Background: In order to kill E. coli, colicins need to cross the bacterial outer membrane.
Results: Neutron scattering data show colicin N at the protein-lipid interface of its receptor OmpF.
Conclusion: Colicins can unfold and penetrate membranes via the outside wall of their receptors.
Significance: The protein-lipid interface may be the route that colicins take into the cell.
Keywords: Bacterial Toxins, Escherichia coli, Lipids, Membrane Biophysics, Neutron Scattering, Colicin, Deuteration, OmpF, Outer Membrane, Lipid Monolayer, SANS, Deuteration, Porin
Abstract
Proteins that translocate across cell membranes need to overcome a significant hydrophobic barrier. This is usually accomplished via specialized protein complexes, which provide a polar transmembrane pore. Exceptions to this include bacterial toxins, which insert into and cross the lipid bilayer itself. We are studying the mechanism by which large antibacterial proteins enter Escherichia coli via specific outer membrane proteins. Here we describe the use of neutron scattering to investigate the interaction of colicin N with its outer membrane receptor protein OmpF. The positions of lipids, colicin N, and OmpF were separately resolved within complex structures by the use of selective deuteration. Neutron reflectivity showed, in real time, that OmpF mediates the insertion of colicin N into lipid monolayers. This data were complemented by Brewster Angle Microscopy images, which showed a lateral association of OmpF in the presence of colicin N. Small angle neutron scattering experiments then defined the three-dimensional structure of the colicin N-OmpF complex. This revealed that colicin N unfolds and binds to the OmpF-lipid interface. The implications of this unfolding step for colicin translocation across membranes are discussed.
Introduction
Secreted proteins cross the ∼3 nm thick cell membrane via dedicated protein translocon complexes, such as Sec (1) or Tat (2), which provide a hydrophilic channel across the lipid bilayer. Protein toxins, on the other hand, enter cells across the lipid bilayer in the opposite direction. Some use protein pores (e.g. cholera toxin uses an endogenous retrograde degradation pathway involving Sec61 (3), whereas anthrax toxin forms its own β barrel channel to translocate large toxic subunits into the cytosol (4)). However, several toxins have been shown to translocate through the lipid bilayer. Adenylate cyclase toxin from Bordetella pertussis, the causative agent of whooping cough, does not require a pore in order to enter cells (5). Diphtheria toxin crosses the lipid bilayer via a translocation subunit (N-terminal translocation domain; T-domain)4 (6), which converts from a membrane-attached to a membrane-inserted state and acts as a chaperone to enable the translocation of toxin subunits in the molten globule state. Finally, pore-forming and nuclease domains of bacterial colicins have been shown to translocate themselves and foreign epitopes across lipid bilayers (7, 8). Thus, two principal modes of protein inward translocation are known: passage through a water filled pore or via the lipid bilayer (with the possible assistance of a protein “chaperone”).
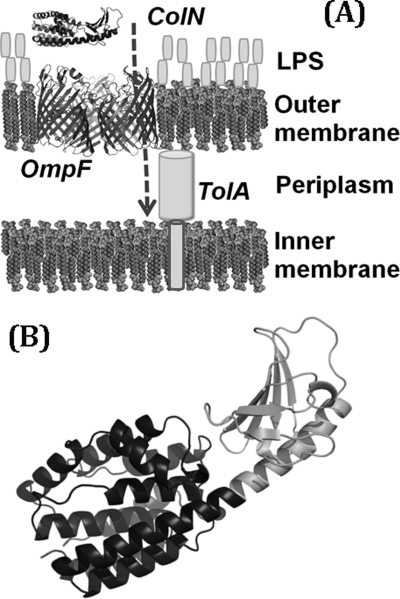
We are investigating the import of Gram-negative bacteriocins, which are large (>40 kDa) water-soluble antibacterial proteins. These antibiotic molecules, produced by bacteria, such as Escherichia coli “colicins” (9) and Yersinia pestis “pesticins” (10), kill competing bacteria, and to do so they must cross the Gram-negative outer membrane. This is highly asymmetric, with dense lipopolysaccharide on its outer face and phospholipids on the inner leaflet. Large bacteriocins only translocate the outer membrane in the presence of specific outer membrane proteins (11), which presumably act as translocons (12) (Fig. 1A), but the benefit of hijacking these proteins is unclear because they normally transport only small molecules and lack obvious protein translocation channels, such as in anthrax toxin (4). As a result, the path that bacteriocins follow to overcome the hydrophobic barrier is still largely undefined, but if understood, it may yet reveal a potential bacterial Achilles' heel useful in antibiotic development.
FIGURE 1.
The route for of colicin N entry into E. coli cells. A, outer leaflet of the outer membrane is composed of LPS, whereas the inner leaflet and inner membrane contain phospholipids. ColN is represented by the x-ray structure (Protein Data Bank code 1A87) (48) in which the 90 N-terminal residues that bind to periplasmic TolA are unresolved. Trimeric OmpF is Protein Data Bank entry 2OMF (52). B, detail of ColN with receptor binding domain in gray.
Colicins comprise three functional domains, the T-domain, a central receptor binding domain (R-domain), and a C-terminal domain that carries the lethal activity (9). Colicin N (ColN) is a pore-forming colicin that depolarizes and kills the target cell by inserting its C-terminal P (pore-forming) domain into the inner membrane (Fig. 1B). ColN uses the outer membrane protein OmpF (11) and cannot kill cells lacking it and its homologues. Thus, OmpF, a trimeric, pore-forming “porin,” is the simple but essential outer membrane translocon for ColN. The interaction with the outer membrane is driven initially by the R-domain (Fig. 1B), with full affinity requiring the entire ColN molecule (11, 13), which then unfolds during translocation (14, 15). The translocation into the periplasm is dependent upon periplasmic receptor proteins. Colicins are divided into group A or B, depending on whether they use the Tol or Ton proteins for this, respectively (9). ColN is a group A colicin that binds, via its unfolded T-domain, the C-terminal domain III of TolA (16–18). Colicin P-domains need to unfold to insert into membranes, and partial denaturation of these stable domains by low pH or detergents greatly increases insertion and pore formation (19–23).
The OmpF pore has a large diameter entrance and exit, but it narrows to a central constriction or “eyelet” region. An OmpF mutation, G119D, situated at this constriction inhibits ColN toxicity and receptor binding (24). Furthermore, experiments in planar lipid bilayers containing OmpF show that, like the R-domain (25), the ColN T-domain can inhibit ion flow through the OmpF channel (26). This agrees with x-ray data, which show parts of the ColE9 T-domain (27) in the lumen of the OmpF channel. These data suggest that colicin translocation could occur via the OmpF pore. Disulfide mutants that locked the OmpF eyelet in place had no effect on toxicity (28) and showed that the colicins would need to fully unfold to enter by this route.
On the other hand, a study by electron microscopy of the OmpF-ColN complex in two-dimensional crystals with phosphatidylcholine unexpectedly revealed that ColN was situated at the rim of the OmpF trimeric complex rather than in the internal water-filled pore (15). It also revealed that ColN displaces tightly bound LPS from the periphery of OmpF. These data suggest that the P domain could also pass through the membrane at the OmpF-LPS interface.
Here we have used complementary neutron scattering methods to unravel the initial steps in ColN translocation. In the first part, we use neutron reflectivity from a lipid monolayer to follow insertion in real time, and in the second part we resolve the structure of the translocon complex in detergent solution. Similar neutron techniques have recently been used to define the dynamic structure of the potassium channel (29) and diphtheria toxin (30).
EXPERIMENTAL PROCEDURES
Materials
Fully hydrogenous and tail-deuterated 1,2-dipalmitoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (DPPG; Avanti) were dissolved in chloroform. 99.8% D2O and d-SDS were from Sigma and CIL, respectively; H2O (18.2 MΩ) was MilliQ-filtered.
Protein Purification
Partially deuterated OmpF was produced at the Bio-Deuteration Laboratory (Oak Ridge National Laboratory) from E. coli BE3000 (31) adapted to D2O minimal medium (32), and 0.5% (w/v) glucose minimal medium in 90% D2O was used for 50 ml precultures and 1-liter fermentations in a Bioflo 3000 fermenter (New Brunswick Scientific) at 35 °C. pD 7.3 was maintained by 10% (w/v) NaOH in 90% D2O. Aeration (0.5–1 liter/min) and stirring (200–800 rpm) maintained a minimum dissolved oxygen level of 30% saturation. Cell growth was monitored at 600 nm, and cells were harvested when glucose limitation caused a sharp rise in dissolved oxygen.
All OmpF was purified as described previously (33), precipitated in cold ethanol, and resuspended in 1% (v/v) octyl-polyoxyethylene (POE) (Enzo), 20 mm sodium phosphate buffer, pH 7.2. ColN(1–387) (ColN) and ColN P-domain (residues 185–387) (ColN-P) were purified as described previously (34), followed by dialysis into 50 mm sodium phosphate, pH 7.6, 300 mm NaCl (buffer A). Contrast variation small angle neutron scattering (SANS) was also carried out, which determined that the neutron scattering length density (nSLD) match point for the h-ColN was at 42% (v/v), and that for d-OmpF was at 87% (v/v) D2O.
Production of OmpF/DPPG Vesicles and Monolayers
Small unilamellar vesicles were produced by sonicating a dry film of DPPG in 20 mm sodium phosphate buffer, pH 7.0 (buffer D) at 40 °C. Mixed OmpF/DPPG vesicles were then produced by adding 0.5 ml of 2 mg/ml OmpF in 1% (v/v) octyl-POE, buffer D to 10 ml of 1 mg/ml vesicles and then dialyzed against buffer D.
A glass slide was inserted at 45° through the surface of 130 ml of buffer D in a Langmuir Trough (model 302M, Nima Ltd.), and 300 μl of an OmpF/DPPG vesicle solution was allowed to roll drop by drop down the slide to the air-liquid interface, where the vesicles ruptured, producing a monolayer film. Isotherms (trough area, 300 cm2; barrier speed, 10 cm2/min) covering a wide pressure range were collected to check film quality (Fig. 2A). DPPG-only monolayers produced by the method described above or by spreading 50 μl of a 1 mg/ml DPPG-chloroform solution onto the interface showed identical properties. ColN (0.31 μm, 1 ml) was added beneath the monolayer held at an initial surface pressure of 35 mN/m, and the interactions were monitored by surface pressure, Brewster angle microscopy (BAM), and neutron reflection (NR).
FIGURE 2.
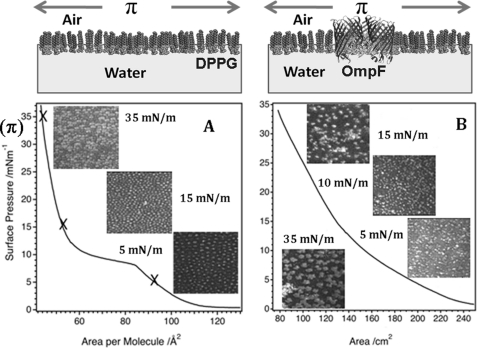
Surface pressure experiments with corresponding schematic diagrams. Surface pressure (π) is the spreading force of the monolayer at the air-water interface, and the area is controlled by movable barriers. A, BAM images (140 × 150 μm) of a DPPG monolayer at surface pressure values of 5, 15, and 35 mN/m with the associated surface pressure versus area isotherm. At low surface pressures, lipid domains are indicated by brighter regions on the image. At high surface pressure (35 mN/m), where the film is in the solid region, the contrast is due to lipid domains of the same thickness within the film but with the lipid tails in each domain having a different orientation relative to the polarized laser incident on the surface. B, BAM images of an OmpF/DPPG monolayer at surface pressure values of 5, 10, 15, and 35 mN/m and the associated surface pressure versus area isotherm of this monolayer. Thicker domains are indicated by brighter regions, whereas areas with disordered structure or lower thickness correspond to darker regions within the image. Because the exact protein lipid composition is unknown, the area per molecule cannot be calculated accurately. From the NR fits, we calculate 1 OmpF to 110 DPPG molecules, which suggests that the OmpF preferentially adsorbed to the interface during the vesicle deposition because the ratio applied was about 1 OmpF to 500 DPPG.
Production of OmpF-ColN Complexes for SANS
Buffer A for the SANS experiment was made at two different H2O/D2O ratios (13% (v/v) and 100% (v/v) D2O). Buffer B was equivalent to buffer A but with 1% (w/v) SDS added. Mixtures of h- and d-SDS were used to match the SDS scattering to the solution; therefore, h-SDS was added to the 13% (v/v) D2O solution, and 95% (w/v) d-SDS plus 5% (w/v) h-SDS were added to the 100% (v/v) D2O solution. Therefore, all mixtures of buffer B contained SDS matched to the solution. Solutions of H2O/D2O ratios (13, 41, 87, and 100% (v/v) D2O) were used. OmpF ethanol precipitation pellets were resuspended in buffer B and mixed with an equal volume of equimolar ColN or ColN-P in buffer A to produce a binary complex in buffer C (50 mm sodium phosphate, pH 7.6, 300 mm NaCl, 0.5% (w/v) h/d-SDS) and equilibrated by dialysis. Dialysis solutions were used for background data collections. Protein concentrations were determined by A280 nm (supplemental Table S2) (18).
Neutron Reflectometry
Specular NR used the white beam INTER (35) or CRISP (36) reflectometers (ISIS; neutron wavelengths 0.5–16 and 0.5–6.5 Å, respectively). The beam was reflected from the air-liquid interface at glancing angles of incidence, 1.5 and 0.8° (CRISP) and 2.3° (INTER). Specular NR probes the scattering length density (nSLD) along the axis normal (37), and isotopic substitution was used to produce a number of reflectometry profiles (supplemental Table S1), which were analyzed to a single physical profile.
The simultaneous constrained fitting of these data sets assists in the determination of complex layer structures. The procedures to obtain and fit protein-lipid profiles were as described (38, 39) and are briefly outlined here. The profiles were analyzed using RasCal (A. V. Hughes, ISIS, Rutherford Appleton Laboratory) which employs the optical matrix formalism, which partitions the interface to a series of slabs, each characterized by its nSLD, thickness, and roughness. In all cases, the simplest possible model (i.e. least number of parameters) that adequately described the data was selected.
Data fitting errors were determined using a “bootstrap” error analysis function within Rascal. This entails resampling the original data set with these new data sets fitted via the same methods outlined above. The distribution of parameter values across these fits is used to estimate fitting errors, and these are propagated through the calculations of the derived parameters according to standard error treatment methods. The lipid, protein, and solution isotopic contrasts used to obtain multiple neutron reflectometry data sets from isostructural monolayers are shown in supplemental Table S1, with the actual contrast mixtures in the respective figure legends. The layer thickness and volume fraction data were used to calculate the surface excess of each component at the interface (40).
Brewster Angle Microscopy
BAM measurements were taken on a BAM2Plus microscope (Nanofilm Technology) mounted on the Langmuir trough, enabling Ångstrom resolution perpendicular to the interface and micrometer scale lateral resolution (38).
Small Angle Neutron Scattering
SANS was conducted at 20 °C in 1 mm path length quartz cuvettes on beamline D22 (Institut Laue-Langevin) at detector distances of 2 and 10 m (Q range = 0.03 < Q < 0.33 Å−1. The radius of gyration (Rg), P(r) distance distribution, and maximum dimension (Dmax) were obtained using the programs PRIMUS and GNOM. All ab initio reconstructions of molecular envelopes were generated using the program MONSA (41), which allows the simultaneous fitting of SANS data collected in different h- and d-solvent contrasts and the assigning of different nSLD to the volumes defined by the model. A two-phase sphere of dummy atoms (42) with P3 symmetry imposed was created by DAMESV with a diameter of 160 Å, as defined by the Dmax determined from the distance distribution functions (43). Resulting models from MONSA were averaged using DAMAVER (43).
RESULTS
OmpF/DPPG Forms Stable Monolayers
DPPG is available in several deuterated forms and was used to simulate the negatively charged interface produced by LPS in vivo. Pure DPPG monolayers behaved as expected (38) (Fig. 2A), and importantly, mixed OmpF/DPPG layers were equally stable (Fig. 2B and supplemental Fig. S1). The surface pressure (π) versus area isotherm and BAM images taken at various surface pressures demonstrate that the mixed OmpF/DPPG monolayers (Fig. 2B) were distinct from a pure DPPG film (Fig. 2A) in both compression behavior and surface topography. Below π = 15 mN/m, the isotherm was curved and resembled that of an insoluble monolayer (44) in the liquid-expanded region. Above 15 mN/m, the pressure response becomes linear and typical of a liquid-condensed monolayer. In this condensed phase, the BAM image of the OmpF/DPPG monolayer showed large distinct domains (see Fig. 2B; π = 15–35 mN/m) compared with the even domain distribution of condensed phase DPPG (Fig. 2A). The resolution of BAM perpendicular to the membrane plane is subnanometer, and we attribute these light regions to thicker protein-rich areas with the dark background consisting essentially of DPPG.
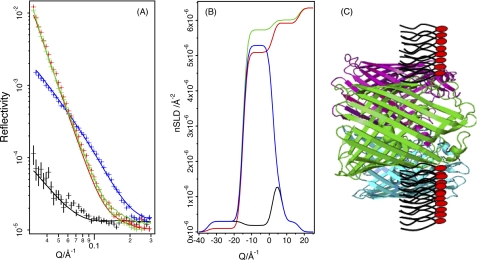
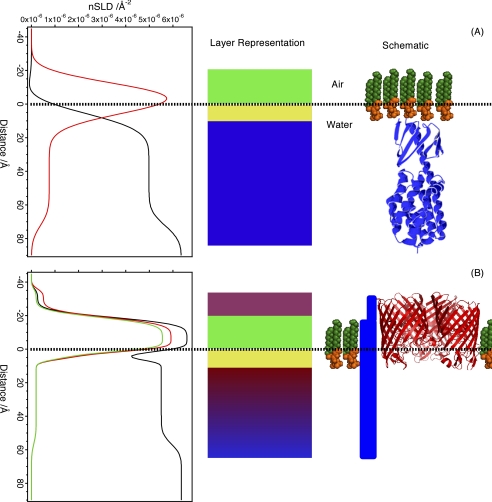
NR analysis of the OmpF/DPPG film used mixtures of d-OmpF/h-DPPG on non-reflecting air contrast-matched water (ACMW), h-OmpF/d-DPPG on D2O, d-OmpF/d-DPPG on D2O, and d-OmpF/h-DPPG on ACMW (Fig. 3A). ACMW and air have the same (zero) nSLD (loosely equivalent to a neutron “refractive index”), whereas deuteration of the components provides contrasting nSLDs, which allow more accurate modeling of the protein lipid system. Omp proteins are known to assemble at the air-water interface in a folded state and oriented with the large extra-cellular loops in the aqueous phase (45, 46). The fitted nSLD (Fig. 3B) demonstrates that the OmpF/DPPG film is a monolayer with this expected structure (Table 1). The reflection data can be fitted as a three-layer model where the upper layer 18 Å (against air) is composed of ∼8% OmpF, the intermediate layer of 17 Å consists of OmpF and DPPG tails, and the lower layer (against the aqueous phase) consists of DPPG headgroups and the extracellular loops of the OmpF (15 Å) (Fig. 3C).
FIGURE 3.
Modeling monolayer structure from NR data. Shown are NR data (symbols) and model data fits (lines) for OmpF/DPPG monolayers (A) and fitted nSLD profiles (B). The isotopic contrasts shown are d-OmpF/h-DPPG on ACMW (black), d-OmpF/d-DPPG on ACMW (blue), h-OmpF/d-DPPG on D2O (red), and d-OmpF/d-DPPG on D2O (green). A schematic diagram depicting the interfacial structure obtained from these data is shown (C), which does not represent the actual porin/lipid ratio within the film. ACMW (air contrast-matched water) is a mixture of D2O and H2O (∼8% D2O) so that the nSLD sums to zero and is therefore equivalent to air. A bare ACMW-air interface does not reflect, ensuring that the only scattering signal is that from the lipids and proteins in the monolayer.
TABLE 1.
Parameters generated by simultaneous fitting of multiple nSLD profiles
Data were collected after 180-min equilibration. d, layer thickness in Å; Г, surface excess of each component at the interface compared with the aqueous bulk. Where the summed percentages for a layer exceed 100%, this is within the overall error for the layer (e.g. OmpF + ColN + DPPG lipid headgroup layer = 101.8 ± 6.7). These results, obtained by not constraining the fits to 100%, emphasize the quality of the models. Where totals are significantly less than 100%, the difference reflects the water content of the layer.
| Layer number | DPPG |
ColN + DPPG |
OmpF + DPPG |
OmpF + ColN +DPPG |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| d | Lipid | Г | d | Lipid | ColN | Г | d | Lipid | OmpF | Г | d | Lipid | OmpF | ColN | Г | |
| Å | % | mg/m2 | Å | % | % | mg/m2 | Å | % | % | mg/m2 | Å | % | % | % | mg/m2 | |
| 1 (protein only/air) | 18.0 ± 2.0 | 8.2 ± 2.3 | 0.34 ± 0.1 | 16.7 ± 0.06 | 8.2 ± 2.4 | 5.3 ± 3.1 | 0.6 ± 0.21 | |||||||||
| 2 (lipid acyl chain) | 17.2 ± 1.77 | 98 ± 5.2 | 1.73 ± 0.09 | 15.8 ± 1.0 | 97.0 ± 3.0 | 3.7 ± 1.6 | 1.64 ± 0.049 | 17.2 ± 0.13 | 69 ± 2.1 | 10.5 ± 1.7 | 1.58 ± 0.10 | 20.4 ± 0.5 | 69 ± 10 | 10.8 ± 4.6 | 11.6 ± 6.0 | 2.26 ± 0.41 |
| 3 (lipid headgroup) | 6.15 ± 0.5 | 74 ± 7.2 | 0.8 ± 0.078 | 8.0 ± 0.01 | 67.0 ± 3.5 | 21 ± 6.0 | 1.31 ± 0.069 | 5 ± 0.5 | 82 ± 0.2 | 8.2 ± 2.1 | 0.8 ± 0.004 | 5.0 ± 0.08 | 82.0 ± 5.0 | 8.2 ± 4.2 | 11.6 ± 1.7 | 1.1 ± 0.3 |
| 4 (protein only/water) | 60.5 ± 2.2 | 40 ± 1.8 | 5.4 ± 0.24 | 10.0 ± 1.0 | 1.0 ± 6.0 | 0.03 ± 0.13 | 50.1 ± 4.1 | 0.5 ± 0.5 | 11.6 ± 1.5 | 1.34 ± 0.17 | ||||||
OmpF-dependent ColN Insertion Revealed by BAM and Surface Pressure Data
Fig. 4, A–C, compares the impact of ColN addition upon DPPG and DPPG-OmpF monolayers as measured by BAM and surface pressure. Using the INTER reflectometer, the variation in the neutron reflection signal (Fig. 4D) could also be observed as a function of time with a resolution of about 3 min (35).
FIGURE 4.
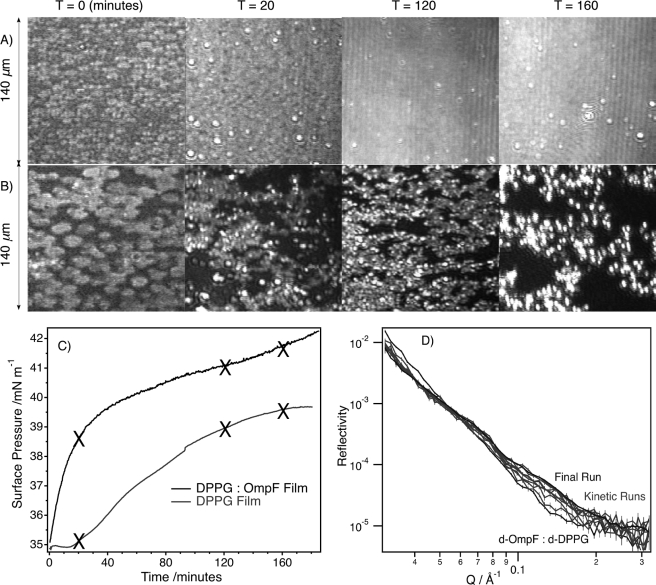
The kinetics of ColN-monolayer interactions at the air-water interface. Shown are BAM images of the monolayer at selected time intervals after the addition of ColN to the subphase beneath pure DPPG film (A) and an OmpF/DPPG film (B). C, time response of the surface pressure after the addition of ColN to the subphase at time zero; crosses indicate where BAM images were acquired. D, neutron reflection data sets with 3 min resolution showing variation in signal due to h-ColN interacting with a d-OmpF/d-DPPG film starting at 35 mN/m.
ColN adsorption to both monolayers resulted in large increases in BAM image intensity, corresponding to large changes in thickness, requiring reduction of the CCD gain to retain sensitivity to the interfacial topography. This revealed distinct differences between the absorption of ColN to condensed phase DPPG (Fig. 4A) and OmpF/DPPG (Fig. 4B) monolayers. ColN adsorption to the condensed phase DPPG monolayer caused a significant but homogeneous increase in the image intensity, which obscured all features of the original film. However, after injection of ColN below the OmpF/DPPG monolayer, circular domains appeared in the BAM image, which then increased in both frequency and size (Fig. 4B) but did not achieve complete coverage of the interface. It is clear from both BAM and surface pressure data that there is lateral reorganization in the OmpF/DPPG layer as a result of ColN interactions (Fig. 4, B and C). The NR signal (Fig. 4B) stabilized after 3 h.
Neutron Reflection Reveals Changes That Accompany Colicin Insertion
The NR data (reflectivity versus Q) are used in model-dependent fits to generate nSLD distributions along the axis perpendicular to the surface. It provides no in plane information. Here the NR data sets were simultaneously refined for multiple nSLD contrasts (combinations of hydrogenous or deuterated lipids, proteins, and water) to determine the structure, normal to the surface, of ColN adsorbed to OmpF/DPPG and DPPG only monolayers (Fig. 5). Fig. 5A contains the NR profiles, best model-to-data fits, and SLD profiles for an equilibrium ColN/DPPG monolayer fitted to a three-layer interfacial model; Table 1 gives the fitted parameters. The notable feature is an adsorbed ColN layer 60.5 Å thick on the aqueous face of the monolayer possibly stabilized by the net positive charge of ColN (47). The volume fraction of the adsorbed layer is 0.4 and correlates with the value of 0.35 for the three-dimensional crystal (48), whereas the thickness of the adsorbed ColN layer is similar to the length of the semimajor axis of the ColN crystal structure (Protein Data Bank code 1A87) (48). In contrast, ColN at pure air-water interfaces creates a sparse 25-Å-thick layer correlating with a few separated molecules at the surface (data not shown)
FIGURE 5.
Different colicin structures in lipid-only and OmpF-containing monolayers. The reflection data from each experiment were fitted to an nSLD profile, where the y axis is the distance from an interface where zero represents the air/liquid boundary; negative distances then are “in air.” The layers in the fits have been represented as blocks, where each color represents a distinct layer, and finally a schematic is drawn, representing the molecular arrangement perpendicular to the interface (protein/lipid ratio not to scale). A, equilibrium ColN-adsorbed DPPG monolayer. The isotopic contrasts shown are ColN-adsorbed d-DPPG on ACMW (red) and h-DPPG on D2O (black). Note that the ColN orientation was not determined, and the schematic shows one orientation only. B, OmpF/DPPG monolayers, h-OmpF/d-DPPG on D2O (black), h-OmpF/d-DPPG on ACMW (green), and d-OmpF/d-DPPG on ACMW (red). There is no suggestion as to the arrangement of ColN in this experiment, so it is drawn as a block where the thickness is proportional to the density of ColN at that height.
The NR-derived model for the ColN/OmpF/DPPG monolayer (Fig. 5B) reveals a very different structure, with ColN inserted across the core of the lipid layer. This resolution is possible because deuteration of the lipid and OmpF protein provides a contrast with the hydrogenous ColN. The colicin still extends 50 Å into the water phase, and thus the total height of the ColN layer is increased to 92 Å (Table 1). The lipid volume fraction in the monolayer was unchanged by ColN interaction (layers 2 and 3 in Table 1); it should be noted that the volume fraction does not add to unity in these tables, with the “missing” material consisting of water. Therefore, prior to ColN interaction, there was ∼20% water in the lipid tail region, which decreased to ∼9% after ColN interaction. There is a concomitant increase in surface excess with ColN penetration (this calculation does not include water). ColN therefore does not alter the composition of the OmpF/DPPG monolayer but increases the lateral pressure by inserting into the film and either directly or indirectly displacing water. Thus, the observed increases in surface pressure, changes in BAM images, and insertion of ColN across the lipid monolayer are contemporary. The surface pressure measurements support a process of insertion and dissociation of ColN from OmpF because there is no saturation of π after ColN addition. If ColN remained bound within the lumen of the OmpF channel, lateral dissociation into the lipid monolayer would not occur, and the increase in π should halt at a defined stoichiometry.
SANS Reveals Three-dimensional Structure and Composition of Translocon Complex
To resolve the three-dimensional structure of the translocon, we studied an OmpF-ColN complex (15) by SANS in detergent solution. The same complex, dependent upon the pore-forming C-terminal domain for stability, has been observed in both SDS and LPS solutions (23). Thus, we believe this to be a biologically relevant complex except that it would be unusual in vivo to observe more than one colicin per OmpF trimer. Crucially, the detergent SDS, which must be present to keep the complex in solution, can be obtained in both hydrogenous and deuterated forms. This means that it can be rendered non-scattering and, therefore, invisible to neutrons by mixing h- and d-SDS so that its nSLD equals that of the buffer solution. Because carbon has a scattering length similar to that of deuterium, the nSLD of h-SDS (C12H25SO4Na) is equivalent to that of 13% D2O. This means that the detergent scatter can be removed from any data collected in the 13–100% D2O range. Subsequently, these mixtures of H2O, D2O, and h/d-SDS were varied to provide three critical conditions: (i) 13% D2O, in which the neutrons were scattered by the entire complex; (ii) 42% D2O, in which the neutrons were scattered by only d-OmpF; or (iii) 87% D2O, in which the neutrons were scattered by only h-ColN.
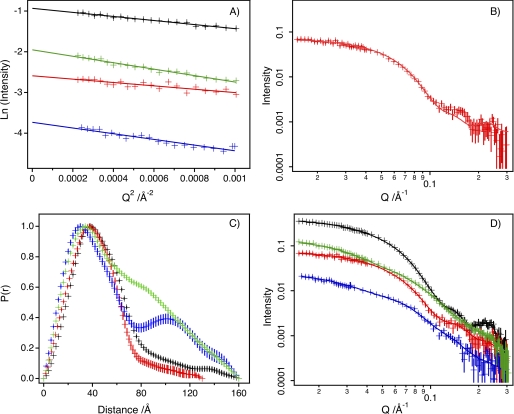
By merging these data to create one model, the independent distributions of OmpF and ColN within the same complex could be modeled. Guinier analysis of the low Q regime of the scattering data (Fig. 6A) indicated that the samples were monodisperse with the expected Rg values (supplemental Table S3).
FIGURE 6.
SANS data for the OmpF-colicin complex. SANS data (symbols) and fitting (lines) from d-OmpF·h-ColN complexes. Black, 13% D2O; red, 41% D2O; blue, 87% D2O; green, 100% D2O. A, Guinier analysis. B, calculated (using Cryson) scattering curve for OmpF trimer (Protein Data Bank code 2OMF) compared with d-OmpF·h-ColN complex with h-ColN contrast-matched (i.e. 41%) D2O. C, distance distribution functions, P(r), calculated using GNOM. D, simultaneous model fitting of data using MONSA. In all graphs, error bars are only shown when larger than data symbols.
We therefore first consider the case where the detergent solution is mixed such that the ColN and detergent are matched (41% D2O), and the scattering signal originates solely from the OmpF trimer in the complex (Fig. 6B). Analysis of the data under these conditions results in an Rg of 35 Å compared with 30 Å calculated from the crystal structure. Compared with previous small angle x-ray scattering data (49–51) on OmpF stabilized by a range of detergents (Rg = 41.0–50.2) our results are much closer to the calculated Rg because the detergent contributes significantly to the small angle x-ray scattering signal. This result gives confidence to apply a constrained fitting approach across all contrasts to reveal the size and shape of the overall complex and the subunits.
Fig. 6, C and D, shows the distance distribution function (P(r)) plots and the data with simultaneous fits for the four different contrast conditions employed. It can be seen from Fig. 6C that the contrast condition with the smallest P(r) was at 41% D2O, which observed OmpF only. At 87% D2O, where OmpF is invisible, a double peak is observed in the P(r) that indicates that the complexed ColN structure is large with some clearly separated centers of mass.
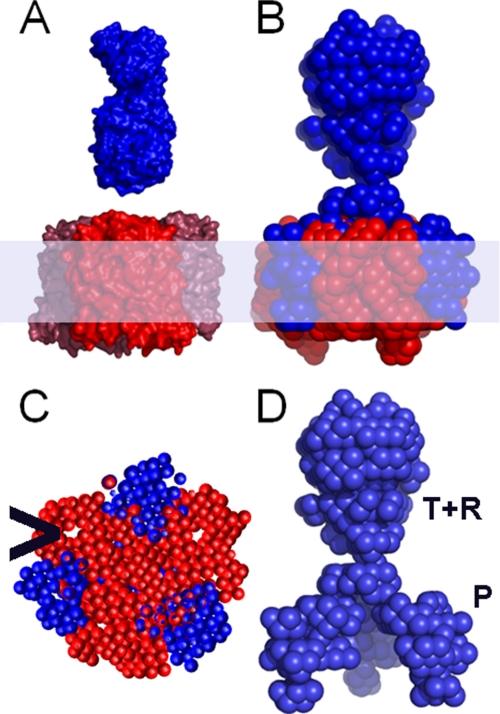
The resulting structural model derived from the SANS data showed that OmpF in the complex resembled the known crystal structure for a native trimer (52) (Fig. 7, A and B, and supplemental Table S3), but the ColN component formed a novel trimer penetrating the hydrophobic region of the membrane via external clefts between the OmpF monomers (Fig. 7, B and C). The pore lumina are resolved and apparently empty (Fig. 7C). The modeled structure of ColN in the complex is extended compared with the x-ray-derived structure (Protein Data Bank code 1A87) (48), and it is not immediately clear where the three domains of ColN are in the complex. The toxic C-terminal P-domain is required for complex formation (23), and when a complex was formed from just P-domains with OmpF, the Rg value obtained from SANS was much smaller, demonstrating that it was the P-domain that inserted into the cleft (supplemental Figs. S2 and S3 and Table S4).
FIGURE 7.
The OmpF-colicin complex modeled from SANS data. A, surface representations of the crystal structures of OmpF trimer (Protein Data Bank code 2OMF) (red) and ColN monomer (Protein Data Bank code 1A87) (blue) OmpF is oriented with the surface loops upward, and the ColN is oriented with the P-domain (Fig. 1) downward. B, the ab initio SANS model of OmpF (red) and ColN (blue) complex is shown with the membrane region shaded. Three molecules of ColN bind one OmpF trimer with the R-domain upward and P-domain on the outside of the OmpF trimer. C, cross-section midway through membrane region to show the position of the P domain in the clefts and the empty lumina (>). D, colicin trimer in the same direction as B with approximate domain distribution labeled.
The SANS-derived structural model indicates that ColN has undergone significant rearrangement to form the trimer. It was previously shown, using engineered disulfide bonds to lock the protein in the x-ray structure, that P-domain unfolding is required for complex formation and in vivo toxicity (15). To test if R-domain unfolding occurs in vivo, we inserted disulfide bridges to lock the R-domain structure in the native form. When the bonds were intact (oxidized), the mutant protein was inactive, but its function was restored upon disulfide reduction (supplemental Fig. S4), thus confirming that R-domain unfolding is required for in vivo toxicity.
DISCUSSION
We have now revealed the structure of the ColN translocon in three environments: a zwitterionic phospholipid bilayer by electron microscopy (15), an anionic phospholipid monolayer by neutron reflection, and anionic detergent micelles by SANS. They all show that OmpF can induce the unfolding and penetration of ColN into the hydrophobic membrane via the protein-lipid interface.
The DPPG and the OmpF/DPPG layers differ in that ColN penetrates the hydrophobic layer of the latter (Fig. 5 and Table 1). Thus, OmpF enhances colicin insertion into membranes, and the lack of π saturation (Fig. 4C) implies that ColN may dissociate from the OmpF trimer. The BAM images of the OmpF-containing monolayer (Fig. 4B) also show distinct regions consistent with both the natural tendency of porins to form crystalline arrays in vivo (53–55) and the two-dimensional crystals that readily form in vitro (15, 56). These OmpF domains become larger upon the addition of ColN. Self-association of an Omp by colicin has also recently been suggested in the case of colicin Ia, which was shown to bind to two copies of its receptor, Cir (57). Furthermore, it has been suggested that colicins that use two different receptors, such as colicin E9, bind first to the high affinity receptor (BtuB) and then recruit the second translocator protein (OmpF) (58).
SANS provides a three-dimensional structural model (Fig. 7) with a resolution comparable with that of electron microscopy but, thanks to selective deuteration, has the advantage of being able to separate the individual components of complexes. A landmark example of this method was the positioning of protein and RNA subunits within the EM-defined envelope of the 70 S E. coli ribosome (41). In membrane proteins, partial deuteration was first used to determine the position of specific deuterated helices of bacteriorhodopsin in crystalline layers (59, 60). Here we use contrast matching to (i) remove any detergent scattering and (ii) separately resolve the ColN and OmpF structures. This combination provides a powerful tool for membrane protein research that is being advanced by improved procedures for membrane protein and lipid deuteration and the introduction of improved neutron beamlines. The OmpF structure within the modeled complex resembled the free trimer with the empty pore lumina resolved (Fig. 7C). However, the complexed ColN formed a novel trimeric structure penetrating the hydrophobic region of the membrane via external clefts between the OmpF monomers (Fig. 7, B and D). ColN is taller in the SANS structural model than in the monolayer. The 3 ColN:3 OmpF ratio complex (11, 15) is used to obtain a homogenous sample for SANS, but the less crowded structure in the monolayer is more physiologically relevant because ColN monomers are unlikely to ever need to share a translocon in vivo. Although the T-domain is not essential for OmpF binding (13, 23, 25), should a single colicin be bound per trimer, the intrinsically unfolded T-domain could insert into a free channel lumen (27). If the protein continues across the outer membrane via the interface, the model presented here removes the need to thread a fully unfolded peptide through a small pore, and if several OmpF proteins associate, then a possible route exists between neighboring proteins.
The evidence for a translocation route via the pore (12, 24, 61, 62) was recently extended by x-ray crystallography data showing an unfolded N-terminal region of colicin E9 (containing OmpF binding sites (26, 58)) in the pore lumen (27). This supports a hypothesis whereby the unfolded translocation domain threads through the OmpF lumen (as in anthrax toxin) and presents the TolB binding site to the correct periplasmic location. If the remainder of the colicin follows, it would need to unfold, and in this respect, destabilization of the P-domain at the lipid interface could help. It was recently shown that some RNase colicins need to be cleaved by proteases during translocation, but pore-forming colicins appear to remain intact (63). However, there is conflicting evidence whether pore-forming colicins cross entirely into the periplasm (64) or continue to span the outer membrane after the inner membrane pore has opened (14). If the latter is true, then the T-domain could remain in the pore with the R-domain outside while the P-domain enters the cell. Interestingly, in both membrane protein folding and the import of mitochondrial membrane proteins, it has been proposed that critical stages of folding or insertion may occur at the protein-lipid interface (1, 65–67).
In summary, the work presented here extends the evidence that colicin N interacts with the protein-lipid interface of OmpF. It also further demonstrates that neutron methods applied to complex membrane systems can obtain structural data that are complementary to x-ray and EM approaches.
Supplementary Material
This work was supported by Wellcome Trust Grant 080342, ISIS Grant RB910077, and the Institut Laue-Langevin. The research at Oak Ridge National Laboratory's Center for Structural Molecular Biology was supported by the Office of Biological and Environmental Research, using facilities supported by the United States Department of Energy, managed by UT- Battelle, LLC under contract DE-AC05–00OR22725.

This article contains supplemental Tables S1–S4 and Figs. S1–S5.
- T-domain
- N-terminal translocation domain
- ColN
- colicin N
- R-domain
- central receptor binding domain
- P-domain
- pore-forming domain
- DPPG
- 1,2-dipalmitoyl-sn-glycero-3-phospho-(1′-rac-glycerol)
- d-
- deuterated
- h-
- hydrogenous
- mN
- millinewtons
- BAM
- Brewster angle microscopy
- NR
- neutron reflection
- nSLD
- neutron scattering length density
- ACMW
- air contrast-matched water
- SANS
- small angle neutron scattering
- POE
- polyoxyethylene.
REFERENCES
- 1. Xie K., Dalbey R. E. (2008) Nat. Rev. Microbiol. 6, 234–244 [DOI] [PubMed] [Google Scholar]
- 2. Berks B. C., Palmer T., Sargent F. (2005) Curr. Opin. Microbiol. 8, 174–181 [DOI] [PubMed] [Google Scholar]
- 3. Schmitz A., Herrgen H., Winkeler A., Herzog V. (2000) J. Cell Biol. 148, 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thoren K. L., Krantz B. A. (2011) Mol. Microbiol. 80, 588–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Osickova A., Masin J., Fayolle C., Krusek J., Basler M., Pospisilova E., Leclerc C., Osicka R., Sebo P. (2010) Mol. Microbiol. 75, 1550–1562 [DOI] [PubMed] [Google Scholar]
- 6. Ren J., Kachel K., Kim H., Malenbaum S. E., Collier R. J., London E. (1999) Science 284, 955–957 [DOI] [PubMed] [Google Scholar]
- 7. Mosbahi K., Lemaître C., Keeble A. H., Mobasheri H., Morel B., James R., Moore G. R., Lea E. J., Kleanthous C. (2002) Nat. Struct. Biol. 9, 476–484 [DOI] [PubMed] [Google Scholar]
- 8. Jakes K. S., Kienker P. K., Slatin S. L., Finkelstein A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 4321–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cascales E., Buchanan S. K., Duché D., Kleanthous C., Lloubès R., Postle K., Riley M., Slatin S., Cavard D. (2007) Microbiol. Mol. Biol. Rev. 71, 158–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vollmer W., Pilsl H., Hantke K., Höltje J. V., Braun V. (1997) J. Bacteriol. 179, 1580–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Evans L. J., Cooper A., Lakey J. H. (1996) J. Mol. Biol. 255, 559–563 [DOI] [PubMed] [Google Scholar]
- 12. Yamashita E., Zhalnina M. V., Zakharov S. D., Sharma O., Cramer W. A. (2008) EMBO J. 27, 2171–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Evans L. J., Labeit S., Cooper A., Bond L. H., Lakey J. H. (1996) Biochemistry 35, 15143–15148 [DOI] [PubMed] [Google Scholar]
- 14. Bénédetti H., Lloubès R., Lazdunski C., Letellier L. (1992) EMBO J. 11, 441–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baboolal T. G., Conroy M. J., Gill K., Ridley H., Visudtiphole V., Bullough P. A., Lakey J. H. (2008) Structure 16, 371–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anderluh G., Hong Q., Boetzel R., MacDonald C., Moore G. R., Virden R., Lakey J. H. (2003) J. Biol. Chem. 278, 21860–21868 [DOI] [PubMed] [Google Scholar]
- 17. Gokce I., Raggett E. M., Hong Q., Virden R., Cooper A., Lakey J. H. (2000) J. Mol. Biol. 304, 621–632 [DOI] [PubMed] [Google Scholar]
- 18. Raggett E. M., Bainbridge G., Evans L. J., Cooper A., Lakey J. H. (1998) Mol. Microbiol. 28, 1335–1343 [DOI] [PubMed] [Google Scholar]
- 19. Lakey J. H., Slatin S. L. (2001) in Pore-forming Toxins (Van Der Goot F. G., ed) pp. 131–161, Springer Verlag, Heidelberg [Google Scholar]
- 20. Lakey J. H., Massotte D., Heitz F., Dasseux J. L., Faucon J. F., Parker M. W., Pattus F. (1991) Eur. J. Biochem. 196, 599–607 [DOI] [PubMed] [Google Scholar]
- 21. van der Goot F. G., González-Mañas J. M., Lakey J. H., Pattus F. (1991) Nature 354, 408–410 [DOI] [PubMed] [Google Scholar]
- 22. Bullock J. O., Cohen F. S. (1986) Biochim. Biophys. Acta. 856, 101–108 [DOI] [PubMed] [Google Scholar]
- 23. Dover L. G., Evans L. J., Fridd S. L., Bainbridge G., Raggett E. M., Lakey J. H. (2000) Biochemistry 39, 8632–8637 [DOI] [PubMed] [Google Scholar]
- 24. Jeanteur D., Schirmer T., Fourel D., Simonet V., Rummel G., Widmer C., Rosenbusch J. P., Pattus F., Pagès J. M. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 10675–10679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stora T., Lakey J. H., Vogel H. (1999) Angew. Chem. Int. Ed. Engl. 38, 389–392 [DOI] [PubMed] [Google Scholar]
- 26. Zakharov S. D., Eroukova V. Y., Rokitskaya T. I., Zhalnina M. V., Sharma O., Loll P. J., Zgurskaya H. I., Antonenko Y. N., Cramer W. A. (2004) Biophys. J. 87, 3901–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Housden N. G., Wojdyla J. A., Korczynska J., Grishkovskaya I., Kirkpatrick N., Brzozowski A. M., Kleanthous C. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 21412–21417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bainbridge G., Armstrong G. A., Dover L. G., Whelan K. F., Lakey J. H. (1998) FEBS Lett. 432, 117–122 [DOI] [PubMed] [Google Scholar]
- 29. Krepkiy D., Mihailescu M., Freites J. A., Schow E. V., Worcester D. L., Gawrisch K., Tobias D. J., White S. H., Swartz K. J. (2009) Nature 462, 473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chenal A., Prongidi-Fix L., Perier A., Aisenbrey C., Vernier G., Lambotte S., Haertlein M., Dauvergne M. T., Fragneto G., Bechinger B., Gillet D., Forge V., Ferrand M. (2009) J. Mol. Biol. 391, 872–883 [DOI] [PubMed] [Google Scholar]
- 31. Garavito R. M., Rosenbusch J. P. (1986) Methods Enzymol. 125, 309–328 [DOI] [PubMed] [Google Scholar]
- 32. Artero J. B., Härtlein M., McSweeney S., Timmins P. (2005) Acta Crystallogr. D Biol. Crystallogr. 61, 1541–1549 [DOI] [PubMed] [Google Scholar]
- 33. Lakey J. H., Watts J. P., Lea E. J. (1985) Biochim. Biophys. Acta 817, 208–216 [DOI] [PubMed] [Google Scholar]
- 34. Fridd S. L., Gökçe I., Lakey J. H. (2002) Biochimie 84, 477–483 [DOI] [PubMed] [Google Scholar]
- 35. Webster J., Holt S., Dalgliesh R. (2006) Physica B Condensed Matter 385, 1164–1166 [Google Scholar]
- 36. Penfold J., Ward R. C., Williams W. G. (1987) J. Phys. E Sci. Instrum. 20, 1411–1417 [Google Scholar]
- 37. Lu J. R., Thomas R. K. (1998) J. Chem. Soc. Faraday Trans. 94, 995–1018 [Google Scholar]
- 38. Clifton L. A., Green R. J., Hughes A. V., Frazier R. A. (2008) J. Phys. Chem. B 112, 15907–15913 [DOI] [PubMed] [Google Scholar]
- 39. Lad M. D., Birembaut F., Clifton L. A., Frazier R. A., Webster J. R., Green R. J. (2007) Biophys. J. 92, 3575–3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Penfold J. (2002) Curr. Opin. Colloid Interface Sci. 7, 139–147 [Google Scholar]
- 41. Svergun D. I., Nierhaus K. H. (2000) J. Biol. Chem. 275, 14432–14439 [DOI] [PubMed] [Google Scholar]
- 42. Svergun D. I. (1999) Biophys. J. 76, 2879–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Volkov V. V., Svergun D. I. (2003) J. Appl. Crystallogr. 36, 860–864 [Google Scholar]
- 44. McConlogue C. W., Vanderlick T. K. (1997) Langmuir 13, 7158–7164 [Google Scholar]
- 45. Schindler H., Rosenbusch J. P. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 2302–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dargent B., Charbit A., Hofnung M., Pattus F. (1988) J. Mol. Biol. 201, 497–506 [DOI] [PubMed] [Google Scholar]
- 47. Evans L. J., Goble M. L., Hales K. A., Lakey J. H. (1996) Biochemistry 35, 13180–13185 [DOI] [PubMed] [Google Scholar]
- 48. Vetter I. R., Parker M. W., Tucker A. D., Lakey J. H., Pattus F., Tsernoglou D. (1998) Structure 6, 863–874 [DOI] [PubMed] [Google Scholar]
- 49. Watanabe Y., Sano Y., Inoko Y. (1999) Jpn. J. Appl. Phys. 38, Suppl. 1, 180–182 [Google Scholar]
- 50. Mo Y., Lee B. K., Ankner J. F., Becker J. M., Heller W. T. (2008) J. Phys. Chem. B 112, 13349–13354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Watanabe Y., Inoko Y. (2005) Protein J. 24, 167–174 [DOI] [PubMed] [Google Scholar]
- 52. Cowan S. W., Schirmer T., Rummel G., Steiert M., Ghosh R., Pauptit R. A., Jansonius J. N., Rosenbusch J. P. (1992) Nature 358, 727–733 [DOI] [PubMed] [Google Scholar]
- 53. Jarosławski S., Duquesne K., Sturgis J. N., Scheuring S. (2009) Mol. Microbiol. 74, 1211–1222 [DOI] [PubMed] [Google Scholar]
- 54. Hoenger A., Ghosh R., Schoenenberger C. A., Aebi U., Engel A. (1993) J. Struct. Biol. 111, 212–221 [DOI] [PubMed] [Google Scholar]
- 55. Spector J., Zakharov S., Lill Y., Sharma O., Cramer W. A., Ritchie K. (2010) Biophys. J. 99, 3880–3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cisneros D. A., Muller D. J., Daud S. M., Lakey J. H. (2006) Angew. Chem. Int. Ed. Engl. 45, 3252–3256 [DOI] [PubMed] [Google Scholar]
- 57. Jakes K. S., Finkelstein A. (2010) Mol. Microbiol. 75, 567–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Housden N. G., Loftus S. R., Moore G. R., James R., Kleanthous C. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13849–13854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Trewhella J., Anderson S., Fox R., Gogol E., Khan S., Engelman D., Zaccai G. (1983) Biophys. J. 42, 233–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Trewhella J., Popot J. L., Zaccaï G., Engelman D. M. (1986) EMBO J. 5, 3045–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nikaido H. (2003) Microbiol. Mol. Biol. Rev. 67, 593–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cao Z., Klebba P. E. (2002) Biochimie 84, 399–412 [DOI] [PubMed] [Google Scholar]
- 63. Chauleau M., Mora L., Serba J., de Zamaroczy M. (2011) J. Biol. Chem. 286, 29397–29407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. El Kouhen R., Pagès J. M. (1996) J. Bacteriol. 178, 5316–5319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hessa T., Kim H., Bihlmaier K., Lundin C., Boekel J., Andersson H., Nilsson I., White S. H., von Heijne G. (2005) Nature 433, 377–381 [DOI] [PubMed] [Google Scholar]
- 66. Rapaport D. (2005) J. Cell Biol. 171, 419–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chacinska A., Koehler C. M., Milenkovic D., Lithgow T., Pfanner N. (2009) Cell 138, 628–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.