Background: Outer membrane protein Lpp of Gram-negative bacteria acts as a receptor for antimicrobial peptide.
Results: We identify and characterize the Lpp, which is responsible for the recognition of cationic antimicrobial peptide.
Conclusion: Lpp is a new target of antimicrobial peptide.
Significance: Lpp may be used as a ligand to develop antimicrobial materials.
Keywords: Antibiotics, Bacterial Pathogenesis, Lipoprotein Receptor, Membrane Proteins, Microbiology, Klebsiella pneumoniae, Antimicrobial Peptide, Enterobacteriaceae, Outer Membrane Protein, Receptor
Abstract
Antimicrobial peptides/proteins (AMPs) are important components of the host innate defense mechanisms. Here we demonstrate that the outer membrane lipoprotein, Lpp, of Enterobacteriaceae interacts with and promotes susceptibility to the bactericidal activities of AMPs. The oligomeric Lpp was specifically recognized by several cationic α-helical AMPs, including SMAP-29, CAP-18, and LL-37; AMP-mediated bactericidal activities were blocked by anti-Lpp antibody blocking. Blebbing of the outer membrane and increase in membrane permeability occurred in association with the coordinate internalization of Lpp and AMP. Interestingly, the specific binding of AMP to Lpp was resistant to divalent cations and salts, which were able to inhibit the bactericidal activities of some AMPs. Furthermore, using His-tagged Lpp as a ligand, we retrieved several characterized AMPs, including SMAP-29 and hRNase 7, from a peptide library containing crude mammalian cell lysates. Overall, this study explores a new mechanism and target of antimicrobial activity and provides a novel method for screening of antimicrobials for use against drug-resistant bacteria.
Introduction
Antimicrobial peptides/proteins (AMPs)2 are produced by all living organisms and are important components promoting innate immunity against invading pathogens (1–3). As but a few examples, human skin is perpetually exposed to microorganisms but is largely free of infection; the biochemical barrier provided by defensins, psoriasin (S100A7), and ribonuclease 7 (hRNase 7) contributes substantially to the natural defense against microbial invasion (4–7). The sheep myeloid antimicrobial peptide 29 (SMAP-29), a leukocyte peptide of the cathelicidin family, reduces the bacterial concentration in both bronchoalveolar lavage fluid and consolidated pulmonary tissues of infected lambs (8). The LL-37, proteolytic fragment from human cathelicidin-like protein hCAP18, is bactericidal for several pathogens including Escherichia coli and methicillin-resistant Staphylococcus aureus (9). Likewise, the CAP-18, C-terminal fragment from rabbit granulocytes, possesses bactericidal activity (10). Protegrin-1, another prominent AMP, is a cysteine-rich, 18-residue β-sheet peptide isolated from porcine leukocytes with antimicrobial activity against a broad range of microorganisms (11). Finally, Polymyxin B is a lipopeptide antibiotic isolated from Bacillus polymyxa and is composed of a polycationic peptide ring and a tripeptide side chain with a fatty acid tail. Both Polymyxin B and colistin (also known as polymyxin E) are used clinically for the therapy of bacterial infection (12).
Although cationic AMPs possess diverse secondary structures, their surfaces are uniformly amphipathic with both hydrophobic and hydrophilic residues, like SMAP-29 and CAP-18 (8, 10). However, some AMP possesses higher percentage of α-helical structure in the presence of hydrophobic environments, like LL37 in trifluoroethanol or lipid A (13). These AMPs have multiple modes of action that are generally perceived as differing from those of conventional antibiotics (1, 7, 14). Most studies have proceeded on the tacit assumption that these AMPs act on bacteria through electrostatic interactions and that lipopolysaccharide (LPS), a component of the outer membrane of most Gram-negative bacteria, serves as the initial AMP binding site. However, recent studies have shown that LPS is not universally associated with susceptibility to the bactericidal effects of AMPs (15, 16). Consistent with this notion, we recently examined the mechanism of antimicrobial action of hRNase 7 against the Gram-negative bacteria, Pseudomonas aeruginosa, which is not mediated through LPS but via the outer membrane protein, OprI (17).
In this report we identify a distinct group of outer membrane lipoprotein (Lpp) of the Enterobacteriaceae family of Gram-negative bacteria that contribute directly to their susceptibilities to α-helical cationic AMPs. The Lpp is one of major outer membrane proteins of E. coli, which anchors to outer membrane and cell wall through the modified N-terminal N-acyl-S-diacylglycerylcysteine residue and C-terminal lysine residue, respectively (18, 19). Lpp is proposed to consist of oligomeric α-helices and act as a barrier against antibiotics and metabolites (20, 21). Here, we provide direct evidence for the cationic α-helical AMP to bind and internalize the surface oligomeric Lpp and present a novel method for screening potential antimicrobials for use against antibiotic-resistant bacteria.
EXPERIMENTAL PROCEDURES
Peptides
The following oligopeptides, sheep SMAP-29 (RGLRRLGRKIAHGVKKYGPTVLRIIRIAG-NH2), rabbit CAP-18 (GLRKRLRKFRNKIKEKLKKIGQKIQGLLPKLAPRTDY), human LL-37 (LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES), porcine Protegrin-1 (RGGRLCYCRRRFCVCVGR-NH2), and R3S1 (RRRSRRRSRRRS) were synthesized by Kelowna International Scientific Inc. (Taipei, Taiwan). Polymyxin B was purchased from Sigma.
Antimicrobial Activity Assay
The bacteria were cultured in Luria-Bertani broth, which contains 0.17 m NaCl, and plated on Luria-Bertani agar for P. aeruginosa Migula (Schroeter) Migula (ATCC BAA-47TM), E. coli K-12 (M61655), Klebsiella pneumoniae (ATCC 13884), Yersinia enterocolitica (ATCC 23715), Serratia marcescens (ATCC 8100), and Salmonella typhimurium. S. aureus subspecies Aureus Rosenbach (ATCC 6538P) was cultured and plated in/on tryptic soy broth/agar (Difco 0369). The yeast Candida albicans (Robin) Berkhout (ATCC 14053) was cultured and plated in/on yeast malt broth/agar, and Pichia pastoris X-33 was cultured and plated in/on yeast extract-peptone-dextrose broth/agar. The microbes were grown overnight, washed, and diluted 1:300 in 10 mm sodium phosphate, pH 7.4. 45 μl of the microbes (5–10 × 104 colony-forming units (cfu)) were mixed with various concentrations of antimicrobial peptide (5 μl), which was dissolved in 20 mm Hepes, pH 7.4, 50 mm NaCl, and incubated at 37 °C for 3 h. Serial dilution of each AMPs-treated bacteria/yeast was prepared and plated for the determination of the remaining cfu.
Preparation of Bacterial Membrane Fraction and Human Cell Lysate
The preparation of membrane fraction was modified and briefly described as follows (22). Overnight culture of K. pneumoniae was resuspended in 20 mm HEPES, pH 7.5, 50 mm NaCl and disrupted by sonication. The pellet of bacterial lysate after centrifugation at 17,000 × g for 60 min at 4 °C was extracted with the buffer (2% Triton X-100, 10 mm Tris-HCl, pH 7.8) at 30 °C for 60 min. The supernatant after ultracentrifugation at 100,000 × g for 60 min at 4 °C was collected for the analysis of native Lpp. The confluent human lung adenocarcinoma cells CL1-0 in a 100-mm plate was washed with phosphate-buffered saline and incubated with 1 ml of ProteoJETTM Mammalian Cell Lysis Reagent (Thermo Scientific, Waltham, MA) at room temperature for 10 min. The supernatant of the total lysate after centrifugation at 16,000 × g for 15 min was collected and stored at −70 °C for use.
Cloning, Expression, and Purification of Lpp
The DNA fragment encoding Lpp (V00302) was cloned from the genomic DNA of E. coli by PCR by the use of two primers 5′-CATATGTGCTCCAGCAACGCTAAAATC3-′ and 5′-GTATACCTTGCGGTATTTAGTAGCC-3′). The PCR product was cloned into the pGEM-T vector (Promega, Madison, WI) then further tagged with a DNA fragment at the 5′ end (CCATGGCTCATCATCATCATCATCATGTCGACAAGCTTATCGAAGGCCGTTGCTCCAGCAACGCTAAA) containing NcoI, His6, SalI-HindIII, and a Factor Xa cleavage site and subcloned into the expression vector pET32a (Novagen) through HindIII and XhoI sites (17). The genes encoding the Lpps of K. pneumoniae and S. typhimurium were made by a QuikChangeTM site-directed mutagenesis kit in pET32a using E. coli Lpp as a template and two pairs of oligonucleotides, 5′-GCAAGCTTATCGAAGGCCGTTGCTCCAGCAACGCTAAAATC-3′/5′CGCTCGAGTTACTTGCGGTATGAATGAGCCTGGTTGTCCAGACGCTGGTTAGC3′ and 5′-GCAAGCTTATCGAAGGCCGTTGCTCCAGCAACGCTAAAATC-3′/5′-CGCTCGAGTTACTTGCGGTATTTAGTAGCCTGGTTGTCCAGACGCTGGTTAGC-3′, respectively, as primers. The OprI and Lpp were expressed in E. coli BL21(DE3) at 26 °C overnight in the presence of 0.5 mm isopropyl-β-d-thiogalactopyranoside. The recombinant fusion protein was purified by use of a nickel nitrilotriacetic acid-agarose gel (Qiagen) column and digested by protease Factor Xa (Ni-NTA Agarose, Novagen). The flow-through from a second nickel-nitrilotriacetic acid column was further purified by fast protein liquid chromatography Superose 12TM column chromatography.
Binding of AMPs to K. pneumoniae
The bacteria (5–10 × 106 cfu in 45 μl of 10 mm sodium phosphate, pH 7.5) were incubated with various AMPs (3 μg in 5 μl) in the presence of 4 mm MgCl2 or 150 mm NaCl for 30 min at 37 °C and subjected to centrifugation at 13,000 × g for 20 min, then washed 5 times with 10 mm sodium phosphate before SDS-PAGE and Coomassie Blue staining.
Assays of Permeability of Bacterial Membrane
The overnight culture of bacteria (107 cfu) was washed and resuspended in 100 μl of water and incubated with 1 μm SYTOX® Green (Molecular Probes) in a dark 96-well plate for 15 min in the dark. After the addition of AMP, the increase of fluorescence due to the binding of the dye to intracellular DNA was measured in the same microplate reader using 485- and 520-nm filters for excitation and emission wavelengths, respectively (17).
Morphological Analysis by Transmission Electron Microscopy
Bacteria (107 cfu in 95 μl) were treated with 40 μm biotinylated SMAP-29 at 37 °C for 0–20 min as indicated. The bacteria were fixed by 4% paraformaldehyde in 0.1 m cacodylate buffer, pH 7.4, and embedded in 2% agarose. Small cubes (1 mm3) were submerged in cryoprotectant (30% glycerol) at room temperature overnight, frozen in nitrogen slurry, transferred to a Leica ASF2 unit at −140 °C, then substituted with methanol at 90 °C, methanol/HM20 (Electron Microscopy Sciences) (1:1), methanol/HM20 (1:2), and pure HM20 at 50 °C. Thin sections were obtained and incubated with rabbit anti-Lpp and mouse anti-biotin antibodies, then with 18-nm and 12-nm gold particle-labeled secondary antibodies, respectively. The morphologies of SMAP-29-treated bacteria were examined under a Hitachi H-7000 transmission electron microscope after double-staining with uranyl acetate and lead citrate (23).
Cross-linking Assay
Small aliquots of recombinant Lpp (0.8 μg in 10 μl) or bacterial suspensions (5–10 × 106 cfu in 45 μl) were incubated with increasing concentrations of EDC and glutaraldehyde in 10 mm sodium phosphate, pH 5.5 and 7.5, respectively, at 37 °C for 30 min. The cross-linked complexes were separated by reducing SDS-PAGE followed by silver staining or Western blotting using anti-Lpp and anti-biotin antibody, respectively.
RESULTS
Broad Antimicrobial Spectra of Cationic AMPs
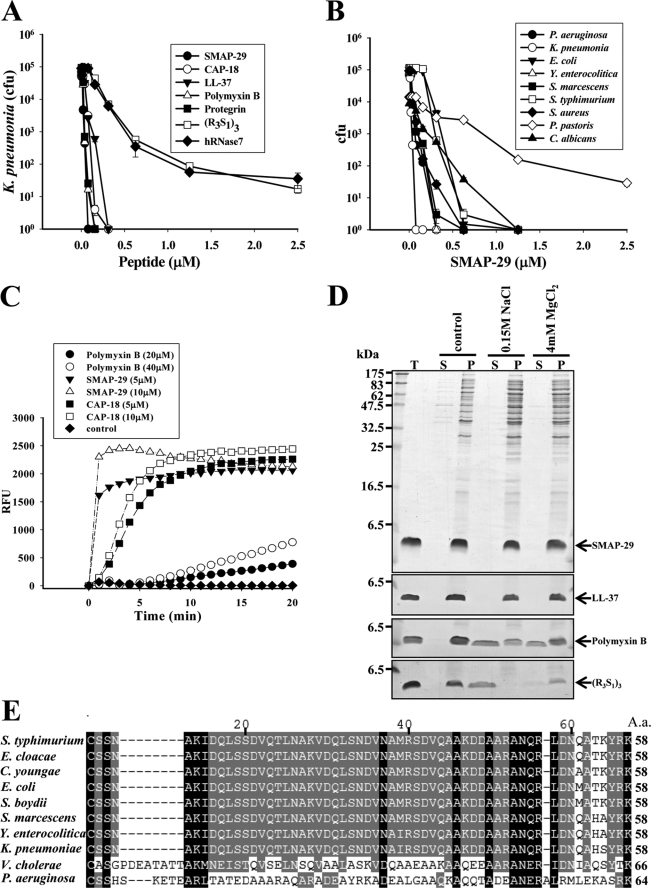
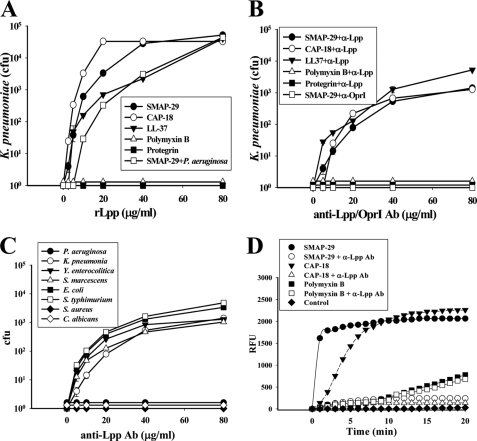
Among the AMPs tested against K. pneumoniae, SMAP-29, Protegrin-1, and Polymyxin B had the highest bactericidal activity with LD99 (the dose that achieves 99% reduction of colony-forming units) less than 0.1 μm. CAP-18 and LL-37 were somewhat weaker with LD99 less than 0.15 μm, whereas the Arg-riched oligopeptide, (R3S1)3, and hRNase 7 were weak, with LD99 0.6 μm (Fig. 1A). SMAP-29 exerted broad spectrum antimicrobial activity against Gram-negative bacteria (P. aeruginosa, K. pneumoniae, Y. enterocolitica, S. marcescens, S. typhimurium, E. coli), Gram-positive bacteria (S. aureus), and fungi (C. albicans) but had minimal activity against P. pastoris X-33) (Fig. 1B). To examine the membrane integrity of K. pneumoniae after AMP treatment, we found that the permeability of membrane markedly increased within 5 min after treatment with 5∼10 μm SMAP-29 or CAP-18, whereas permeability resulting from 20∼40 μm Polymyxin B increased slowly (Fig. 1C).
FIGURE 1.
Antimicrobial spectra of cationic AMPs and their target protein on Gram-negative bacteria. A, bactericidal activities of AMPs on K. pneumoniae are shown. B, antimicrobial activity of SMAP-29 on various microbes is shown. Microbes (5–10 × 104 cfu) were treated with AMPs at 37 °C in 10 mm sodium phosphate, pH 7.5, for 3 h and then plated on agar plates for the determination of remaining cfu. C, increase of membrane permeability of K. pneumoniae after AMP treatment is shown. Bacteria (107 cfu) were incubated with 1 μm SYTOX® Green in a dark 96-well plate for 15 min before the addition of AMP. The increase of fluorescence was measured using 485- and 520-nm filters for excitation and emission wavelengths, respectively. RFU, relative fluorescence units. D, binding of AMPs to K. pneumoniae is shown. Overnight cultures of bacteria (107 cfu) were incubated with SAMP-29, LL-37, Polymyxin B, or (R3S1)3 (3 μg each) in 50 μl at 37 °C for 30 min, then spun at 10,000 × g for 10 min followed by SDS-PAGE and Coomassie Blue staining. NaCl or MgCl2 was added as indicated. T represents total AMPs employed, S is for supernatant, and P is for pellet. E, amino acid sequence alignment of Lpp-homologous proteins by the software ClustalX is shown. The Lpp sequences of S. typhimurium (NP_460342), Enterobacter cloacae (YP_003612855), Citrobacter youngae (ZP_06353081), E. coli (NP_416192), Shigella boydii (YP_001880438), S. marcescens (AAA26566), Y. enterocolitica (YP_001006405), K. pneumoniae (YP_001335792), V. cholerae (ZP_01972724), and OprI sequence of P. aeruginosa (NP_251543) were obtained from National Center for Biotechnology Information.
The Lpp Family Proteins in Gram-negative Bacteria
Among the cationic AMPs tested, SMAP-29, and LL-37 were able to bind susceptible bacteria K. pneumoniae even in the presence of 0.15 m NaCl or 4 mm MgCl2, at which the binding abilities of Polymyxin B and (R3S1)3 were markedly reduced (Fig. 1D). These results suggest that the bindings of cationic SMAP-29 and LL-37 to the susceptible bacteria are mediated through a specific receptor rather than nonspecific ionic interaction, as that occurs between cationic AMP and anionic LPS in most Gram-negative bacteria. A search of the National Center for Biotechnology Information protein data base with the primary amino acid sequence of P. aeruginosa OprI protein, a known binding protein for the AMP, human RNase 7 (24), we identified a homologous lipoprotein, Lpp, which is expressed in all the Enterobacteriaceae and Vibrio cholerae. Lpp protein sequence from these bacterial species is 23 and 25% identical, respectively, to P. aeruginosa OprI but interestingly was not detected in Gram-positive bacteria or fungi (Fig. 1E); these microbes clearly have other targets for AMP binding that remain to be explored.
Oligomerization of Recombinant Lpp/OprI
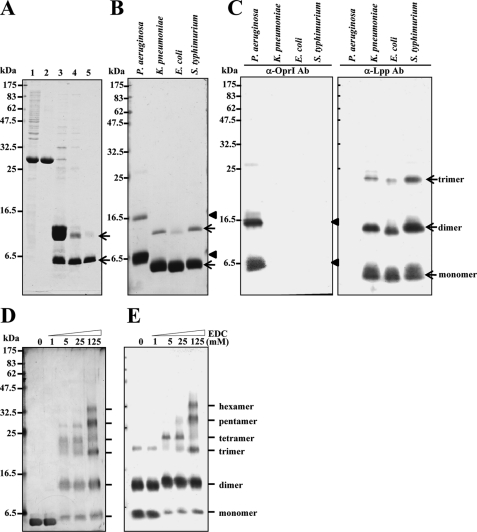
The primary sequences of Lpp family proteins in the Enterobacteriaceae were highly homologous to one another with minor variations primarily at C terminus. We cloned Lpp genes of K. pneumoniae, E. coli, and S. typhimurium, expressed them in E. coli BL21-DE3 as thioredoxin-fused proteins, and purified them to high homogeneity after cleavage with protease Factor Xa (Fig. 2, A and B). Both recombinant OprI and Lpp proteins (shown by arrowhead and arrow, respectively) were detected predominantly in monomer form and with minor amounts in dimer conformation on reducing SDS-PAGE visualized by Coomassie Blue staining (Fig. 2B). However, three major bands were observed by Western blot using antibodies raised against recombinant OprI from P. aeruginosa and Lpp from E. coli (Fig. 2C). The antibody raised against E. coli Lpp could react with the Lpps of E. coli, K. pneumoniae, and S. typhimurium but not with the OprI of P. aeruginosa and vice versa. To investigate the stronger antigenicity of dimeric Lpp than monomeric Lpp to anti-Lpp antibody as shown in Fig. 2C, a His-tagged Lpp of K. pneumoniae was contact-transferred onto duplicate PVDF membranes and detected by antibodies against His6 and Lpp, respectively. The antigenicity of dimeric His6-tagged Lpp to anti-Lpp antibody was stronger than that to anti-His6 antibody (see supplemental Fig. S1). The preference of anti-Lpp antibody to dimeric Lpp indicates that the Lpp used as immunogen to raise antibody may exist in the form of dimer or more linked by a disulfide bond at the N terminus. To further examine the status of polymerization, the recombinant Lpp of K. pneumoniae was cross-linked with an increasing amount of EDC and analyzed by silver staining and Western blot (Fig. 2, D and E). The presence of oligomers ranging dimer to hexamer suggests that the recombinant Lpp exists in hexamer.
FIGURE 2.
Production of oligomeric Lpps. A, shown is analysis of recombinant Lpp of K. pneumoniae at different purification steps by 14% SDS-PAGE and Coomassie Blue staining. Lane 1, crude lysate from E. coli BL21(DE3) transformed with thioredoxin/Lpp-fused gene; lane 2, eluate of nickel column; lane 3, protease Factor Xa-digested product; lane 4, flow-through of second nickel column; lane 5, eluate of SuperoseTM 12 column chromatography. B, purity of recombinant OprI/Lpp is shown. Equal amounts of recombinant OprI of P. aeruginosa and Lpps of K. pneumoniae, E. coli, and S. typhimurium (3 μg each) were taken for 14% SDS-PAGE separation and Coomassie Blue staining. C, a Western blot analysis of recombinant OprI and Lpp is shown. The recombinant proteins as mentioned in panel B (0.2 μg each) were taken for Western blot analysis using anti-OprI (left panel) and anti-Lpp (right panel) antibody (Ab), respectively. D and E, cross-linking of Lpp by EDC is shown. The recombinant Lpp of K. pneumoniae (0.8 μg each) was incubated with increasing amounts of EDC as indicated at pH 5.5 for 30 min and subjected to 14% SDS-PAGE and silver staining (panel D) and Western blot analysis (panel E).
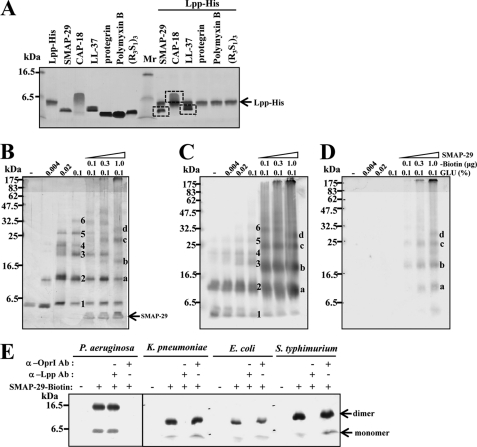
Specific Binding of Cationic α-Helical AMPs to Recombinant Lpp Family Proteins
Only cationic α-helical AMPs, like SMAP-29, CAP-18, and LL-37, were recognized by His-tagged Lpp and pulled down by Ni-NTA Agarose, whereas the non-α-helical cationic AMPs, like Protegrin-1, Polymyxin B, and (R3S1)3, were not (Fig. 3A). The specific interaction between Lpp and biotinyl-SMAP-29 was further validated by glutaraldehyde cross-linking followed by SDS-PAGE, silver staining, and Western blot using anti-Lpp and anti-biotin antibodies, respectively (Fig. 3, B–D). Oligomers of Lpp from monomer to hexamer were observed upon glutaraldehyde cross-linking (marked with numbers 1–6). Interestingly, the Lpp·biotinyl-SMAP-29 complexes were detected in the presence of biotinyl-SMAP-29 (marked with a–d). In addition to K. pneumoniae Lpp, the Lpp of E. coli and S. typhimurium was also recognized by biotinylated SMAP-29 and pulled down by streptavidin-conjugated beads detected by Western blot using anti-Lpp antibody. The formation of Lpp·biotinyl-SMAP-29 complex were abolished by anti-Lpp antibody but not by anti-OprI antibody (Fig. 3E). Similarly, the P. aeruginosa OprI was also bound to biotinyl-SMAP-29 and abolished by anti-OprI antibody but not by anti-Lpp antibody. It is interesting to note that the signal of dimer is stronger than that of monomer probably due to the preference of biotinyl-SMAP-29 to recognize and bind disulfide bond-linked oligomer to monomer. These results indicate that cationic α-helical AMP could specifically recognize both OprI of P. aeruginosa and Lpp of Enterobacteriaceae bacteria.
FIGURE 3.
Specific binding of cationic α-helical AMPs to recombinant Lpp. A, binding of cationic AMPs to recombinant Lpp is shown. Cationic AMPs as indicated (4 μg each) were incubated with 1.5 μg of His-tagged Lpp in 10 mm sodium phosphate buffer, pH 7.5, containing 0.15 m NaCl and 40 mm imidazole, precipitated by Ni-NTA Agarose, and analyzed by SDS-PAGE and Coomassie Blue staining. B–D, specific interaction between Lpp and SMAP-29 is shown. Recombinant Lpp of K. pneumoniae (0.8 μg each) was incubated with increasing amounts of biotinylated SMAP-29 for 30 min, cross-linked with glutaraldehyde for 20 min, separated by 14% SDS-PAGE, and analyzed by silver staining (B), Western blot analysis with anti-Lpp antibody (C), or anti-biotin antibody(D). Numbers 1–6 represent the status of Lpp polymerization, and a–d indicate the positions of biotinylated SMAP-29·Lpp complexes. E, binding of recombinant Lpp/OprI by biotinylated SMAP-29 is shown. The recombinant OprI of P. aeruginosa and Lpps of K. pneumoniae, E. coli, and S. typhimurium (2 μg each) were incubated with 2 μg of biotinylated SMAP-29 and analyzed by Western blot with anti-Lpp antibody (Ab).
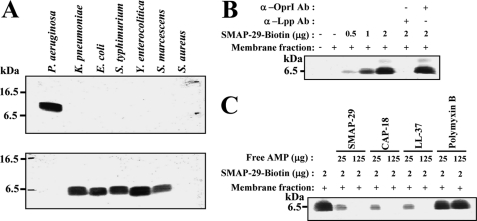
Specific Recognition of Native K. pneumoniae Lpp by Cationic α-Helical AMPs
In contrast to recombinant Lpp/OprI, only one signal with slower mobility than that of monomeric recombinant Lpp/OprI was observed for the crude total lysates of tested Gram-negative bacteria by Western blot as shown in Fig. 4A. The native Lpp of membrane fraction of K. pneumoniae was recognized by biotinyl-SMAP-29 and pulled down by streptavidin-conjugated beads detected by Western blot. The specific interaction was abolished by anti-Lpp antibody but not by anti-OprI antibody (Fig. 4B). Furthermore, the interaction was competed by excess amounts of free SMAP-29 and other cationic α-helical AMPs, like CAP-18 and LL-37, but not by Polymyxin B (Fig. 4C).
FIGURE 4.
Specific recognition of native Lpp of K. pneumoniae by cationic α-helical AMPs. A, shown is antigenicity of native OprI and Lpp to anti-OprI and anti-Lpp antibodies. The total lysates of indicated bacteria (0.5 μg each) were separated by 14% SDS-PAGE and detected by Western blot using anti-OprI (top) and anti-Lpp (bottom) antibody, respectively. B, shown is binding of native Lpp by biotinylated SMAP-29. The membrane fraction of K. pneumoniae (2 μg each) was incubated with increasing amounts of biotinylated SMAP-29 in 10 mm sodium phosphate buffer, pH 7.5, containing 0.15 m NaCl and 0.15% Triton X-100, precipitated with streptavidin-conjugated beads, and analyzed by Western blot using anti-Lpp antibody (Ab). Anti-Lpp or anti-OprI antibodies (4 μg each) were added to the mixture as control. C, competition of the formation of Lpp·SMAP-29 complex by free cationic α-helical AMPs is shown. The membrane fraction of K. pneumoniae (2 μg each) was incubated with biotinylated SMAP-29 in the presence of free SMAP-29, CAP-18, LL-37, and Polymyxin B, precipitated, and analyzed as panel B.
Role of Lpp in Susceptibility of Bacteria to Cationic α-Helical AMPs
To determine whether Lpp is the target of cationic α-helical AMP in vivo, increasing amounts of recombinant Lpp were employed for the competition of AMP action in a colony-forming unit assay. We found that the antimicrobial activities of cationic α-helical AMPs, like SMAP-29, CAP-18, and LL37, against K. pneumoniae were blocked by exogenous Lpp, but those of Polymyxin B and Protegrin-1 were not. As a control, we found that the antimicrobial activity of SMAP-29 against P. aeruginosa was also inhibited by exogenous Lpp (Fig. 5A). These results indicate that the actions of SMAP-29 were blocked by exogenous Lpp regardless of the nature of the specific susceptible target bacteria, as this is a distinct property of the ligand. Furthermore, the surface Lpp of K. pneumoniae was protected from the actions of these cationic α-helical AMPs, including SMAP-29, by anti-Lpp antibody but not by anti-OprI antibody. However, the actions of Polymyxin B and Protegrin-1 were not blocked by the anti-Lpp antibody (Fig. 5B). In addition to K. pneumoniae, several bacteria of the Enterobacteriaceae family (Y. enterocolitica, S. marcescens, S. typhimurium, and E. coli) were also protected by the anti-Lpp antibody, but the P. aeruginosa, S. aureus, and C. albicans not belonging to Enterobacteriaceae were not protected by the anti-Lpp antibody (Fig. 5C). The increase in membrane permeability caused by SMAP-29 or CAP-18 was inhibited by anti-Lpp antibody, but that caused by Polymyxin B was not (Fig. 5D). These results demonstrate that surface Lpp plays a key role in the susceptibility of Enterobacteriaceae bacteria, but not that of P. aeruginosa, to cationic α-helical AMPs.
FIGURE 5.
Protection of bacteria from AMP-mediated toxicity. A and B, repression of antimicrobial activity of AMP by the addition of exogenous recombinant Lpp or anti-Lpp antibody is shown. The viability of K. pneumoniae or P. aeruginosa (5–10 × 104 cfu) treated with 0.1 μm SMAP-29, 0.2 μm CAP-18, 0.2 μm LL-37, 0.1 μm Protegrin-1, or 0.1 μm Polymyxin B was measured in the presence of increasing amounts of recombinant Lpp (A) or anti-Lpp antibody (B) as indicated. C, protection of various microbes from the action of SMAP-29 by the pretreatment with anti-Lpp antibody is shown. D, repression of AMP-induced increase of membrane permeability by anti-Lpp antibody is shown. Bacteria (107 cfu) containing 1 μm SYTOX® Green were incubated with or without anti-Lpp antibody in a dark 96-well plate for 15 min before the treatment of 5 μm SMAP-29, 5 μm CAP-18, and 40 μm Polymyxin B and assayed for fluorescence as described in Fig. 1C.
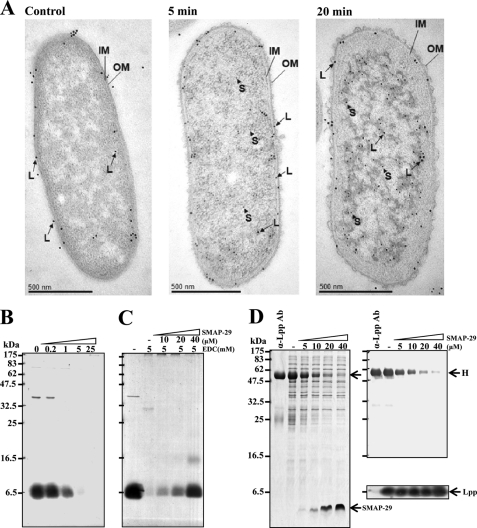
Morphological Changes and Entry of Surface Lpp Caused by SMAP-29
Morphologically, the outer membrane of K. pneumoniae detached from inner cytoplasmic membrane and blebbed outward 5 min after SMAP-29 treatment. The cytosol condensed into a heavily stained body, leaving behind a lightly stained space beneath the membrane 20 min after SMAP-29 treatment (Fig. 6A). Most Lpp as well as biotinylated SMAP-29 (large and small gold particles shown as L and S, respectively) relocated from outer membrane to the condensed cytosolic area. To further investigate whether Lpp is depleted from the outer membrane, we found that the surface Lpp was accessible to EDC and cross-linked into a large complex without SMAP-29 treatment because the complex was unable to be dissociated into monomer by SDS-PAGE analysis (Fig. 6B); however, the Lpp became inaccessible to EDC after SMAP-29 treatment probably due to internalization into cytosol (Fig. 6C). In addition, the anti-Lpp antibody could not recognize and bind the SMAP-29-treated bacteria anymore, although the total amount of Lpp remained constant (Fig. 6D). These results indicate that the abundant surface Lpp was targeted and internalized into cytosol by SMAP-29.
FIGURE 6.
Morphological changes and entry of surface Lpp caused by SMAP-29. A, internalization of Lpp and SMAP-29 is shown. Bacteria (1 × 107) were incubated with 60 μg of biotinyl-SMAP-29 at 37 °C for 5 and 20 min, respectively. Thin sections of bacteria were incubated with both anti-Lpp antibody from rabbit and anti-biotin antibody from mouse then with secondary antibody conjugated with large gold particles (18 nm, for Lpp (L)), for rabbit antibody and small gold particles (12 nm, SMAP-29 (S)), and for mouse antibody and examined under transmission electron microscopy. OM and IM represent outer membrane and inner membrane, respectively. B, cross-linking of bacterial surface Lpp by EDC is shown. Small aliquots of K. pneumoniae were incubated with EDC as indicated in 10 mm sodium phosphate, pH 5.5, and analyzed by Western blot using anti-Lpp antibodies. C, shown is protection of surface Lpp from EDC cross-linking by SMAP-29 treatment. The bacteria were treated with SMAP-29 for 10 min before cross-linking with 5 mm EDC and analyzed as described above. D, masking of surface Lpp by SMAP-29 treatment is shown. The binding capacities of SMAP-29-treated bacteria to anti-Lpp antibody (4 μg) were assayed by SDS-PAGE and Coomassie Blue staining (left panel) and Western blot ( of protein loaded to left panel) using anti-rabbit immunoglobulin antibody (right top panel) and anti-Lpp antibody (right bottom panel), respectively. Anti-Lpp antibody at left lane acts as a control, and H represents the heavy chain of immunoglobulin.
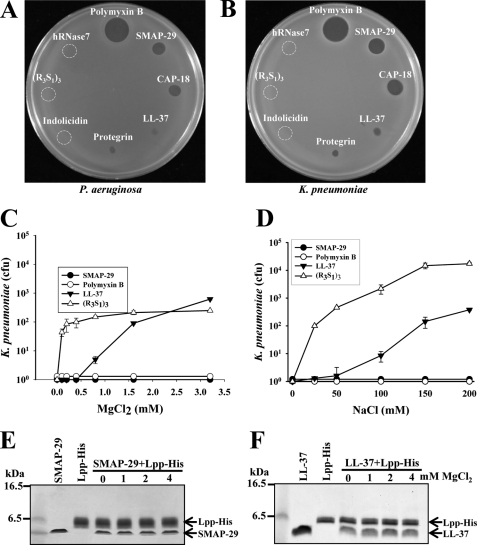
Factors Interfere with Bactericidal Activity of AMPs
To explore and identify factors that could reduce the efficacy of AMPs in clinical trials and the possible bactericidal pathway of AMP, we found that the bactericidal activities, analyzed by the diameter of inhibition zone on Luria Broth agar plate, of hRNase 7, (R3S1)3, and Indolicidin (4 μg each) against E. coli and K. pneumoniae were reduced, whereas those of Polymyxin B and SMAP-29 were only slightly reduced (Fig. 7m, A and B). To investigate the possible factors involved in Luria Broth culture medium, which contains divalent cations and salts (0.17 m NaCl), we found the bactericidal activities of LL-37 and (R3S1)3 against K. pneumoniae were markedly repressed by 1.5 mm MgCl2 or 0.15 m NaCl, but those of SMAP-29 and Polymyxin B were not (Fig. 7, C and D). To investigate whether the inhibitory effects of ions and salts are mediated through the reduced binding of AMP to Lpp, we found that the binding ability of LL-37 as well as SMAP-29 to His-tagged Lpp was not inhibited by MgCl2 (0∼4 mm) in the presence of 0.15 m NaCl, although the bactericidal activity of LL-37 was susceptible to the inhibition by 1.5 mm MgCl2 or 0.15 m NaCl. Thus, other downstream events in the bactericidal pathway or the structure of LL-37 are suspected to be affected by divalent cations and salts because the structure of LL-37 varies with the hydrophobicities of solvents (13).
FIGURE 7.
Factors interfere with bactericidal activity of AMPs. A and B, bactericidal activities of AMPs assayed by the diameter of inhibition zone on Luria-Bertani agar plates are shown. P. aeruginosa (panel A) and K. pneumoniae (panel B) were plated on Luria-Bertani agar plates in which AMPs were dotted (4 μg each) as indicated. C and D, effects of MgCl2 and NaCl on the bactericidal activity of AMPs are shown. The bactericidal activities of 10 μm SMAP-29, 10 μm LL-37, 10 μm Polymyxin B, and 10 μm (R3S1)3 against K. pneumoniae were determined in the presence of MgCl2 and NaCl as indicated. E and F, shown is the effect of MgCl2 on the binding of AMP to Lpp. SMAP-29 and LL-37 (4 μg each) were incubated with 1.5 μg Lpp-His in the presence of MgCl2 as indicated before pulling down by Ni-NTA Agarose and visualized by Coomassie blue staining.
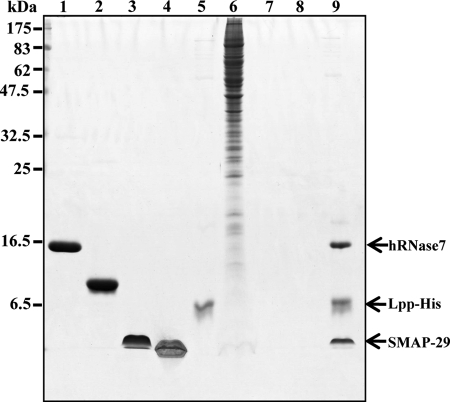
Screening of AMPs by His-tagged Lpp
Because Lpp/OprI is the target of cationic α-helical AMP, we intended to screen for AMPs from natural source using the His-tagged Lpp as a ligand. For proteins in the RNase A superfamily, human hRNase 7 possesses bactericidal activity, whereas the oocytic ribonuclease from bullfrog, Rana Catesbeiana (RC-RNase) does not. For bactericidal cationic peptides, SMAP-29 is an α-helical AMP, whereas Polymyxin B is not. To mimic the complicate environments of natural source of AMPs including ions, salts, and other cellular components, we retrieved hRNase 7 and SMAP-29 from the above-mentioned RNase/peptide mixture in the presence of crude lysate of human lung adenocarcinoma cell CL1-0 by the His-tagged Lpp (Fig. 8). The result provides a novel method for screening of antimicrobials from natural sources for use against drug-resistant bacteria.
FIGURE 8.
Screening of bactericidal AMPs by His-tagged Lpp. The mixture of human RNase 7 (2 μg, lane 1), RC-RNase from bullfrog, Rana Catesbeiana (2 μg, lane 2), SMAP-29 (4 μg, lane 3), and Polymyxin B (4 μg, lane 4) were incubated with His-tagged Lpp (1.5 μg, lane 5) in 10 mm sodium phosphate, pH 7.5, containing 0.15 m NaCl and 40 mm imidazole in the presence of crude cell lysates of human lung adenocarcinoma cell CL1-0 (30 μg, lane 6), then pulled down by Ni-NTA Agarose and analyzed by SDS-PAGE and Coomassie Blue staining. The human RNase 7 and SMAP-29 were pulled down as shown on the right (lane 9). No peptide/protein was pulled down by Ni-NTA Agarose from the two peptide/two protein mixture (lane 7) and the crude cell lysate (lane 8) without the His-tagged Lpp ligand.
DISCUSSION
The widespread use of antibiotics in both medicine and agriculture plays a significant role in the emergence of drug-resistant bacteria. Conventional antibiotics typically inhibit the synthesis of nucleic acids, proteins, or cell wall components. Microorganisms develop resistance to these antibiotics through their inactivation/modification by enzymes, modification of target site, alteration of metabolic pathway, and reduction of drug accumulation by decreasing drug permeability or increasing active efflux (25–28). Thus, development of new antimicrobials with unique targets and action mechanisms is an acute and urgent need. Natural AMP that fill this need have been isolated from multiple sources (1, 2, 14, 16). Although these AMPs possess different secondary structures like α-helix, β-strand, and random coils, as a group they exert antimicrobial activity by increasing membrane permeability, ostensibly via mechanisms that are different from those exerted by conventional antibiotics (1, 7, 14).
Although primary studies on the action mechanism of AMPs on bacterial membrane concluded that LPS was the initial and direct binding site of cationic oligopeptides, it is now clear that the susceptibility of a given bacterial strain to AMPs is not strictly correlated with the presence of LPS on bacteria or divalent cations in the assay environments (15, 16). In our previous study we determined that the susceptibility of P. aeruginosa to hRNase 7, a lysine-enriched cationic protein from human skin, was not via LPS but by the outer membrane protein, OprI (17). Here, we identify another AMP-binding protein, Lpp, found in Enterobacteriaceae and V. cholerae that is likewise responsible for their susceptibilities to cationic α-helical AMPs. These proteins are highly homologous to each other except for minor variations at the C termini. Direct binding of cationic α-helical AMP to Lpp was shown by the pulldown experiment and cross-linking of Lpp·SMAP-29 complex followed by Western blot analysis (Fig. 3). The formation of biotinylated SMAP-29·Lpp complex was specifically competed by free cationic α-helical AMPs and inhibited by anti-Lpp antibody (Fig. 4). In addition, the role of Lpp on the susceptibility of bacteria to these AMPs was further shown by the blocking of bactericidal activity with anti-Lpp antibody (Fig. 5). These results clearly demonstrated the Lpp interacts with and promotes susceptibilities of Enterobacteriaceae bacteria to the bactericidal activities of cationic α-helical AMPs.
Hundreds of natural AMPs have been isolated, although only few AMPs exhibit promising antimicrobial activity in animal models or human clinical trials because components in serum and/or body fluids, such as divalent cations and salts, inhibit their bioactivities. For instance, the bactericidal activity of human skin-derived psoriasin, a CaCl2-binding protein, is mediated by the depletion of zinc from bacteria, but its activity is inhibited by divalent cations (4). Thus, AMPs like psoriasin and hRNase 7 exert bactericidal activity only in environments deficient in divalent cations, and they cannot be employed as therapeutic agents (4, 6, 17). Here, we find that the bactericidal activity, the binding abilities to the whole bacteria and Lpp protein of SMAP-29, were resistant to 4 mm MgCl2 and 0.15 m NaCl. However, the bactericidal activity of LL-37 was labile to the above-mentioned divalent cations and salts, although its binding abilities to the whole bacteria and Lpp protein were resistant. With respect to the coiled Arg-riched oligopeptide, (R3S1)3, both bactericidal activity and binding ability to the whole bacteria were inhibited by these two compounds (Figs. 1 and 7). SMAP-29 contains two major domains, the N-terminal amphipathic α-helical region (residues 8–17) for antimicrobial activity and the C-terminal hydrophobic region (residues 20–28) for hemolytic activity. It shows disordered structure in aqueous buffers and significant helicity in membrane-like environments (29, 30). In addition, the secondary structure (percentage of α-helix) of cationic α-helical LL-37 varies with the concentrations of trifluoroethanol or lipid A in the environments (13). Therefore, we suggest that cationic α-helical AMP may be composed of two domains, one for Lpp-binding and the other for bactericidal activity. The structure of the individual domain is critical for its function and labile to the components in environments, like divalent cations, salts, and hydrophobicity.
It is interesting to find the role of Lpp on the bactericidal pathway caused by AMP. Lpp is known to be one of major outer membrane proteins of E. coli (7.2 × 105 molecules/cell), which exists in two major forms, one bound form linked to the peptidoglycan of cell wall through C-terminal lysine residue and the other free form without anchoring, but both contain modified N-acyl-S-diacylglycerylcysteine at the N terminus for the outer membrane integration (18, 19, 31, 32). E. coli mutants deficient in Lpp form large blebs on their surface, leak periplasmic enzymes into the medium, and have difficulty forming septa during cell division (33). The structure of Lpp is proposed to form a tubular channel through the outer membrane with a hydrophilic interior and hydrophobic outer surface, which act as a barrier against antibiotics and metabolites (20). The Lpp assembly is stabilized in the outer membrane not only by the hydrophobic interaction between its hydrophobic outer surface and the membrane lipid bilayer but also by three lipid acids linked to N-terminal cysteine. Furthermore, the Lpp is also linked to the peptidoglycan of cell wall by C-terminal lysine (19). By contrast, the Lpp is proposed to be composed of trimeric polypeptides from studies using recombinant Lpp proteins devoid of the N-terminal cysteine. The structure of one Lpp-hybrid protein, which contains the signal peptide, and the Ala-Glu residue of the OmpF N terminus and the Cys-1-deficient Lpp, is determined to be trimeric by the cross-linking with dithiobis (succinimidyl propionate) and dimethyl suberimidate, respectively (34). The structure of a truncated Lpp, Lpp56, deficient in Cys-1 and Lys-58 at both ends, has been determined to be a trimeric α-helix with a diameter of ∼21 Å and a length of ∼83 Å on x-ray crystallography (21). With respect to native Lpp of E. coli in the addition to monomer, dimer, and trimer, a fourth band of free form Lpp, probably tetramer or more, is observed on SDS-PAGE after cross-linking with increasing concentrations of disuccinimidyl suberate (32). In this study the recombinant Lpp possessed an N-terminal Cys-1 residue at which lipid acids were added for outer membrane integration and a C-terminal Lys-58 residue for cell wall anchoring. Based on the cross-linking experiments of recombinant Lpp with EDC and Lpp·SMAP-29 complex with glutaraldehyde, we suggest that the functional recombinant Lpp is composed of six α-helical polypeptides (hexamer). In addition, pairs of these oligopeptides may be tethered by disulfide bonds as well as intermolecular interactions because some dimeric Lpps are still tightly associated and preferentially recognized by anti-Lpp antibody, even reduced by 2-mercaptoethanol and separated by SDS-PAGE (Figs. 2, B–E, and 3, B–E).
In this report we found that interaction of SMAP-29 with Lpp resulted in the outward blebbing of the outer membrane, membrane permeability markedly increased, and the Lpp relocated from outer membrane to cytosol in K. pneumoniae (Fig. 6A). The dramatic damages to cellular structure caused by AMP occurring within minutes of primary challenge would not most likely allow bacteria to survive nor permit them to generate drug-resistant progeny. Although Lpp and OprI belong to different bacterial families, they possess three identical residues (Cys-Ser-Ser) at the N termini and could be recognized by SMAP-29, EDC cross-linking agent, and respective antibodies on bacterial surface. However, the Lpp was internalized into cytosol and not detectable on the bacterial surface in minutes through an unknown mechanism after SMA-29 treatment (17) (Figs. 1E and 6, B–D). Thus, a specific motif near N terminus of Lpp/OprI is exposed to outer environments of bacteria and is suggested to be a target site for cationic AMP. Screening or design of drugs against the specific motif of Lpp/OprI would provide a good chance to circumvent the current emergence of drug-resistance in Gram-negative bacteria. In this report, we retrieved cationic α-helical SMAP-29 and cationic α-helix-containing hRNase 7 from a library that contains crude lysate of human cancer cell CL1-0 to mimic the natural source of materials (Fig. 8).
Supplementary Material
Acknowledgments
We thank Dr. Helene Rosenberg for critical reading and suggestions on how to improve the manuscript. We also thank the core facility of the Institute of Cellular and Organismic Biology (Academia Sinica) for assistance in immunohistochemistry.
This work was supported by Academia Sinica and National Science Council Grant NSC 98-2311-B-001-017-MY3.

This article contains supplemental Fig. 1.
- AMP
- antimicrobial peptide/protein
- cfu
- colony-forming unit(s)
- hRNase 7
- human ribonuclease 7
- SMAP-29
- sheep myeloid antimicrobial peptide 29
- EDC
- 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride
- Lpp
- murein lipoprotein.
REFERENCES
- 1. Zasloff M. (2002) Nature 415, 389–395 [DOI] [PubMed] [Google Scholar]
- 2. Brown K. L., Hancock R. E. (2006) Curr. Opin. Immunol. 18, 24–30 [DOI] [PubMed] [Google Scholar]
- 3. Bowdish D. M., Davidson D. J., Hancock R. E. (2005) Curr. Protein Pept. Sci. 6, 35–51 [DOI] [PubMed] [Google Scholar]
- 4. Gläser R., Harder J., Lange H., Bartels J., Christophers E., Schröder J. M. (2005) Nat. Immunol. 6, 57–64 [DOI] [PubMed] [Google Scholar]
- 5. Harder J., Schroder J. M. (2002) J. Biol. Chem. 277, 46779–46784 [DOI] [PubMed] [Google Scholar]
- 6. Harder J., Bartels J., Christophers E., Schroder J. M. (2001) J. Biol. Chem. 276, 5707–5713 [DOI] [PubMed] [Google Scholar]
- 7. Schröder J. M., Harder J. (2006) Cell. Mol. Life Sci. 63, 469–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brogden K. A., Kalfa V. C., Ackermann M. R., Palmquist D. E., McCray P. B., Jr., Tack B. F. (2001) Antimicrob. Agents Chemother. 45, 331–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zaiou M., Nizet V., Gallo R. L. (2003) J. Invest. Dermatol. 120, 810–816 [DOI] [PubMed] [Google Scholar]
- 10. Travis S. M., Anderson N. N., Forsyth W. R., Espiritu C., Conway B. D., Greenberg E. P., McCray P. B., Jr., Lehrer R. I., Welsh M. J., Tack B. F. (2000) Infect Immun. 68, 2748–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Steinberg D. A., Hurst M. A., Fujii C. A., Kung A. H., Ho J. F., Cheng F. C., Loury D. J., Fiddes J. C. (1997) Antimicrob. Agents Chemother. 41, 1738–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zavascki A. P., Goldani L. Z., Li J., Nation R. L. (2007) J. Antimicrob. Chemother. 60, 1206–1215 [DOI] [PubMed] [Google Scholar]
- 13. Turner J., Cho Y., Dinh N. N., Waring A. J., Lehrer R. I. (1998) Antimicrob. Agents Chemother. 42, 2206–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Boix E., Nogués M. V. (2007) Mol. Biosyst. 3, 317–335 [DOI] [PubMed] [Google Scholar]
- 15. Peterson A. A., Fesik S. W., McGroarty E. J. (1987) Antimicrob. Agents Chemother. 31, 230–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Epand R. M., Vogel H. J. (1999) Biochim. Biophys. Acta 1462, 11–28 [DOI] [PubMed] [Google Scholar]
- 17. Lin Y. M., Wu S. J., Chang T. W., Wang C. F., Suen C. S., Hwang M. J., Chang M. D., Chen Y. T., Liao Y. D. (2010) J. Biol. Chem. 285, 8985–8994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Inoyye S., Takeishi K., Lee N., DeMartini M., Hirashima A., Inouye M. (1976) J. Bacteriol. 127, 555–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sankaran K., Wu H. C. (1994) J. Biol. Chem. 269, 19701–19706 [PubMed] [Google Scholar]
- 20. Inouye M. (1974) Proc. Natl. Acad. Sci. U.S.A. 71, 2396–2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shu W., Liu J., Ji H., Lu M. (2000) J. Mol. Biol. 299, 1101–1112 [DOI] [PubMed] [Google Scholar]
- 22. Mizushima S., Yamada H. (1975) Biochim. Biophys. Acta 375, 44–53 [DOI] [PubMed] [Google Scholar]
- 23. Pilhofer M., Ladinsky M. S., McDowall A. W., Jensen G. J. (2010) Methods Cell Biol. 96, 21–45 [DOI] [PubMed] [Google Scholar]
- 24. Huang Y. C., Lin Y. M., Chang T. W., Wu S. J., Lee Y. S., Chang M. D., Chen C., Wu S. H., Liao Y. D. (2007) J. Biol. Chem. 282, 4626–4633 [DOI] [PubMed] [Google Scholar]
- 25. Shepard B. D., Gilmore M. S. (2002) Microbes Infect. 4, 215–224 [DOI] [PubMed] [Google Scholar]
- 26. De Pascale G., Wright G. D. (2010) Chembiochem 11, 1325–1334 [DOI] [PubMed] [Google Scholar]
- 27. Mahamoud A., Chevalier J., Alibert-Franco S., Kern W. V., Pagès J. M. (2007) J. Antimicrob. Chemother. 59, 1223–1229 [DOI] [PubMed] [Google Scholar]
- 28. Pagès J. M., James C. E., Winterhalter M. (2008) Nat. Rev. Microbiol. 6, 893–903 [DOI] [PubMed] [Google Scholar]
- 29. Tack B. F., Sawai M. V., Kearney W. R., Robertson A. D., Sherman M. A., Wang W., Hong T., Boo L. M., Wu H., Waring A. J., Lehrer R. I. (2002) Eur. J. Biochem. 269, 1181–1189 [DOI] [PubMed] [Google Scholar]
- 30. Shin S. Y., Park E. J., Yang S. T., Jung H. J., Eom S. H., Song W. K., Kim Y., Hahm K. S., Kim J. I. (2001) Biochem. Biophys. Res. Commun. 285, 1046–1051 [DOI] [PubMed] [Google Scholar]
- 31. Inouye M., Shaw J., Shen C. (1972) J. Biol. Chem. 247, 8154–8159 [PubMed] [Google Scholar]
- 32. Cowles C. E., Li Y., Semmelhack M. F., Cristea I. M., Silhavy T. J. (2011) Mol. Microbiol. 79, 1168–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fung J., MacAlister T. J., Rothfield L. I. (1978) J. Bacteriol. 133, 1467–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Choi D. S., Yamada H., Mizuno T., Mizushima S. (1986) J. Biol. Chem. 261, 8953–8957 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.