Background: Deactivation of the JAK1/STAT3 pathway is tightly controlled in cells.
Results: CUEDC2 inhibits JAK1/STAT3 signaling through binding to SOCS3.
Conclusion: CUEDC2 is a novel regulator of JAK1/STAT3 signaling.
Significance: Our study identified a novel potential mechanism of SOCS3-mediated suppression on the JAK/STAT signaling pathway and provided important insight into the critical roles of CUEDC2 in the complex signal transduction network.
Keywords: JAK Kinase, Phosphorylation, Signal Transduction, STAT3, Ubiquitination, CUEDC2, Interaction, SOCS3
Abstract
Janus kinase 1/signal transducers and activators of transcription 3 (JAK1/STAT3) pathway is one of the recognized oncogenic signaling pathways that frequently overactivated in a variety of human tumors. Despite rapid progress in elucidating the molecular mechanisms of activation of JAK/STAT pathway, the processes that regulate JAK/STAT deactivation need to be further clarified. Here we demonstrate that CUE domain-containing 2 (CUEDC2) inhibits cytokine-induced phosphorylation of JAK1 and STAT3 and the subsequent STAT3 transcriptional activity. Further analysis by a yeast two-hybrid assay showed that CUEDC2 could engage in a specific interaction with a key JAK/STAT inhibitor, SOCS3 (suppressors of cytokine signaling 3). The interaction between CUEDC2 and SOCS3 is required for the inhibitory effect of CUEDC2 on JAK1 and STAT3 activity. Additionally, we found CUEDC2 functions collaboratively with SOCS3 to inhibit JAK1/STAT3 signaling by increasing SOCS3 stability via enhancing its association with Elongin C. Therefore, our findings revealed a new biological activity for CUEDC2 as the regulator of JAK1/STAT3 signaling and paved the way to a better understanding of the mechanisms by which SOCS3 has been linked to suppression of the JAK/STAT pathway.
Introduction
The Janus kinase/signal transducers and activators of transcription (JAK/STAT) pathway was originally described as a signal-transducing pathway induced by interferons (1). However, it has now been demonstrated that the JAK/STAT pathway mediates a multitude of distinct biological signals. JAK kinases are associated with cell surface receptors, such as cytokine and tyrosine kinase receptors. Binding of ligand triggers JAK-mediated phosphorylation of specific tyrosine residues in the cytoplasmic portion of the receptor. Cytoplasmic STAT proteins are recruited to the membrane by phosphorylated receptor and then phosphorylated by JAKs. Phosphorylated STATs then dimerize via their Src homology 2 (SH2)4 domains, translocate to the nucleus, and transactivate target genes (2, 3). JAK/STAT pathway regulates many cellular processes critical for hematopoiesis, immune response, and allelotaxy, including innate and adaptive immune function, development, proliferation, differentiation, apoptosis, and inflammation. Duration and degree of JAK/STAT activation are tightly controlled, and any deregulation will lead to disease, including tumor development (4).
The STAT protein family is composed of multiple members, termed STAT1–6. STAT3 was first described as a DNA-binding protein activated by epidermal growth factor and interleukin-6 (IL-6) capable of interacting with an enhancer element in the promoters of acute phase genes (5, 6). Later studies demonstrated that STAT3 is activated in response to several cytokines and growth factors, such as interferon (IFN) and leptin. In normal cells, STAT3 activation is transient like other STAT family members; however, in a large number of primary tumors and cancer-derived cell lines, it remains persistently activated, which may be caused by impairment of negative regulation or mutation of STAT3 itself (7, 8). Evidence indicates that constitutive activation of STAT3 may contribute to cellular transformation, induce tumor angiogenesis, and suppress anti-tumor immune responses, further enhancing tumor progression (9, 10). Therefore, STAT3 proteins are emerging as ideal targets for cancer therapy (9, 11, 12).
A large body of literature has been generated on the regulation of the JAK/STAT pathway. After well defining of the positive regulators of this signaling pathway, three main classes of proteins that negatively control the JAK/STAT pathway were concluded, including SOCS (suppressors of cytokine signaling) proteins, PIAS (protein inhibitors of activated stats) family proteins, and protein-tyrosine phosphatases (13). The SOCS family consists of eight members, including cytokine-inducible SH2 protein and SOCS-1–7. The SOCS proteins contain a central SH2 domain and a conserved carboxyl-terminal region called the SOCS box. Expression of SOCS proteins is rapidly induced by cytokine-mediated STAT activation (14) and subsequently leads to down-regulation of cytokine-induced signal transduction. Therefore, SOCS proteins act in a classic negative feedback loop to regulate JAK/STAT signal transduction. Despite the structural similarity of SOCS family members, they inhibit JAK/STAT signaling through different mechanisms. For example, cytokine-inducible SH2 protein, which was the first family member identified, was shown to compete with STAT5 for binding sites within the erythropoietin receptor (EpoR), thereby attenuating the JAK2/STAT5 signaling pathway (15). SOCS1 binds to the activation loop of JAKs via its SH2 domain and inhibits JAK kinase activity (16). The detailed mechanism by which SOCS3 functions to inhibit JAK/STAT signaling has not been well characterized. Some studies have reported that SOCS3 could also bind to and directly inhibit JAKs (17), whereas other reports suggest that it is necessary for SOCS3 to associate with the activated cytokine receptors to attenuate JAK/STAT signaling (18–22).
CUEDC2 is a CUE domain-containing protein. Previous work from our laboratory showed that CUEDC2 interacts with the progesterone receptor and promotes progesterone-induced proteasomal degradation of the progesterone receptor (23). More importantly, we found CUEDC2 is a crucial determinant of resistance to endocrine therapies in breast cancer through affecting estrogen receptor α protein stability (24). These functions provide insight into the mechanism by which CUEDC2 regulates breast cancer cells. In this study, we demonstrate that CUEDC2 inhibits JAK1/STAT3 activation by attenuating their phosphorylation. In addition, we identify CUEDC2 as a novel SOCS3 binding partner that stabilizes SOCS3 protein, resulting in suppression of JAK1/STAT3 signaling. Therefore, our novel findings suggest that CUEDC2 cooperates with SOCS3 to suppress the JAK/STAT signaling pathway.
EXPERIMENTAL PROCEDURES
Plasmid Constructions
HA-CUEDC2 full-length and truncated mutants were described as before (25). To generate bacterial expression vector for GST-CUEDC2 and the mutants, the corresponding CUEDC2 cDNAs (1–287, 1–133, 1–180,133–287, and 180–287 aa) were cloned in-frame into pGEX-KG vector (Amersham Biosciences). FLAG-SOCS3 and FLAG-SOCS3 (ΔSH2) were amplified by PCR from mammary library and cloned into pCDNA3.0 vector; other FLAG-SOCS3 deletion mutants were kindly provided by Dr. Fred Schaper (26). PACT-Luc and m67-Luc were, respectively, provided by Dr. Tarik Moroy (27) and Dr. Yong-Yun Kong (28). FLAG-STAT3 constructs were obtained from Dr. Darnell Jr., and GST-STAT3 was from Dr. XinMin Cao. IFN-γ activating sequence reporter was gift from Dr. Geoffrey L. Greene. JAK1 expression plasmid was kindly provided from Dr. Claude Haan. To generate Gp130 expression construct, a DNA fragment containing the intracytoplasmic domain of gp130 was amplified by PCR and inserted into PXJ40-HA vector.
Cell Culture, Transfection, and Reporter Gene Analysis
HEK293T cells and HeLa cells were cultured in DMEM containing 10% newborn calf serum at 37 °C in a humidified atmosphere of 5% CO2. HepG2 cells were maintained in MEM containing 10% fetal bovine serum. Cells were transfected using Lipofectamine 2000 (Invitrogen) following the manufacturer's instruction. Luciferase assays were carried out using the luciferase kit (Promega, Madison, WI) as described by the manufacturer's instructions, and luciferase activities were determined using a Dual-Luciferase Reporter Assay System (Promega). All experiments were repeated at least three times.
Immunoprecipitation, Immunoblotting, and Antibodies
For immunoprecipitation experiments, cells were lysed in E1A buffer (50 mm Hepes, pH 7.6, 250 mm NaCl, 0.1% Nonidet P-40, 5 mm EDTA) containing a mixture of protease inhibitors. Immunoprecipitations were performed by incubating whole cell extracts with the indicated antibody and rocking at 4 °C for 6 h after the preincubating with protein A/G-Sepharose (Santa Cruz Biotechnology),. Immunoprecipitates were washed 3 times and resuspended in 40 μl of 1× SDS sample buffer, then resolved by SDS-PAGE. All the samples for phosphorylation assays were prepared in M2 buffer. Mouse anti-HA antibody, rabbit anti-SOCS3 (sc-9023), and anti-STAT3 (sc-7179) antibodies were purchased from Santa Cruz Biotechnology. Anti-FLAG (M2) (F3165) monoclonal antibody was from Sigma. Rabbit anti-pSTAT3 (Tyr-705) (#9131), anti-JAK1 (#3332), and anti-pJAK1 (#1022/1023) (#3331) were purchased from cell signaling technology. CUEDC2 monoclonal antibody and GAPDH and GFP polyclonal antibodies were prepared in our laboratory.
GST Pulldown Assay
GST and GST fusion proteins were expressed in DH5α and purified according to the manufacturer's instructions (GE Healthcare). FLAG-SOCS3 protein, obtained from the whole cell lysates of HEK293T cells, which were transfected with FLAG-SOCS3 and/or its mutants plasmids, were incubated with GST and GST-CUEDC2 or its truncate fusion protein bound to agarose beads in 1 ml of binding buffer (20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA, 2% glycerol, and 0.1% Nonidet P-40) containing a protease inhibitor mixture at 4 °C for 6 h. Beads were then washed 3 times and resuspended in 30 μl of 1× SDS-PAGE sample buffer and detected by immunoblotting.
RNA Interference
The two small interfering RNAs (siRNA) that target CUEDC2 were purchased, respectively, from Invitrogen (#1, HSS149051) and Dharmacon (#2, J-014272-20); target sequences were, respectively, 5′-CCAAGAUGAGGCAACUGGCGCUGAG-3′ (#1) and 5′-CAUCAGAGGAGAACUUCGA-3′ (#2). For control siRNA against Photinus pyralis luciferase gene (Invitrogen), the target sequence was 5′-GGAUUUCGAGUCGUCUUAAUGUAUA-3′. Relative expression of endogenous CUEDC2 was detected by anti-CUEDC2 (from our laboratory). SOCS3 was selectively suppressed by using the RNA interference method, and the siRNA used for targeting human SOCS3 were: 5′-CCAAGAACCUGCGCATCCA-3′ and 5′-TGGATGCGCAGGTTCTTGG-3′ (29).
Stable Cell Lines Construction
The pSUPER retro shRNA retrovirus vector expressing CUEDC2 siRNA (target sequence 5′-GAAGCTGATCCGATACATC-3′; 5′- GTACATGATGGTGGATAGC-3′) were constructed by recombinant DNA technology. The packaging cells Phoenix (from ATCC) were transfected with these combinant plasmids using a liposome-based transfection method, and virus supernatant was collected and then infected into HeLa cells. The stable integrant was selected using G418 for 2 weeks.
For HepG2 cell lines stably expressing CUEDC2, CUEDC2 cDNA was inserted into pBabe-retro-puro retrovirus vector. HepG2 cells were infected with virus supernatant from Phoenix cells transfected with pBabe-GFP or pBabe-CUEDC2, and stable integrant was selected with puromycin for 2 weeks. HeLa cells stably expressing CUEDC2 were described as before (25).
RESULTS
CUEDC2 Inhibits STAT3 Transcriptional Activity
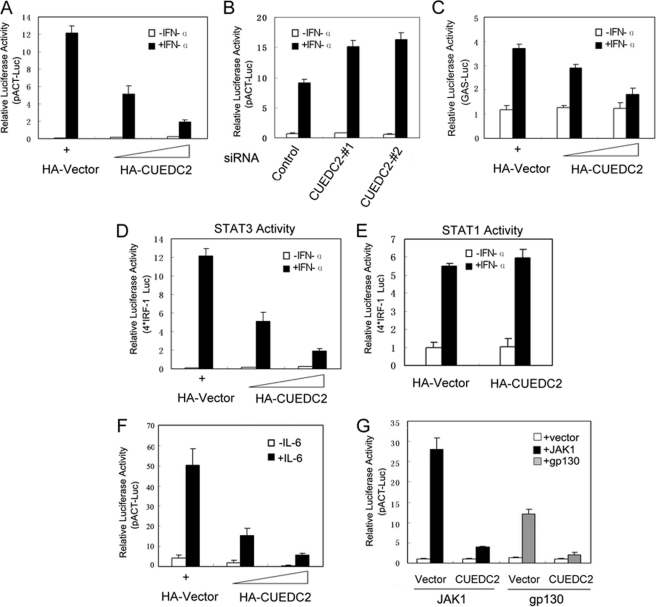
CUEDC2 interacts with progesterone receptor and estrogen receptor, leading to the ubiquitination and proteasome-dependent degradation of these two proteins (23, 24). To gain further insight into the function of CUEDC2 and elucidate other potential roles of CUEDC2 in cytokine-mediated signal transduction, we investigated whether CUEDC2 regulates other transcription factors that are also ubiquitinated. To investigate this possibility, reporter gene assays were used to determine the effect of CUEDC2 on the transcriptional activity of various transcription factors. STAT3 activity was shown to be affected by CUEDC2 in a screening assay.5 As shown in Fig. 1A, HEK293T cells were transfected with pACT-Luc (a luciferase reporter plasmid containing the promoter of α1-antichymotrypsin that is has two STAT3 binding sites) (27) and an increasing dose of HA-CUEDC2 constructs. Results show that CUEDC2 clearly inhibits IFN-α-induced STAT3 transcriptional activity in a dose-dependent manner. Similar results were also obtained in HeLa cells (supplemental Fig. 1A). To further confirm the role of endogenous CUEDC2 in STAT3 transcriptional activation, CUEDC2 expression was knocked down using two different siRNA in HEK293T cells. As anticipated, IFN-α-induced STAT3 activation was higher in CUEDC2 knockdown cells compared with control cells. Western blot analysis demonstrates that both of the two CUEDC2 siRNA, but not control siRNA, specifically reduced the expression of endogenous CUEDC2 (Fig. 1B and supplemental Fig. 1B). To exclude the nonspecific effect that may be caused by the α1-antichymotrypsin reporter gene, the same experiments were performed by using other STAT3-responsive luciferase reporter constructs of GAS (IFN-γ-activating sequence) reporter and m67 reporter (a synthetic STAT3-responsive promoter) (31, 32), and the results showed that CUEDC2 also inhibits the expression of these two reporters in a dose-dependent manner (Fig. 1C and supplemental Fig. 1C).
FIGURE 1.
CUEDC2 inhibits STAT3 transcriptional activity. A, HEK293T cells were transiently transfected in a 12-well plate with pACT luciferase reporter (200 ng), FLAG-STAT3 (200 ng), and increasing amounts of HA-CUEDC2 vectors (0, 200, and 500 ng) as indicated. 24 h after transfection, cells were stimulated with IFN-α (50 ng/ml) for an additional 6 h, and luciferase activity was measured. Renilla reporter pRL-TK (20 ng/well) vectors were used as an internal control for transfection efficiency. B, HEK 293T cells were transfected with control siRNA or the two different CUEDC2 siRNAs (20 nm) (#1 and #2). 24 h later pACT-Luc and FLAG-STAT3 plasmids were cotransfected as in A. After another 24 h, cells were treated with IFN-α for 6 h, and luciferase reporter assays were performed. C, HEK293T cells were transfected the same as in A, except the luciferase reporter gene was replaced by STAT3 responsive IFN-γ activating sequence reporter (500 ng/well). D and E, HEK293T cells were transfected with 4× IRF-1 luciferase reporter (500 ng/well) construct and STAT3 or STAT1(200 ng/well) together with or without CUEDC2 as indicated. Twenty-four hours after transfection, cells were left untreated (open columns) or treated with IFN-α (50 ng/ml) for 6 h, and luciferase activity was determined. F, HepG2 cells were transiently transfected with pACT-Luc (200 ng/well) together with increasing amounts of CUEDC2 vectors; 24 h after transfection cells were starved for 16–18 h in MEM with 0.5% serum then treated with IL-6 (100 ng/ml) for another 6 h, and luciferase activity was measured.G, JAK1 or glycoprotein 130 (gp130) expression vectors (200 ng/well) were co-transfected with pACT-Luc, FLAG-STAT3, and HA-CUEDC2 (or HA-vector) into HEK293T cells; 24 h later cells were harvested and detected for luciferase activity. All the results are the means ± S.E. of three independent experiments.
To test the specificity of CUEDC2 on STAT3 activation, a luciferase reporter construct (4× interferon regulatory factor-1 (IRF-1)) containing four copies of the STAT binding sequence from the IRF-1 gene were used (33). Cells cotransfected with STAT3 and (4× IRF-1) reporter showed a 200-fold increase of luciferase expression upon IFN-α treatment. In keeping with the above data, expression of CUEDC2 strongly inhibited IFN-α-induced STAT3-dependent gene expression. However, CUEDC2 had no such inhibitory effect on STAT1-mediated transcriptional activation in response to IFN-α (Fig. 1, D and E), indicating that CUEDC2 specifically inhibits STAT3 transcriptional activity. Because STAT3 is also the major mediator of IL-6 signaling, it was next investigated whether CUEDC2 plays a role in IL-6-mediated STAT3 activation. As shown in Fig. 1F, CUEDC2 expression decreases IL-6-induced STAT3 activation in HepG2 cells. Notably, our results also demonstrate that CUEDC2 can inhibit STAT3 activation triggered by forced expression of JAK1 or the intracytoplasmic domain of cell surface receptor glycoprotein 130 (gp130) (Fig. 1G). These data suggest that CUEDC2-mediated suppression of STAT3 activation is not restricted to a particular stimulation but may be a general mechanism of STAT3 regulation.
CUEDC2 Inhibits Phosphorylation of JAK1 and STAT3
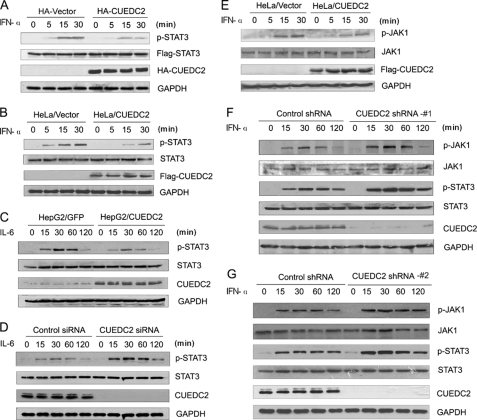
Phosphorylation of STAT3 at specific residues, particularly Tyr-705, is necessary for its activation. Therefore, it was next determined whether CUEDC2 inhibits STAT3 activity through affecting its phosphorylation. To this end, either HA-CUEDC2 or control vectors were co-transfected with FLAG-STAT3 into HEK293T cells. Time course analysis of STAT3 phosphorylation demonstrates that, in response to IFN-α treatment, the amount of phosphorylated STAT3 increased substantially in the cells transfected with a control vector (HA vector); in contrast, this phosphorylation of STAT3 was markedly decreased in cells transfected with the CUEDC2 vector (HA-CUEDC2) (Fig. 2A). To analyze the phosphorylation status of endogenous STAT3, HeLa cells stably expressing FLAG-CUEDC2 (HeLa/CUEDC2) or the control vector (HeLa/Vector) were treated with or without IFN-α for the indicated times, and cell lysate was resolved by SDS-PAGE and subjected to Western blot analysis. The results show that IFN-α-induced phosphorylation of STAT3 was much less in cells expressing CUEDC2 compared with control cells. Levels of total STAT3 protein were similar in both samples (Fig. 2B). Next we examined the influence of CUEDC2 on IL-6-induced STAT3 phosphorylation. In keeping with the luciferase reporter assay results, IL-6-induced phosphorylation of STAT3 at Tyr-705 was also reduced in HepG2 cells stably expressing CUEDC2 (HepG2/CUEDC2) compared with control cells expressing GFP (HepG2/GFP) (Fig. 2C). However, inhibition of CUEDC2 by siRNA in HepG2 cells led to an elevated level of phosphorylated STAT3 (Fig. 2D). Furthermore, the impact of CUEDC2 on JAK1 phosphorylation was also investigated in HeLa cells because of its major role in STAT3 activation. The results showed that, just as STAT3, IFN-α-induced JAK1 phosphorylation was lower in cells stably expressing CUEDC2 compared with the control cells (Fig. 2E). Consistent with these results, phosphorylation levels of JAK1 and STAT3 were elevated when CUEDC2 was knocked down by shRNA(#1) in HeLa cells (Fig. 2F). To rule out the off-target effect, we tested the kinetics of JAK1 and STAT3 phosphorylation with a different CUEDC2 shRNA(#2), and the same results were also obtained (Fig. 2G). Thus, these data indicate that CUEDC2 inhibits the phosphorylation of JAK1 and STAT3 and thus attenuates STAT3 transcriptional activation.
FIGURE 2.
CUEDC2 decreases STAT3 and JAK1 tyrosine phosphorylation. A, HEK293T cells in 6-well plates were transfected with FLAG-STAT3 (500 ng/well) together with HA-CUEDC2 or control vectors (1.5 μg/well). 24 h after transfection cells were stimulated by IFN-α (50 ng/ml) for the indicate periods, and total cell lysates were immunoblotted using anti-p-STAT3 (Tyr-705), anti-FLAG and anti-HA antibodies. B, HeLa/vector and HeLa/CUEDC2 cells were stimulated for 0–30 min with IFN-α (50 ng/ml), and endogenous STAT3 phosphorylation at Tyr-705 was detected by immunoblotting. C, HepG2 cells infected with retrovirus construction of pBabe-GFP(HepG2/GFP) or pBabe-CUEDC2(HepG2/CUEDC2) were starved for 16–18 h in MEM with 0.5% serum and then treated with IL-6 (100 ng/ml) for the indicated times. Cell lysates were analyzed by immunoblotting with indicated antibodies. D, HepG2 cells transfected with CUEDC2 siRNA(#1) or control siRNA were starved for 16–18 h in MEM with 0.5% serum and then treated with IL-6 (100 ng/ml) for the indicated times. Cell lysates were analyzed by immunoblotting, and STAT3 phosphorylation at Tyr-705 was detected. E, HeLa/vector and HeLa/CUEDC2 cells were treated the same as in B, phosphorylated JAK1 was detected by immunoblotting with anti-p-JAK1 (Tyr-1022/1023). F and G, cell extracts were prepared from HeLa cells with control or CUEDC2 shRNA (#1 or #2), then treated by IFN-α (50 ng/ml) for the indicated periods, and immunoblot analysis was performed.
CUEDC2 Associates with SOCS3
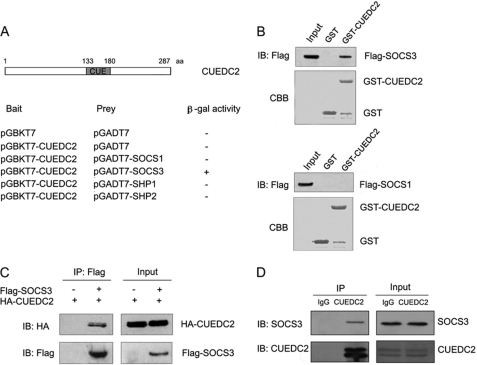
Because CUEDC2 inhibits JAK1 and STAT3 phosphorylation and the subsequent STAT3 transcriptional activity, was next determined whether CUEDC2 directly interacts with JAK1 and/or STAT3. However, we found that no association was observed between CUEDC2 and either JAK1 or STAT3, although the positive controls (JAK1 interacts with SOCS1 and STAT3 interacts with PIAS3) in the same assay worked well (supplemental Fig. 2) (33, 34). This indicates that CUEDC2 inhibits JAK1/STAT3 signaling by recruiting some other essential molecules. To further explore how CUEDC2 mediates JAK1 and STAT3 inhibition, it was determined whether SOCS or SHP (the SH2 domain-containing protein-tyrosine phosphatase) family members, which are key negative regulators of JAK/STAT signaling, interact with CUEDC2 in a yeast strain, AH109, by performing β-galactosidase assays. Interestingly, out of all the proteins tested, CUEDC2 only interacts with SOCS3 (Fig. 3A). The specificity of the interaction between CUEDC2 and SOCS3 was further confirmed by GST pulldown assays. Results show that substantial amounts of SOCS3 were pulled down by GST-CUEDC2 but not by GST alone (Fig. 3B, upper panel). Additionally, SOCS1, another member of the SOCS family proteins, could bind neither GST nor GST-CUEDC2 (Fig. 3B, lower panel). These results suggest that CUEDC2 specifically associates with SOCS3 in vitro.
FIGURE 3.
CUEDC2 interacts with SOCS3 in vitro and in vivo. A, the yeast strain AH109 was cotransformed with pGADT7-Gal4-AD, pGADT7-Gal4-AD-SOCS1'SOCS3, SHP1, or SHP2 plus the pGBKT7-Gal4-DBD or pGBKT7-Gal4-DBD-CUEDC2 as indicated. A cotransformant from each plate was streaked on SD/−Trp/−Leu/−His/−Ade medium with β-galactosidase (+, positive interaction; −, no interaction). At the top of the graph is a schematic diagram of the CUEDC2 protein. B, cell lysates of HEK293T cells expressing FLAG-tagged SOCS3 (up panel) or SOCS1 (down panel) were incubated with agarose beads coupled to GST alone or a fusion protein of GST and CUEDC2 (GST-CUEDC2), and the interacting proteins were detected with anti-FLAG. IB, immunoblot; CBB, Coomassie Brilliant Blue staining. C, HEK293T cells were transfected with vectors expressing HA-tagged CUEDC2(3 μg/well) and FLAG-tagged SOCS3(1 μg/well) as indicated. Whole cell lysates were then immunoprecipitated (IP) with anti-FLAG monoclonal antibodies and immunoblotted with anti-HA antibody. D, HeLa cell lysates were immunoprecipitated with mouse IgG or anti-CUEDC2 monoclonal antibodies, and immunoprecipitates were determined by immunoblotting with anti-SOCS3 and anti-CUEDC2 antibodies.
To further verify this interaction in eukaryotic cells, HA-CUEDC2 and FLAG-SOCS3 were co-expressed into HEK293T cells, and co-immunoprecipitation experiments were performed with anti-FLAG antibody and followed by Western blot analysis. Again, data show that CUEDC2 associates with SOCS3 specifically (Fig. 3C). As data from Fig. 3, A–C, demonstrate that these two proteins interact was performed with exogenous protein expression, we next examined whether CUEDC2 and SOCS3 interact under physiological conditions, To do so, cell extracts from HeLa cells were prepared, and immunoprecipitations were performed with anti-CUEDC2 antibody or mouse immunoglobulin G (IgG) control. Precipitates were resolved by SDS-PAGE followed by Western blot analysis using SOCS3 antibody. As shown in Fig. 3D, SOCS3 was detected in the immunoprecipitates obtained from cell extracts with antibody to CUEDC2 (anti-CUEDC2) but not with control IgG. Collectively, these data indicate that CUEDC2 interacts with SOCS3 in vitro and in vivo.
Interaction between CUEDC2 and SOCS3 Is Essential for CUEDC2-mediated Inhibition of STAT3 Activity
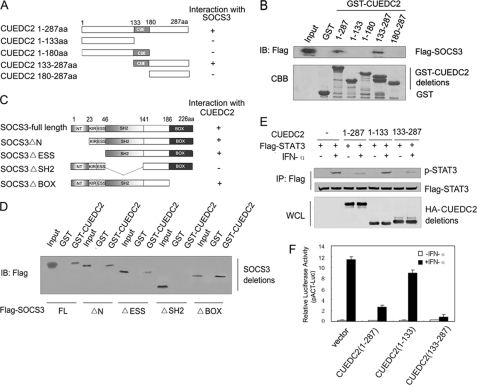
To delineate which region of CUEDC2 was responsible for association with SOCS3, a series of GST-CUEDC2 deletion mutants was constructed (Fig. 4A), and GST pulldown assays were performed in HEK293T cells. As indicated in Fig. 4B, only wild type CUEDC2 and the carboxyl-terminal region of CUEDC2 (133–287 aa) interacted with SOCS3; none of the other CUEDC2 mutants (1–133, 1–180, and 180–287aa) was enough to bind SOCS3. Therefore, these results revealed that association of CUEDC2 and SOCS3 requires the carboxyl-terminal region including the CUE domain. To analyze the binding domains of SOCS3 in better detail, SOCS3 deletion mutants, respectively, lacking the 23 amino-terminal amino acids (SOCS3 ΔN), the amino-terminal region identified as an extension of the SH2 domain (SOCS3 ΔESS), the SH2 domain (SOCS3 ΔSH2) and the carboxyl-terminal SOCS-box (SOCS3 ΔBox) were generated (Fig. 4C). These mutants were transiently expressed in HEK293T cells, and pulldown assays were performed. The results demonstrated that CUEDC2 associates with all the SOCS3 deletion mutants except SOCS3 ΔSH2 (Fig. 4D), suggesting that the SH2 domain of SOCS3 is essential for its interaction with CUEDC2.
FIGURE 4.
Interaction between CUEDC2 and SOCS3 is essential for inhibition of CUEDC2 on STAT3 activity. A, shown are deletion mutants of CUEDC2 used in domain-mapping experiments; numbers indicate amino acids included in constructs. B, purified GST or truncated GST-CUEDC2 fusion proteins were incubated with cell lysates from HEK293T cells transiently transfected with FLAG-SOCS3 vector. After extensive washes, bound proteins were analyzed by immunoblotting (IB) with anti-FLAG antibody. The GST fusion proteins were resolved by SDS-PAGE and stained with Coomassie Blue. C, shown is a schematic diagram depicting different SOCS3 deletion mutants used in the domain-mapping experiments. D, HEK 293T cells were transiently transfected with FLAG-SOCS3 or its truncated vectors, cell lysates were prepared and incubated with GST or GST-CUEDC2, and bound proteins were analyzed by immunoblotting with anti-FLAG antibody. E, HEK 293T cells were transfected with FLAG-STAT3 vectors (500 ng/well) and plasmids expressing full-length CUEDC2 or CUEDC2 deletion mutants (1.5 μg/well) and then treated with or without IFN-α (50 ng/ml) for 20 min. Lysates immunoprecipitated (IP) with anti-FLAG and whole-cell lysates were analyzed by immunoblotting with indicated antibodies. F, full-length CUEDC2 or CUEDC2 mutant expression vectors (500 ng/well) were cotransfected with pACT-Luc (200 ng/well) and FLAG-STAT3 (200 ng/well) into HEK293T cells and left treated with or without IFN-α (50 ng/ml) for 6 h, then luciferase reporter assays were performed. Data are representative of at least three independent experiments and are shown as means ± S.E.
Based on the results that CUEDC2 binds SOCS3, it was next determined whether interaction between CUEDC2 and SOCS3 was essential for CUEDC2 inhibition on STAT3 activity. To this end, the effect of each CUEDC2 mutant on IFN-α induced STAT3 phosphorylation was determined. As shown in Fig. 4E, there were much lower levels of STAT3 phosphorylation induced by IFN-α in cells overexpressing wild type CUEDC2 or CUEDC2 (133–287 aa) than the control vector, whereas robust phosphorylation of STAT3 was still observed in cells transfected with CUEDC2 (1–133 aa), which could not bind SOCS3. These results were further confirmed by luciferase reporter assays. Only those deletion mutants of CUEDC2 that can interact with SOCS3 were able to inhibit STAT3 transcriptional activity (Fig. 4F). These data suggests that negative regulation of STAT3 activation by CUEDC2 requires its interaction with SOCS3.
CUEDC2 Cooperates with and Requires SOCS3 to Inhibit STAT3 Activation
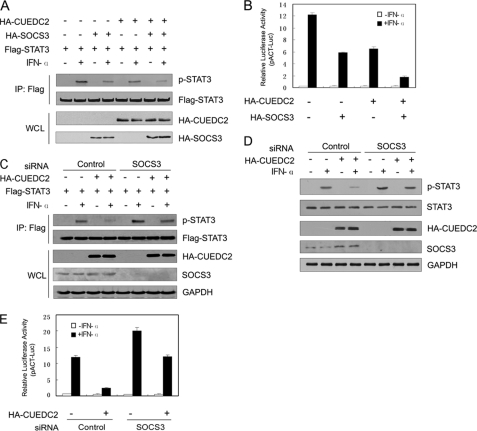
Given that the interaction with SOCS3 is essential for the ability of CUEDC2 to inhibit STAT3 activity, further characterization of this relationship was required. To define the requirement of SOCS3 in CUEDC2-mediated inhibition of STAT3 activity, HEK293T cells were co-transfected with FLAG-STAT3 and either SOCS3 and CUEDC2 alone or together. Cell lysates were immunoprecipitated with anti-FLAG, and Western blot analysis was performed. As shown in Fig. 5A, the levels of phosphorylated STAT3 were lower in cells expressing either CUEDC2 or SOCS3 (fourth lane and sixth lane, compared with the second lane). Notably, co-expression of both CUEDC2 and SOCS3 almost completely abrogated STAT3 phosphorylation (eighth lane compared with the second lane). Similar results were also obtained in luciferase reporter assays (Fig. 5B). These data indicate that CUEDC2 enhances SOCS3-mediated inhibition of STAT3 activation. Therefore, it is probable that SOCS3 mediates the repression of CUEDC2 on STAT3 activity. To test this possibility, SOCS3 was knocked down by siRNA, and the effect of CUEDC2 on STAT3 activity was examined. We found that the inhibition of CUEDC2 on STAT3 phosphorylation (either exogenous or endogenous) was partially recovered by SOCS3 knockdown (Fig. 5, C and D). Luciferase reporter assay further confirmed these data (Fig. 5E). Taken together, these results suggest that CUEDC2 cooperates with and requires SOCS3 to attenuate STAT3 activation.
FIGURE 5.
CUEDC2 cooperates with and requires SOCS3 to inhibit STAT3 activation. A, FLAG-STAT3 expression vectors (500 ng/well) were transfected alone or together with expression constructs of HA-CUEDC2 (500 ng/well) and/or HA-SOCS3 (100 ng/well) into HEK293T cells. 24 h after transfection, cells were stimulated with or without IFN-α (50 ng/ml) for 20 min, lysates were immunoprecipitated (IP) with anti-FLAG, and the immunoprecipitates and whole-cell lysates were analyzed by immunoblot as indicated. B, HEK 293T cells were transfected as in Fig. 1 with HA-CUEDC2 (100 ng/well) and/or HA-SOCS3 (20 ng/well) vectors. 24 h later cells were treated with IFN-α (50 ng/ml) for 6 h, and luciferase activity was determined and normalized for transfection efficiency. Data are shown as means ± S.E. C, HEK293T cells were transfected with either control siRNA or SOCS3 siRNA 24 h before the cotransfection CUEDC2 vectors (1.5 μg/well) and FLAG-STAT3 vectors (500 ng/well). After another 24 h, cells were treated with IFN-α (50 ng/ml) for 20 min and immunoprecipitated with anti-FLAG, and whole cell lysates (WCL) were analyzed by immunoblotting with indicated antibodies. D, HEK293T cells were transfected the same as in C but not with FLAG-STAT3, and phosphorylation of endogenous STAT3 was analyzed by immunoblotting. E, HEK293T cells were transfected with control siRNA or SOCS3 siRNA 24 h before the cotransfection CUEDC2 vectors (200 ng/well) and FLAG-STAT3 vectors (200 ng/well), and luciferase activity was determined and normalized for transfection efficiency. Data are shown as the means ± S.E.
CUEDC2 Stabilizes SOCS3 Protein by Protecting It from Degradation via Enhancing SOCS3-Elongin C Interaction
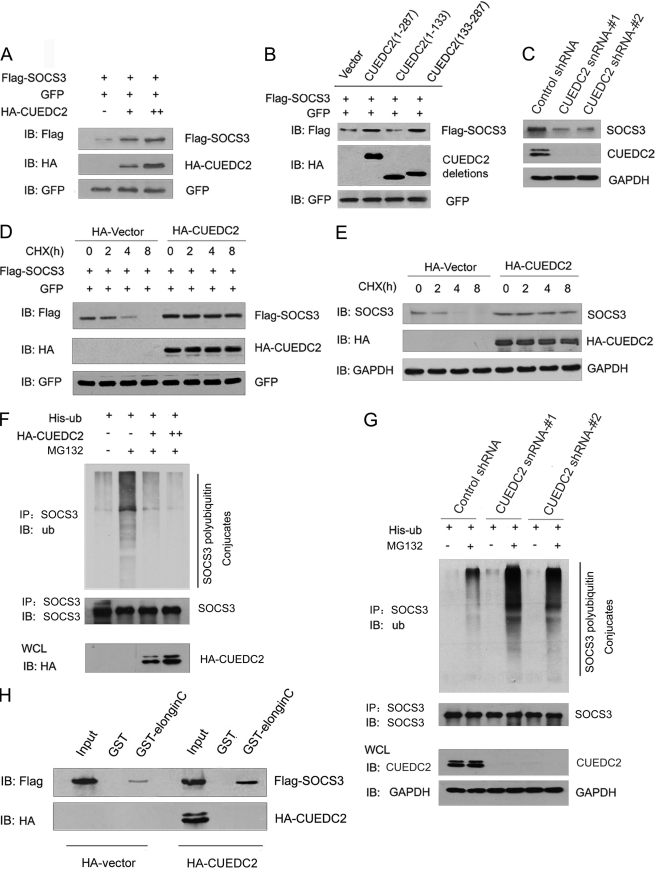
Because CUEDC2 inhibits JAK1/STAT3 signaling by forming a complex with SOCS3, how CUEDC2 and SOCS3 cooperated was next explored. We first detected the impact of CUEDC2 on SOCS3 protein level. As shown in Fig. 6A, CUEDC2 and SOCS3 expression vectors were cotransfected into HEK293T cells. GFP expression vector was included in these experiments as a control for transfection efficiency. The results show that overexpression of CUEDC2 increases SOCS3 protein levels in a dose-dependent manner. To investigate whether this up-regulation of CUEDC2 on SOCS3 protein level depends on their interaction, wild type and different deletion mutants of CUEDC2 constructs were transfected with SOCS3 and GFP vectors into HEK293T cells. We found that only the full-length CUEDC2 and CUEDC2 (133–287 aa), which can bind SOCS3, were able to increase SOCS3 levels. This indicates that the interaction of CUEDC2 and SOCS3 was essential for the effect of CUEDC2 on SOCS3 protein level (Fig. 6B). To further substantiate these findings, we examined whether loss of CUEDC2 affects the protein level of endogenous SOCS3. As shown in Fig. 6C, a decrease of SOCS3 protein levels was observed in HeLa cells with two different CUEDC2 shRNAs compared with the cells with control shRNA. As known, an increase in protein level can be a result of increased production or decreased degradation. To distinguish between these two possibilities, protein levels of SOCS3 were monitored after the addition of cycloheximide to block protein synthesis. The results show that the half-life of SOCS3 protein was significantly prolonged when CUEDC2 was overexpressed (Fig. 6, D and E). Therefore, it is most likely that CUEDC2 regulates the SOCS3 protein level through increasing its stability.
FIGURE 6.
CUEDC2 stabilizes SOCS3 by protecting it from degradation via enhancing SOCS3-Elongin C interaction. A, HEK293T cells were transfected with equal amounts of FLAG-SOCS3 (200 ng), GFP expression vector (50 ng), and increasing amounts of HA-CUEDC2 expression vectors (0, 0.5, and 1.5 μg). Cell lysates were subjected to immunoblotting (IB) using antibodies as indicated. Levels of GFP were shown as equal transfection efficiency. B, wild type or different deletions of CUEDC2 constructs were transfected into HEK293T cells, and the same assays were performed as in A. C, lysates from HeLa cells with control or CUEDC2 shRNA (#1 and #2) were subjected to immunoblotting using antibodies as indicated. D and E, HA-CUEDC2 or HA vectors were transfected into HeLa cells. 16 h after transfection, cells were treated with the protein synthesis inhibitor cycloheximide (CHX, 50 μg/ml), harvested at the indicated times, and then exogenous (D) and endogenous SOCS3 (E) protein levels were analyzed by immunoblotting. F, HeLa cells were transfected as indicated and treated with or without the proteasome inhibitor MG132 (10 μm) for an additional 6 h. Cell lysates were immunoprecipitated (IP) with anti-SOCS3 and detected with antibodies as indicated. ub, ubiquitin, G, HeLa cells with control or CUEDC2 shRNA (#1 and #2) were transfected and treated as indicated, and cell lysates were immunoprecipitated with anti-SOCS3 and detected with antibodies as indicated. H, HEK293T cells were cotransfected with HA-CUEDC2 (or HA-vectors) and FLAG-SOCS3, cell lysates were prepared, and input was normalized. After incubation with purified GST or GST-Elongin C fusion protein and extensive washes, bound proteins were analyzed by immunoblotting with anti-FLAG and anti-HA antibody. WCL, whole cell lysates.
Although the detailed mechanism of SOCS3 degradation and its regulation are still not fully known, it is generally accepted that SOCS3 is degraded by the proteasome-mediated pathway. A ubiquitylation assay showed that the CUEDC2 overexpression attenuated the level of ubiquitinated species of SOCS3 in the presence of MG132 (Fig. 6F), and knock down of CUEDC2 led to an increasing level of SOCS3 ubiquitination (Fig. 6G). Thus, these data suggest that CUEDC2 stabilizes SOCS3 protein by blocking its degradation via decreasing its ubiquitination. It has been shown that the interaction of SOCS3 with Elongin B and C can limit its turnover. Any factors that can block the interaction between SOCS family members and Elongin could promote SOCS degradation (35–37), so we next determined whether CUEDC2 affects the interaction between SOCS3 and Elongin B/C. To this end, SOCS3 was co-expressed in cells with CUEDC2 or control vector. Twenty-four hours after transfection, cell lysates were incubated with either GST or a GST-Elongin C fusion protein. As shown in Fig. 6H, more SOCS3 was pulled down in the presence of CUEDC2 as normalized to input. Taken together, our data demonstrate that CUEDC2 enhances the SOCS3-Elongin C interaction, attenuates SOCS3 ubiquitination, and facilitates its stabilization.
DISCUSSION
Recent studies have shown that the JAK/STAT signaling pathway can be regulated at multiple steps through distinct mechanisms. Suppressors of JAK/STAT signaling can be summarized as the following. First, cofactors of STATs, such as PIAS family proteins (33), GRIM-19, Daxx, and LMW-DSP2 (38–40), were reported to interact with STATs and repress their activation. Inhibition occurs either through blocking of STATs-DNA binding, preventing their nuclear import, or through undefined mechanisms. Second, protein-tyrosine phosphatase, including SHP1, SHP2, CD45, protein-tyrosine phosphatase-1b and so on, inhibit the JAK/STAT pathway by dephosphorylating cytokine receptors, JAKs, or some other essential pathway components (41, 42). Discovery of the SOCS proteins unveiled another mechanism of negative regulation of the JAK/STAT signaling. Distinct SOCS family members inhibit cytokine signaling through several different mechanisms. Many studies suggest that SOCS1 inhibits cytokine signaling by binding to JAKs and inhibiting their catalytic activity (34, 43). However, in the case of SOCS3, the detailed mechanisms are not mutually exclusive, and whether there are any other components involved in SOCS3-mediated inhibition remains poorly understood. In this study we found that CUEDC2 impairs JAK1 and STAT3 phosphorylation and inhibits STAT3 transcriptional activation. However, it seems that CUEDC2 does not act as a cofactor of STAT3-like PIAS3 or GRIM-19, as no interaction was observed between CUEDC2 and STAT3. Furthermore, results from β-galactosidase assays in yeast showed no association between CUEDC2 and SHP1.
Here, we demonstrate that CUEDC2 specifically interacts with SOCS3 and acts as a novel SOCS3 co-operator required for inhibition of JAK1/STAT3 signaling. Some studies have shown that SOCS3 inhibits the activity of JAKs by direct binding. However, compared with SOCS1, SOCS3 binds JAKs with much lower affinity, and significantly higher levels of SOCS3 must be expressed compared with SOCS1 for equivalent inhibition of JAK kinase activity (44, 45). In this study we show that CUEDC2 associates with and stabilizes SOCS3 protein; this up-regulation of SOCS3 via protein stabilization may increase the possibility of interaction between SOCS3 and JAKs, thus leading to JAK inhibition. This indicates a novel potential mechanism of SOCS3-mediated suppression of the JAK/STAT signaling. Notably, the interaction between CUEDC2 and SOCS3 is essential for CUEDC2-mediated deactivation of JAK1/STAT3 signaling. Loss of SOCS3 expression counteracts CUEDC2-induced inhibition on STAT3 activity and suggests that inhibition of CUEDC2 on JAK1/STAT3 signaling is SOCS3-dependent.
Although the exact mechanism and the E3-ligase by which SOCS3 protein is degraded is not well understood, evidence has been provided that Elongin B/C complex plays important roles in SOCS protein regulation. The Elongin B/C complex was initially identified as a positive regulator of RNA polymerase II elongation factor Elongin A (46, 47) and, subsequently, as a component of the multiprotein von Hippel-Lindau disease tumor-suppressor complex (48, 49). Recently, Hilton et al. (51) and Kile et al. (50) identified Elongin C as a component of ubiquitin ligases that include Elongin B, the ring finger protein Roc1, and one of the scaffold proteins Cul2 or Cul5. It has been reported that the SOCS box mediates the interactions with Elongin C, and this interaction to Elongin C stabilizes SOCS protein; disruption of this interaction leads to proteasome-mediated SOCS degradation (35, 36). Our results demonstrate that CUEDC2 enhances the interaction of SOCS3 and Elongin C, thus inhibiting SOCS3 degradation. Additionally, we did not observe direct interaction between CUEDC2 and Elongin C (Fig. 6H). Therefore, it appears as though CUEDC2 does not recruit SOCS3 to Elongin C, and one possibility is that binding of CUEDC2 induces a conformational change of SOCS3 that may facilitate its tighter association with Elongin C.
As known, appropriate activation of JAK1/STAT3 signaling is necessary for cell proliferation and differentiation, but persistent STAT3 activation may result in disease, even tumor. Recent studies also demonstrated that hyperactivation of STAT3 in tumor cells negatively regulates induction of adaptive immunity and mediates immune evasion by blocking the production and inhibiting the sensing of inflammatory signals (52, 53). NF-κB is another critical transcription factor in both immunologic and inflammation response. An important role of IκB kinase-dependent NF-κB activation has been documented both during pathogen infection and in cancers caused by chronic inflammation and other stimuli (54). Our earlier studies demonstrated that CUEDC2 inhibits NF-κB activation by recruitment of protein phosphatase 1, leading to dephosphorylation of IκB kinase. Recently, Lee et al. (30) reported that persistently activated STAT3 maintains constitutive NF-κB activity in tumors. Based on the cross-talk between NF-κB and STAT3, CUEDC2 might play critical roles in controlling chronic inflammation and preventing immune evasion by inhibiting NF-κB and STAT3 activation, which then negatively regulates tumor development or progression.
In conclusion, we established CUEDC2 as an important inhibitor of JAK1/STAT3 signaling that exerts its inhibitory role by attenuating JAK1 and STAT3 phosphorylation via interacting and cooperating with SOCS3. CUEDC2 enhances SOCS3-Elongin C association, thereby preventing SOCS3 degradation by the proteasome. Loss of CUEDC2 expression leads to decreased levels of SOCS3 protein and increased JAK1/STAT3 activation. Therefore, our study not only identified a novel SOCS3 cofactor and a potential mechanism of SOCS3-mediated suppression of the JAK/STAT signaling pathway but also provided important insight into the critical roles of CUEDC2 in the complex signal transduction network.
Supplementary Material
Acknowledgments
We thank Dr. Darnell Jr., Dr. Fred Schaper, Dr. Tarik Moroy, Dr. Claude Haan, Dr. Yong-Yun Kong and Dr. xinmin Cao for generously providing materials.
This work was supported by National Basic Research Program of China Grant 2012CB9100700, National Natural Science Foundation of China Grants 81130037, 30900754, and 31100978, and National High Technology Research and Development Program of China Grant 2009AA02Z103.

This article contains supplemental Figs. S1 and S2.
W.-N. Zhang, L. Wang, Q. Wang, X. Luo, D.-F. Fang, Y. Chen., X. Pan, J.-H. Man, Q. Xia, B.-F. Jin, W.-H. Li, T. Li, B. Liang, L. Chen, W.-L. Gong, M. Yu, A.-L. Li, T. Zhou, and H.-Y Li, unpublished data.
- SH2
- Src homology 2
- SOCS
- suppressor of cytokine signaling
- PIAS
- protein inhibitor of activated stats
- CUE
- coupling of ubiquitin conjugation to endoplasmic reticulum degradation
- CUEDC2
- CUE domain-containing 2
- EpoR
- erythropoietin receptor
- MEM
- minimum essential medium
- aa
- amino acids
- IRF-1
- interferon regulatory factor-1.
REFERENCES
- 1. Kishimoto T., Taga T., Akira S. (1994) Cell 76, 253–262 [DOI] [PubMed] [Google Scholar]
- 2. Levy D. E., Darnell J. E., Jr. (2002) Nat. Rev. Mol. Cell Biol. 3, 651–662 [DOI] [PubMed] [Google Scholar]
- 3. Levy D. E., Lee C. K. (2002) J. Clin. Invest. 109, 1143–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Igaz P., Tóth S., Falus A. (2001) Inflamm Res. 50, 435–441 [DOI] [PubMed] [Google Scholar]
- 5. Zhong Z., Wen Z., Darnell J. E., Jr. (1994) Science 264, 95–98 [DOI] [PubMed] [Google Scholar]
- 6. Bowman T., Garcia R., Turkson J., Jove R. (2000) Oncogene 19, 2474–2488 [DOI] [PubMed] [Google Scholar]
- 7. Bromberg J. (2002) J. Clin. Invest. 109, 1139–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lesina M., Kurkowski M. U., Ludes K., Rose-John S., Treiber M., Klöppel G., Yoshimura A., Reindl W., Sipos B., Akira S., Schmid R. M., Algül H. (2011) Cancer Cell 19, 456–469 [DOI] [PubMed] [Google Scholar]
- 9. Yu H., Jove R. (2004) Nat. Rev. Cancer 4, 97–105 [DOI] [PubMed] [Google Scholar]
- 10. Lee H., Deng J., Kujawski M., Yang C., Liu Y., Herrmann A., Kortylewski M., Horne D., Somlo G., Forman S., Jove R., Yu H. (2010) Nat. Med. 16, 1421–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sinibaldi D., Wharton W., Turkson J., Bowman T., Pledger W. J., Jove R. (2000) Oncogene 19, 5419–5427 [DOI] [PubMed] [Google Scholar]
- 12. O'Shea J. J., Gadina M., Schreiber R. D. (2002) Cell 109, S121–S131 [DOI] [PubMed] [Google Scholar]
- 13. Greenhalgh C. J., Hilton D. J. (2001) J. Leukoc. Biol. 70, 348–356 [PubMed] [Google Scholar]
- 14. Krebs D. L., Hilton D. J. (2000) J. Cell Sci. 113, 2813–2819 [DOI] [PubMed] [Google Scholar]
- 15. Matsumoto A., Masuhara M., Mitsui K., Yokouchi M., Ohtsubo M., Misawa H., Miyajima A., Yoshimura A. (1997) Blood 89, 3148–3154 [PubMed] [Google Scholar]
- 16. Yasukawa H., Misawa H., Sakamoto H., Masuhara M., Sasaki A., Wakioka T., Ohtsuka S., Imaizumi T., Matsuda T., Ihle J. N., Yoshimura A. (1999) EMBO J. 18, 1309–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sasaki A., Yasukawa H., Suzuki A., Kamizono S., Syoda T., Kinjyo I., Sasaki M., Johnston J. A., Yoshimura A. (1999) Genes Cells 4, 339–351 [DOI] [PubMed] [Google Scholar]
- 18. Nicholson S. E., De Souza D., Fabri L. J., Corbin J., Willson T. A., Zhang J. G., Silva A., Asimakis M., Farley A., Nash A. D., Metcalf D., Hilton D. J., Nicola N. A., Baca M. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6493–6498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sasaki A., Yasukawa H., Shouda T., Kitamura T., Dikic I., Yoshimura A. (2000) J. Biol. Chem. 275, 29338–29347 [DOI] [PubMed] [Google Scholar]
- 20. Bjorbak C., Lavery H. J., Bates S. H., Olson R. K., Davis S. M., Flier J. S., Myers M. G., Jr. (2000) J. Biol. Chem. 275, 40649–40657 [DOI] [PubMed] [Google Scholar]
- 21. Hörtner M., Nielsch U., Mayr L. M., Johnston J. A., Heinrich P. C., Haan S. (2002) J. Immunol. 169, 1219–1227 [DOI] [PubMed] [Google Scholar]
- 22. Hörtner M., Nielsch U., Mayr L. M., Heinrich P. C., Haan S. (2002) Eur. J. Biochem. 269, 2516–2526 [DOI] [PubMed] [Google Scholar]
- 23. Zhang P. J., Zhao J., Li H. Y., Man J. H., He K., Zhou T., Pan X., Li A. L., Gong W. L., Jin B. F., Xia Q., Yu M., Shen B. F., Zhang X. M. (2007) EMBO J. 26, 1831–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pan X., Zhou T., Tai Y. H., Wang C., Zhao J., Cao Y., Chen Y., Zhang P. J., Yu M., Zhen C., Mu R., Bai Z. F., Li H. Y., Li A. L., Liang B., Jian Z., Zhang W. N., Man J. H., Gao Y. F., Gong W. L., Wei L. X., Zhang X. M. (2011) Nat. Med. 17, 708–714 [DOI] [PubMed] [Google Scholar]
- 25. Li H. Y., Liu H., Wang C. H., Zhang J. Y., Man J. H., Gao Y. F., Zhang P. J., Li W. H., Zhao J., Pan X., Zhou T., Gong W. L., Li A. L., Zhang X. M. (2008) Nat. Immunol. 9, 533–541 [DOI] [PubMed] [Google Scholar]
- 26. Schmitz J., Weissenbach M., Haan S., Heinrich P. C., Schaper F. (2000) J. Biol. Chem. 275, 12848–12856 [DOI] [PubMed] [Google Scholar]
- 27. Rödel B., Tavassoli K., Karsunky H., Schmidt T., Bachmann M., Schaper F., Heinrich P., Shuai K., Elsässer H. P., Möröy T. (2000) EMBO J. 19, 5845–5855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kwon M. C., Koo B. K., Moon J. S., Kim Y. Y., Park K. C., Kim N. S., Kwon M. Y., Kong M. P., Yoon K. J., Im S. K., Ghim J., Han Y. M., Jang S. K., Shong M., Kong Y. Y. (2008) EMBO J. 27, 642–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Komyod W., Böhm M., Metze D., Heinrich P. C., Behrmann I. (2007) Mol. Cancer Res. 5, 271–281 [DOI] [PubMed] [Google Scholar]
- 30. Lee H., Herrmann A., Deng J. H., Kujawski M., Niu G., Li Z., Forman S., Jove R., Pardoll D. M., Yu H. (2009) Cancer Cell 15, 283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leong H., Mathur P. S., Greene G. L. (2009) Breast Cancer Res. Treat. 117, 505–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bromberg J. F., Wrzeszczynska M. H., Devgan G., Zhao Y., Pestell R. G., Albanese C., Darnell J. E., Jr. (1999) Cell 98, 295–303 [DOI] [PubMed] [Google Scholar]
- 33. Chung C. D., Liao J., Liu B., Rao X., Jay P., Berta P., Shuai K. (1997) Science 278, 1803–1805 [DOI] [PubMed] [Google Scholar]
- 34. Endo T. A., Masuhara M., Yokouchi M., Suzuki R., Sakamoto H., Mitsui K., Matsumoto A., Tanimura S., Ohtsubo M., Misawa H., Miyazaki T., Leonor N., Taniguchi T., Fujita T., Kanakura Y., Komiya S., Yoshimura A. (1997) Nature 387, 921–924 [DOI] [PubMed] [Google Scholar]
- 35. Kamura T., Sato S., Haque D., Liu L., Kaelin W. G., Jr., Conaway R. C., Conaway J. W. (1998) Genes Dev. 12, 3872–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Haan S., Ferguson P., Sommer U., Hiremath M., McVicar D. W., Heinrich P. C., Johnston J. A., Cacalano N. A. (2003) J. Biol. Chem. 278, 31972–31979 [DOI] [PubMed] [Google Scholar]
- 37. Hanada T., Yoshida T., Kinjyo I., Minoguchi S., Yasukawa H., Kato S., Mimata H., Nomura Y., Seki Y., Kubo M., Yoshimura A. (2001) J. Biol. Chem. 276, 40746–40754 [DOI] [PubMed] [Google Scholar]
- 38. Lufei C., Ma J., Huang G., Zhang T., Novotny-Diermayr V., Ong C. T., Cao X. (2003) EMBO J. 22, 1325–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Muromoto R., Nakao K., Watanabe T., Sato N., Sekine Y., Sugiyama K., Oritani K., Shimoda K., Matsuda T. (2006) Oncogene 25, 2131–2136 [DOI] [PubMed] [Google Scholar]
- 40. Sekine Y., Tsuji S., Ikeda O., Sato N., Aoki N., Aoyama K., Sugiyama K., Matsuda T. (2006) Oncogene 25, 5801–5806 [DOI] [PubMed] [Google Scholar]
- 41. Irie-Sasaki J., Sasaki T., Matsumoto W., Opavsky A., Cheng M., Welstead G., Griffiths E., Krawczyk C., Richardson C. D., Aitken K., Iscove N., Koretzky G., Johnson P., Liu P., Rothstein D. M., Penninger J. M. (2001) Nature 409, 349–354 [DOI] [PubMed] [Google Scholar]
- 42. Myers M. P., Andersen J. N., Cheng A., Tremblay M. L., Horvath C. M., Parisien J. P., Salmeen A., Barford D., Tonks N. K. (2001) J. Biol. Chem. 276, 47771–47774 [DOI] [PubMed] [Google Scholar]
- 43. Naka T., Narazaki M., Hirata M., Matsumoto T., Minamoto S., Aono A., Nishimoto N., Kajita T., Taga T., Yoshizaki K., Akira S., Kishimoto T. (1997) Nature 387, 924–929 [DOI] [PubMed] [Google Scholar]
- 44. Suzuki R., Sakamoto H., Yasukawa H., Masuhara M., Wakioka T., Sasaki A., Yuge K., Komiya S., Inoue A., Yoshimura A. (1998) Oncogene 17, 2271–2278 [DOI] [PubMed] [Google Scholar]
- 45. Pezet A., Favre H., Kelly P. A., Edery M. (1999) J. Biol. Chem. 274, 24497–24502 [DOI] [PubMed] [Google Scholar]
- 46. Bradsher J. N., Jackson K. W., Conaway R. C., Conaway J. W. (1993) J. Biol. Chem. 268, 25587–25593 [PubMed] [Google Scholar]
- 47. Aso T., Lane W. S., Conaway J. W., Conaway R. C. (1995) Science 269, 1439–1443 [DOI] [PubMed] [Google Scholar]
- 48. Duan D. R., Pause A., Burgess W. H., Aso T., Chen D. Y., Garrett K. P., Conaway R. C., Conaway J. W., Linehan W. M., Klausner R. D. (1995) Science 269, 1402–1406 [DOI] [PubMed] [Google Scholar]
- 49. Kibel A., Iliopoulos O., DeCaprio J. A., Kaelin W. G., Jr. (1995) Science 269, 1444–1446 [DOI] [PubMed] [Google Scholar]
- 50. Kile B. T., Schulman B. A., Alexander W. S., Nicola N. A., Martin H. M., Hilton D. J. (2002) Trends Biochem. Sci. 27, 235–241 [DOI] [PubMed] [Google Scholar]
- 51. Hilton D. J., Richardson R. T., Alexander W. S., Viney E. M., Willson T. A., Sprigg N. S., Starr R., Nicholson S. E., Metcalf D., Nicola N. A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 114–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang T., Niu G., Kortylewski M., Burdelya L., Shain K., Zhang S., Bhattacharya R., Gabrilovich D., Heller R., Coppola D., Dalton W., Jove R., Pardoll D., Yu H. (2004) Nat. Med. 10, 48–54 [DOI] [PubMed] [Google Scholar]
- 53. Yang X. P., Ghoreschi K., Steward-Tharp S. M., Rodriguez-Canales J., Zhu J., Grainger J. R., Hirahara K., Sun H. W., Wei L., Vahedi G., Kanno Y., O'Shea J. J., Laurence A. (2011) Nat. Immunol. 12, 247–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Naugler W. E., Sakurai T., Kim S., Maeda S., Kim K., Elsharkawy A. M., Karin M. (2007) Science 317, 121–124 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.