Abstract
Nature's strategies for evolving catalytic functions can be deciphered from the information contained in the rapidly expanding protein sequence databases. However, the functions of many proteins in the protein sequence and structure databases are either uncertain (too divergent to assign function based on homology) or unknown (no homologs), thereby limiting the utility of the databases. The mechanistically diverse enolase superfamily is a paradigm for understanding the structural bases for evolution of enzymatic function. We describe strategies for assigning functions to members of the enolase superfamily that should be applicable to other superfamilies.
Keywords: Bacterial Metabolism, Carbohydrate Metabolism, Computation, Enzyme Kinetics, Enzyme Mechanisms, Enzymes, Evolution
Introduction
Nature continues to evolve an extraordinarily diverse collection of enzymes.2 The reactions they catalyze and the metabolic pathways in which they participate provide organisms with the ability to thrive in different metabolic niches; they also provide potential for biomedical and chemical applications. To fully understand the importance and implications of this diversity, the challenges are both to provide descriptions of the full range of functional diversity, i.e. molecular and biological functions, and to use those descriptions to understand and exploit natural design principles. However, the number of protein sequences2 is growing much faster than the number of experimentally based annotations (ExPASy UniProtKB/Swiss-Prot Database). Therefore, novel approaches for reliable functional annotation are required to maximize the utility of the data.
In genome sequencing projects, automated methods identify the sequences and then typically annotate their functions by transfer from a homolog using simple pairwise comparisons (1), despite the availability of more sophisticated and orthogonal tools (2). A recent analysis of the annotation error rate revealed that it can be very high, sometimes exceeding 80% for particular monofunctional families (3). Even now, this error rate prevents reliable use of annotations to infer in vitro molecular and in vivo biological functions; the problem will become even more critical as the errors in functional annotation are propagated.
Assignment of correct functions to homologous enzymes is confounded because they need not catalyze the same reaction. From a survey of structurally characterized superfamilies, almost 40% are functionally diverse, i.e. different members catalyze reactions with different EC numbers (4). Thus, trivial annotation transfer by sequence homology is often not sufficient to assign function. In mechanistically diverse enzyme superfamilies, all of the homologous enzymes catalyze different reactions using a conserved partial reaction; in functionally distinct enzyme suprafamilies, homologous enzymes catalyze different reactions that do not share any mechanistic attribute (5).
This minireview focuses on approaches for functional assignment in the mechanistically diverse enolase superfamily, a paradigm for understanding relationships among sequence, structure, and function in homologous enzymes. The current challenge is to devise a general strategy for assignment of functions to members discovered in genome projects. This challenge is not restricted to the enolase superfamily, so the approaches developed for the enolase superfamily should be applicable to other superfamilies.
Sequence-Structure-Function Relationships in Enolase Superfamily
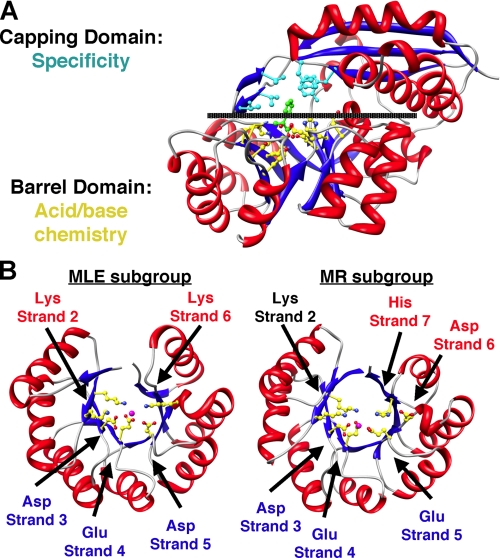
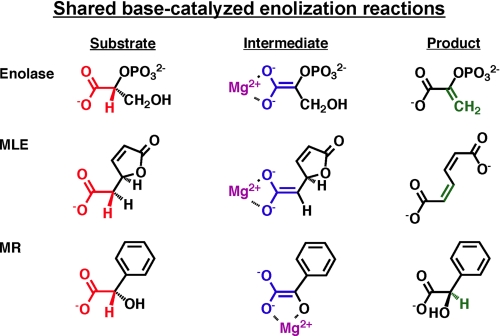
The first mechanistically diverse superfamily was discovered when the structural similarity of muconate lactonizing enzyme (MLE)3 and mandelate racemase (MR) was recognized (Fig. 1A). These proteins share 1) a (β/α)7β-barrel domain (a modified (β/α)8-barrel (TIM barrel)) containing the active site functional groups that support chemistry and 2) an α+β-capping domain formed from polypeptide segments at the N and C termini that provide many of the determinants for substrate recognition (6). Later, the structural similarity to enolase was recognized,4 establishing the “enolase superfamily” (7). The substrates for MLE, MR, and enolase are carboxylate anions with a proton on the α-carbon. In each reaction, the proton is abstracted to form an enolate anion intermediate, although the overall reactions are different (Fig. 2).
FIGURE 1.
A, structure of MR showing the bidomain structure described in text. B, (β/α)7β-barrel domains of MLE and MR showing the conserved and divergent metal ion ligands and general basic/acidic catalysts.
FIGURE 2.
Substrates, enolate anion intermediates, and products for reactions catalyzed by MLE, MR, and enolase.
The active sites are located at the interface between the barrel and capping domains, sequestered from solvent by a flexible loop in the capping domain that allows access of the substrate. The active sites contain an essential Mg2+ ion coordinated to conserved side chain functional groups in the barrel domain (Fig. 1B) and to at least one carboxylate oxygen of the substrate. A basic catalyst abstracts the α-proton to generate the intermediate that is stabilized by coordination to the Mg2+ ion. An acidic catalyst usually directs the intermediate to the product; the acidic catalyst can be the conjugate acid of the basic catalyst or another catalyst.
Unlike the ligands for the essential Mg2+ ion, the basic/acidic catalysts are not conserved across the superfamily. The differences allow the superfamily to be partitioned into several subgroups that provide structure-based restrictions on the mechanism and therefore the overall reaction: enolase, MLE, MR, and d-glucarate dehydratase (31); d-mannonate dehydratase (23); β-methylaspartate ammonia lyase (32, 33); and galactarate dehydratase (28). The structures of the barrel domains in the MLE and MR subgroups are compared in Fig. 1B.
The Mg2+ ion and the basic/acidic catalysts are “hard-wired” features of the active sites. The capping domain, including its flexible loop, defines the volume, shape, and polarity of the active site, thereby selecting the substrate that is presented to the hard-wired catalytic residues. This dissection of reactivity mediated by the barrel domain and substrate specificity by the capping domain may appear simplistic. However, substitutions of specificity-determining residues are sufficient to interchange the substrate specificities of “monofunctional” members of the enolase superfamily (8–10). Remarkably, in this laboratory evolution experiment, the mechanism was determined by the identity of the substrate, e.g. an enzyme catalyzing a 1,1-proton transfer using two basic/acidic catalysts (l-Ala-D/l-Glu epimerase (AEE)) was evolved to an enzyme catalyzing syn-dehydration using a single basic/acidic catalyst (o-succinylbenzoate synthase (OSBS)). These experiments suggest that Nature could use the same strategy for divergent evolution, i.e. “new” reactions can be catalyzed if volume, shape, and/or polarity of the active site is changed.
Enolase Superfamily
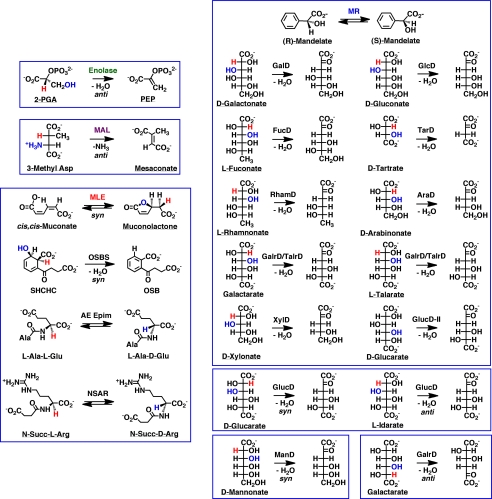
The enolase superfamily now includes >8000 members5; the size is increasing rapidly as microbial genome projects are completed (ExPASy UniProtKB/Swiss-Prot Database). The superfamily can be partitioned into “clusters” using sequence similarity networks (11); in these networks, each sequence (node) is connected to all other sequences that share a maximum BLASTP E value (Fig. 3). The network was constructed using an E value threshold of <10−90, corresponding to ≥35% sequence identity. Most of the discrete clusters are isofunctional families. Each node is colored according to reaction specificity, with gray indicating unknown function; ∼50% of the members have functions that are currently unknown. (The accompanying minireview by Brown and Babbitt (12) provides additional examples of the use of sequence similarity networks to discover functional divergence in enzyme superfamilies.) More than 20 different reactions have been identified in the enolase superfamily by in vitro enzymatic assays (Fig. 4); many more remain to be discovered.
FIGURE 3.
Sequence similarity network for enolase superfamily members (excluding enolase subgroup). The various sequences (nodes) are shown as circles; the lines (edges) connecting the nodes are BLASTP connections with E values ≤10−90. The nodes colored gray have unknown functions; those with other colors have assigned functions.
FIGURE 4.
Reactions catalyzed by enolase superfamily members. The various boxes enclose reactions catalyzed by the seven subgroups that have been recognized. 2-PGA, 2-phosphoglycerate; PEP, phosphoenolpyruvate; MAL, β-methylaspartate ammonia lyase; SHCHC, 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylic acid; OSB, o-succinylbenzoic acid; AE Epim, l-Ala-d/l-Glu epimerase; GalD, d-galactonate dehydratase; GlcD, d-gluconate dehydratase; FucD, l-fuconate dehydratase; TarD, d-tartrate dehydratase; RhamD, l-rhamnonate dehydratase; AraD, d-arabinonate dehydratase; GalrD/TalrD, galactarate dehydratase/l-talarate dehydratase; XylD, d-xylonate dehydratase; GlucD, d-glucarate dehydratase; ManD, d-mannonate dehydratase.
In the following sections, three strategies for assigning functions are described. A single general strategy with “high throughput” capabilities is desirable, given the rapidly increasing membership of the superfamily.
Assigning Function
Operon Context
The majority of the members of the enolase superfamily are found in microbes.6 This offers advantages for annotating members with unknown functions. 1) Enzymes in metabolic pathways are often encoded by operons; and 2) many bacterial species are genetically tractable, so genetics can be used to construct knock-out mutants, allowing assessment of the relevance of an in vitro enzymatic activity to physiology (or vice versa).
In some cases, genome context alone provides sufficient information to assign function. The gene encoding a member of previously unknown function in Escherichia coli (ycjG) is clustered with genes encoding enzymes that hydrolyze murein peptides and also a periplasmic binding protein specific for l-Ala-d-Glu-meso-diaminopimelic acid, a component of the murein peptide (13). Assuming that it could catalyze a 1,1-proton transfer reaction (Lys catalysts on opposite faces of the active site) (14) and use a substrate derived from the murein peptide, YcjG was assigned as AEE, a previously unknown reaction.
Members of another family of previously unknown function (“unknowns”) are encoded by operons that also encode orthologs of enzymes in the β-ketoadipate pathway. The MLE from Pseudomonas putida, a “founding member” of the superfamily, catalyzes the syn-cycloisomerization of cis,cis-muconate to (4S)-muconolactone in the pathway; this MLE shares ∼30% sequence identity with members of the unknown family. A member of the unknown family catalyzes the MLE reaction (15), as expected from its operon context; however, the reaction is an anti-cycloisomerization, i.e. the proton is delivered to the opposite face of the proximal double bond of the substrate. Structures for product-liganded complexes revealed that the unanticipated (and biologically irrelevant) distinct stereochemical behavior of this second family of MLEs is explained by different spatial arrangements of the substrate specificity determinants relative to the conserved Lys acidic catalyst (16). Supporting the notion that new functions evolve readily in mechanistically diverse superfamilies, phylogenetic analysis suggested that the new MLEs evolved from a different intermediate ancestor than the previously recognized MLEs.
We have encountered many examples of functional promiscuity, which further complicates functional annotation, including enzymes that catalyze both the OSBS reaction in menaquinone biosynthesis and the racemization of N-succinylamino acids (N-succinylamino acid racemase (NSAR)) (17). OSBSs are highly divergent in sequence; however, the genes encoding OSBSs can be recognized by their co-localization with genes encoding orthologs of enzymes in the menaquinone biosynthetic pathway (18). Some promiscuous OSBSs/NSARs are encoded by operons that encode a succinyl-CoA:d-amino acid N-succinyltransferase and a N-succinyl-l-amino acid hydrolase, both previously unknown reactions, with NSAR equilibrating the N-succinyl d-amino acid and its enantiomer (19). The common substrate specificities provide unequivocal evidence for a previously unknown metabolic pathway that irreversibly converts a d-amino acid to an l-amino acid; however, its physiological context is unknown. This example emphasizes a further “complication” in functional assignment: the relationship of in vitro enzymatic reactions to in vivo physiological function. As novel in vitro reactions are discovered, biological experiments are required to establish their in vivo context.
A more common situation is that genome context provides clues of reaction type but not substrate specificity. Many unknown members are encoded by operons that encode aldose 1-epimerases, dehydrogenases, aldolases, and/or sugar kinases in catabolic pathways for carbohydrates. In such cases, the unknown members almost certainly are acid-sugar dehydratases, but the identities of the substrates cannot be specified. In some cases, the specificity of one enzyme in the catabolic pathway can be confidently assigned by sequence homology, e.g. a protein annotated as a “fucose permease” provided sufficient information to assign the l-fuconate dehydratase function (20). However, operon context usually is not sufficient to specify the identity of the substrate for either the operon or the dehydratase.
Library Screening
An alternative approach is to screen a physical library of possible substrates, e.g. a library of acid sugars for potential acid-sugar dehydratases. This approach has been successful for identifying substrates for dipeptide epimerases. However, for the putative dehydratases, this approach has been less successful (21–24). We have assembled a library of all mono- and diacids derived from d- and l-hexoses, d- and l-pentoses, and d- and l-tetroses; a partial set of hexuronic acids; and the (very few) commercially available phosphorylated acid sugars (20). Many members of the library are not included in the BRENDA and KEGG databases of known metabolites.
We identified an acid-sugar dehydratase from Salmonella typhimurium LT2 that catalyzes dehydration of both l-talarate and meso-galactarate as well as their interconversion by epimerization (22). The identification of l-talarate as a substrate illustrates a challenge for functional assignment: l-talarate is not in the databases of metabolites. The gene encoding the dehydratase is adjacent to a gene encoding a permease, suggesting that l-talarate may be a previously unrecognized carbon source for S. typhimurium LT2. We determined that l-talarate is a carbon source; when the gene encoding the l-talarate dehydratase is disrupted, l-talarate no longer serves as a carbon source. Galactarate is catabolized by another pathway, so the dehydration of l-talarate appears to be both the in vitro enzymatic function and the in vivo physiological function. Library screening not only allowed a physiologically relevant function assignment but also discovered an unknown metabolite.
However, for many putative dehydratases, this library has been ineffective. The explanation for failure is uncertain: if the function of an enzyme is not known, it is impossible to determine whether the protein is catalytically active as isolated. A more troubling explanation is that the library is incomplete and does not contain the substrate. Elaboration of a physical library requires extensive resources.
A corollary of the fact that many of the proteins/enzymes in public databases do not have experimentally based functions is that the identities of many metabolites are not known. Thus, even if all known metabolites were available, the library would not be useful for all enzymes with unknown functions. We have concluded that in vitro physical library screening, by itself, is not a general solution for functional assignment in the enolase superfamily.
In Silico Ligand Docking
The identity and spatial arrangement of the amino acids in the active site of an enzyme determine its substrate specificity; in the enolase superfamily, the specificity-determining residues are usually located in the capping domain. Computational ligand docking methods, more commonly used for computer-aided drug design, provide an approach for inferring substrate specificity from the structure of a binding site. Specifically, we and others (25–27) have demonstrated the feasibility of using in silico libraries of known or potential metabolites in combination with various docking methods to suggest substrates for experimental testing.
This approach has resulted in the assignment of novel functions to members of the enolase superfamily (and the amidohydrolase superfamily, e.g. Ref. 25); it has also suggested further developments in the strategy both to increase its success rate and to provide a connection between in vitro enzymatic functions and in vivo physiological functions.
The Protein Structure Initiative (PSI) provided structures for many enzymes of unknown function in meeting its goal of “increasing structural coverage of sequenced genes” (PSI-2, www.nigms.nih.gov/research/featuredprograms/psi/centers/, and PSI:Biology, www.nigms.nih.gov/research/featuredprograms/psi/psi_biology/). In the case of Protein Data Bank code 2OQY, a divergent member of the enolase superfamily, in silico ligand docking was used to focus in vitro enzymatic assays (28). Unlike other members, the structure of 2OQY revealed the presence of two Mg2+ ions in the active site: one in the canonical position and a second 10.6 Å away in the capping domain. Also, the basic/acidic residues differed from those previously observed in the superfamily. In silico docking of the KEGG library identified diacid sugars and phosphorylated acid sugars as likely ligands, with each of the anionic groups interacting with one of the Mg2+ ions. 2OQY was screened with our acid sugar library; only galactarate was identified as a substrate.
However, in silico ligand docking to experimental structures is not always successful: conformational changes frequently accompany ligand binding, so a structure determined in the absence of a ligand may not be useful for inferring substrate specificity. In the enolase superfamily, this is a major challenge because the flexible loop in the capping domain, which contains residues important for selectivity, is almost always “open” or disordered in the absence of a ligand. An alternative approach is to use homology modeling to obtain “dockable” structures using an experimentally determined liganded structure as the template. Residue substitutions in the binding site provide information about differences in specificity, which can be captured using docking methods.
Using the substrate/product-liganded structure of the AEE from Bacillus subtilis (14) as the template to generate homology models, we have successfully predicted a novel enzymatic function as well as the substrate specificities for many members of the specificity diverse dipeptide epimerase family. The members of a family of previously uncharacterized enzymes do not possess the specificity determinants in the AEE template, so we expected a novel function (29). N-Succinyl-Arg was predicted to be the “best” substrate, followed by dipeptides with C-terminal Arg/Lys residues and also N-succinyl-Lys. Assays using N-succinylamino acids and dipeptides confirmed that N-succinyl-Arg is the best substrate for racemization (kcat = 71 s−1 and kcat/Km = 1.4 × 105 m−1 s−1). N-Succinyl-Lys also is a good substrate; various dipeptides with Arg/Lys in the C-terminal epimerized position are substrates with inferior kinetic constants. A subsequent x-ray structure with N-succinyl-Arg confirmed the predicted structure of the liganded complex, thereby explaining the successful prediction of this novel enzymatic function. However, the physiological function of the in vitro N-succinyl-Arg racemase is unknown: the genomic context does not provide any clues about its function.
This approach has also been applied with excellent success to diverse dipeptide epimerases; these are identified by a conserved Asp-X-Asp motif at the end of the eighth β-strand in the barrel domain that provides the specificity determinant for the α-ammonium group of the dipeptide. Although most are aees, some have widely varying specificities, such as a dipeptide epimerase from Thermotoga maritima with specificity for dipeptides such as l-Ala-d/l-Phe (30). The physiological roles of these specificity diverse epimerases are unknown.
Summary
The mechanistically diverse enolase superfamily continues to serve as a paradigm that provides novel insights into the structural basis for divergent evolution. The current challenge is to develop strategies that allow definition of the full set of reactions catalyzed by its members. Our experience is that this can be best (only?) accomplished by multidisciplinary efforts that include bioinformatics, structural enzymology, computational enzymology, and experimental enzymology to establish in vitro enzymatic functions and genetics/physiology/metabolomics to establish in vivo physiological functions. To help meet this challenge, the Enzyme Function Initiative (supported by National Institutes of Health Grant U54GM093342) has been assembled to develop and disseminate integrated sequence/structure-based strategies for predicting the substrates for unknown enzymes.
This work was supported, in whole or in part, by National Institutes of Health Grants P01GM071790-07 and U54GM093342-01. This is the fourth article in the Thematic Minireview Series on Enzyme Evolution in the Post-genomic Era.
The (public) TrEMBL non-redundant protein sequence database contains 13,499,622 sequences (Release 2011_02, February 8, 2011), primarily the result of microbial genome projects (1457 completed, 1775 in assembly stage, and 2881 incomplete; www.ncbi.nlm.nih.gov/genomes/lproks.cgi). The number of bases in the GenBankTM Data Bank has doubled every 18 months (ftp.ncbi.nih.gov/genbank/gbrel.txt), so the content of protein sequence databases is expected to continue to increase.
Although enolase shares the characteristic bidomain structure with MLE and MR (and all other structurally characterized members), the barrel domain in enolase has an unusual βααβ(β/α)5β-structure.
The membership of the enolase superfamily is frequently updated and made publically available by the Structure-Function Linkage Database (SFLD; supported by National Institutes of Health Grants R01GM60595 and P41RR01081 and National Science Foundation Grant DBI-0234768) (34). SFLD facilitates investigation of sequence, structure, and function in mechanistically diverse superfamilies by providing tools that allow sequence relationships to be explored.
l-Fuconate dehydratases are found in humans and other chordates, acid- sugar dehydratases with diverse substrate specificities are found in fungi, and a dipeptide epimerase family is found in plants. Of course, enolase is ubiquitous because of its role in glycolysis and gluconeogenesis.
- MLE
- muconate lactonizing enzyme
- MR
- mandelate racemase
- AEE
- l-Ala-d/l-Glu epimerase
- OSBS
- o-succinylbenzoate synthase
- NSAR
- N-succinylamino acid racemase.
REFERENCES
- 1. Frishman D. (2007) Chem. Rev. 107, 3448–3466 [DOI] [PubMed] [Google Scholar]
- 2. Loewenstein Y., Raimondo D., Redfern O. C., Watson J., Frishman D., Linial M., Orengo C., Thornton J., Tramontano A. (2009) Genome Biol. 10, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schnoes A. M., Brown S. D., Dodevski I., Babbitt P. C. (2009) PLoS Comput. Biol. 5, e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Almonacid D. E., Babbitt P. C. (2011) Curr. Opin. Chem. Biol. 15, 435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gerlt J. A., Babbitt P. C. (2001) Annu. Rev. Biochem. 70, 209–246 [DOI] [PubMed] [Google Scholar]
- 6. Neidhart D. J., Kenyon G. L., Gerlt J. A., Petsko G. A. (1990) Nature 347, 692–694 [DOI] [PubMed] [Google Scholar]
- 7. Babbitt P. C., Hasson M. S., Wedekind J. E., Palmer D. R., Barrett W. C., Reed G. H., Rayment I., Ringe D., Kenyon G. L., Gerlt J. A. (1996) Biochemistry 35, 16489–16501 [DOI] [PubMed] [Google Scholar]
- 8. Schmidt D. M., Mundorff E. C., Dojka M., Bermudez E., Ness J. E., Govindarajan S., Babbitt P. C., Minshull J., Gerlt J. A. (2003) Biochemistry 42, 8387–8393 [DOI] [PubMed] [Google Scholar]
- 9. Vick J. E., Schmidt D. M., Gerlt J. A. (2005) Biochemistry 44, 11722–11729 [DOI] [PubMed] [Google Scholar]
- 10. Vick J. E., Gerlt J. A. (2007) Biochemistry 46, 14589–14597 [DOI] [PubMed] [Google Scholar]
- 11. Atkinson H. J., Morris J. H., Ferrin T. E., Babbitt P. C. (2009) PLoS ONE 4, e4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brown S. D., Babbitt P. C. (2012) J. Biol. Chem. 287, 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schmidt D. M., Hubbard B. K., Gerlt J. A. (2001) Biochemistry 40, 15707–15715 [DOI] [PubMed] [Google Scholar]
- 14. Gulick A. M., Schmidt D. M., Gerlt J. A., Rayment I. (2001) Biochemistry 40, 15716–15724 [DOI] [PubMed] [Google Scholar]
- 15. Murakami S., Kohsaka C., Okuno T., Takenaka S., Aoki K. (2004) FEMS Microbiol. Lett. 231, 119–124 [DOI] [PubMed] [Google Scholar]
- 16. Sakai A., Fedorov A. A., Fedorov E. V., Schnoes A. M., Glasner M. E., Brown S., Rutter M. E., Bain K., Chang S., Gheyi T., Sauder J. M., Burley S. K., Babbitt P. C., Almo S. C., Gerlt J. A. (2009) Biochemistry 48, 1445–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Palmer D. R., Garrett J. B., Sharma V., Meganathan R., Babbitt P. C., Gerlt J. A. (1999) Biochemistry 38, 4252–4258 [DOI] [PubMed] [Google Scholar]
- 18. Glasner M. E., Fayazmanesh N., Chiang R. A., Sakai A., Jacobson M. P., Gerlt J. A., Babbitt P. C. (2006) J. Mol. Biol. 360, 228–250 [DOI] [PubMed] [Google Scholar]
- 19. Sakai A., Xiang D. F., Xu C., Song L., Yew W. S., Raushel F. M., Gerlt J. A. (2006) Biochemistry 45, 4455–4462 [DOI] [PubMed] [Google Scholar]
- 20. Yew W. S., Fedorov A. A., Fedorov E. V., Rakus J. F., Pierce R. W., Almo S. C., Gerlt J. A. (2006) Biochemistry 45, 14582–14597 [DOI] [PubMed] [Google Scholar]
- 21. Yew W. S., Fedorov A. A., Fedorov E. V., Wood B. M., Almo S. C., Gerlt J. A. (2006) Biochemistry 45, 14598–14608 [DOI] [PubMed] [Google Scholar]
- 22. Yew W. S., Fedorov A. A., Fedorov E. V., Almo S. C., Gerlt J. A. (2007) Biochemistry 46, 9564–9577 [DOI] [PubMed] [Google Scholar]
- 23. Rakus J. F., Fedorov A. A., Fedorov E. V., Glasner M. E., Vick J. E., Babbitt P. C., Almo S. C., Gerlt J. A. (2007) Biochemistry 46, 12896–12908 [DOI] [PubMed] [Google Scholar]
- 24. Rakus J. F., Fedorov A. A., Fedorov E. V., Glasner M. E., Hubbard B. K., Delli J. D., Babbitt P. C., Almo S. C., Gerlt J. A. (2008) Biochemistry 47, 9944–9954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hermann J. C., Marti-Arbona R., Fedorov A. A., Fedorov E., Almo S. C., Shoichet B. K., Raushel F. M. (2007) Nature 448, 775–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Macchiarulo A., Nobeli I., Thornton J. M. (2004) Nat. Biotechnol. 22, 1039–1045 [DOI] [PubMed] [Google Scholar]
- 27. Tyagi S., Pleiss J. (2006) J. Biotechnol. 124, 108–116 [DOI] [PubMed] [Google Scholar]
- 28. Rakus J. F., Kalyanaraman C., Fedorov A. A., Fedorov E. V., Mills-Groninger F. P., Toro R., Bonanno J., Bain K., Sauder J. M., Burley S. K., Almo S. C., Jacobson M. P., Gerlt J. A. (2009) Biochemistry 48, 11546–11558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Song L., Kalyanaraman C., Fedorov A. A., Fedorov E. V., Glasner M. E., Brown S., Imker H. J., Babbitt P. C., Almo S. C., Jacobson M. P., Gerlt J. A. (2007) Nat. Chem. Biol. 3, 486–491 [DOI] [PubMed] [Google Scholar]
- 30. Kalyanaraman C., Imker H. J., Fedorov A. A., Fedorov E. V., Glasner M. E., Babbitt P. C., Almo S. C., Gerlt J. A., Jacobson M. P. (2008) Structure 16, 1668–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gulick A. M., Hubbard B. K., Gerlt J. A., Rayment I. (2001) Biochemistry 40, 10054–10062 [DOI] [PubMed] [Google Scholar]
- 32. Asuncion M., Blankenfeldt W., Barlow J. N., Gani D., Naismith J. H. (2002) J. Biol. Chem. 277, 8306–8311 [DOI] [PubMed] [Google Scholar]
- 33. Levy C. W., Buckley P. A., Sedelnikova S., Kato Y., Asano Y., Rice D. W., Baker P. J. (2002) Structure 10, 105–113 [DOI] [PubMed] [Google Scholar]
- 34. Pegg S. C., Brown S. D., Ojha S., Seffernick J., Meng E. C., Morris J. H., Chang P. J., Huang C. C., Ferrin T. E., Babbitt P. C. (2006) Biochemistry 45, 2545–2555 [DOI] [PubMed] [Google Scholar]