Background: PHF20 is a methyl lysine binding protein that is a component of the MOF histone acetyltransferase protein complex.
Results: PHF20 knockout mouse die just after birth but display normal H4K16Ac levels.
Conclusion: Promoters that are marked with high H4K16Ac levels display reduced gene expression when PHF20 is lost.
Significance: PHF20 works downstream of the H4K16Ac mark to regulate transcription.
Keywords: Epigenetics, Protein Domains, Protein Methylation, Transcription Coactivators, Transgenic Mice, Lysine Acetylation, Effector Protein, Lysine Methylation, Tudor Domain
Abstract
In epigenetic signaling pathways, histone tails are heavily modified, resulting in the recruitment of effector molecules that can influence transcription. One such molecule, plant homeodomain finger protein 20 (PHF20), uses a Tudor domain to read dimethyl lysine residues and is a known component of the MOF (male absent on the first) histone acetyltransferase protein complex, suggesting it plays a role in the cross-talk between lysine methylation and histone acetylation. We sought to investigate the biological role of PHF20 by generating a knockout mouse. Without PHF20, mice die shortly after birth and display a wide variety of phenotypes within the skeletal and hematopoietic systems. Mechanistically, PHF20 is not required for maintaining the global H4K16 acetylation levels or locus specific histone acetylation but instead works downstream in transcriptional regulation of MOF target genes.
Introduction
After reading epigenetic signals, effector molecules elicit influences on transcription, DNA repair, and replication by virtue of intrinsic enzymatic activity or by acting as a scaffolding molecule that recruits a protein complex harboring enzymatic activity. The list of known effector molecules is constantly growing (1). One approach utilized to identify these histone code readers is the screening of protein domain microarrays. Using this approach, we identified a handful of new reader domains, including a Tudor domain within an uncharacterized protein, plant homeodomain finger protein 20 (PHF20, also called C20orf104 or GLEA2) (2). At the time of this discovery, literature concerning PHF20 was limited but did include findings of autoantibodies against PHF20 in patients suffering from various cancers, including glioblastoma, hepatocellular carcinoma, and medulloblastoma (3). Recently, protein biochemistry has revealed the presence of PHF20 and a close ortholog, PHF20L1, in the NSL2 complex (4–7). Besides PHF20, the NSL complex contains the primary H4K16 histone acetyltransferase MOF, the O-linked N-acetylglucosamine transferase enzyme OGT, the scaffolding molecule WDR5, HCF-1, and NSL1–3 (4), suggesting that it is a major player in epigenetic gene regulation. Indeed, in flies, the NSL complex is a powerful transcriptional activation complex (8, 9). Although PHF20 is known to be in the NSL complex, its function both within and outside of this context is unknown. Therefore, we generated PHF20 knockout mice to start uncovering the biological roles of PHF20. We set out to answer the following questions. Where is PHF20 important during mouse development, and how is MOF activity (H4K16ac deposition and transcriptional regulation) affected by loss of PHF20?
EXPERIMENTAL PROCEDURES
Southern Blot Genotyping
Approximately 800 bp of intronic PHF20 sequence downstream of the trap cassette insertion site was cloned out to use as a template for probe generation by PCR. The genomic DNA from WT and targeted mouse embryonic stem cell (mES) clones was isolated and digested with a panel of restriction enzymes with identified sites external to the trap insertion. Southern blot analyses were performed on the panel of differentially digested DNA to screen for the restriction enzymes (RE) that produced different size bands for the WT and KO alleles. After the trap insertion was verified, the targeted mES line(s) were given to the Genetically Engineered Mouse Core Facility of the M. D. Anderson Cancer Center for blastocyst injection and chimeric mouse production.
Alcian/Alizarin Stain
The bone and cartilage stain protocol used is in accordance with Ref. 22. Newborn pups or E18.5 embryos from heterozygous mating crosses were euthanized by placing them on ice for 30 min, and then the bodies were scalded in hot tap water (65–70 °C) for 20–30 s, which aids in skin removal with forceps. The body cavity was cleaned out, and the carcasses were fixed in 95% ethanol overnight. The next day they were transferred to acetone to remove the fat and left there overnight at room temperature. The mice were then rinsed with double distilled H2O, followed by a 24-hour soak in Alcian blue stain (for cartilage, 5% acetic acid, 0.05% Alcian blue 8GX). The stain was washed out with two to three changes of 70% ethanol over a 6- to 8-h time period. To clear the tissues, the mice were then soaked in 1% KOH until tissue clearing was complete, around 5 h for newborns. The Alizarin bone stain was then added for an overnight soak (50 mg alizarin red (Sigma-Aldrich) in 1L 1% KOH). The mouse skeleton was finally cleared by soaking in 1% KOH, 20% glycerol for 1–2 days. The samples were then moved to a permanent storage 1:1 solution of glycerol:ethanol.
MicroCT Bone Scans
Adult (day 25) femurs were fixed in formalin for 48 h and then transferred to 70% ethanol and stored at 4 °C until analysis. The bones were sent to the Baylor College of Medicine MicroCT Core, where they were scanned at a 16-micron resolution.
Protein Domain Arrays and Peptide Pull-downs
Experiments were performed as described in Ref. 2.
Flow Cytometry Analysis
Timed pregnancies were set up to harvest embryos at E17.5-E18.5. The embryos were euthanized on ice for 10 min, and then the thymuses were removed and placed on ice in RMPI + 5% FCS. The thymuses were pressed through a 70-micron cell strainer using 25 ml of cold PBS + 5% FCS to wash the thymocytes through the mesh. This was spun down to pellet cells and then resuspended in 5 ml of PBS + 5% FCS for cell counts. 1–2 million cells were used per embryo for immunolabeling. All antibodies were allowed to bind the respective cell surface proteins for 15 min on ice, followed by a single wash in PBS + 5% FCS. The following antibodies were used: CD4-PE (BD Biosciences L3T4, catalog no. 553049), CD8-FITC (eBiosciences, catalog no. 11-0081-85), CD3-Biotin (eBiosciences, catalog no. 13003185), CD45-PerCP Cy5.5 (eBiosciences, catalog no. 450451-82). For the CD3 label, secondary SA-Qdot655 (Invitrogen, catalog no. Q10121MP) was added for 15 min and then washed out as above. Analysis was performed at the Cell and Tissue Analysis Core of the M. D. Anderson Cancer Center, Science Park, TX, using the BD Biosciences LSR Fortessa machine.
Acid Extraction of Histones
Embryonic mouse tissue was harvested for histone acid extraction. The following protocol is for ∼100 mg of tissue. The tissue was minced with fine scissors in cold PBS containing protease inhibitors and then spun down at a low speed to pellet the tissue. The cell pellet was suspended in 800 μl of ice-cold RSB buffer (10 mm Tris·HCl (pH 7.4)/10 mm NaCl/3 mm MgCl2) and then centrifuged at a low speed (2500 rpm) for 5 min at 4 °C. The pellet was resuspended in 800 μl of RSB plus 0.5% Nonidet P-40 and homogenized, and the cellular membrane was broken with 50 pulses in a loose-fitting glass homogenizer and then placed on ice for 10 min. The lysed cells were then centrifuged again (2500 rpm) for 5 min at 4 °C. The pellet (nuclei) was resuspended in 400 μl of 5 mm MgCl2. Then 400 μl of 0.8 m HCl was added, and histones were extracted on ice for 20 min. This was spun down at 14,000 rpm for 10 min at 4 °C, and the supernatant (containing the histones) was removed and carefully transferred to a new tube. The histones were precipitated with 800 μl of 50% (w/v) TCA acid and then centrifuged at 14,000 rpm for 20 min at 4 °C. The pelleted histones, which collect on the tube walls, were washed once in acetone:0.3 M HCl, then twice with cold acetone, and allowed to air-dry completely. The purified and dried histones were resuspended in 100 μl of deionized water and 2 μl of 1.0 m Tris-HCL (pH 8.8).
Tissue ChIP
Timed pregnancies were used to obtain E14.5 embryos. The whole head of each embryo was used for ChIP. The head (brain tissue) was minced with fine scissors and then cross-linked with 10 ml of 1% formaldehyde in PBS per gram of tissue for 10 min at room temperature with constant rocking. To stop the reaction, glycine was added for a final concentration of 0.125 m for an additional 5 min. The tissue was spun down at 2500 rpm for 5 min, and the fixing solution removed. Then the tissue was washed two times with PBS. From this point on, the tissue was kept on ice, and protease and phosphatase inhibitors were added to buffers. The fixed tissue was resuspended in 10 ml of PBS per gram of tissue, then homogenized with a loose-fitting glass homogenizer (50 strokes), and spun down at 2500 rpm for 5 min. The tissue was then resuspended in FA lysis buffer (8 ml/gram of tissue, 50 mm HEPES-KOH (pH7.5), 140 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS) and sonicated on ice in 400-μl aliquots five times at 40% output for 20 s each and then cleared by high-speed centrifugation. 50 μl of the sonicated material was used to generate the input DNA and to check shearing efficiency, aiming for most of the DNA being between 100 and 500 bp after shearing. Using the input DNA to estimate the chromatin concentration, 25 μg of DNA-chromatin was used per ChIP, diluted 1:10 in radioimmune precipitation assay buffer (50 mm Tris-HCl (pH8.0), 150 mm NaCL, 2 mm EDTA, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS). 4 μl of H4K16Ac antibody (Active Motif, catalog no. 39167) or 4 μl of IgG antibody (Millipore, catalog no. 12-370) was added to this and allowed to bind overnight at 4 °C. The next day, 40 μl of ChIP beads (Millipore, catalog no. 16-157) were added to each ChIP and allowed to bind the complexes for 2 h at 4 °C. The beads-chromatin complexes were then subjected to the following washes: once in low-salt wash buffer (0.1% SDS, 1% Triton-X-100, 2 mm EDTA, 150 mm NaCl, 20 mm Tris-HCl pH 8.0) for 10 min at 4 °C, once in high-salt wash buffer (0.1% SDS, 1% Triton-X-100, 2 mm EDTA, 500 mm NaCl, 20 mm Tris-HCl (pH 8.0)), once in Li-Cl buffer (Millipore, catalog no. 20-156), and finally twice in TE buffer (10 mm Tris-Cl, pH 8.0, 1 mm EDTA). The chromatin was eluted into two 15-min washes of 250 μl of fresh elution buffer (1% SDS, 100 mm NaHCO3), and the eluates were combined. To this, 20 μl of 5 m NaCl was added, and this was incubated at 65 °C overnight to reverse the cross-links. The next day, 10 μl of 0.5 m EDTA, 20 μl of 1 m Tris-HCl (pH 6.5), and 20 μg of proteinase K was added to each sample and incubated at 45 °C for 1 h. Finally, phenol:chloroform extraction followed by ethanol precipitation was used to extract and purify the DNA from each ChIP. The DNA was then subjected to deep sequencing on the Illumina Solexa Genome Analyzer II at the Center for Cancer Epigenetics Solexa Sequencing Core of the M. D. Anderson Cancer Center. For ChIP-sequence confirmation, the primer sets shown in the supplemental data were used in a traditional PCR reaction, templated with DNA obtained from the above protocol. FLAG-PHF20 or FLAG-MOF expression vectors were a gift from Dr. Yali Dou (University of Michigan).
ChIP-Seq Analysis
In total, 36,312,750 and 37,585,722 raw reads were generated from the wild-type and knockout samples, of which 61.17% and 61.26% passed quality filtering and were uniquely mapped to the mouse reference genome version NCBI37/mm9. The uniquely mapped and high-quality reads were then subjected to model-based analysis for ChIP-sequence (MACS) (10) for H4K16ac intensity calculation at each base pair and peak calling. Briefly, the MACS parameter “mfold” was set to 3 to robustly estimate DNA fragment size (d) in each sample. Then each read was shifted for a distance of d/2 toward the 3′ direction and extended to be 100 bp long. H4K16ac intensity at each base pair was then calculated as coverage of these reads and further normalized by fold change to ensure that whole genome average intensity equaled 1 in each sample. A p value cutoff, 1e-5, was used to call H4K16ace intensity peaks. Finally, 393 and 581 peaks were identified from wild-type and knockout samples, respectively. 28405 refSeq genes (refGene) were obtained from the UCSC genome browser (11) to examine H4K16ac distribution around transcription start sites.
Real-time qPCR
The whole heads of E14.5 embryos were harvested and stored in RNAlater (Qiagen, catalog no. 76104) until RNA extraction, which was performed following the Qiagen RNeasy kit instructions, including a DNase digestion step (Qiagen, catalog no. 79254). The RNA (3 μg per embryo) was converted into 60 μl of first-strand cDNA using the Invitrogen SuperScript III First-Strand synthesis kit (catalog no. 18080-051). 2 μl of cDNA was used per real-time qPCR reaction using the IQ SYBR SuperMix from Bio-Rad (catalog no. 170-8880 mix. Traditional PCR using each primer set was performed first to ensure the specific amplification of one band per primer set. Then the real-time PCR reactions carried out on a 7900HT real-time PCR analyzer. The ΔCt method was used to calculate the relative expression of selected genes in the knockout embryos as compared with the wild-type embryos, using GAPDH as an internal calibrator. The gene expression analysis of three sets of embryos was performed, each in triplicate. One representative embryo pair was used to generate Fig. 4E. Primer sets shown in the supplemental data were used in the RT-qPCR reactions.
FIGURE 4.
PHF20 is not required for global or locus-specific H4K16Ac, but loss of PHF20 negatively affects expression of H4K16Ac target genes. A, co-IP analysis confirms a strong interaction between endogenous PHF20 and MOF in embryonic brain tissue. WB, Western blot. However, loss of PHF20 in mice does not affect global levels of H4K16Ac, H4K8Ac, or H4K5Ac (B). C, to determine the requirement for PHF20 in locus-specific targeting of MOF as measured by H4K16Ac levels, we performed a ChIP-seq of H4K16Ac from WT and KO embryonic brain tissue. The H4K16Ac peaks identified in this experiment reside at the 5′ end of genes and closely flank the transcription start site. There was no difference in the average peak size or location with or without PHF20. D, snapshots of the UCSC mouse genome browser are shown to highlight H4K16Ac profiles for four selected genes, Ing1, Actb, Cdk4, and Morf4L1. E, RT-qPCR analysis of these four genes in E14.5 brain tissue revealed a striking decrease in expression levels when PHF20 is absent.
Antibodies
The characterization of the anti-PHF20 antibodies is shown in supplemental Fig. S1. Other antibodies used include anti-H4K16Ac (Active Motif, catalog no. 39167), anti-H4K8Ac (Active Motif, catalog no. 61104), anti-H4K5Ac (Active Motif, catalog no. 39584), anti-actin (Sigma, catalog no. A5441), anti-MOF (Bethyl, catalog no. A300–992A), and anti-FLAG (Rockland, catalog no. 600-401-383).
RESULTS
In a previous report, we demonstrated the use of protein microarrays as an approach to identify methyl-dependent protein domains (2) and identified the Tudor domain of PHF20 (here referred to as Tudor2′). The domain composition of PHF20 includes another Tudor-like domain, Tudor1, Tudor2, an AT hook, a Zn finger, a coiled-coil region, and a plant homeodomain (Fig. 1A), and hints toward a role for PHF20 in chromatin binding and regulation. We constructed a condensed domain array and probed this simultaneously with the histone tails of H3 and H4, dimethylated at lysines 9 and 20, respectively. Using two different fluorescent tags, we can visualize the binding to one or both of these methylated peptides. The Tudors of PHF20 and 53BP1 behave similarly on our array, binding both H3K9me2 and H4K20me2 (Fig. 1B). By reversing this experiment and using recombinant effector molecules to probe a library of arrayed peptides representing histone modifications, we consistently observe a striking overlap in the binding profile between the Tudor domains of DNA damage repair protein 53BP1 and that of PHF20 (12). Therefore, we initially predicted that PHF20 may also have a role in DNA damage response and repair. However, as will be described in the subsequent paragraphs, PHF20 knockout mice and 53BP1 knockout mice display very different phenotypes (13). Additionally, we looked for the presence of PHF20 at sites of DNA damage and were unable to visualize PHF20 at break foci following UV or ionizing radiation (data not shown). Therefore, we believe the biological functions of 53BP1 and PHF20 are distinct. To validate and describe the methyl lysine binding of PHF20 Tudor2, the critical residues Trp-97 and Tyr-103 within the aromatic cage were mutated. The sites were predicted on the basis of homology to the published structure of the tandem Tudor of 53BP1 (14). As expected, mutation of these two residues abolishes the dimethyl lysine binding abilities of a GST-PHF20 construct containing the Tudor1 and 2 domains (Fig. 1C).
FIGURE 1.
Analysis of the PHF20 Tudor2 domain and generation of PHF20 null mice. A, PHF20 harbors a number of protein domains. PHD, plant homeodomain. B, a condensed protein array was constructed to compare the binding profiles of PHF20 Tudor2 to other methyl lysine binding domains. PHF20 Tudor2 and the tandem Tudors of 53BP1 bound both H3K9me2 and H4K20me2 peptides. The Tudors of JMJD2A only bound H4K20me2, whereas the chromo of HP1β only bound H3K9me2. C, peptide pull-down assays reveal that the mutation of key residues (Trp-97 Tyr-103) within the predicted methyl-binding cage of PHF20 Tudor2 block dimethyl lysine binding. D, PHF20 knockout mice were generated from targeted mES clones obtained from BayGenomics. The gene trap (TRAP) insertion site is depicted and verified by Southern blot analysis with a probe external to the trap. ATG, initiator methionine. E, IP-Western analysis (WB) confirms the absence of PHF20 protein in the null mice. F, the murine PHF20 expression pattern was assessed using IP-Western.
Although it is clear that, in vitro, PHF20 can bind methylated histone tails, the in vivo role of full-length PHF20 remains unknown. We therefore generated PHF20 knockout mice, beginning with a gene trap-targeted mES cell line obtained from Bay Genomics (clone XN131). The location of the gene trap was confirmed by Southern blot analysis and resides between exons two and three (Fig. 1D, bottom panel). A splice acceptor within the gene trap hijacks the splicing machinery, resulting in the production of a fusion protein between the first 24 amino acids of the Tudor1 domain and β-galactosidase (trap disruption point in protein shown in Fig. 1A). To facilitate subsequent experiments we generated a panel of PHF20 antibodies, which were characterized in detail (supplemental Fig. S1). Using these reagents, we confirmed by IP-Western analysis that the KO mice do not express PHF20 (Fig. 1E). Using the same IP-Western analysis approach, tissues from newborn mice were analyzed for PHF20 protein expression. The expression pattern is not ubiquitous, but is primarily in the brain, thymus, and bone (Fig. 1F). In agreement, immunohistochemistry analysis of E18.5 brain sections shows strong PHF20 expression in the developing cerebral cortex (supplemental Fig. S1E). In mouse brain and thymus tissue, PHF20 appears in two bands (Fig. 1, E and F). However, in bone marrow, skeletal muscle, mouse embryonic fibroblasts, and mES cells, PHF20 is present as just one band around 150 kDa (Fig. S1, C and D). It is likely that the 120-kDa band observed in the brain and thymus is a low-abundant splice variant of PHF20, as is the 130-kDa band observed in heart tissue.
Full necropsy revealed many phenotypes upon loss of PHF20 in vivo, summarized in Fig. 2A. The most striking is perinatal lethality. 95% of the PHF20 knockout mice are born alive but die within the first day of life (Fig. 2C). The null pups are runted, weighing only 50% (Avg. 51% ± 9%, p = 0.007) that of their heterozygous and WT littermates. This size difference is retained in the few null mice that do survive the first day (Fig. 2B). The exact cause of death is currently unknown. Other major abnormalities were found in the skeleton and hematopoietic system. X-ray examination revealed the PHF20 knockouts as having a frail skeleton and a missing lumbar vertebra (Fig. S2, A and B). In agreement with this finding, we observed strong PHF20 expression in the developing ventral neural tube (supplemental Fig. S2C). Alcian/Alizarin bone and cartilage stains of newborn mice clearly show the reduction from six to five lumbar vertebra in the PHF20 KO mice (Fig. 2D). A delay in bone formation is also evident in the KO mouse. There are many fewer areas of calcified bone (red) in the KO as compared with its WT littermate. Concerning the hematopoietic system, the cellularity within bone marrow of day 25 adult mouse vertebrae is reduced in the PHF20 knockout mice (Fig. S2D). The spleen and thymus of PHF20 knockout E18.5 embryos and surviving day 25 adults are smaller (after normalization to total body weight), with the spleen demonstrating disorganization between the white and red pulp regions (supplemental Fig. S2E). Together, we observe defects in many different hematopoietic tissues, including the hematopoietic stem/progenitor containing bone marrow and tissues populated with cells derived from both lymphoid and erythroblast progenitors. This suggests a defect early in hematopoietic stem/progenitor cell differentiation or proliferation. Although this possibility is currently under investigation, for this study we performed a more detailed analysis focusing on thymic-dependent hematopoiesis, as there is a marked decrease in the number of thymocytes in PHF20-null embryos (Fig. 2E).
FIGURE 2.
Loss of PHF20 in vivo results in a wide variety of developmental phenotypes. Those observed to date are listed in A. B, PHF20 KO embryos and surviving adults are smaller than normal, with 95% of PHF20 KO mice dying before weaning (C). D, newborn mice (left panel) and E18.5 embryos (right panel) were subjected to Alcian/Alizarin bone (red) and cartilage (blue) staining. The PHF20 KO newborn and embryo clearly display a missing lumbar vertebra and a reduction in the bone-to-cartilage ratio. E, the total number of thymocytes within E18.5 PHF20 KO thymi are significantly reduced.
We asked if PHF20 knockout mice are defective in T cell development. Thymocytes were obtained from PFH20-deficient or wild-type embryos at E17.5 or E18.5. The frequency and number of thymocyte subsets defined by CD4 and CD8 expression was determined by FACS analysis. The most immature thymocyte precursors do not express CD4 or CD8 and are thus termed double negative (DN) thymocytes. DN thymocytes that undergo successful TCRβ gene rearrangement express pre-TCR molecules that promote survival and differentiation to the CD4+CD8+ double positive (DP) maturation stage. Subsequent rearrangement of the TCRα locus in DP thymocytes results in expression of αβ-TCRs. DP thymocytes that express αβ-TCRs with moderate affinity for self peptide-MHC complexes are positively selected for continued maturation. Positively selected DP thymocytes down-regulate either CD4 or CD8 to become single positive (SP) thymocytes that migrate into the medulla where they are subjected to a negative selection process that deletes autoreactive cells. PHF20-deficient mice have a significant increase in the percentage and number of DN thymocytes compared with littermate controls (Fig. 3, A and C). There is a corresponding decrease in DP thymocyte subsets, indicating that PHF20 knockout mice have a partial block in thymocyte maturation at the DN stage. Since the DN population is heterogeneous, additional FACS analyses are required to precisely identify the DN subset(s) affected by loss of PHF20. Nevertheless, it is clear that PHF20 plays a role in maturation of thymocyte precursors. CD3 expression is up-regulated when DP thymocytes are positively selected and differentiate to the single positive stage. Interestingly, we find that CD4+CD8-SP thymocytes from PHF20 deficient mice fail to up-regulate CD3 expression (Fig. 3B). This observation suggests that loss of PHF20 impairs thymocyte differentiation at relatively mature, as well as immature, stages of thymocyte maturation.
FIGURE 3.
PHF20 knockout mice display impaired thymocyte development. A, thymocyte subsets defined by CD4 and CD8 expression. FACS plots show an increased percentage of DN thymocytes and a corresponding decreased percentage of DP thymocytes in the PHF20 KO mice (C). B, the expression of another marker of thymocyte maturation, CD3, was also evaluated. Within the mature CD4+ population, the levels of CD3 in the PHF20 knockout thymocytes is markedly reduced (dashed lines) compared with WT littermates (solid lines), also demonstrating a block in thymocyte development in the absence of PHF20. When considering the same number of events for all the embryos, the total number of thymocytes within each subset is graphed in C, showing the significant differences between WT and KO thymocyte distribution within the DN, DP, and CD8+ single positive populations. These data suggest that PHF20 knockout mice have a block at the DN stage of development.
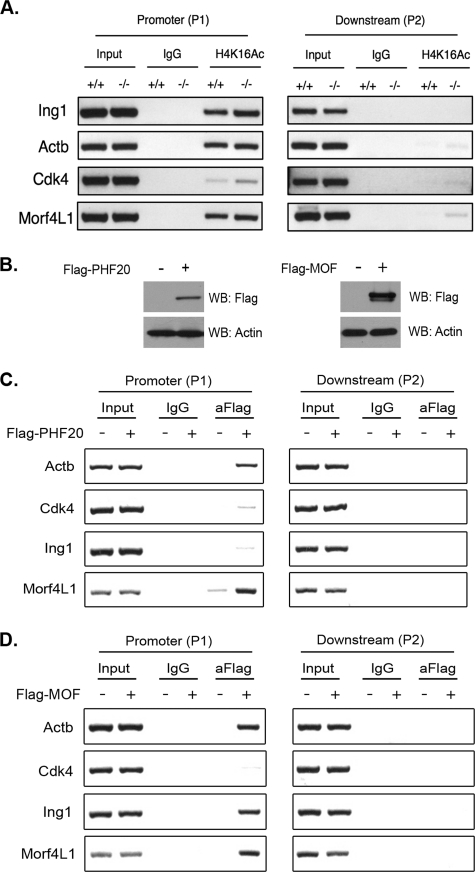
PHF20 is a key member of the NSL complex in flies and mammals (4, 5, 9). The NSL complex is one of two protein complexes that contain the primary H4K16 acetyltransferase MOF (MYST1, KAT8). H4K16 acetylation is a powerful histone modification. This single alteration to the histone tail can decondense chromatin by disrupting internucleosomal and inter-30-nm fiber binding to promote transcriptional activation (15, 16). Furthermore, MOF activity in Drosophila allows for the 2-fold increase in transcription of the male X chromosome, providing dosage compensation and transcriptional output equivalent to that of two female X chromosomes (17, 18). Previous studies have shown that loss or reduction of NSL complex components, either the enzymatic component MOF or non-enzymatic components MRCS2 or MSL1, results in global decreases of H4K16 acetylation (5, 9, 19). Therefore, we wondered if loss of PHF20 in our mouse model also resulted in global decreases in H4K16Ac. First, we confirmed the interaction between PHF20 and MOF in embryonic brain tissues. In agreement with the previous studies, we could strongly coimmunoprecipitate the two proteins (Fig. 4A). Second, we purified histones from E18.5 brain, as this organ expresses high amounts of PHF20 and MOF (Fig. 1F and supplemental Fig. S2F) and tested for changes in the bulk levels of various acetyl marks contributing to NSL complex activity. We could not detect a decrease in H4K16Ac, H4K8Ac, or H4K5Ac levels in the PHF20 KO tissues (Fig. 4B). This analysis was also extended to purified histones from E18.5 thymus (data not shown), primary MEFs derived from E14.5 embryos (data not shown), as well as cell culture-based experiments where PHF20 protein levels were reduced by RNAi (supplemental Fig. S2G). All of these experiments confirmed that loss of PHF20 does not affect global levels of H4K16Ac. However, as PHF20 contains many chromatin-binding domains, including the methyl-binding Tudor2 domain, we speculated that PHF20 was critical for correctly targeting MOF and the NSL complex to specific genomic locations and target genes. The targets of PHF20 itself are not known, and the PHF20 antibodies we raised do not ChIP, so we performed a ChIP-seq analysis of H4K16Ac peaks in WT and PHF20 KO embryonic mouse brains to mark global locations of NSL complex activity. In this way, we hoped to identify a subset of NSL complex target genes that also require PHF20 for proper levels of MOF activity and thus H4K16ac. Consistent with public H4K16Ac ChIP-seq data from mammalian cells (20), H4K16Ac peaks were located in the 5′ region of genes, flanking the transcription start site. Global genome comparison, however, revealed no difference in the location or intensity of H4K16Ac peaks between PHF20 WT and KO E14.5 brains (Fig. 4C). Recent work on chromatin recruitment and transcriptional activation of the Drosophila NSL complex is in agreement with our findings here. Raja et al. (9) found that RNAi knockdown of the Drosophila ortholog of PHF20, MBD-R2, did not affect NSL complex integrity or localization in the nucleus. Additionally, the depletion of another NSL complex component, MCRS2, reduced MBD-R2 occupancy on chromatin and H4K16Ac, suggesting that MRCS2 is the “ recruiter” of the NSL complex (9). Interestingly however, this same report, as well as a subsequent study from Prestel et al. (8) identified MBD-R2 as the key factor affecting the transcriptional activation ability of the NSL complex, especially on active autosomal gene targets. Thus, we compiled a short list of genes that display strong H4K16Ac peaks in their 5′ region and asked if their transcriptional output was hindered in the PHF20-null situation. The following genes were selected: Ing1, Actb, Cdk4, and Morf4L1. Displayed in Fig. 4D are the screen shots of the selected gene loci from the H4K16Ac ChIP-seq results. The brains from three independent pairs of E14.5 embryos were harvested to generate cDNA and perform an expression analysis by qPCR. In all three cases, we saw a marked reduction in the expression of target genes Ing1, Actb, and Morf4L1, with Cdk4 showing a less dramatic reduction in expression (Fig. 4E). Using traditional ChIP-PCR from E14.5 embryos brain tissue, we confirmed the enrichment of the H4K16Ac mark at these four loci (Fig. 5A). In NIH-3T3 cells, both FLAG-tagged PHF20 and MOF are enriched at the promoters of these selected genes (Fig. 5, B–D), demonstrating that they are indeed direct targets of the NSL complex. Thus, we conclude that PHF20 is acting downstream of MOF chromatin recruitment to aid in transcriptional activation.
FIGURE 5.
Recruitments of PHF20 and MOF to the promoters of genes with elevated H4K16Ac levels. A, traditional ChIP-PCR analysis was performed on four selected genes that showed strong H4K16Ac peaks in the ChIP-seq analysis, using WT and KO embryonic brain tissue. In agreement, H4K16Ac marked the promoter but not downstream regions of these genes, and there was no drop-in signal as a result of PHF20 absence. B, NIH-3T3 cells were transient transfected with either FLAG-PHF20 or FLAG-MOF in a ratio of 10 μg/10-cm plate. Cells were harvested 48 h after transfection, and the expressions of both proteins were detected through Western blot analysis using anti-FLAG antibody. C, NIH-3T3 cells were transfected as in B with FLAG-PHF20, and traditional ChIP-PCR assays were performed to test the enrichments of PHF20 at the promoters but not downstream regions of target genes. D, similar experiments were performed as in C, except that FLAG-MOF was transfected as indicated.
DISCUSSION
Our initial interest in PHF20 function manifested after discovering the Tudor2 domain as a major reader of dimethyl lysine and, therefore, likely a critical molecule involved in epigenetic-based mechanisms. In agreement with this prediction, PHF20 deletion in vivo results in a slew of developmental defects across a variety of tissue types. PHF20-null mice die in the perinatal period, are runted, show a delay in bone formation or defects in skeletal composition, and display hematopoietic defects including aberrant thymocyte development. The wide array of defects in the PHF20 KO mice suggests a broad role of PHF20 in proper and timely transcriptional activation. Indeed, investigation into the function of PHF20 within the NSL protein complex revealed that PHF20 is not required for global or locus-specific MOF activity, as gauged by H4K16Ac. However, loss of PHF20 does prohibit full transcriptional activation of selected NSL complex target genes, suggesting that it is a potent coactivator. The PHF20 mouse model, therefore, is a valuable tool for investigating the transcriptional roles of MOF and the NSL complex while avoiding the early lethality associated with MOF depletion (21). Initially, we assumed that the primary function of the Tudor2 of PHF20 was to bind methylated lysine residues on the histone tails to target its complex components. Although methyl-histone binding by PHF20 may provide stability for the NSL complex on chromatin, the results here suggest that PHF20 works downstream of NSL and MOF targeting. It is likely that many transcription factors and basal transcription machinery proteins are modified in ways that mimic the histone code (i.e. combinations of acetylation and methylation), generating a similar docking platform for effector molecules such as Tudor2 of PHF20. The list of identified methylated non-histone proteins is growing, but many more remain to be discovered. Currently, we are identifying proteins that bind PHF20 through the Tudor2 domain in a methyl-dependent fashion. In this way, we aim to identify methylated non-histone proteins linked to the NSL complex through PHF20 that will aid in further understanding of our findings here that PHF20 is required for normal development, viability, and MOF target gene expression.
Supplementary Material
Acknowledgments
We thank A. Espejo for PHF20 protein microarray work and the Center for Cancer Epigenetics and Center for Environmental and Molecular Carcinogenesis Centers at the M. D. Anderson Cancer Center for assistance.
This work was supported, in whole or in part, by National Institutes of Health Training Grant 5T32ES007247-19 (to A. B.) and NIEHS National Institutes of Health Institutional Grants ES007784 and DA023449 (to M. T. B.). This work was also supported by Cancer Prevention and Research Institute of Texas Grants RP110471 (to M. T. B.) and RP110471 (to W. L.).
Deep-sequencing data can be accessed with GEO accession number GSE29306.

This article contains supplemental Figs. S1 and S2 and data.
- NSL
- nonspecific lethal
- E18.5
- embryonic day 18.5
- qPCR
- quantitative PCR
- IP
- immunoprecipitation
- DN
- double negative
- DP
- double positive
- MOF
- H4K16 histone acetyltransferases
- mES
- mouse embryonic stem cells
- RE
- restriction enzymes.
REFERENCES
- 1. Yun M., Wu J., Workman J. L., Li B. (2011) Cell Res. 21, 564–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kim J., Daniel J., Espejo A., Lake A., Krishna M., Xia L., Zhang Y., Bedford M. T. (2006) EMBO Rep. 7, 397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Heisel S. M., Ketter R., Keller A., Klein V., Pallasch C. P., Lenhof H. P., Meese E. (2008) PloS ONE 3, e2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cai Y., Jin J., Swanson S. K., Cole M. D., Choi S. H., Florens L., Washburn M. P., Conaway J. W., Conaway R. C. (2010) J. Biol. Chem. 285, 4268–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li X., Wu L., Corsa C. A., Kunkel S., Dou Y. (2009) Mol. Cell 36, 290–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mendjan S., Taipale M., Kind J., Holz H., Gebhardt P., Schelder M., Vermeulen M., Buscaino A., Duncan K., Mueller J., Wilm M., Stunnenberg H. G., Saumweber H., Akhtar A. (2006) Mol. Cell 21, 811–823 [DOI] [PubMed] [Google Scholar]
- 7. Smith E. R., Cayrou C., Huang R., Lane W. S., Côté J., Lucchesi J. C. (2005) Mol. Cell. Biol. 25, 9175–9188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Prestel M., Feller C., Straub T., Mitlöhner H., Becker P. B. (2010) Mol. Cell 38, 815–826 [DOI] [PubMed] [Google Scholar]
- 9. Raja S. J., Charapitsa I., Conrad T., Vaquerizas J. M., Gebhardt P., Holz H., Kadlec J., Fraterman S., Luscombe N. M., Akhtar A. (2010) Mol. Cell 38, 827–841 [DOI] [PubMed] [Google Scholar]
- 10. Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., Liu X. S. (2008) Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fujita P. A., Rhead B., Zweig A. S., Hinrichs A. S., Karolchik D., Cline M. S., Goldman M., Barber G. P., Clawson H., Coelho A., Diekhans M., Dreszer T. R., Giardine B. M., Harte R. A., Hillman-Jackson J., Hsu F., Kirkup V., Kuhn R. M., Learned K., Li C. H., Meyer L. R., Pohl A., Raney B. J., Rosenbloom K. R., Smith K. E., Haussler D., Kent W. J. (2011) Nucleic Acids Res. 39, D876–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu H., Galka M., Iberg A., Wang Z., Li L., Voss C., Jiang X., Lajoie G., Huang Z., Bedford M. T., Li S. S. (2010) J. Proteome Res. 9, 5827–5836 [DOI] [PubMed] [Google Scholar]
- 13. Ward I. M., Minn K., van Deursen J., Chen J. (2003) Mol. Cell. Biol. 23, 2556–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Botuyan M. V., Lee J., Ward I. M., Kim J. E., Thompson J. R., Chen J., Mer G. (2006) Cell 127, 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shia W. J., Pattenden S. G., Workman J. L. (2006) Genome Biol. 7, 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shogren-Knaak M., Ishii H., Sun J. M., Pazin M. J., Davie J. R., Peterson C. L. (2006) Science 311, 844–847 [DOI] [PubMed] [Google Scholar]
- 17. Kind J., Vaquerizas J. M., Gebhardt P., Gentzel M., Luscombe N. M., Bertone P., Akhtar A. (2008) Cell 133, 813–828 [DOI] [PubMed] [Google Scholar]
- 18. Smith E. R., Pannuti A., Gu W., Steurnagel A., Cook R. G., Allis C. D., Lucchesi J. C. (2000) Mol. Cell. Biol. 20, 312–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Taipale M., Rea S., Richter K., Vilar A., Lichter P., Imhof A., Akhtar A. (2005) Mol. Cell. Biol. 25, 6798–6810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Z., Zang C., Cui K., Schones D. E., Barski A., Peng W., Zhao K. (2009) Cell 138, 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thomas T., Dixon M. P., Kueh A. J., Voss A. K. (2008) Mol. Cell. Biol. 28, 5093–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nagy A., Gertsenstein M., Vintersten K., Behringer R. (2003) Manipulating the Mouse Embryo, 3rd Ed., CSHL Press, Cold Spring Harbor, NY [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.