Background: Fructooligosaccharides as prebiotics are a preferred carbon source of bifidobacteria.
Results: A potential fructose-specific transporter was analyzed in B. longum NCC2705.
Conclusion: The identified transporter is a functional ABC transporter with high affinity for fructose and is regulated by its substrate.
Significance: This transporter represents the first fructose-specific sugar transporter identified in bifidobacteria.
Keywords: ABC Transporter, Metabolism, Microbiology, Protein-Protein Interactions, Proteomics
Abstract
Recently, a putative ATP-binding cassette (ABC) transport system was identified in Bifidobacterium longum NCC2705 that is highly up-regulated during growth on fructose as the sole carbon source. Cloning and expression of the corresponding ORFs (bl0033–0036) result in efficient fructose uptake by bacteria. Sequence analysis reveals high similarity to typical ABC transport systems and suggests that these genes are organized as an operon. Expression of FruE is induced by fructose, ribose, or xylose and is able to bind these sugars with fructose as the preferred substrate. Our data suggest that BL0033–0036 constitute a high affinity fructose-specific ABC transporter of B. longum NCC2705. We thus suggest to rename the coding genes to fruEKFG and the corresponding proteins to FruE (sugar-binding protein), FruK (ATPase subunit), FruF, and FruG (membrane permeases). Furthermore, protein-protein interactions between the components of the transporter complex were determined by GST pulldown and Western blot analysis. This revealed interactions between the membrane subunits FruF and FruG with FruE, which in vivo is located on the external side of the membrane, and with the cytoplasmatic ATPase FruK. This is in line with the proposed model for bacterial ABC sugar transporters.
Introduction
Bifidobacteria are Gram-positive, high G+C content bacteria commonly present in human and animal intestines. The potential health-promoting or probiotic activities of bifidobacteria are well known and include reduction of symptoms of irritable bowel disease (1), stimulation of the immune response (2), and inhibition or competitive exclusion of pathogenic bacteria (3). Carbohydrates represent one of the major structural building blocks of all organisms. Colonization and survival of bifidobacteria in the large intestine are dependent on utilization and uptake of fermentable carbohydrates not absorbed or metabolized by the host. As saccharolytic organisms, bifidobacteria are able to utilize a wide range of catabolic pathways that confer a growth advantage over other intestinal bacteria (4, 5). Several nondigestible carbohydrates such as oligofructose, inulin, and raffinose have been identified as bifidogenic compounds and are used as food additives (prebiotics) to selectively promote growth of bifidobacteria in the gut (5, 6).
Transport of sugars across the cytoplasmatic membrane is a prerequisite for the subsequent carbohydrate metabolism of bacteria. More than 8.5% of the predicted proteins encoded on the genome of Bifidobacterium longum NCC2705 are involved in carbohydrate transport and metabolism, ∼30% more than observed in other organisms (5). Bifidobacteria are reported to possess only one metabolic pathway for the fermentation of hexoses: the bifidus shunt. The first central intermediate of the bifidus shunt, fructose-6-phosphate, links this pathway to several other pathways including N-acetyl hexose fermentation, galactose catabolism, and peptidoglycan biosynthesis (7). Although B. longum is able to grow on a wide variety of sugars as sole carbon source including glucose, fructose, and ribose, the specificity of the majority of transporter systems that feed these sugars into the catabolic pathways have not been defined to date.
Three energy-dependent sugar uptake mechanisms have been characterized in bacteria. The major facilitator superfamily transporters are used for the uptake of galactose, xylose, and lactose in Escherichia coli and operate by proton symport. A second system is the phosphoenolpyruvate:sugar phosphortransferase system (PTS)5 for transporting glucose, fructose, mannose, and sucrose found in many Gram-negative bacteria. A third mechanism is the periplasmic binding protein-dependent ATP-binding cassette (ABC) transporter, which are ubiquitous membrane protein complexes that use the energy generated from ATP hydrolysis for uptake or export of a large variety of solutes across biological membranes (8, 9).
Most ABC transporters share a modular architecture containing two transmembrane domains or subunits and two cytosolic nucleotide-binding domains or subunits, also known as ATP-binding cassettes. The substrate specificity is accomplished by the transmembrane domains, which display basically no sequence homologies and feature varying numbers of transmembrane helices among different ABC transporters. Nucleotide-binding domains with the ability to bind and hydrolyze ATP can provide the energy required for substrate translocation. In addition to the classical nucleotide-binding Walker A and B motifs, each nucleotide-binding domain also contains a conserved LSGG(N)QQ signature motif that is diagnostic of ABC ATPases (10). One of the best functionally characterized ABC transporters is the maltose uptake system (MalFGK2-E) of E. coli and Salmonella typhimurium, which is composed of a periplasmic maltose-binding protein (MalE), two integral membrane proteins (MalF and MalG), and two copies of the cytoplasmic ATP-binding cassette (MalK). Thus, this system serves as a model for studying the molecular mechanism by which ABC importers exert their functions (11–14). In bacteria, the ABC superfamily transports a wide range of substrates, including a variety of monosaccharides such as arabinose or ribose, as well as di- and tri-saccharides or higher oligosaccharides (15).
A total of 19 carbohydrate uptake systems were predicted in the genome sequence of B. longum NCC2705 in GenBankTM data base provided by the National Center for Biotechnology Information, of which 13 are putative ABC transporters (5). Although carbohydrate utilization is critical to understanding bifidobacterial survival and colonization in the gut, only a few sugar transport systems in this genus have been characterized to date. A phosphoenolpyruvate:glucose PTS has been characterized in Bifidobacterium breve NCBF 2275. Activities of a lactose/proton symporter, a glucose/potassium symport, and an unsaturable galactose permease have been detected in Bifidobacterium bifidum DSM 20082 (7, 16). In B. longum NCC2705, glucose PTS activity and an inducible glucose/proton symport subject to lactose repression were detected (17, 18).
The transport of fructose in bacteria is usually performed by a fructose-specific PTS, which upon transport generates fructose-1-phosphate or major facilitator superfamily transporters (9). Fructose-specific PTSs show several features that are not found in the PTS of other sugars. Although the PTSs for other sugars use a common histidine-containing phosphocarrier protein to transfer phosphate between enzyme I and the sugar-specific enzyme II, the fructose-specific PTS has its own fructose-specific phosphocarrier protein (19).
Bioinformatics analysis of B. longum NCC2705 genome sequence revealed no fructose-specific PTS (5). We recently reported on a sugar-binding protein (BL0033) and ATP-binding protein (BL0034), which are part of a putative ABC transport system with similarity to the ribose-specific ABC transporter of E. coli (17) and are induced when bacteria are grown on fructose, ribose, and xylose as sole carbon sources (20). We thus speculated that BL0033–0036 form an ABC transporter system involved in the uptake of fructose, ribose, and/or xylose. This transport system could play an important role because fructose and fructooligosaccharides are preferred substrates of bifidobacteria. These fructose-containing sugars are components of human breast milk and frequently used as prebiotics. It is speculated that they might be responsible for the early colonization of infants. In 2011, Fukuda et al. (21) showed that B. longum NCC2705 and other bifidobacteria-harboring homologues of BL0033–0036 exert a protective effect against E. coli O157:H7 infections in mice, whereas a mutant of B. longum NCC2705 and strains that do not encode homologues of BL0033–0036 had no protective effect. Moreover, compared with the mutant, B. longum NCC2705 was more efficient in the conversion of fructose to acetate in vitro, and the amount of acetate in feces of mice treated with the wild type strains was significantly higher (21). Both the protective effect and efficient acetate conversion could be transferred to strains that did not encode homologues of BL0033–0036 by heterologous expression of the corresponding genes bl0033-0036 (21).
We thus propose to rename the genes bl0033–0036 into fruEKFG and the encoded proteins into FruE, FruK, FruF, and FruG. Here, we report the molecular characterization of a fructose-specific ABC transporter in B. longum NCC2705. In addition to the preferred substrate fructose, ribose and xylose might be also transported by this system, albeit with lower affinity. GST pulldown and Western blot experiments showed interactions of the different components of the ABC transporter. Identification and characterization of carbohydrate transporter systems of bifidobacteria will help to further understand the nutritional lifestyle and their contribution to the protective effects of this important group of intestinal bacteria.
EXPERIMENTAL PROCEDURES
Strains, Medium, and Growth Conditions
The strains, plasmids, and oligonucleotides used for this study are listed in Table 1. B. longum DCP-18, deficient in fructose-transport and fermentation (see supplemental Table S1), was used as host for homologous complementation with bl0033. To analyze expression of the ABC transporter system, B. longum strain NCC2705 (kindly provided by the Nestlé Research Center, Lausanne, Switzerland) was grown anaerobically at 37 °C in 400 ml of De Man-Rogosa-Sharpe broth (22) containing 0.05% l-cysteine or modified Garches medium (MGM) containing d-glucose, fructose, xylose, or d-ribose as the sole carbon source as described previously (20) for 8 h (early exponential phase), 12 h (midexponential phase), 16 h (end of exponential phase), and 24 h (stationary growth phase). In the concentration series experiments, B. longum NCC2705 cells were grown in MGM supplemented with either fructose, glucose, xylose, or d-ribose at 1, 2, 3, or 4 g/liter, respectively. The cultures were grown in 500-ml flasks, and anaerobic conditions were maintained by sparging the cultures with O2-free N2 gas (5 ml/min). For experiments, the bacteria were harvested in midexponential phase at an A600 of 0.9 corresponding to 1.5 × 108 colony-forming units/ml.
TABLE 1.
Strains, plasmids, and oligonucleotides used for this study
| Strain, plasmid, or oligonucleotides | Relevant characteristics or nucleotide sequence (5′ → 3′) | Reference or purpose |
|---|---|---|
| Strains | ||
| B. longum NCC2705 | Type strain, genome sequenced | Ref. 5 |
| B. longum DCP-18 | Fructose-negative strain | This study |
| E. coli mutant LR2–177 | galP manA nagE glcA fruA ptsI glk+ mak° (deficient for mannofructokinase activity) | Ref. 23 |
| E. coli DH5α | hsdR17 (rk− mk−) recA1 endA1 gyrA96 thi-1 relA1 | Commercial strain |
| E. coli BL21-CondonPlus(DE3)-R-IL | dcm hsdSB(rB− mB−) gal(λDE3[lacI lacUV5-T7 gene1 ind1 Sam7 nin5] | Commercial strain |
| E. coli Rosetta (DE3) | BL21 derivatives designed to alleviate codon bias when expressing heterologous proteins in E. coli | Commercial strain |
| Plasmids | ||
| pET-32a | Ampr lacI; expression vector; 5.4 kb | Commercial plasmid |
| pGEX-4T-1 | Ampr lacI; expression vector; 4.9 kb | Commercial plasmid |
| pET32a-3 | pET-32a containing the gene fruE | This study |
| pET32a-4 | pET-32a containing the gene fruK | This study |
| pGEX-4T-1-3 | pGEX-4T-1 containing the gene fruE | This study |
| pGEX-4T-1-5 | pGEX-4T-1 containing the gene fruF | This study |
| pGEX-4T-1-6 | pGEX-4T-1 containing the gene fruG | This study |
| pGEX-FruEKFG | pGEX-4T-1 containing the full fruEKFG operon | This study |
| pDG7 | E. coli-Bifidobacterium shuttle vector | Ref. 27 |
| pDG7-FruE | pDG7 vector containing the fruE gene | This study |
| Oligonucleotides for reverse transcription PCR, gene cloning, and expression | ||
| 16SF | TCCAGTTGATCGCATGGTC | RT-PCR |
| 16SR | GGGAAGCCGTATCTCTACGA | |
| fruE-F | AGGTGGCAGCTCGGACTCC | RT-PCR |
| fruE-R | GCGGCGTCGAAGGTCTTGG | |
| fruE-F′ | GGCGCGGATCCATGAAGAATTGGAAGAAGGC | Cloning of fruEKFG in pGEX-4T-1 |
| fruE-R′ | ATTACCGCTCGAGTCAGTAGGCGCGGGTGTT | |
| Ebl33F | CGGAATTCCTCGGTGCGCAGTAGATGGT | Cloning of fruE in pDG7 |
| Ebl33R | CCCAAGCTTTCAGTAGGCGCGGGTGTTGT | |
| fruK-F | GCGCGGATCCATGACAGATAAAAACCCC | Cloning of fruK constructs |
| fruK-R | TACCGCTCGAGTCATGCCTCCTTTCCGGT | |
| fruF-F | CGGGATCCATGACAACAGCTACGGCAA | Cloning of fruF constructs |
| fruF-R | CCGCTCGAGCTATTTTTTATCTCCGCCG | |
| fruG-F | GCGCGGATCCATGGCTGAAAAGGCAAAAGC | Cloning of fruG constructs |
| fruG-R | AACCGCTCGAGTCATGCTGCCACCGCCTTA | |
To test specificity and reversibility of relative gene expression induced by fructose, the cultures were grown in 400 ml of MGM-containing fructose or glucose for 6 h as described above. Then each bacterial culture was washed twice with prewarmed PBS, and the cells were split in two and resuspended in 400 ml of fresh medium with either fructose or glucose as control at a final concentration of 2 g/liter. The two cultures were then incubated further for 6 h.
E. coli DH5α, Rosetta (DE3) and BL21-CodonPlus (DE3)-R-IL were used as cloning and expression hosts. E. coli mutant LR2–177 was used for heterologous complementation experiments. E. coli strains were routinely grown at 37 °C in LB medium with shaking. Ampicillin was added to a final concentration of 100 μg/ml for selection of transformed bacteria.
Sugar Transport Assays
The radiolabeled sugars [U-14C]fructose (10.2 GBq/mmol) and [U-14C] glucose (10.9 GBq/mmol) were purchased from GE Healthcare. Sugar transport assays were performed as described (9). Cells grown in MGM were harvested at A600 of 0.5 to 0.7, washed twice in PBS, and resuspended in carbon-free MGM at a final A600 of 0.5. Uptake was determined by rapid filtration through glass microfiber filters (Whatman GF/F) that were rinsed with 3 ml of MGM. The radioactivity remaining on the filters was detected with a liquid scintillation spectrometer (Beckman Instruments, Villepinte, France). For competition experiments, cold unlabeled fructose, ribose, xylose, mannose, galactose, and glucose were added at a final concentration of 4.5 mm into a 50 μm [U-14C]fructose solution for 1min before filtration. All of the assays were conducted in triplicate on two independent cultures at 37 °C with 1 ml of cell suspension and radioactive substrate (100,000 dpm) at final concentrations of 0.5–500 μm for 1–10 min.
Determination of Km and Vmax was carried out in B. longum NCC2705, E. coli LR2–177 (pGEX-FruEKFG), and B. longum DCP-18 (pDG7-FruE). For heterologous expression, pGEX-FruEKFG was induced in E. coli LR2–177 (23) for 1 h (A600 of 0.5) with 1 mm IPTG. The cells were harvested and washed twice with chilled PBS. Fructose uptake was conducted at 37 °C as described above.
Specific Substrate Binding of FruE
GST-fused FruE and GST alone were purified by affinity chromatography using glutathione-Sepharose 4B beads (GE Healthcare). After washing with buffer B (10 mm Tris-HCl, pH 8.0, 14 mm β-mercaptoethanol, 0.1% Tween 20, 20% (v/v) ethanol, and 0.5 m NaCl), proteins were eluted with buffer C (10 mm reduced glutathione, 50 mm Tris-HCl, pH 7.5). Fructose, ribose, xylose, or glucose (final concentration, 1 g/liter) was separately added into 10 ml of purified protein solution and incubated for 1 h at room temperature. The mixture was transferred into ultrafiltration tubes, which can retain macromolecules. After centrifugation at 1000 × g for 30 min, both retentate and filtrate were collected and subjected to capillary electrophoresis (P/ACE MDQ capillary electrophoresis system; Beckman Coulter). Furthermore, the capacity of BL0033 to bind substrates was assayed by an enzymatic test using a glucose/fructose kit (BioSenTec Co.) at different substrate concentrations and incubation times according to the manufacturer's instructions. In all of the experiments, nonfused GST was used as a negative control, and purified protein was used as a positive control.
Preparation of Whole Cell Protein Extracts
Preparation of whole cellular protein extracts was performed as described previously (24) with modifications. Briefly, the cells were centrifuged for 10 min at 8,000 × g in a Sigma 3K12 centrifuge and washed four times with 40 ml of ice-cold low salt washing buffer (3 mm KCl, 1.5 mm KH2PO4, 68 mm NaCl, 9 mm NaH2PO4) (24). Bacterial pellets were resuspended in 5 ml of lysis buffer (7 m urea, 2 m thiourea, 4% (w/v) CHAPS, and 50 mm DTT) containing complete protease inhibitors (Roche Applied Science) at 1.25 mm. The cells were disrupted by sonication for 5 min (cycles of 2 s of sonication followed by a 3-s rest) on ice with a Sonifier 750 (Branson Ultrasonics Corp., Danbury, CT) set at 25% duty cycle. After adding 2.5 mg of RNase A (Promega, Madison, WI) and 100 units of RQ1 DNase (Promega), the bacterial lysate was incubated for 1 h at 15 °C to solubilize proteins and then centrifuged for 45 min at 20,000 × g to pellet the insoluble components. The supernatants were collected, and protein concentrations were measured using the PlusOne two-dimensional Quant kit (GE Healthcare). The samples were stored at −70 °C in 1-mg aliquots until further use. The experiments were performed at least six times.
Two-dimensional Polyacrylamide Gel Electrophoresis
Isoelectric focusing was carried out as described previously (24) by loading 1 mg of protein onto ImmobilineTM DryStrip gels in three pH ranges (pH 3–10, nonlinear/linear, 18 cm; pH 4–7 and pH 4.5–5.5, linear, 18 cm; Amersham Biosciences). Isoelectric focusing was conducted at 20 °C for 65,000 Vh in the IPGphor system (GE Healthcare). For the second dimension, vertical slab SDS-PAGE (12.5% gel) was performed using the Bio-Rad Protean II Xi apparatus, with runs for about 4 h at 30 mA/gel. Following electrophoresis, gels were stained with Coomassie Brilliant Blue G-250 (Amresco, Solon, OH) and then scanned with an ImageScanner (GE Healthcare). Image analysis was carried out using ImageMaster two-dimensional Platinum software (GE Healthcare). To facilitate the discrimination between true spots and artifacts, the spot detection parameters of the software were set to: smooth, 3; minimum area, 50; and saliency, 6. The relative volume of each spot was quantified by determining the spot intensity in pixel units and normalizing that value to the sum of the intensities of all the spots in the gel. Proteins were considered differentially expressed if their relative intensity differed more than 3-fold between the two conditions compared. Each experiment was performed at least three times.
In-gel Protein Digestion and MALDI-TOF/TOF MS/MS and Protein Identification
The Coomassie-stained protein spots were excised, and the protein was digested as previously described (24, 25). MALDI-TOF/TOF MS/MS measurements were performed on a Bruker Ultraflex III TOF/TOF-MS (Bruker Daltonics GmbH, Bremen, Germany) equipped with a 337-nm wavelength nitrogen laser (model LSI 337i; Bruker) working in reflectron mode. The mass range of peptides detected (m/z) was from 800 to 4500 Da. The Mascot search engine uses mass spectrometry data to identify proteins from primary sequence databases as previously described (25). Peptide mass fingerprinting searches were performed by using the program Mascot v2.2.06 (Matrix Science Ltd.) licensed in-house against the publically available NCBI nonredundant protein databases (NCBInr v20100616, 11,205,16 sequences; 3,821,460,163 residues). For those proteins identified in the NCBInr data base, the proteins of B. longum spp. were selected as the best hits from the lists of homologue proteins.
Nanospray Electrospray Ionization MS/MS
The peptide solution collected after in-gel protein digestion was dried, reconstituted in 30 μl of 0.1% TFA in 30% acetonitrile/water, and then desalted with ZipTip C18 pipette tips (Millipore, Bedford, MA). Electrospray ionization MS/MS was carried out with a hybrid quadrupole orthogonal acceleration tandem mass spectrometer (Q-TOF2; Micromass, Manchester, UK). Glufibrinopeptide was used to calibrate the instrument in the MS/MS mode, and internal calibration was carried out using enzyme autolysis peaks. MS/MS peak lists were created by MaxEnt3 (MassLynx v3.5; Micromass), and amino acid sequences were interpreted manually using MassSeq (Micromass). All of the MS/MS ion data base searches were performed using Mascot v2.2.06 on the freely accessible internet website against protein databases of NCBInr (v20100430, 10,927,723 sequences; 3,720,794,783 residues of all bacteria). We checked the fragment ion intensity patterns because the peak intensities should be matched by the search.
Gene Expression Analysis by Semi-quantitative RT-PCR
The relative abundance of mRNA of genes of interest was analyzed by semi-quantitative RT-PCR. Total RNAs were extracted using the EPICENTRE MasterPure RNA purification kit (Epicenter Technologies, Madison, WI) as recommended by the manufacturer. The RNA concentrations were determined by an ND-1000 Spectrophotometer (Nanodrop Technologies) at 260 nm. Reverse transcription was performed using the Omniscript reverse transcription kit (Qiagen) with 2 μg of total RNA as a template. Primers used for RT-PCR assays were designed to generate PCR products of comparable sizes (Table 1). The PCR amplification was performed with the following conditions: 94 °C for 5 min initial denaturation followed by 25–30 cycles at 94 °C for 20 s, 60 °C for 30 s, and 72 °C for 1 min and a final extension at 72 °C for 5 min to complete the reaction. To control for RNA quantity, RT-PCR was performed targeting the 16S rRNA as described elsewhere (26). Negative control PCRs were performed without primers, reverse transcriptase, or Taq polymerase (Promega) to confirm the absence of contaminating DNA in the 16S rRNA preparations. PCR products were analyzed on 1% agarose gels and visualized by ethidium bromide staining. Signal intensities were quantified using Quantity One quantitation software (Bio-Rad).
Cloning, Expression, and Purification of the ABC Transport System and Antibody Preparation
Total cellular DNA was prepared from B. longum NCC2705 as previously described (24). Based on the gene sequence of the component of the identified ABC transporter system (bl0033, bl0034, bl0035, and bl0036), primers were designed (Table 1) to amplify these genes by PCR.
The amplified PCR products were cloned into plasmids pET32a and/or pGEX-4T-1 or pDG7 (27) using BamHI and XhoI restriction sites (underlined in the primer sequences in Table 1). All of the constructed expression plasmids were verified by DNA sequencing (AUGC Company, Beijing, China). Transformants were cultured in LB broth with ampicillin (100 mg/liter) to an A600 of 0.6. Expression of the inserted gene was then induced by the addition of IPTG (0.05 mm; Invitrogen), and the culture was grown for an additional 10 h at 16 °C. To determine the subcellular localization of the expressed protein, the supernatant and pellet were analyzed on a 12% SDS-PAGE using a Mini-Protein II gel apparatus (Bio-Rad).
The GST fusions of the two membrane permeases FruF and FruG were purified using a protocol for the preparation of insoluble proteins (28). E. coli Rosetta strains containing the pGEX-constructs harboring fruF or fruG were grown at 37 °C in LB medium with 100 μg/ml of ampicillin to an A600 of 0.6 and induced with 0.5 mm of IPTG for 4 h. The cells were harvested and washed twice with chilled STE (10 mm Tris, pH 8.0, 150 mm NaCl, 1 mm EDTA). The pellet was solubilized in ice-cold STE containing 100 μg/ml of lysozyme and incubated on ice for 15 min. Just before sonication, 5 mm DTT, 1.25 mm protease inhibitors (Roche Applied Science), and 0.7% sarkosyl (Sigma) were added. After being mixed thoroughly for 5 s, the cells were sonicated on ice for 1 min. After centrifugation at 16,000 × g for 20 min, the supernatant was transferred to a new Eppendorf tube, Triton X-114 was added to the final concentration (2%) from a 10% stock in STE, and the lysate was incubated at room temperature for 30 min.
GST fusion proteins were purified by adding glutathione-Sepharose 4B beads (GE Healthcare) to bacterial lysates and incubation at room temperature for 1 h. The beads were washed once with PBS, and protein was eluted by GST elution butter (50 mm Tris-HCl, 10 mm reduced glutathione, pH 8.0).
In the case of the His6-tagged constructs, the coding DNA fragments were amplified from B. longum NCC2705 genomic DNA and fused to an His6 tag in pET32a at the N terminus. Polyhistidine has a high affinity for nickel-nitrilotriacetic acid resin. The His6-tagged protein was purified batch-wise under native conditions using nickel-nitrilotriacetic acid-agarose and 1 mg/ml total protein from bacterial pellets following the manufacturer's instructions (Qiagen). The washing buffer contained 20 mm imidazole and the elution buffer contained 500 mm imidazole. The protein was concentrated using an Amicon Ultra-4 centrifugal filter unit equipped with a 50-kDa cut-off membrane (Millipore), transferred to 100 mm Tris-HCl buffer, pH 7.4, and stored on ice. Protein concentrations were determined by the Bradford method. All of the protein samples were subjected to SDS-PAGE and analyzed by Western immunoblotting as described below.
To generate polyclonal antibodies, purified His-tagged BL0033, BL0034, BL0035, and BL0036 proteins were injected subcutaneously into BALB/c female mice. Sera from the immunized mice were collected and purified using an immobilized protein A kit (Pierce) according to the manufacturer's instructions.
Resistance of FruF and FruG to proteinase K was determined as described (29) by adding 100 mg/ml of proteinase K to E. coli spheroblasts expressing either FruF or FruG and subsequent analysis by SDS-PAGE and Western blot using specific antibodies.
Protein-Protein Interaction by GST Pulldown and Western Blotting
In GST pulldown experiments, the GST fusions were incubated with bacterial lysates of E. coli BL21 cells expressing different His6-tagged proteins in equimolar ratios for 2 h in PBS buffer (140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4, pH 7.5) at 4 °C. Glutathione-Sepharose 4B beads were added, and the bound proteins were precipitated by centrifugation.
Total cell proteins or periplasmic proteins were separated by PAGE and transferred to nitrocellulose membrane (Hybond protein; 0.2-mm pore size) by electroblotting. Immunoblotting was performed using a 1/5000 dilution of a polyclonal serum raised against FruE or FruK of B. longum NCC2705. In direct binding assays, the adsorbates were washed with lysis buffer and then subjected to SDS-PAGE and immunoblot analysis with anti-His, anti-GST, and anti-FruE or anti-FruK antibodies. In all GST pulldown and Western blotting experiments, GST vector or His vector was used as negative control.
Bioinformatic Analysis
Protein sequences were analyzed for conserved domains/motifs, transmembrane helices, etc., using the web-based InterProScan Sequence Search (30) and the ScanProsite software of the Swiss Institute of Bioinformatics. The signal peptide of BL0033/FruE was verified using the SignalP version 4.0 (31).
Statistical Analysis
Each experiment was performed at least three times. Statistical significance was determined using the Student's t test.
RESULTS AND DISCUSSION
Fructose Uptake Activity in B. longum NCC2705
To analyze the parameters of fructose transport, B. longum NCC2705 was grown in MGM with 1 g/liter fructose as the sole carbon source to midexponential growth phase. Bacteria were then washed, and uptake of fructose was measured using [U-14C]fructose concentrations ranging from 0.5 to 500 mm (data not shown). The apparent Km of fructose uptake by B. longum NCC2705 was found to be 8 ± 1 μm with a Vmax of 45 ± 5 nmol/min/mg of protein, indicating the presence of a high affinity fructose transport system. Growth in presence of 2 g/liter fructose did not significantly modify the Km but resulted in a doubling of the Vmax (96 ± 4 nmol/min/mg of protein), indicating that the substrate induces its own transport.
Analysis of bl0033–0036 in Fructose-negative Strains
Analysis of the fermentation pattern of 45 B. longum strains isolated from human feces revealed that one of the isolates, B. longum DCP-18, was unable to grow on fructose as a sole carbon source (i.e. it is fructose-negative; supplemental Table S1). We thus performed a time course analysis of fructose uptake with a substrate concentration of 10 μm, which clearly demonstrated that B. longum DCP-18 is not able to transport fructose (Fig. 1A). Similar results were obtained for substrate concentrations up to 100 μm. In contrast, transport assays using glucose showed that the uptake of the sugar was not altered in B. longum DCP-18 (data not shown), which is in agreement with the growth phenotype. These results clearly suggested that in B. longum NCC2705 (and other strains), fructose is transported by a unique high affinity system, which is absent or inactivated in the fructose-negative strain B. longum DCP-18.
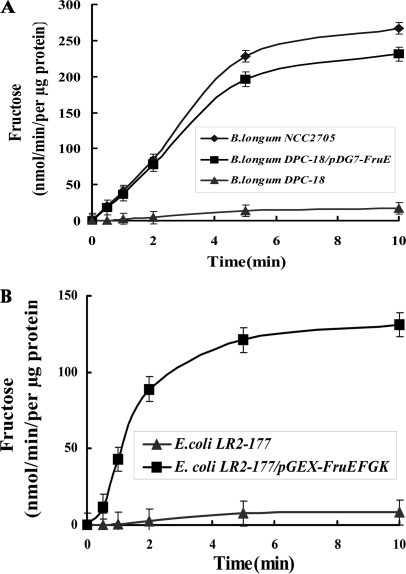
FIGURE 1.
Fructose uptake activity in B. longum and E. coli strains, binding assays of FruE with different substrates and quantitative assay of FruE sugar binding. A, fructose uptake activity in B. longum NCC2705 wild type (♦), DCP-18 (▴), and DCP-18/pDG7-FruE (■). Bacteria were grown in MGM with glucose (1 g/liter) as the sole carbon source, harvested at an A600 of 0.5, and assayed for uptake of [U-14C]fructose, which was added to the assay at a final concentration of 10 mm. B, fructose uptake of E. coli LR2–177 (▴) and LR2–177/pGEX-FruEKFG (■). E. coli strains were grown in LB medium with glucose (1 g/liter), and expression of FruEKFG was induced with 0.05 mm IPTG. All of the values are the means of three independent cultures. The error bars indicate the standard deviation.
The presence of all four genes of the operon encoding our putative fructose-specific ABC transporter (fruEKFG) was analyzed by PCR on isolated genomic DNA. All strains harbored homologues of fruK, fruF, and fruG. By contrast, B. longum DCP-18 was the only strain that did not yield a PCR product for fruE (supplemental Table S1), suggesting that this strain lacks a functional fruE gene. To probe whether this activity is related to the putative sugar-binding protein FruE, fruE was cloned into pDG7 under the control of its native promoter and transformed into B. longum DCP-18. Interestingly, the recombinant strain B. longum DCP-18 (pDG7-FruE) was able to grow on fructose as sole carbon source and, more importantly, was able to transport fructose with almost the same efficiency as B. longum NCC2705 (Fig. 1A).
Additionally, the full-length operon fruEKFG was cloned into pGEX-4T-1, and the resulting plasmid transformed into E. coli LR2–177, a strain that is not able to transport fructose (23). This successfully complemented the fructose-negative phenotype of E. coli LR2–177 (Fig. 1B). Collectively these data strongly suggest that fruEKFG encode for a functional ABC transporter specific for fructose.
Sequence Analysis of the Fructose Transporter Components
The putative ABC transporter is encoded by the four open reading frames fruEKFG preceded by conserved ribosome-binding sites. Transcriptional coupling is very likely in the case of fruK, fruF, and fruG, which are overlapping by two and three nucleotides, whereas fruE and fruK are separated by an intergenic region of 141 bp. In silico analysis of this intergenic region between fruE and fruK did not detect putative transcriptional terminator structures in this intergenic region. This suggests that fruEKFG form an operon. The deduced amino acid sequence of the four gene products showed significant similarity to the E. coli proteins RbsB (FruE), RbsA (FruK), and RbsC (FruF and FruG), which form a high affinity ribose-specific ABC transporter (supplemental Fig. S1, A–C).
A search for conserved domains/motifs revealed that FruE contains a conserved periplasmatic sugar-binding domain (PFAM PF00532, amino acid residues 40–268) and a Gram-positive lipoprotein precursor signal peptide with a predicted cleavage site between amino acid residues Gly-24 and Ser-25 (supplemental Fig. S1A).
The FruK protein contains a duplicate ATP-binding cassette domain (PFAM PF00005, residues 47–173 and 305–433) and the ATP-binding and hydrolysis motifs Walker A (GXXGXGKS, residues 40–47) and Walker B (X4DEPT, residues 166–173 and 426–433) at the end of the ATP-binding cassettes, as well as the conserved signature motif (LSGGNQ(Q/R)Q, residues 406–412), which are all specific features of the ATP-binding subunits of ABC transporters (supplemental Fig. S1B) (15, 32).
Both FruF and FruG display a ABC transporter permease domain (PFAM PF02653, FruF: residues 60–325; FruG: residues 55–326) and seven and eight transmembrane helices, respectively (supplemental Fig. S1C). Moreover, by aligning the amino acid sequence of FruF to other known membrane permeases of ABC sugar transport systems identified in of Gram-positive bacteria, a conserved EAA loop was identified (supplemental Fig. S1D). The resistance of soluble FruF and FruG against proteinase K was tested, revealing that both proteins were resistant both at 37 and 42 °C as described previously for MalF of E. coli (29).
Thus, FruE, FruK, FruG, and FruF are the sugar-binding periplasmic protein, the ATP-binding cytoplasmic protein, and the integral membrane proteins (permease) of the putative ABC sugar transporter. We propose to rename this fructose transporter into FruEKFG according to the name of the maltose uptake system (MalEFGK) of E. coli and S. typhimurium.
Sugar Substrate Binding Specificity of FruE
Based on the similarity to the ribose ABC transporter of E. coli, Parche et al. (17) suggested that this system may transport ribose or ribose-containing oligosaccharides in B. longum NCC2705. However, at the same time, the authors observed that this operon was induced in the presence of fructooligosaccharide, which is in contrast to the predicted substrate specificity (17). Recently, we identified FruE as one of the proteins that are induced when B. longum NCC2705 is grown in the presence of fructose, ribose, and xylose (20). We thus analyzed the substrate specificity of FruE in more detail.
To demonstrate FruE sugar binding activity, a GST-FruE fusion protein was incubated with fructose, ribose, xylose, or glucose and analyzed by capillary electrophoresis. Furthermore, the binding intensity of the substrate was assayed, and unfused GST was used as a negative control in all experiments as described in “Experimental Procedures.” A specific peak of FruE-fructose, FruE-ribose, or FruE-xylose was observed at ∼2 min of retention time when GST-FruE was incubated with a 5 mm excess of fructose, ribose, and xylose, respectively (Fig. 2A). This peak was absent when GST-FruE was incubated with glucose and GST alone, suggesting that the binding of fructose, ribose, and xylose by FruE was specific. However, the binding peaks were slightly weaker when GST-FruE was incubated with ribose and xylose compared with fructose. The binding intensity of FruE with fructose was 2.5 mmol (0.45 mg) fructose/g of GST-FruE (Fig. 2B) as assayed quantitatively using the glucose/fructose kit. Glucose was not bound by FruE under any condition tested (Fig. 2B).
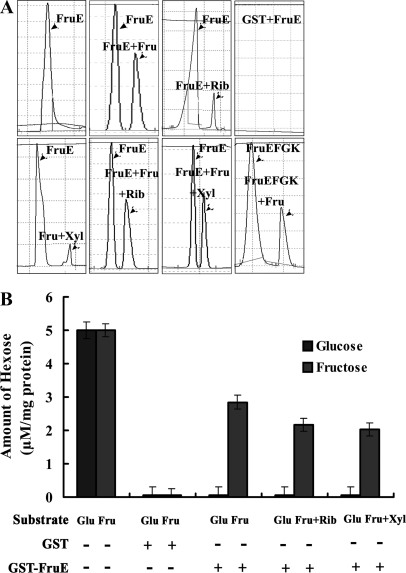
FIGURE 2.
Sugar binding of GST-FruE. A, specific sugar binding of purified GST-FruE with glucose, fructose, ribose, and xylose measured by capillary electrophoresis. Peaks corresponding to FruE, FruE + glucose, FruE + fructose, FruE + ribose, FruE + xylose, FruE + fructose, and FruEFGK + fructose with the excess of ribose or xylose are indicated. B, quantitative assay of glucose (dark gray bars) and fructose (light gray bars) bound to purified recombinant GST-FruE protein. 5 μm glucose or fructose were used in all assays. To test the accuracy of the assay, 5 μm glucose or fructose were assayed without any protein. GST alone was not bound by any of the two sugars. For the competitive binding, 5 μm ribose or xylose were added to the assays additionally to fructose/glucose.
Because binding of ribose and xylose to FruE suggests that these sugars might also transported by the FruEKFG system, we tested binding of fructose to FruE in the presence of competing ribose or xylose. The peak of FruE-fructose in the presence of an excess of ribose or xylose was similar to the FruE-fructose without the presence of other sugars (Fig. 2A). However, a slight reduction of the amount of fructose bound to FruE was observed in the presence of ribose or xylose as a competitor (Fig. 2B). We thus quantitatively assessed the inhibition of fructose binding activity of FruE by different sugars using [U-14C]fructose. The addition of cold ribose and xylose inhibited fructose binding activity of FruE by 52 and 46%, respectively, when using a 50-fold excess of competitor (Table 2). The addition of glucose, galactose, and mannose as competitors at the same excess ratio only had minor effects on [U-14C]fructose binding. These results suggest that B. longum NCC2705 uses the FruEKFG ABC transport system for uptake of fructose, ribose, and xylose with fructose as the preferred substrate.
TABLE 2.
Effect of various competitors on fructose uptake
| Competitor | Mean inhibition of uptakea |
|---|---|
| % | |
| Negative control | 0 |
| d-Glucose | 12 |
| d-Mannose | 10 |
| d-Galactose | 12 |
| d-Ribose | 52 |
| d-Xylose | 46 |
| d-Fructose | 99 |
a The results are expressed as percentages of inhibition of fructose uptake and are means of five measurements from two independent experiments, with a variation of less than 5%. Uptake was realized with [U-14C]fructose at 50 μm and 50-fold excess of unlabeled competitors. The control value was 26 nmol of fructose transported/min/mg of protein.
FruE Is Induced Specifically and Reversibly by Fructose
FruE is a probable sugar-binding protein of an ABC transporter system that mediates the first step of sugar uptake. To further investigate whether the FruE expression is induced specifically and reversibly by its substrates, we compared its expression both at the protein and transcriptional levels in cells grown on fructose, ribose, xylose, or glucose in the early exponential phase (8 h), the midexponential phase (12 h), the end of exponential phase (16 h), and the stationary phase (24 h). The different time points were determined by recording bacterial growth curves. To assess the protein abundance, two-dimensional PAGE was performed. Using our previously published proteome reference map of B. longum NCC2705 (25), the spots representing FruE were searched on the two-dimensional gel (Fig. 3), and their relative intensities were quantified. To verify that these spots really represent FruE, the spots were then excised, and protein was extracted and analyzed by MALDI-TOF/TOF MS/MS and electrospray ionization MS/MS. This identified all spots nonambiguously as FruE by several internal peptides (supplemental Table S2).
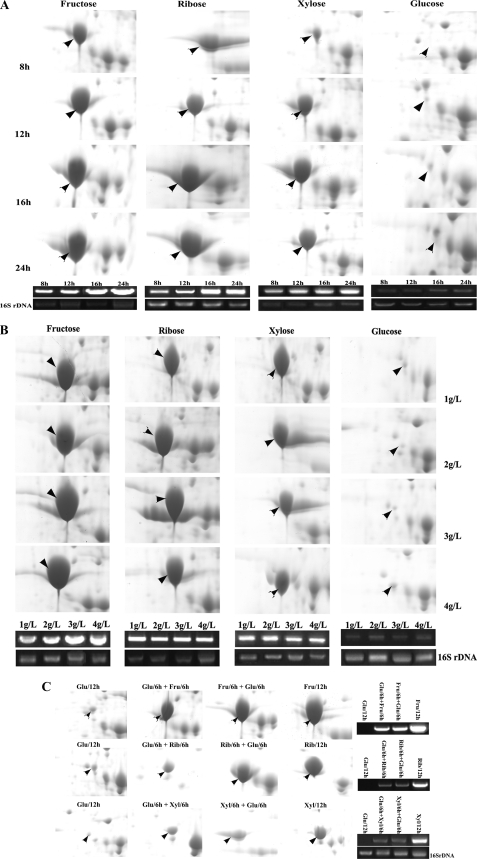
FIGURE 3.
Specificity and reversibility of induction of FruE by fructose, ribose, and xylose. A, spots corresponding to FruE in two-dimensional proteome maps (upper panels) and the RT-PCR-amplified fragments of fruE and 16 S rDNA (lower panels) of B. longum NCC2705 grown for 8, 12, 16, and 24 h in MGM with fructose (Fru), ribose (Rib), xylose (Xyl), or glucose (Glu) as the sole carbon source. B, two-dimensional proteome maps zoomed in on the FruE spots and RT-PCR targeting fruE of B. longum NCC2705 grown to midexponential growth phase in MGM with 1, 2, 3, or 4 g/liter of fructose, ribose, or xylose, respectively. C, FruE spots on two-dimensional proteome maps and RT-PCR targeting fruE of B. longum NCC2705 grown on MGM containing any of the substrates for 6 h and subsequently changed to a medium containing another sugar.
Clearly, expression of FruE was increased over time during growth on MGM containing fructose, ribose, or xylose as the sole carbon source, whereas in glucose-grown cells FruE is barely detectable (Fig. 3A, upper panels). To corroborate these results, RT-PCR analysis was performed targeting the fruE mRNA. This confirmed the results obtained in two-dimensional PAGE analysis on the transcriptional level (Fig. 3A, lower panels). This suggests that expression of FruE is either repressed in the presence of glucose or induced by fructose, ribose, and xylose.
We then performed experiments with B. longum NCC2705 cells grown to midexponential growth phase in MGM supplemented with different concentrations of fructose, ribose, and xylose (1, 2, 3, or 4 g/liter). Interestingly, both protein and transcript levels of FruE increased with increasing fructose concentration, whereas no concentration-dependent increase was observed for ribose, xylose, and glucose (Fig. 3B). This suggests that expression of FruE is induced in the presence of fructose in a dose-dependent manner.
Finally, we tested the reversibility of the FruE induction by fructose, ribose, and xylose. B. longum NCC2705 cells cultured on glucose, fructose, ribose, or xylose for 6 h were harvested, washed by PBS, split in two, and resuspended in fresh medium with the previous or any of the other three substrates. The two cultures were then further incubated for 6 h and analyzed for FruE protein or fruE transcript levels by two-dimensional PAGE and RT-PCR. As observed in our previous experiments, expression of FruE was strongly induced when bacteria were grown in the presence of its substrate fructose, ribose, and xylose but not on glucose. Furthermore, expression of FruE was greatly reduced both on the transcript and protein level when these inductors were withdrawn from the culture medium (Fig. 3C), indicating a reversible mechanism of regulation by these sugars.
Protein-Protein Interactions of the FruEKFG ABC Transporter Complex
In most bacteria, typical ABC sugar transporters are composed of the following components: a secreted or membrane-anchored substrate-binding protein, a membrane-integral permease, and an ATPase subunit (9, 10, 15). Within the fruEKFG operon, all of the genes required for a fully functional ABC transport system are present. Both the sugar-binding protein and the ATPase subunit must interact with the membrane-spanning permease(s) to transmit conformational changes evoked by hydrolysis of ATP and subsequently leading to import of the substrate. However, whether or not the proteins encoded by fruE–G constitute a functional ABC transporter has yet to be demonstrated.
Thus, we cloned and expressed GST fusions of all proteins of the system. Additionally, His6-tagged FruE and FruK proteins were obtained. All of the fusion proteins were found to be preferentially expressed in soluble form in the cytoplasm of the E. coli strain used for expression (Fig. 4A). All of the proteins were purified by standard affinity chromatography using their respective tags (Fig. 4A).
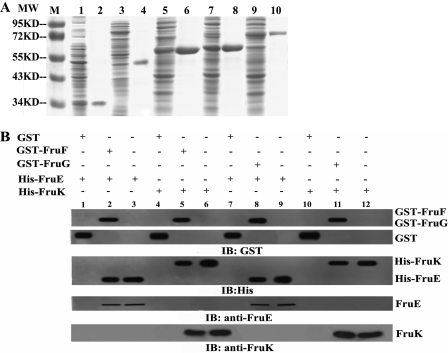
FIGURE 4.
SDS-PAGE and GST pulldown assays to analyze protein-protein interactions of the FruEKFG ABC transporter subunits. A, Coomassie-stained SDS-PAGE of crude extracts (lanes 1, 3, 5, 7, and 9; 30 μg of protein were loaded per sample) and purified proteins (lanes 2, 4, 6, 8, and 10; 5–10 μg of purified protein was loaded per sample) of IPTG-induced E. coli BL21(DE3) containing pET32a-FruE (lanes 1 and 2), pET32a-FruK (lanes 3 and 4), pGEX-4T-1-FruF (lanes 5 and 6), pGEX-4T-1-FruG (lanes 7 and 8), and pGEX-4T-1-FruE (lanes 9 and 10). Lane M, molecular weight marker. The proteins were purified by Ni+ affinity column (His-FruE and His-FruK) or GST beads (GST-FruF, GST-FruG, and GST-FruE). B, GST pulldown assays probing interactions between FruE or FruK with the membrane permeases FruF and FruG. For pulldown, 25 μg of GST fusion protein was incubated with 5 μl of glutathione-Sepharose 4B beads for 2 h in PBS at 4 °C. Then 200 μl of lysate containing a total of 25 μg of protein of an E. coli BL21 strain expressing the respective His6-tagged protein were added, and the bound proteins were precipitated by centrifugation. Lane 1, GST + His-FruE (negative control); lane 2, GST-FruF + His-FruE; lane 3, His-FruE (positive control); lane 4, GST + His-FruK (negative control); lane 5, GST-FruF + His-FruK; lane 6, His-FruK (positive control); lane 7, GST + His-FruE (negative control); lane 8, GST-FruG + His-FruE; lane 9, His-FruE (positive control); lane 10, GST + His-FruK (negative control); lane 11, GST-FruF + His-FruK; Lane 12, His-FruK (positive control). IB, immunoblot.
These proteins were then tested for interaction with each other using GST pulldown experiments in which GST-tagged proteins were incubated with cellular lysates of E. coli BL21 expressing His6 fusions of the respective interaction partner. Adsorbates were probed with anti-His, anti-GST, anti-FruE, or anti-FruK antibodies. GST alone was used as a negative control, and purified protein was used as a positive control. As expected, both the sugar-binding protein FruE and the ATPase subunit FruK showed an interaction with FruF and FruG (Fig. 4B).
Conclusion
The ability of the Bifidobacterium species to survive and persist in the competitive environment of the intestinal tract is correlated with their capacity to utilize fermentable carbohydrates not absorbed and metabolized by the host. Although bifidobacteria have been studied for over a century, the lack of genetic tools has prevented a comprehensive and coherent view of their biosynthetic capabilities. Until now, very little was known about fructose uptake in bifidobacteria. Basically, glucose transport in B. longum was characterized as an active nonphosphorylating process (16). We recently identified BL0033 as a protein that is highly expressed in B. longum NCC2705 grown on fructose, ribose, or xylose (20), which is part an operon with similarity to a ribose-specific ABC transporter of E. coli (17).
In this study, we have characterized the fruEKFG operon and the functionality of the encoded proteins as an ABC sugar transporter. This system displays many of the characteristics of binding protein-dependent ABC transporters with a soluble periplasmic substrate-binding protein (BL0033/FruE), integral membrane proteins (permeases, BL0035/FruF and Bl0036/FruG), and an energy-transducing ATP-binding cassette protein (BL0034/FruK). These proteins display the invariably conserved motifs found in bacterial ABC transporters.
The high homology of the components of the FruEKFG proteins of B. longum NCC2705 with the ribose ABC transporter of E. coli suggests a common origin. This hypothesis is strengthened by (i) the presence of all classical components of an ABC transport system in the fruE-G locus and (ii) the apparent ability of the B. longum system to transport ribose and xylose, as shown by uptake and binding competition experiments.
To show that FruEKFG constitutes a functional fructose-specific ABC transporter, we screened a large number of B. longum strains for the inability to grow on fructose as the sole carbon source. Of 45 strains screened a single strain (B. longum DCP-18) exhibited a fructose-negative phenotype. In addition to the inability to ferment fructose, this strains also lacks a functional fruE gene and is deficient for fructose transport across the cytoplasmatic membrane. Complementation of this strain with a plasmid-based fruE gene restored fructose transport and fermentation. Similar results were obtained with E. coli LR2–177, a strain that cannot transport fructose and thus does not grow on this sugar.
We further could show that FruE is able to bind to fructose, ribose, and xylose. By using competition assays, we observed that fructose binding of FruE is inhibited only partially in the presence of an excess of ribose or xylose, suggesting that FruE has affinity to all three sugars with a preference of fructose over the other two substrates.
Expression analysis of FruE by two-dimensional SDS-PAGE and RT-PCR revealed that FruE is induced specifically and reversibly by fructose both in a time- and dose-dependent manner. Collectively, our data suggest that FruEKFG constitute a functional ABC transporter with high affinity to fructose. We thus propose to rename the genes bl0033–0036 and the encoded proteins BL0033–0036 as fruEKFG and FruEKFG, respectively.
Furthermore, we found that both the sugar-binding protein FruE and the ATPase subunit FruK interacted with the two membrane permeases. Thus, the interactions observed support a conformation of the FruEKFG holocomplex in which FruE is located on the cell surface and interacts with the permeases. Intracellularly FruK would then be bound to the permeases and upon ATP hydrolysis evokes conformational changes leading to fructose import.
Most described fructose transporters belong to the PTS family. The only known fructose-specific ABC transporter was identified in Sinorhizobium meliloti (33). To our knowledge, the system described here is the first report on a fructose-specific ABC transport system in Gram-positive bacteria. Our analysis extends previous studies on the physiological characteristics and supports the hypothesis regarding the adaptation to the human gastrointestinal tract, as formulated by Schell et al. (5). More importantly, our data confirmed that the Fru-ABC transporter system could clearly serve to import fructose and with lower affinity of ribose and xylose. Our study on the transporter and metabolism of fructose and other sugars will hopefully facilitate and stimulate further in-depth research to elucidate the nutritional lifestyle of B. longum NCC2705 and other bifidobacteria.
Supplementary Material
Acknowledgments
We are indebted to the Nestlé Research Center for kindly providing B. longum NCC2705 and helpful information. We are grateful to Feng Liu for technical assistance and helpful discussions.
This work was supported by National Natural Science Foundation of China Grant 81071321, Mega-projects of Science and Technology Research of China Grant 2009ZX10004-205, and National High Technology Research and Development Program of China 863 Program Grant 2007AA02Z118.

This article contains supplemental Tables S1 and S2 and Fig. S1.
- PTS
- phosphotransferase system
- ABC
- ATP-binding cassette
- MGM
- modified Garches medium
- IPTG
- Isopropyl β-d-1-thiogalactopyranoside
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate.
REFERENCES
- 1. Brigidi P., Vitali B., Swennen E., Bazzocchi G., Matteuzzi D. (2001) Res. Microbiol. 152, 735–741 [DOI] [PubMed] [Google Scholar]
- 2. Young S. L., Simon M. A., Baird M. A., Tannock G. W., Bibiloni R., Spencely K., Lane J. M., Fitzharris P., Crane J., Town I., Addo-Yobo E., Murray C. S., Woodcock A. (2004) Clin. Diagn. Lab. Immunol. 11, 686–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zimmermann M. B., Chassard C., Rohner F., N'goran E. K., Nindjin C., Dostal A., Utzinger J., Ghattas H., Lacroix C., Hurrell R. F. (2010) Am. J. Clin. Nutr. 92, 1406–1415 [DOI] [PubMed] [Google Scholar]
- 4. He T., Roelofsen H., Alvarez-Llamas G., de Vries M., Venema K., Welling G. W., Vonk R. J. (2007) J. Microbiol. Methods 69, 364–370 [DOI] [PubMed] [Google Scholar]
- 5. Schell M. A., Karmirantzou M., Snel B., Vilanova D., Berger B., Pessi G., Zwahlen M. C., Desiere F., Bork P., Delley M., Pridmore R. D., Arigoni F. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tannock G. W., Munro K., Bibiloni R., Simon M. A., Hargreaves P., Gopal P., Harmsen H., Welling G. (2004) Appl. Environ. Microbiol. 70, 2129–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Caescu C. I., Vidal O., Krzewinski F., Artenie V., Bouquelet S. (2004) J. Bacteriol. 186, 6515–6525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Henderson P. J., Baldwin S. A., Cairns M. T., Charalambous B. M., Dent H. C., Gunn F., Liang W. J., Lucas V. A., Martin G. E., McDonald T. P. (1992) Int. Rev. Cytol. 137, 149–208 [PubMed] [Google Scholar]
- 9. Saier M. H., Jr (2000) Mol. Microbiol. 35, 699–710 [DOI] [PubMed] [Google Scholar]
- 10. Bordignon E., Grote M., Schneider E. (2010) Mol. Microbiol. 77, 1354–1366 [DOI] [PubMed] [Google Scholar]
- 11. Grote M., Polyhach Y., Jeschke G., Steinhoff H. J., Schneider E., Bordignon E. (2009) J. Biol. Chem. 284, 17521–17526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jacso T., Grote M., Daus M. L., Schmieder P., Keller S., Schneider E., Reif B. (2009) Biochemistry 48, 2216–2225 [DOI] [PubMed] [Google Scholar]
- 13. Oldham M. L., Khare D., Quiocho F. A., Davidson A. L., Chen J. (2007) Nature 450, 515–521 [DOI] [PubMed] [Google Scholar]
- 14. Orelle C., Ayvaz T., Everly R. M., Klug C. S., Davidson A. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12837–12842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Higgins C. F. (1992) Annu. Rev. Cell Biol. 8, 67–113 [DOI] [PubMed] [Google Scholar]
- 16. Briczinski E. P., Phillips A. T., Roberts R. F. (2008) Appl. Environ. Microbiol. 74, 6941–6948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Parche S., Amon J., Jankovic I., Rezzonico E., Beleut M., Barutçu H., Schendel I., Eddy M. P., Burkovski A., Arigoni F., Titgemeyer F. (2007) J. Mol. Microbiol. Biotechnol. 12, 9–19 [DOI] [PubMed] [Google Scholar]
- 18. Parche S., Beleut M., Rezzonico E., Jacobs D., Arigoni F., Titgemeyer F., Jankovic I. (2006) J. Bacteriol. 188, 1260–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kornberg H. L., Prior T. I. (1989) FEMS Microbiol. Rev. 5, 193–200 [DOI] [PubMed] [Google Scholar]
- 20. Liu D., Wang S., Xu B., Guo Y., Zhao J., Liu W., Sun Z., Shao C., Wei X., Jiang Z., Wang X., Liu F., Wang J., Huang L., Hu D., He X., Riedel C. U., Yuan J. (2011) Proteomics 11, 2628–2638 [DOI] [PubMed] [Google Scholar]
- 21. Fukuda S., Toh H., Hase K., Oshima K., Nakanishi Y., Yoshimura K., Tobe T., Clarke J. M., Topping D. L., Suzuki T., Taylor T. D., Itoh K., Kikuchi J., Morita H., Hattori M., Ohno H. (2011) Nature 469, 543–547 [DOI] [PubMed] [Google Scholar]
- 22. De Man J. C., Rogosa M., Sharpe M. E. (1960) J. Appl. Microbiol. 23, 130–135 [Google Scholar]
- 23. Aulkemeyer P., Ebner R., Heilenmann G., Jahreis K., Schmid K., Wrieden S., Lengeler J. W. (1991) Mol. Microbiol. 5, 2913–2922 [DOI] [PubMed] [Google Scholar]
- 24. Yuan J., Zhu L., Liu X., Li T., Zhang Y., Ying T., Wang B., Wang J., Dong H., Feng E., Li Q., Wang J., Wang H., Wei K., Zhang X., Huang C., Huang P., Huang L., Zeng M., Wang H. (2006) Mol. Cell Proteomics 5, 1105–1118 [DOI] [PubMed] [Google Scholar]
- 25. Yuan J., Wang B., Sun Z., Bo X., Yuan X., He X., Zhao H., Du X., Wang F., Jiang Z., Zhang L., Jia L., Wang Y., Wei K., Wang J., Zhang X., Sun Y., Huang L., Zeng M. (2008) J. Proteome Res. 7, 375–385 [DOI] [PubMed] [Google Scholar]
- 26. Matsuki T., Watanabe K., Tanaka R., Fukuda M., Oyaizu H. (1999) Appl. Environ. Microbiol. 65, 4506–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rossi M., Brigidi P., Matteuzzi D. (1998) Lett. Appl. Microbiol. 26, 101–104 [DOI] [PubMed] [Google Scholar]
- 28. Frangioni J. V., Neel B. G. (1993) Anal. Biochem. 210, 179–187 [DOI] [PubMed] [Google Scholar]
- 29. Ehrle R., Pick C., Ulrich R., Hofmann E., Ehrmann M. (1996) J. Bacteriol. 178, 2255–2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zdobnov E. M., Apweiler R. (2001) Bioinformatics 17, 847–848 [DOI] [PubMed] [Google Scholar]
- 31. Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011) Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 32. Saurin W., Dassa E. (1994) Protein Sci. 3, 325–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lambert A., Østerås M., Mandon K., Poggi M. C., Le Rudulier D. (2001) J. Bacteriol. 183, 4709–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.