, STAT3, STAT6, or unspecific control siRNA 48 h before stimulation showed that silencing of STAT6 completely abolished IL-27-mediated cell proliferation. Silencing of STAT3 had a less pronounced but still significant effect (Fig. 4B).
FIGURE 4.
IL-27 induces cell proliferation and cell migration in IEC that are mediated by STAT signaling as well as p38. A, after stimulation with 50 ng/ml IL-27 for 48 h, cell proliferation was significantly higher in IL-27-treated cells in comparison with unstimulated cells. Pretreatment with the p38 inhibitor SB203580 abolished IL-27-mediated increase of cell proliferation. *, p < 0.01; **, p < 0.0005 versus unstimulated in the same inhibitor group. B, transfection with STAT3 or STAT6 siRNA inhibited IL-27-mediated enhancement of cell proliferation. In all experiments, proliferation in unstimulated control cells was arbitrarily set as 1.0. *, p < 0.005; **, p < 10−6 versus unstimulated in the same siRNA group. C, wounding assays were performed to analyze cell migration. Presented are representative images of unstimulated and IL-27-stimulated cells. D, analysis of siRNA-transfected cells stimulated with IL-27 or left unstimulated. IL-27 significantly increased the area overgrown by cells after 24 h (t = 0 h, 0%), which is dependent on STAT3 and STAT6. Data are derived from four independent experiments. *, p < 0.01; **, p < 10−5 versus unstimulated in the same siRNA group.
We then performed wounding assays analyzing the influence of IL-27 on IEC restitution (Fig. 4C). In control and STAT1 siRNA-transfected cells, stimulation with IL-27 significantly increased cell restitution in comparison with unstimulated cells (Fig. 4, C and D). IL-27-mediated cell migration could be completely abolished by the silencing of STAT3 or STAT6 (Fig. 4D).
Microarray Analysis of IL-27-induced Gene Expression Patterns in IEC
To analyze the IL-27-mediated gene expression pattern in IEC, we performed microarray experiments in IL-27-stimulated DLD-1 cells. After correction for multiple testing, 428 genes remained significantly regulated more than two-fold by IL-27 (316 up and 112 down; p < 0.05). The 20 most up-regulated genes are depicted in Table 1.
TABLE 1.
Overview of 20 genes whose expression was most strongly induced by IL-27 in DLD-1 cells after 6 h of stimulation (p < 0.05 versus unstimulated and adjusted for multiple testing)
| Gene ID | Gene symbol | Description | -Fold increase |
|---|---|---|---|
| NM_003955 | SOCS3 | Suppressor of cytokine signaling 3 | 17.34 |
| NM_002164 | IDO1 | Indoleamine-pyrrole 2,3-dioxygenase | 13.83 |
| NM_018664 | BATF3 | Basic leucine zipper transcription factor, ATF-like 3 | 11.88 |
| NM_007329 | DMBT1 | Deleted in malignant brain tumors 1 | 8.13 |
| NM_018027 | FRMD4A | FERM domain containing 4A | 7.74 |
| NM_000014 | A2M | α2-Macroglobulin | 7.58 |
| NM_182920 | ADAMTS9 | ADAM metallopeptidase with thrombospondin type 1 motif, 9 | 7.24 |
| X94553 | FOXE1 | HFKH4 mRNA for forkhead-like protein | 6.66 |
| NM_144586 | LYPD1 | LY6/PLAUR domain containing 1 | 6.56 |
| NM_000246 | CIITA | Class II, major histocompatibility complex, transactivator | 6.52 |
| NM_152486 | SAMD11 | Sterile α motif domain containing 11 | 6.06 |
| THC2559929 | THC2559929 | SNC73 protein, partial (54%) | 5.84 |
| NM_002089 | CXCL2 | Chemokine (CXC motif) ligand 2 | 5.82 |
| NM_006161 | NEUROG1 | Neurogenin 1 | 5.77 |
| NM_006169 | NNMT | Nicotinamide N-methyltransferase | 5.74 |
| NM_138700 | TRIM40 | Tripartite motif-containing 40 | 5.70 |
| NM_006851 | GLIPR1 | GLI pathogenesis-related 1 (glioma) | 5.56 |
| NM_013271 | PCSK1N | Proprotein convertase subtilisin/kexin type 1 inhibitor | 5.27 |
| NM_006705 | GADD45G | Growth arrest and DNA damage-inducible, γ | 5.24 |
| NM_014069 | PSORS1C2 | Psoriasis susceptibility 1 candidate 2 | 4.80 |
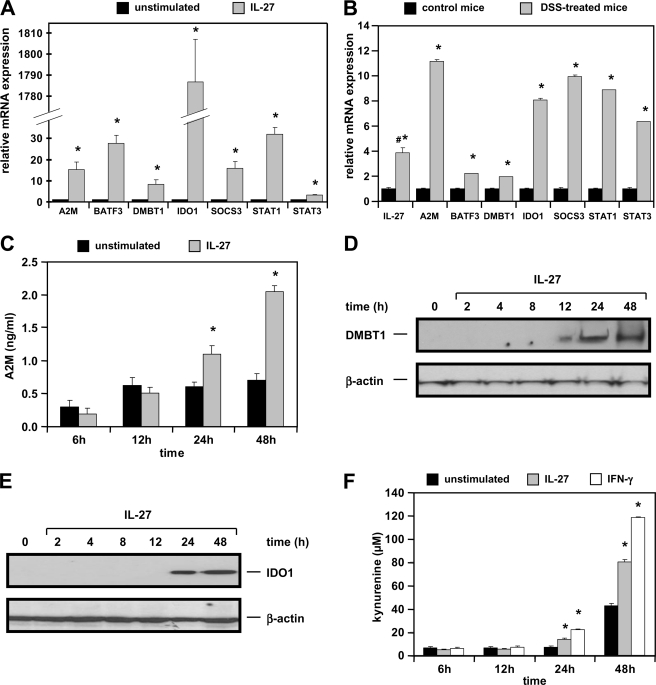
To verify the IL-27-induced gene expression in DLD-1 cells, we performed qPCR in three independent sets of cDNA samples choosing some of the most strongly up-regulated genes from the microarray analysis. Expression of α2-macroglobulin (A2M), basic leucine zipper transcriptional factor ATF-like 3 (BATF3), deleted in malignant brain tumor (DMBT1), IDO1, and suppressor of cytokine signaling 3 (SOCS3) were increased between 8.2- and 1785-fold in IL-27-stimulated cells as determined by qPCR (Fig. 5A). Moreover, we observed that STAT1 and STAT3 genes are additional targets up-regulated 32- and 3.2-fold, respectively, by IL-27.
FIGURE 5.
Target genes and proteins induced by IL-27 in human IEC are also up-regulated in the DSS colitis model in primary murine IEC. A, qPCR analysis confirms induction of A2M, BATF3, DMBT1, IDO1, SOCS3, STAT1, and STAT3 mRNA expression by IL-27 in DLD-1 cells. Expression in unstimulated cells was arbitrarily set to 1.0 for each gene. The very strong increase in IDO1 expression is a result of very low basal expression in unstimulated cells. *, p < 0.005 versus unstimulated. B, IL-27 target genes are up-regulated in isolated primary IEC from BALB/c mice treated with DSS for 5 days. #, IL-27 expression was determined in whole colonic tissue as it is not expressed in IEC. *, p < 0.05 versus control mice. C, A2M protein concentration in the cell culture supernatant as determined by ELISA is increased by stimulation with IL-27. *, p < 0.05 versus unstimulated. D, DMBT1 protein expression is activated by IL-27. E, IDO1 protein expression is induced by IL-27 after 24 h. F, IDO1 enzymatic activity is stimulated by IL-27 as determined by measuring the kynurenine concentration in the cell culture supernatant. IFN-γ (1000 units/ml) was used as a positive control. *, p < 0.05 versus unstimulated.
IL-27 and IL-27 Target Genes Are Up-regulated in Murine DSS Colitis
Having shown that IL-27 is up-regulated in human colitis, we next analyzed expression levels of Il27 and its target genes in murine DSS colitis. Il27 expression (measured in whole colonic tissue as IEC do not express IL-27 protein) was significantly higher in DSS-treated mice compared with control mice (Fig. 5B). In IEC isolated from DSS-treated mice, expression of the IL-27 target genes A2m, Batf3, Dmbt1, Ido1, Socs3, Stat1, and Stat3 was significantly up-regulated in comparison with control mice (Fig. 5B) suggesting that the detected increased IL-27 cytokine levels in the colon may induce those target genes.
Validation of Protein Expression of IL-27 Target Genes by ELISA, Western Blot, and Enzymatic Activity Assay
To analyze if the IL-27-induced gene expression results in increased protein levels, A2M protein was measured by ELISA. A2M, for which increased fecal expression reflecting disease activity has been shown in IBD patients (17), was significantly up-regulated after 48 h of stimulation with IL-27 as determined from the cell culture supernatants of DLD-1 cells (Fig. 5C). However, overall concentrations were very low and close to the detection limit of the ELISA. In Western blot experiments, we confirmed induction of DMBT1 and IDO1 protein by IL-27 beginning 12 and 24 h after stimulation, respectively (Fig. 5, D and E).
Next, we analyzed IDO1 enzymatic activity by determining the concentration of its metabolite kynurenine spectrophotometrically in the cell culture supernatants. Kynurenine concentration was very low in the supernatant of unstimulated cells, whereas IL-27 induced a significant increase in kynurenine concentration up to 82 μm (Fig. 5F). This effect was comparable with that of IFN-γ, which is a known IDO1 inducer (18).
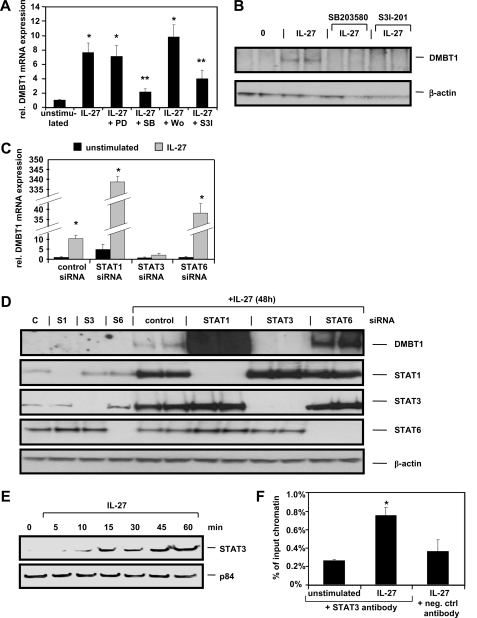
IL-27-induced DMBT1 mRNA and Protein Expression Is Mediated by p38 and STAT3 and Is Negatively Regulated by STAT1 and STAT6
We then aimed to determine the signal transduction pathways that regulate expression of DMBT1, a scavenger receptor involved in bacterial recognition (19). Therefore, we pretreated the cells with inhibitors for MEK1 (PD98059), p38 (SB203580), PI3K (wortmannin), or STAT3 (S3I-201). Following stimulation with IL-27 for 6 h, induction of DMBT1 mRNA expression was negatively regulated by the p38 inhibitor SB203580 and the STAT3 inhibitor S3I-201 (Fig. 6A), whereas expression of other genes (A2M, BATF3, IDO1, and SOCS3) was not negatively affected by these inhibitors (data not shown). The dependence of IL-27-induced DMBT1 protein expression on p38 and STAT3 was confirmed by Western blot experiments (Fig. 6B). siRNA transfection experiments confirmed that the IL-27-induced transcriptional up-regulation of DMBT1 is mediated by STAT3 as silencing of STAT3 nearly completely inhibited DMBT1 mRNA as well as DMBT1 protein expression (Fig. 6, C and D). Silencing of STAT6 and STAT1 resulted in a strong increase of DMBT1 mRNA and protein expression following IL-27 stimulation (Fig. 6, C and D), identifying STAT1 and STAT6 as negative regulators of IL-27-mediated DMBT1 up-regulation. Moreover, an up-regulation of STAT1 and STAT3 protein following IL-27 stimulation was observed (Fig. 6D) confirming the data derived from qPCR analysis (Fig. 5A).
FIGURE 6.
DMBT1 induction through IL-27 is mediated by p38 and STAT3 and negatively regulated by STAT1 and STAT6. A, DMBT1 mRNA is induced by IL-27 and can be inhibited by treatment with p38 inhibitor SB203580 and STAT3 inhibitor S3I-201. DMBT1 expression in unstimulated control cells was arbitrarily set to 1.0. *, p < 0.01 versus control; **, p < 0.05 versus IL-27 only. PD, PD98059; SB, SB203580; Wo, wortmannin; S3I, S3I-201. B, DMBT1 protein is induced by IL-27 after 48 h. This effect is abolished by the p38 inhibitor SB203580 and the STAT3 inhibitor S3I-201. C, DMBT1 mRNA is induced 10-fold by IL-27 after 6 h in control siRNA-transfected cells. Silencing of STAT3 inhibited this effect, and silencing of STAT6 and even stronger silencing of STAT1 increased expression of DMBT1 mRNA induced by IL-27. D, DMBT1 protein is induced by IL-27 in control siRNA-transfected cells. STAT3 silencing abolished and STAT1 and STAT6 silencing increased expression of DMBT1 protein mediated by IL-27. Representative STAT1, STAT3, and STAT6 blots show silencing of the respective genes by the respective specific siRNA (C, control; S1, STAT1; S3, STAT3; S6, STAT6). E, Western blots analyzing nuclear extracts from IL-27-treated cells show a transfer of STAT3 protein into the nucleus. The nuclear protein p84 was used as loading control. F, ChIP analysis with an antibody against STAT3 demonstrates increased STAT3 binding to the DMBT1 promoter region (between bp −266 and −144 relative to the translation start site) in IL-27-stimulated cells. The negative control antibody (normal rabbit IgG antibody) did not produce any significant signal. *, p < 0.05 versus unstimulated.
Using nuclear extracts, we next demonstrated in Western blot experiments that STAT3 protein is transferred to the nucleus upon IL-27 stimulation (Fig. 6E). To prove that STAT3 directly binds to the DMBT1 promoter, we performed ChIP analyses with a STAT3-specific antibody followed by quantitative PCR using cells stimulated with IL-27 for 30 min, and we compared the results with unstimulated cells. All primer pairs were chosen to amplify regions with putative STAT3-binding sites in the pulled down DNA and are listed in supplemental Table S2. Our experiments revealed that STAT3 binds within the DMBT1 promoter region between bp −266 and −144 relative to the transcription start site. This region includes a recently described STAT3-binding site identified in IL-22-stimulated IEC (20). STAT3 binding in the DMBT1 promoter resulted in a 3-fold increased PCR signal in IL-27-stimulated cells in comparison with unstimulated cells in our experiments (Fig. 6F).
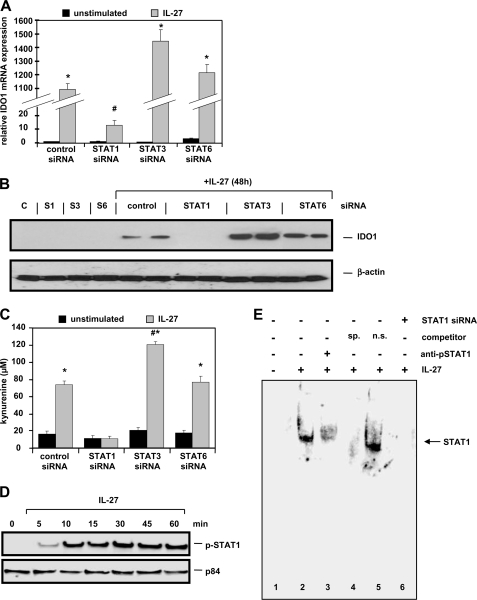
IL-27-induced IDO1 Expression and Enzymatic Activity Is STAT1-dependent
In siRNA transfection experiments, we next investigated the pathways that regulate IDO1 activation. Following STAT1 expression silencing, IL-27-induced IDO1 mRNA expression was nearly completely inhibited (Fig. 7A). In Western blot experiments, we verified induction of IDO1 expression by IL-27 in control siRNA-transfected cells (Fig. 7B). Silencing of STAT1 completely abolished IDO1 expression, whereas silencing of STAT3 had a weak positive effect on IDO1 mRNA and protein expression (Fig. 7, A and B). Western blots with antibodies against STAT1, STAT3, and STAT6 confirmed silencing of the respective genes (data not shown). IL-27-mediated IDO1 enzymatic activity was also strongly dependent on STAT1 as determined by the IL-27-induced kynurenine levels (Fig. 7C). Transfection with STAT3 siRNA resulted in a slightly stronger kynurenine production following IL-27 stimulation compared with control transfected cells (Fig. 7C). However, silencing of STAT6 did not significantly influence IL-27-activated IDO1 expression and enzymatic activity (Fig. 7, A–C). IL-27 stimulation results in an enrichment of STAT1 protein in the nucleus as shown by Western blot analysis with nuclear extracts (Fig. 7D). The direct binding of STAT1 to the IDO1 promoter was then analyzed in EMSA experiments using different oligonucleotides with sequences from the IDO1 promoter containing putative STAT1-binding sites (for sequences see supplemental Table S3). We show that STAT1 binds to an oligonucleotide (IDO1_-1007, supplemental Table S3) containing the −1007- to −994-bp promoter region of IDO1 in IL-27-stimulated cells but not in unstimulated cells (Fig. 7E, lanes 1 and 2). Including a phospho-STAT1-specific antibody in the reaction inhibited STAT1 DNA binding (Fig. 7E, lane 3). The addition of 100-fold excess of unlabeled IDO1_-1007 oligonucleotide confirmed the specificity of binding, whereas the addition of 100-fold excess of an unrelated oligonucleotide sequence (containing a mutant STAT1-binding site) did not influence the STAT1 binding to the IDO1_-1007 oligonucleotide (Fig. 7E, lanes 5 and 6). Together, these data confirm that STAT1 directly binds to the IDO1 promoter thereby inducing IDO1 expression.
FIGURE 7.
IL-27-induced expression and enzymatic activity of IDO1 is dependent on STAT1. A, IDO1 mRNA is strongly induced by IL-27 stimulation. Whereas silencing of STAT6 did not significantly influence IDO1 mRNA increase, silencing of STAT1 nearly completely abolished IDO1 mRNA induction. Silencing of STAT3 had a weak positive effect. IDO1 mRNA expression was arbitrarily defined as 1.0 in unstimulated control transfected cells. *, p < 0.01 versus unstimulated; #, p < 0.01 versus control siRNA + IL-27. B, IDO1 protein is induced by IL-27 in control siRNA-transfected cells. IDO1 induction is inhibited by STAT1 silencing and slightly increased by STAT3 silencing. C, IL-27-mediated up-regulation of IDO1 enzymatic activity as determined by the formation of kynurenine after 48 h is completely inhibited by silencing of STAT1. *, p < 10−9 versus unstimulated; #, p < 10−8 versus control siRNA + IL-27. D, increased levels of phospho-STAT1 protein can be found in nuclear extracts from IL-27-stimulated cells as demonstrated by Western blot. The nuclear protein p84 shows equal loading of the protein samples. E, EMSA experiments demonstrate binding of STAT1 to the promoter region of the IDO1 gene in IL-27-stimulated cells after 30 min (lane 2 versus lane 1). Preincubation of the binding reaction with a phospho-STAT1-specific antibody (anti-pSTAT1) inhibited STAT1 DNA binding (lane 3) as well as did preincubation with a 100-fold excess of unlabeled specific probe (lane 4, sp, specific). Preincubation with a 100-fold excess of unspecific competitor oligonucleotide (5′-CATGTTATGCATATTGGAGTAAGTG-3′; n.s., nonspecific) did not influence STAT1 binding (lane 5). Nuclear extracts from IL-27-stimulated cells with siRNA-mediated knock-out of STAT1 show no binding of protein to the IDO1 probe (lane 6). The sequence (sense orientation) of the double-stranded labeled probe included in all lanes was 5′-TTTTCCTGTAAAAT-3′ (IDO1_-1007). The STAT1 binding site is underlined.
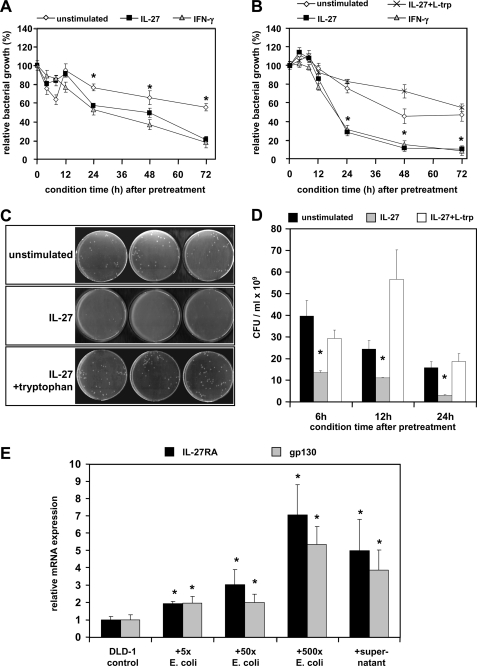
IL-27-induced IDO1 Inhibits Bacterial Growth and IL-27RA and gp130 Expressions Are Induced by Intestinal Bacteria
IDO1 activity has been related to decreased microbial growth (21). We therefore next analyzed the influence on IL-27-induced IDO1 on the growth of intestinal bacteria. Conditioned RPMI medium from IL-27-treated or untreated DLD-1 cells was prepared as described under “Experimental Procedures.” E. coli strains K12 and WP2 (the latter deficient in tryptophan synthesis) were incubated for 6–8 h in conditioned RPMI from DLD-1 cells (collected 4, 8, 12, 24, 48, and 72 h after IL-27 removal and the addition of fresh RPMI). Medium from IL-27-pretreated cells conditioned for 24, 48, or 72 h exhibited a significant growth inhibition on E. coli K12 (similar to IFN-γ) in comparison with conditioned medium from cells without IL-27 pretreatment (Fig. 8A), whereas shorter time intervals had no significant effect. This IL-27-mediated effect was even more pronounced in the E. coli strain WP2 (Fig. 8B) suggesting the lack of tryptophan as a major reason for growth inhibition.
FIGURE 8.
IL-27 inhibits the growth of intestinal bacteria through tryptophan depletion via the induction of IDO1 enzymatic activity. A, E. coli K12 bacteria were incubated for 6–8 h in conditioned medium from IL-27 pretreated or untreated DLD-1 cells, and bacterial growth was determined photometrically at 600 nm. Medium from IL-27 pretreated cells conditioned for 24 h or longer exhibited a significant growth inhibiting effect on E. coli K12. IFN-γ (1000 units/ml) was used as a positive control. Bacterial growth rates were normalized to growth in fresh, untreated RPMI medium. *, p < 0.01 unstimulated versus IL-27 or versus IFN-γ. B, growth of E. coli WP2 (tryptophan auxotroph) was determined as in A. The IL-27-mediated significant bacteriostatic effect was more pronounced in strain WP2 in comparison with strain K12. The addition of l-tryptophan (10 μg/ml) completely reversed the bacterial growth-inhibiting effect of conditioned medium from IL-27-pretreated DLD-1 cells. *, p < 0.001 versus unstimulated or versus IL-27 + l-tryptophan. C and D, number of colony-forming units (cfu) per ml of bacterial cell suspension prepared as in A was significantly lower in conditioned medium from IL-27-pretreated cells in comparison with conditioned medium from untreated cells or when additional l-tryptophan was added. C is a representative picture of LB agar plates with a 10−9 dilution of the bacterial suspension grown in medium conditioned for 24 h. *, p < 0.01 versus unstimulated or versus IL-27 + l-tryptophan. E, expression of IL-27RA and gp130 in DLD-1 cells increased significantly when cells were co-cultured with E. coli bacteria for 6 h. The number of E. coli bacteria added was 5-, 50-, or 500-fold higher than the number of DLD-1 cells per well. A similar effect was achieved when sterile-filtered supernatant from an E. coli overnight culture was added to DLD-1 cells. Expression levels were normalized to the expression in untreated DLD-1 cells. *, p < 0.05 versus unstimulated.
To analyze if a depletion of tryptophan was responsible for the IL-27-related bacterial growth inhibition, E. coli WP2 bacteria were incubated with or without supplemental l-tryptophan in conditioned RPMI. The addition of l-tryptophan completely reversed the inhibiting effect of IL-27-conditioned medium on bacterial growth demonstrating that the lack of tryptophan but not the presence of kynurenine causes decreased bacterial growth (Fig. 8B). Western blots and photometric measurements confirmed IDO1 induction and increased kynurenine production in DLD-1 cells up to 72 h after the removal of IL-27 and the addition of fresh medium (data not shown). Together, these data confirm that the IL-27-induced IDO1 enzymatic activity leading to tryptophan depletion is responsible for bacterial growth inhibition mediated by IL-27.
To determine the influence of intestinal bacteria on IL-27 signaling, we co-cultured DLD-1 cells with increasing amounts of E. coli bacteria and determined the expression of both IL-27 receptor subunits. E. coli bacteria induced a significant, dose-dependent increase in IL-27RA and gp130 expression in DLD-1 cells. Similarly, the diluted, sterile supernatant from an E. coli bacteria overnight culture induced expression of IL-27RA and gp130 suggesting that some soluble factor produced by the bacteria is involved in increasing IL-27 receptor expression in IEC.
DISCUSSION
We here present for the first time a comprehensive analysis of the expression, signal transduction, and biological functions of IL-27 and its receptors in IEC, which has not been analyzed previously. We show that IEC express both receptor subunits, IL-27RA and gp130. Their expression is increased in IEC under proinflammatory conditions in vitro and in intestinal inflammation in CD patients in vivo pointing to an important role of this cytokine receptor system in IBD. Given that our analyses demonstrated IL-27 receptor expression also in colorectal cancer cells, we hypothesize that IL-27 may additionally play a role in colorectal cancer cell migration, tumor growth, and metastasis as well as in the pathogenesis of colitis-induced colorectal cancer.
Stimulation of IEC with IL-27 results in the activation of the MAPK signaling pathways p38 and ERK as well as of the PI3K-Akt pathway. IL-27 also activates the transcription factors STAT1, STAT3, and STAT6. STAT1 and STAT3 activation by IL-27 has been shown in other studies in T cells, monocytes, and keratinocytes (22, 23). However, this is the first study reporting phosphorylation of STAT6 by IL-27. Most important, we identified here a novel signaling pathway responsible for IEC proliferation and migration. We have shown by STAT6 silencing in siRNA-transfected cells that STAT6 signaling is largely responsible for the IL-27-induced cell proliferation and cell migration. In line with our data, it has been reported that STAT6 knock-out results in decreased proliferation and increased apoptosis in IEC and in an exacerbated DSS colitis (24, 25).
In contrast to cell proliferation and migration, the expression of the IL-27-induced target genes A2M, BATF3, DMBT1, IDO1, and SOCS3 was not negatively influenced by STAT6 silencing suggesting differential functions of this transcription factor in IL-27-mediated effects on IEC. Furthermore, we have shown that STAT3 but not STAT1 is crucial for IL-27-induced cell proliferation and cell restitution. This is in agreement with previous studies by us and others showing STAT3 as an important signaling molecule for cell proliferation and migration (26–28).
To identify potential target genes of IL-27 in IEC, we performed microarray analysis in IL-27-stimulated DLD-1 cells. We found a total number of 316 genes that were significantly up-regulated more than two-fold by IL-27. In murine DSS colitis experiments, IL-27 and selected IL-27 target genes identified in the microarray experiments were up-regulated accordingly.
We then focused on a more detailed analysis of two genes with potential roles in IBD pathogenesis, DMBT1 and IDO1. The DMBT1 protein acts as a pattern recognition and scavenger receptor and has been shown to influence DSS colitis (19, 29). It is up-regulated by proinflammatory stimuli and in IBD patients (20, 30) and inhibits LPS and muramyl dipeptide-induced NF-κB activation as well as Salmonella enterica cytoinvasion (31). Recently, an association of a DMBT1 deletion allele with CD has been demonstrated (30).
We here show for the first time that DMBT1 protein is expressed following IL-27 stimulation in IEC. This was dependent on p38 and STAT3 signaling, which is in agreement with a recent study describing STAT3 signaling as a prerequisite for DMBT1 induction by IL-22 in IEC (20). However, silencing of STAT6 or STAT1 protein resulted in a massive induction of DMBT1 expression suggesting that DMBT1 expression is negatively regulated by STAT6 and even stronger by STAT1 signaling. Interestingly, DMBT1 is a target gene of the pattern recognition receptor NOD2 (31), which is encoded by the first identified CD susceptibility gene NOD2/CARD15 (32). Given that DMBT1 and other antibacterial genes are major transcriptional targets of IL-27, an important role of IL-27 following bacterial infection is likely. This is further supported by microarray analyses in which IEC were infected with the Salmonella strain Salmonella dublin (33). The IL-27 subunit EBI3 was found among the most up-regulated genes following bacterial infection (33). Considering the increased IL-27 receptor expression in intestinal inflammation and after bacterial infection demonstrated in our experiments, the IL-27-IL-27RA axis might serve as autocrine amplifier of antibacterial and barrier protective responses in IEC. Interestingly, a CD-associated IL27 risk variant results in lower IL-27 expression (14). Therefore, one could suggest that IEC of CD patients exhibit less antibacterial activity resulting in a weakened intestinal barrier and increased bacterial transmigration, which is typically observed in CD patients.
Moreover, we analyzed the IL-27 target gene IDO1 that encodes an enzyme involved in tryptophan metabolism catalyzing the first step in the conversion from tryptophan to kynurenine. IDO1 has been shown to be overexpressed in CD patients (34) with a specific epithelial expression pattern flanking ulcers or crypt abscesses (35). It limits the severity of DSS and 2,4,6-trinitrobenzene sulfonic acid colitis (36) suggesting an important role for this enzyme in IBD. The increased expression of IDO1 mRNA, protein, and enzymatic activity in IEC following IL-27 stimulation observed here was completely dependent on STAT1 signaling. This ability of IL-27 to activate IDO1 expression via STAT1 was comparable with that of IFN-γ from which it is known to induce IDO1 in a STAT1- and IRF1-dependent manner (37). Moreover, the STAT1 DNA-binding site in the IDO1 promoter identified here is identical to a described STAT1- binding site following IFN-γ stimulation (38).
We show here that IL-27-mediated stimulation of IDO1 expression and enzymatic activity effectively inhibits the growth of intestinal bacteria. This function is mediated via an increased degradation of tryptophan to kynurenine resulting in local tryptophan depletion. Our data are consistent with other studies demonstrating that the induction of IDO1 and the subsequent tryptophan deprivation is one of the antibacterial and antiviral key mechanisms of IFN-γ action (18, 39, 40).
Aside from being a substrate for IDO1 and the kynurenine pathway, l-tryptophan is also the precursor molecule for the synthesis of serotonin via tryptophan hydroxylase 1 (THP1). Interestingly, it has been shown that THP1 knock-out mice with reduced serotonin levels in the gastrointestinal tract are less susceptible to DSS or 2,4,6-trinitrobenzene sulfonic acid colitis (41). The increased tryptophan catabolism via the IDO1 pathway points to another anti-inflammatory function of IL-27.
Moreover, IDO1 is not only linked to antibacterial functions but also plays an important role in T cell differentiation. It has been shown that kynurenines and IDO1 activity stimulate the development of anti-inflammatory regulatory T cells but prevent Th17 cell differentiation by inhibiting the expression of the Th17 cell-specific transcription factor RORγt (42–44). Kynurenine production in IEC induced by IL-27 might help to create a microenvironment for regulatory T cell generation and for limiting intestinal inflammation. Altogether, our data suggest an anti-inflammatory, antibacterial, and epithelial protective role for IL-27 signaling in IEC. An overview of the IL-27-mediated effects in IEC is depicted in supplemental Fig. S3.
In summary, this is the first report on the expression and functions of IL-27 and its receptors in IEC and in patients with IBD. Here, we identify IEC as a novel target of IL-27 signaling. IL-27 activates MAPK and STAT signal transduction pathways in IEC resulting in strong epithelial barrier protective effects characterized by increased cell proliferation and wound healing. Those properties of IL-27 are differentially mediated via p38, STAT3, and STAT6 but not STAT1 signaling. Moreover, IL-27 induces STAT-dependent expression of the antibacterial proteins DMBT1 and IDO1 resulting in decreased microbial growth. In conclusion, in addition to its known functions as an inhibitor of Th17 cells and as an inducer of regulatory T cell differentiation, our results demonstrate that IL-27 is an epithelial barrier protective factor, suggesting beneficial functions of this cytokine in IBD. Functional studies in IEC-specific Il27ra−/− mice will be important to further characterize the antibacterial role of IL-27 in IEC in vivo.
Supplementary Material
Acknowledgment
We thank A. Bedynek (Ludwig-Maximilians University Munich) for excellent technical support.
This work was supported, in whole or in part, by National Institutes of Health Grants DK44319 and DK51362 (to R. S. B.).

This article contains supplemental methods, Figs. S1–S3, Tables S1–S4, and additional references.
- IBD
- inflammatory bowel disease
- CD
- Crohn's disease
- IEC
- intestinal epithelial cell
- IDO1
- indoleamine 2,3-dioxygenase
- Treg
- regulatory T cell
- DSS
- dextran sulfate sodium
- qPCR
- quantitative PCR
- A2M
- α2-macroglobulin.
REFERENCES
- 1. Brand S. (2009) Gut 58, 1152–1167 [DOI] [PubMed] [Google Scholar]
- 2. Pflanz S., Timans J. C., Cheung J., Rosales R., Kanzler H., Gilbert J., Hibbert L., Churakova T., Travis M., Vaisberg E., Blumenschein W. M., Mattson J. D., Wagner J. L., To W., Zurawski S., McClanahan T. K., Gorman D. M., Bazan J. F., de Waal Malefyt R., Rennick D., Kastelein R. A. (2002) Immunity 16, 779–790 [DOI] [PubMed] [Google Scholar]
- 3. Diveu C., McGeachy M. J., Boniface K., Stumhofer J. S., Sathe M., Joyce-Shaikh B., Chen Y., Tato C. M., McClanahan T. K., de Waal Malefyt R., Hunter C. A., Cua D. J., Kastelein R. A. (2009) J. Immunol. 182, 5748–5756 [DOI] [PubMed] [Google Scholar]
- 4. El-behi M., Ciric B., Yu S., Zhang G. X., Fitzgerald D. C., Rostami A. (2009) J. Immunol. 183, 4957–4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Murugaiyan G., Mittal A., Lopez-Diego R., Maier L. M., Anderson D. E., Weiner H. L. (2009) J. Immunol. 183, 2435–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Owaki T., Asakawa M., Morishima N., Hata K., Fukai F., Matsui M., Mizuguchi J., Yoshimoto T. (2005) J. Immunol. 175, 2191–2200 [DOI] [PubMed] [Google Scholar]
- 7. Yoshimoto T., Yoshimoto T., Yasuda K., Mizuguchi J., Nakanishi K. (2007) J. Immunol. 179, 4415–4423 [DOI] [PubMed] [Google Scholar]
- 8. Pflanz S., Hibbert L., Mattson J., Rosales R., Vaisberg E., Bazan J. F., Phillips J. H., McClanahan T. K., de Waal Malefyt R., Kastelein R. A. (2004) J. Immunol. 172, 2225–2231 [DOI] [PubMed] [Google Scholar]
- 9. Nieuwenhuis E. E., Neurath M. F., Corazza N., Iijima H., Trgovcich J., Wirtz S., Glickman J., Bailey D., Yoshida M., Galle P. R., Kronenberg M., Birkenbach M., Blumberg R. S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 16951–16956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wirtz S., Billmeier U., McHedlidze T., Blumberg R. S., Neurath M. F. (2011) Gastroenterology 141, 1875–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Troy A. E., Zaph C., Du Y., Taylor B. C., Guild K. J., Hunter C. A., Saris C. J., Artis D. (2009) J. Immunol. 183, 2037–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Honda K., Nakamura K., Matsui N., Takahashi M., Kitamura Y., Mizutani T., Harada N., Nawata H., Hamano S., Yoshida H. (2005) Inflamm. Bowel Dis. 11, 1044–1052 [DOI] [PubMed] [Google Scholar]
- 13. Villarino A. V., Artis D., Bezbradica J. S., Miller O., Saris C. J., Joyce S., Hunter C. A. (2008) Int. Immunol. 20, 739–752 [DOI] [PubMed] [Google Scholar]
- 14. Imielinski M., Baldassano R. N., Griffiths A., Russell R. K., Annese V., Dubinsky M., Kugathasan S., Bradfield J. P., Walters T. D., Sleiman P., Kim C. E., Muise A., Wang K., Glessner J. T., Saeed S., Zhang H., Frackelton E. C., Hou C., Flory J. H., Otieno G., Chiavacci R. M., Grundmeier R., Castro M., Latiano A., Dallapiccola B., Stempak J., Abrams D. J., Taylor K., McGovern D., Silber G., Wrobel I., Quiros A., Barrett J. C., Hansoul S., Nicolae D. L., Cho J. H., Duerr R. H., Rioux J. D., Brant S. R., Silverberg M. S., Taylor K. D., Barmuda M. M., Bitton A., Dassopoulos T., Datta L. W., Green T., Griffiths A. M., Kistner E. O., Murtha M. T., Regueiro M. D., Rotter J. I., Schumm L. P., Steinhart A. H., Targan S. R., Xavier R. J., Libioulle C., Sandor C., Lathrop M., Belaiche J., Dewit O., Gut I., Heath S., Laukens D., Mni M., Rutgeerts P., Van Gossum A., Zelenika D., Franchimont D., Hugot J. P., de Vos M., Vermeire S., Louis E., Cardon L. R., Anderson C. A., Drummond H., Nimmo E., Ahmad T., Prescott N. J., Onnie C. M., Fisher S. A., Marchini J., Ghori J., Bumpstead S., Gwillam R., Tremelling M., Delukas P., Mansfield J., Jewell D., Satsangi J., Mathew C. G., Parkes M., Georges M., Daly M. J., Heyman M. B., Ferry G. D., Kirschner B., Lee J., Essers J., Grand R., Stephens M., Levine A., Piccoli D., Van Limbergen J., Cucchiara S., Monos D. S., Guthery S. L., Denson L., Wilson D. C., Grant S. F., Daly M., Silverberg M. S., Satsangi J., Hakonarson H. (2009) Nat. Genet. 41, 1335–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Franke A., McGovern D. P., Barrett J. C., Wang K., Radford-Smith G. L., Ahmad T., Lees C. W., Balschun T., Lee J., Roberts R., Anderson C. A., Bis J. C., Bumpstead S., Ellinghaus D., Festen E. M., Georges M., Green T., Haritunians T., Jostins L., Latiano A., Mathew C. G., Montgomery G. W., Prescott N. J., Raychaudhuri S., Rotter J. I., Schumm P., Sharma Y., Simms L. A., Taylor K. D., Whiteman D., Wijmenga C., Baldassano R. N., Barclay M., Bayless T. M., Brand S., Büning C., Cohen A., Colombel J. F., Cottone M., Stronati L., Denson T., De Vos M., D'Inca R., Dubinsky M., Edwards C., Florin T., Franchimont D., Gearry R., Glas J., Van Gossum A., Guthery S. L., Halfvarson J., Verspaget H. W., Hugot J. P., Karban A., Laukens D., Lawrance I., Lemann M., Levine A., Libioulle C., Louis E., Mowat C., Newman W., Panés J., Phillips A., Proctor D. D., Regueiro M., Russell R., Rutgeerts P., Sanderson J., Sans M., Seibold F., Steinhart A. H., Stokkers P. C., Torkvist L., Kullak-Ublick G., Wilson D., Walters T., Targan S. R., Brant S. R., Rioux J. D., D'Amato M., Weersma R. K., Kugathasan S., Griffiths A. M., Mansfield J. C., Vermeire S., Duerr R. H., Silverberg M. S., Satsangi J., Schreiber S., Cho J. H., Annese V., Hakonarson H., Daly M. J., Parkes M. (2010) Nat. Genet. 42, 1118–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Becker K., Niederau C., Frieling T. (1999) Z. Gastroenterol. 37, 597–605 [PubMed] [Google Scholar]
- 18. Thomas S. M., Garrity L. F., Brandt C. R., Schobert C. S., Feng G. S., Taylor M. W., Carlin J. M., Byrne G. I. (1993) J. Immunol. 150, 5529–5534 [PubMed] [Google Scholar]
- 19. Edwards A. M., Manetti A. G., Falugi F., Zingaretti C., Capo S., Buccato S., Bensi G., Telford J. L., Margarit I., Grandi G. (2008) Mol. Microbiol. 68, 1378–1394 [DOI] [PubMed] [Google Scholar]
- 20. Fukui H., Sekikawa A., Tanaka H., Fujimori Y., Katake Y., Fujii S., Ichikawa K., Tomita S., Imura J., Chiba T., Fujimori T. (2011) Inflamm. Bowel Dis. 17, 1177–1188 [DOI] [PubMed] [Google Scholar]
- 21. MacKenzie C. R., Hadding U., Däubener W. (1998) J. Infect. Dis. 178, 875–878 [DOI] [PubMed] [Google Scholar]
- 22. Guzzo C., Che Mat N. F., Gee K. (2010) J. Biol. Chem. 285, 24404–24411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kanda N., Watanabe S. (2008) Eur. J. Immunol. 38, 1287–1296 [DOI] [PubMed] [Google Scholar]
- 24. Elrod J. W., Laroux F. S., Houghton J., Carpenter A., Ando T., Jennings M. H., Grisham M., Walker N., Alexander J. S. (2005) Inflamm. Bowel Dis. 11, 883–889 [DOI] [PubMed] [Google Scholar]
- 25. Zhang M., Zhou Y., Xie C., Zhou F., Chen Y., Han G., Zhang W. J. (2006) Cancer Lett. 243, 38–46 [DOI] [PubMed] [Google Scholar]
- 26. Brand S., Dambacher J., Beigel F., Zitzmann K., Heeg M. H., Weiss T. S., Prüfer T., Olszak T., Steib C. J., Storr M., Göke B., Diepolder H., Bilzer M., Thasler W. E., Auernhammer C. J. (2007) Am. J. Physiol. Gastrointest. Liver Physiol. 292, G1019–G1028 [DOI] [PubMed] [Google Scholar]
- 27. Kida H., Mucenski M. L., Thitoff A. R., Le Cras T. D., Park K. S., Ikegami M., Müller W., Whitsett J. A. (2008) Am. J. Pathol. 172, 1542–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brand S., Beigel F., Olszak T., Zitzmann K., Eichhorst S. T., Otte J. M., Diepolder H., Marquardt A., Jagla W., Popp A., Leclair S., Herrmann K., Seiderer J., Ochsenkühn T., Göke B., Auernhammer C. J., Dambacher J. (2006) Am. J. Physiol. Gastrointest. Liver Physiol. 290, G827–G838 [DOI] [PubMed] [Google Scholar]
- 29. Loimaranta V., Hytönen J., Pulliainen A. T., Sharma A., Tenovuo J., Strömberg N., Finne J. (2009) J. Biol. Chem. 284, 18614–18623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Renner M., Bergmann G., Krebs I., End C., Lyer S., Hilberg F., Helmke B., Gassler N., Autschbach F., Bikker F., Strobel-Freidekind O., Gronert-Sum S., Benner A., Blaich S., Wittig R., Hudler M., Ligtenberg A. J., Madsen J., Holmskov U., Annese V., Latiano A., Schirmacher P., Amerongen A. V., D'Amato M., Kioschis P., Hafner M., Poustka A., Mollenhauer J. (2007) Gastroenterology 133, 1499–1509 [DOI] [PubMed] [Google Scholar]
- 31. Rosenstiel P., Sina C., End C., Renner M., Lyer S., Till A., Hellmig S., Nikolaus S., Fölsch U. R., Helmke B., Autschbach F., Schirmacher P., Kioschis P., Hafner M., Poustka A., Mollenhauer J., Schreiber S. (2007) J. Immunol. 178, 8203–8211 [DOI] [PubMed] [Google Scholar]
- 32. Hugot J. P., Chamaillard M., Zouali H., Lesage S., Cézard J. P., Belaiche J., Almer S., Tysk C., O'Morain C. A., Gassull M., Binder V., Finkel Y., Cortot A., Modigliani R., Laurent-Puig P., Gower-Rousseau C., Macry J., Colombel J. F., Sahbatou M., Thomas G. (2001) Nature 411, 599–603 [DOI] [PubMed] [Google Scholar]
- 33. Eckmann L., Smith J. R., Housley M. P., Dwinell M. B., Kagnoff M. F. (2000) J. Biol. Chem. 275, 14084–14094 [DOI] [PubMed] [Google Scholar]
- 34. Wolf A. M., Wolf D., Rumpold H., Moschen A. R., Kaser A., Obrist P., Fuchs D., Brandacher G., Winkler C., Geboes K., Rutgeerts P., Tilg H. (2004) Clin. Immunol. 113, 47–55 [DOI] [PubMed] [Google Scholar]
- 35. Ferdinande L., Demetter P., Perez-Novo C., Waeytens A., Taildeman J., Rottiers I., Rottiers P., De Vos M., Cuvelier C. A. (2008) Int. J. Immunopathol. Pharmacol. 21, 289–295 [DOI] [PubMed] [Google Scholar]
- 36. Ciorba M. A., Bettonville E. E., McDonald K. G., Metz R., Prendergast G. C., Newberry R. D., Stenson W. F. (2010) J. Immunol. 184, 3907–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chon S. Y., Hassanain H. H., Gupta S. L. (1996) J. Biol. Chem. 271, 17247–17252 [DOI] [PubMed] [Google Scholar]
- 38. Chon S. Y., Hassanain H. H., Pine R., Gupta S. L. (1995) J. Interferon Cytokine Res. 15, 517–526 [DOI] [PubMed] [Google Scholar]
- 39. Mackenzie C. R., Willberg C. B., Däubener W. (1998) J. Neuroimmunol. 89, 191–197 [DOI] [PubMed] [Google Scholar]
- 40. Mao R., Zhang J., Jiang D., Cai D., Levy J. M., Cuconati A., Block T. M., Guo J. T., Guo H. (2011) J. Virol. 85, 1048–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ghia J. E., Li N., Wang H., Collins M., Deng Y., El-Sharkawy R. T., Côté F., Mallet J., Khan W. I. (2009) Gastroenterology 137, 1649–1660 [DOI] [PubMed] [Google Scholar]
- 42. Fallarino F., Grohmann U., You S., McGrath B. C., Cavener D. R., Vacca C., Orabona C., Bianchi R., Belladonna M. L., Volpi C., Santamaria P., Fioretti M. C., Puccetti P. (2006) J. Immunol. 176, 6752–6761 [DOI] [PubMed] [Google Scholar]
- 43. De Luca A., Montagnoli C., Zelante T., Bonifazi P., Bozza S., Moretti S., D'Angelo C., Vacca C., Boon L., Bistoni F., Puccetti P., Fallarino F., Romani L. (2007) J. Immunol. 179, 5999–6008 [DOI] [PubMed] [Google Scholar]
- 44. Baban B., Chandler P. R., Sharma M. D., Pihkala J., Koni P. A., Munn D. H., Mellor A. L. (2009) J. Immunol. 183, 2475–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.