Background: We have examined the effect of PDI knockdown in pancreatic β-cells.
Results: Upon knockdown, proinsulin oxidation to form native disulfide bonds is enhanced and accompanied by improved exit from the ER with increased total insulin secretion.
Conclusion: We hypothesize that PDI exhibits unfoldase activity for proinsulin.
Significance: Unexpectedly, PDI increases retention of proinsulin within the ER of pancreatic β-cells.
Keywords: Endoplasmic Reticulum (ER), Insulin, Protein Export, Protein Folding, Protein Isomerase
Abstract
For insulin synthesis, the proinsulin precursor is translated at the endoplasmic reticulum (ER), folds to include its three native disulfide bonds, and is exported to secretory granules for processing and secretion. Protein disulfide isomerase (PDI) has long been assumed to assist proinsulin in this process. Herein we have examined the effect of PDI knockdown (PDI-KD) in β-cells. The data establish that upon PDI-KD, oxidation of proinsulin to form native disulfide bonds is unimpaired and in fact enhanced. This is accompanied by improved proinsulin exit from the ER and increased total insulin secretion, with no evidence of ER stress. We provide evidence for direct physical interaction between PDI and proinsulin in the ER of pancreatic β-cells, in a manner requiring the catalytic activity of PDI. In β-cells after PDI-KD, enhanced export is selective for proinsulin over other secretory proteins, but the same effect is observed for recombinant proinsulin trafficking upon PDI-KD in heterologous cells. We hypothesize that PDI exhibits unfoldase activity for proinsulin, increasing retention of proinsulin within the ER of pancreatic β-cells.
Introduction
Protein disulfide isomerase (PDI,3 gene name: P4HB) is an evolutionarily conserved protein folding catalyst that influences disulfide bond formation of substrate proteins in the endoplasmic reticulum (ER) (1, 2). PDI is part of a large family of ER oxidoreductases (3) and is one of the more abundant transcripts (see entry for P4HB in the T1DBase) in the human pancreatic islet transcriptome (4) and proteome (5). In principle, disulfide isomerization (reshuffling of intramolecular disulfide bonds in substrate proteins) involves both reduction of improper cystine pairings and reoxidation of substrates to make proper disulfide bonds, so the catalytic capabilities of PDI are, at the very minimum, “bidirectional” (6). PDI can also influence substrate protein folding via its peptide-binding domain, as a molecular chaperone (7).
We are interested in the folding of proinsulin, which is essential for efficient insulin production in pancreatic β-cells (8, 9). Some in vitro studies have implicated PDI in proinsulin folding (10, 11), whereas other studies have implied a direct role for PDI in proinsulin oxidation in the ER of β-cells (12). ER-oxidoreductin 1-β (ERO1β) in pancreatic β-cells has recently been found important for proinsulin oxidation, which is linked to enhanced proinsulin export from the ER (13, 14), and ERO1β has been shown to oxidize PDI in vitro (13).
By contrast in some cases, PDI can retain proteins within the ER, a phenomenon that has been called “anti-chaperone activity” (15) but might be more appropriately considered “unfoldase activity” (16). Interestingly, even in native proinsulin or insulin, the C(B7)–C(A7) disulfide bond is nearly fully exposed at the surface of the folded polypeptide (17), rendering it potentially susceptible to attack by PDI (18). Indeed, overexpression of PDI in pancreatic β-cells actually lowers glucose-stimulated insulin secretion and induces ER stress (19). Therefore, despite current assumptions, it is far from clear that PDI is required to promote net oxidation of proinsulin and to facilitate its export from the ER. We now report that knockdown of PDI expression does not perturb proinsulin oxidation and actually promotes its export, increasing insulin secretion. The results indicate that PDI acts as a retention factor limiting proinsulin egress from the ER in pancreatic β-cells.
EXPERIMENTAL PROCEDURES
Materials
Anti-PDI was from Dr. P. Kim (University of Cincinnati) and from StressGen; anti-ERO1β from Proteintech; anti-IAPP from Dr. B. Verchere (University of British Columbia); anti-KDEL from StressGen; anti-ERp72 from Enzo Life Sciences; anti-P5 from Thermo Scientific; anti-ERp57 from Dr. D. Williams (University of Toronto); anti-phospho-eIF2α from Cell Signaling; anti-γ-tubulin and mAb M2 anti-FLAG from Sigma; and guinea pig anti-insulin from Millipore. Wild-type human α1-antitrypsin (AAT) cDNA in pCDNA3 was from Dr. R. Sifers (Baylor College of Medicine); rabbit anti-AAT was from DAKO.
Cell Culture and Transfection
293 and HepG2 cells were grown in DMEM (containing 4.5 g/liters d-glucose, 2.5 mm l-glutamine, and 110 mg/liters sodium pyruvate) plus 10% FBS; Min6 medium was the same and supplemented with 140 μm 2-mercaptoethanol. INS1 cells were grown in RPMI 1640 (11 mm glucose) supplemented with 10% FBS, and 28 μm 2-mercaptoethanol.
PDI knockdown in INS1 and 293 cells employed transfection with 40 nm siRNA using RNAiMax Lipofectamine reagent (Invitrogen); plasmid transfection in these cells used Lipofectamine 2000 (Invitrogen), both following the manufacturer's instructions. The sequences of ERO1β siRNA were as follows: GCGCUCAAUUGUUGAUCUUTT and GCUAAGUAACGAAAGCAAATT. The sequences of PDI siRNA were as follows: for INS1, GGGAGAGACAUACAAGGAUTT and GCGCAUACUUGAGUUCUUUTT, and for 293 and HepG2, GACCUCCCCUUCAAAGUUGUU and CCGACAGGACGGUCAUUGAUUACAA. siRNA duplex controls for GFP or luciferase knockdown were from Invitrogen.
Metabolic Labeling
At 72 h after transfection, cells were labeled with 100 μCi of [35S]Cys/Met (MP Biomedicals) in Cys/Met-free DMEM. When indicated, cells were washed and incubated with 10 mm N-ethylmaleimide in ice-cold PBS and then lysed in 1% Nonidet P-40, 0.1% SDS, 150 mm NaCl, 2 mm EDTA, 10 mm Tris-HCl (pH 7.4) containing 10 mm NEM and a protease inhibitor mixture. Lysates were immunoprecipitated overnight with anti-insulin, boiled in SDS sample buffer ± 0.1 m DTT, and analyzed by Tris-Tricine-urea-SDS-PAGE.
RIA and ELISA
An RIA that detects proinsulin+insulin of all species (Millipore) was used in all experiments. Media were collected for 24 h, at which time total insulin levels were assessed in cell lysates and media. Secretion of IAPP was measured by ELISA cross-reacting with rat IAPP (Millipore).
Expression of WT and Mutant PDI
An expression plasmid encoding mouse PDI (in pcDNA3.1; from Dr. B. Tsai, University of Michigan) was mutated by site-directed mutagenesis (Stratagene) such that all catalytic Cys residues (Cys-58, Cys-62, Cys-398, Cys-402) were changed to Ala to encode the “PDI-DEAD” protein or to a single C62A to encode the “PDI-TRAP” protein (20, 21). In the experiments in Fig. 1E, 3 × 106 cells were transfected using the AMAXA nucleoporator (14). In addition to PDI-KD, plasmids encoding FLAG-tagged WT-PDI, PDI-DEAD, PDI-TRAP, or GFP (control) were transfected. At 72 h after transfection, cells were labeled for 20 min with 500 μCi of [35S]Cys/Met. The cells were washed once with ice-cold Hanks' buffer with 20 mm NEM and then lysed in buffer containing 15 mm NEM. Then lysates were split in half; one-half was immunoprecipitated with anti-insulin, and the other half was immunoprecipitated with mAb M2 anti-FLAG. Immunoprecipitates were analyzed by 4–12% gradient SDS-PAGE (Invitrogen) followed by autoradiography.
FIGURE 1.
PDI-KD enhances proinsulin export from ER. A, at 72 h after transfection with either scrambled or ERO1β (panels at left) or PDI siRNA duplexes (panels at right) (40 nm), INS1 cells were pulse-labeled for 15 min with [35S]Cys/Met and lysed in SDS buffer with 10 mm NEM. Lysates were immunoprecipitated in duplicate with anti-insulin and analyzed by Tris-Tricine-urea-SDS-PAGE under nonreduced or reduced (100 mm DTT) conditions. These data were replicated in three experiments. Recovery of the native proinsulin disulfide isomer (N) divided by total proinsulin recovery under reduced conditions (P) was quantified in these experiments with mean ± S.D. shown below; the asterisks signify p < 0.05. B, INS1 cells treated with siRNA duplexes (100 nm) designed for knockdown of the luciferase mRNA (a negative control, C1), GFP mRNA (negative control, C2), or PDI-KD were lysed at 72 h after siRNA transfection. Upper eight set of bands: Western blotting for the antigens indicated, normalized to protein (10 or 20 μg, depending on antigen), with γ-tubulin serving as a loading control. BiP and GRP94 were identified with anti-KDEL antibodies. From four such experiments, no significant differences were observed in PDI-KD cells except for the PDI protein itself. Bottom row: XBP1 mRNA was measured by RT-PCR using primers that amplify both unspliced (u) and spliced (s) forms (19). As a positive control for ER stress, cells were treated with thapsigargin (downward arrowhead in first lane: 1 μm for 3 h). From three such experiments, no significant differences were observed in PDI-KD cells. C, at 72 h after transfection with either scrambled (Con) or PDI siRNA (KD) duplexes (100 nm), INS1 cells were pulse-labeled for 20 min with [35S]Cys/Met and either lysed immediately or chased for 1 h, and both media (M) and cell lysates (C) were collected. Samples were immunoprecipitated with anti-insulin and analyzed by reducing Tris-Tricine-urea-SDS-PAGE. Note that two-thirds of the radioactivity of insulin is lost upon reducing SDS-PAGE because the A-chain is not recovered. Upper panel: Western blotting of PDI in the cells used in this experiment. D, at 48 h after transfection of INS1 cells with either scrambled (light bars) or PDI siRNA duplexes (dark bars), the media were changed, and fresh growth media (11 mm glucose) were applied and collected after an additional 24 h. At that time, the cells were lysed, and total insulin (i.e. insulin plus proinsulin) in both cell lysates and media was measured by RIA. The data are the mean of three experiments with six independent replicates; control values were set to 100%. When comparing control versus PDI-KD, either cells or media, the asterisks signify p < 0.05. E, Min6 cells were transiently transfected to express empty vector (Control), FLAG-PDI-AGHA,AGHA′ (PDI-DEAD), FLAG-WT-PDI, or FLAG-PDI-CGHA (PDI-TRAP). At 72 h after transfection, the Min6 cells were pulse-labeled for 20 min with [35S]Cys/Met and lysed in SDS-containing lysis buffer with 15 mm NEM. The lysates were immunoprecipitated with either anti-FLAG or anti-insulin as indicated. The anti-FLAG immunoprecipitates (IP) were analyzed on the same SDS gel with phosphorimaging (i.e. identical exposure) under nonreducing or reducing conditions as indicated; the anti-insulin immunoprecipitates come from an independent gel analysis of the same samples. The positions of FLAG-PDI, PDI-proinsulin adduct, and newly synthesized proinsulin (under reducing conditions) are indicated.
Statistical Analysis
Data are presented as mean value ± S.D. Statistical significance was analyzed by the Student's t test; a p value < 0.05 was deemed statistically significant.
RESULTS
Proinsulin Disulfide Bond Formation
In the case of genetic deficiency of ERO1β, at least in part as a consequence of incomplete proinsulin thiol oxidation, insulin production and consequent insulin secretion are diminished (13, 14). If proinsulin oxidation from ERO1β is mediated by PDI, then loss of PDI expression would be expected to phenocopy ERO1β knockdown. We therefore examined the effect of ERO1β or PDI knockdown (ERO1β-KD or PDI-KD) in INS1 (rat) β-cells. We monitored the recovery after pulse-labeling of newly synthesized native proinsulin (nonreducing conditions) relative to total proinsulin (reducing conditions) as analyzed by Tris-Tricine-urea-SDS-PAGE (22). As expected (13), ERO1β-KD decreased initial oxidation of proinsulin to the native monomer (Fig. 1A, lanes 3 and 4 versus lanes 1 and 2; quantitation in graph below). By contrast, PDI-KD did not perturb proinsulin oxidation and actually augmented native proinsulin (Fig. 1A, lanes 11 and 12 versus lanes 9 and 10; with a 20% increase in recovery of native proinsulin quantitated in the graph below). Similar results were obtained at pulse-labeling times of 2, 5, and 20 min; also, the same results were observed in Min6 (mouse) β-cells (not shown). Importantly, PDI-KD did not result in up-regulation of other β-cell ER oxidoreductases (Fig. 1B). Moreover, PDI-KD in INS1 cells did not up-regulate levels of ER molecular chaperones BiP and GRP94, nor did it activate net phosphorylation of eIF2α (Fig. 1B) or activate splicing of XBP1 (unlike thapsigargin treatment, Fig. 1B, extra lane). Thus, we found no discernible evidence that PDI-KD triggers ER stress in β-cells.
Proinsulin Egress through Secretory Pathway
The perturbation of proinsulin oxidation upon ERO1β-KD (Fig. 1A) is accompanied by inefficient proinsulin transport through the secretory pathway with decreased insulin production (13, 14). However, by pulse-chase analysis after PDI-KD, we actually observed faster maturation of newly synthesized proinsulin (Fig. 1C, left) with an increased insulin:proinsulin ratio at 1 h of chase (Fig. 1C, right). To estimate the consequences of this acceleration on the steady-state distribution of proinsulin+insulin in INS cells, we used RIA to measure total output and found that PDI-KD shifted the distribution of proinsulin+insulin from cells to media; the redistribution shown in Fig. 1D was obtained in standard INS1 growth media (11 mm glucose), but an even greater shift from cells to media was observed at 25 mm glucose (not shown). These data suggested that PDI might function to actively retain proinsulin within the ER.
To determine whether PDI physically interacts with proinsulin, INS1 cells were transfected with either FLAG-tagged PDI, a catalytically dead PDI-AGHA,AGHA′ (PDI-DEAD) mutant, or a PDI-CGHA mutant (PDI-TRAP) that can make a mixed disulfide bond with PDI substrates but is defective for resolving the mixed disulfide (20, 21). Upon pulse-labeling cells with 35S-amino acids for 20 min, comparable amounts of proinsulin (Fig. 1E, right panel) and FLAG-PDI constructs (middle panel) were newly synthesized. The expression of PDI-TRAP, followed by immunoprecipitation with anti-FLAG and analysis by nonreducing SDS-PAGE and phosphorimaging, revealed recovery of a disulfide-linked adduct that ran just above the position of the PDI-TRAP mutant itself (Fig. 1E, left panel). Upon analysis of the same sample under reducing conditions, PDI-TRAP was fully reduced, and the adduct disappeared; with this, co-precipitated endogenous proinsulin clearly appeared (Fig. 1E, middle panel). Catalytically dead PDI neither formed the disulfide-linked adduct (left panel) nor co-precipitated endogenous proinsulin (middle panel). Wild-type PDI expressed in INS1 cells also co-precipitated endogenous proinsulin, although the stability (amount) of the mixed disulfide adduct was clearly less than that obtained for the PDI-TRAP mutant designed to covalently capture PDI substrates (20, 21). These data establish direct PDI-proinsulin interaction via a mechanism requiring the catalytic activity of PDI.
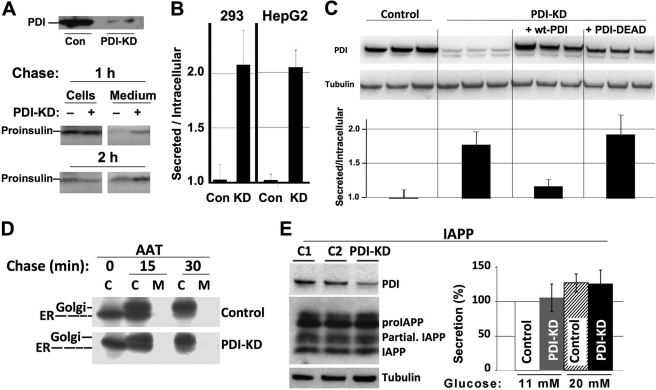
Upon steady-state radiolabeling, immunoprecipitation, and analysis by nonreducing Tris-Tricine-urea-SDS-PAGE (not shown), we observed that most insulin immunoreactivity secreted from PDI-KD cells was composed of mature insulin molecules, i.e. those exhibiting proper disulfide bond formation, ER exit, and proteolytic processing in secretory granules. To confirm that augmented insulin production in PDI-KD cells was not mediated by indirect effects enhancing prohormone convertase activity, we also examined the effect of PDI-KD on recombinant proinsulin trafficking in both 293 cells and HepG2 cells. Neither cell line has either secretory granules or processing enzymes, but it has been reported that in HepG2 cells, PDI-KD actually delays disulfide bond formation and ER exit of secretory proteins such as transferrin (21). Even without prohormone convertases, PDI-KD (Fig. 2A, upper panel) still led to accelerated proinsulin transport (lower panel). Moreover, in both 293 cells and HepG2 cells, PDI-KD shifted the steady-state distribution of proinsulin from cells to media (Fig. 2B). To confirm that intracellular proinsulin retention was a specific effect of PDI, we re-expressed siRNA-resistant WT-PDI or PDI-DEAD in PDI-KD cells. As judged by immunoblotting (Fig. 2C, upper panel), re-expressed PDI was not overexpressed, yet WT-PDI restored the intracellular retention of proinsulin in PDI-KD cells (lower panel). By contrast, PDI-DEAD was unable to confer intracellular retention of proinsulin (Fig. 2C, lower panel), again indicating that PDI action on proinsulin requires the catalytically active thioredoxin motifs of PDI.
FIGURE 2.
PDI-KD effects on enhanced ER export are selective for proinsulin as substrate. A, at 48 h after transfection with either scrambled (Con) or PDI siRNA duplexes (100 nm), 293 cells were pulse-labeled for 30 min with [35S]Cys/Met and chased for 1 or 2 h; chase medium was then collected, and cells were lysed. Proinsulin was immunoprecipitated with anti-insulin and analyzed by Tris-Tricine-urea-SDS-PAGE under reducing conditions. Upper panel: Western blotting of PDI in the cells used in this experiment. B, at 48 h after transfection with either scrambled (Con) or PDI siRNA duplexes (KD), the media bathing either 293 cells or HepG2 cells as indicated were changed, and fresh media were collected between 48 and 72 h. At that time, the cells were lysed, proinsulin contained in both media and cell lysates was measured by RIA, and the extracellular/intracellular ratio of proinsulin was reported. The data are the mean of three experiments with six independent replicates; control ratio was set to 1.0. Control versus PDI-KD cells: p < 0.05 for both sets of cells. C, PDI-KD 293 cells were transfected to express empty vector, WT-PDI, or PDI-AGHA,AGHA′ (PDI-DEAD), and these lanes all compared with control cells that did not have PDI knockdown (triplicates at left). Upper panel: Western blotting of PDI in the cells used in this experiment. Lower panel: analysis by insulin RIA identical to that shown in panel B. Each experiment was performed in triplicate and repeated twice (six independent replicates); control ratio was set to 1.0. Control versus PDI-KD cells or PDI-KD cells expressing PDI-DEAD: p < 0.05. The difference between control and PDI-KD cells expressing WT-PDI was not statistically significant. C, cell lysates; M, media. D, INS1 cells were transfected to express AAT. The cells were then split and treated with either scrambled or PDI siRNA duplexes (40 nm, PDI knockdown was confirmed by Western blotting (not shown)). The cells were pulse-labeled for 20 min with [35S]Cys/Met and chased for the times indicated; media were collected, and cells were lysed as in Fig. 1C. The samples were immunoprecipitated with anti-AAT and analyzed by SDS-PAGE. ER exit of AAT was evaluated by the change in gel migration from the ER-glycosylated form of AAT to the Golgi-glycosylated form, as indicated. E, INS1 cells treated with siRNAs duplexes (100 nm) designed for knockdown of luciferase mRNA (C1), GFP mRNA (C2), or PDI-KD exactly as in Fig. 1B, for 72 h. Left panel: Western blotting for the antigens indicated, normalized to protein (10 μg), with γ-tubulin serving as a loading control. proIAPP, IAPP precursor; Partial IAPP, partially processed IAPP. The result is representative of three independent experiments. Right panel: INS1 cells transfected with siRNAs (100 nm) were grown for 3 days in RPMI media at lower (11 mm) or higher (20 mm) glucose. The media were collected, and IAPP secretion was analyzed by ELISA (see “Experimental Procedures”). The differences from control versus PDI-KD cells at either glucose concentration were not statistically significant.
Importantly, in INS1 cells, PDI-KD did not accelerate the acquisition of Golgi glycosylation of recombinant AAT (Fig. 2D), nor did it affect the level of IAPP processing (Fig. 2E, left) or IAPP secretion over 72 h at low or high glucose (Fig. 2E, right). Thus, the data indicate that PDI exhibits selective retention activity for proinsulin in the ER.
DISCUSSION
Although it is widely assumed that PDI participates in oxidation of proinsulin disulfide bonds in pancreatic β-cells, which in turn facilitates proinsulin export, we find in fact that PDI is an ER retention factor for proinsulin. Upon PDI-KD, proinsulin transit along the secretory pathway is enhanced: accelerated as measured by pulse-chase and with a net increase in secreted insulin as measured by RIA. Although compensatory changes in gene expression, especially those of other ER oxidoreductases, could account for enhanced proinsulin transport, there are no discernible changes in protein levels of other major ER oxidoreductases (ERp72, P5, or ERp57) upon PDI-KD.
The argument is compelling that proinsulin egress from the ER is linked to the kinetics and efficiency of disulfide bond formation; specifically, loss of ERO1β activity is linked not only to decreased formation of the three native proinsulin disulfide bonds but also to inefficient export of newly synthesized proinsulin with delayed and decreased production of newly synthesized insulin (13). Given the current finding that in PDI-KD β-cells, native proinsulin disulfide bond formation is not impaired, it seems safe to surmise that PDI is not the primary oxidase responsible for receiving reducing equivalents from proinsulin thiols for transfer to ERO1β.
This does not preclude a role for PDI to oxidize secretory protein substrates in other cell types or in β-cells in special situations. Certainly, proinsulin oxidation by PDI can be reconstituted in vitro (10, 11). However, even in HepG2 cells, where PDI has been suggested to be a primary oxidase for secretory proteins such as transferrin (21), PDI-KD significantly enhances (recombinant) proinsulin export. This effect of PDI is absolutely dependent upon the catalytic activity of the enzyme; a PDI-TRAP mutant makes an unresolved mixed disulfide with proinsulin, and a catalytically dead PDI cannot confer intracellular proinsulin retention. For these reasons, we consider it plausible that in the ER of β-cells, PDI may act as a net reductase for proinsulin, providing unfoldase activity (16) that lengthens the ER residence time of proinsulin. Substrate unfolding has been implicated in the activity of PDI to promote polypeptide retrotranslocation for ER-associated degradation of selected substrates (23) that may include proinsulin. PDI-mediated unfoldase activity for proinsulin could account for diminished insulin production and induction of ER stress upon increased expression of PDI in β-cells (19). Certainly, we have no evidence that PDI-KD leads to either proinsulin or generalized protein misfolding in INS1 β-cells. Indeed, there is no net increase in phosphorylation of eIF2α, no increased splicing of XBP1, and no increase of the hsp70 and hsp90 family members of the ER, BiP, and GRP94. Thus, all evidence points to the idea that PDI-KD in β-cells is nontoxic. With this in mind, we find interesting the recent speculation that PDI could be a therapeutic target to actually prevent ER stress and neuronal cell degeneration (24).
Altogether, our accumulated evidence favors that PDI is a retention factor for proinsulin in the ER of pancreatic β-cells. Additional studies are needed to directly analyze the contributions of PDI to proinsulin unfolding and to limiting insulin production in type 2 diabetes.
Acknowledgment
We acknowledge NIH5P60-DK20572 Diabetes Center Molecular Biology and Sequencing Cores.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-DK48280 (to P. A.). This work was also supported by Canadian Institutes of Health Research Grant MOP-86641 (to A. V.).
- PDI
- protein disulfide isomerase
- PDI-KD
- PDI knockdown
- ER
- endoplasmic reticulum
- ERO1β
- ER-oxidoreductin 1-β
- IAPP
- islet amyloid polypeptide
- NEM
- N-ethylmaleimide
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Frand A. R., Cuozzo J. W., Kaiser C. A. (2000) Pathways for protein disulphide bond formation. Trends Cell Biol. 10, 203–210 [DOI] [PubMed] [Google Scholar]
- 2. Tu B. P., Ho-Schleyer S. C., Travers K. J., Weissman J. S. (2000) Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science 290, 1571–1574 [DOI] [PubMed] [Google Scholar]
- 3. Kozlov G., Määttänen P., Thomas D. Y., Gehring K. (2010) A structural overview of the PDI family of proteins. FEBS J. 277, 3924–3936 [DOI] [PubMed] [Google Scholar]
- 4. Cras-Méneur C., Inoue H., Zhou Y., Ohsugi M., Bernal-Mizrachi E., Pape D., Clifton S. W., Permutt M. A. (2004) An expression profile of human pancreatic islet mRNAs by Serial Analysis of Gene Expression (SAGE). Diabetologia 47, 284–299 [DOI] [PubMed] [Google Scholar]
- 5. Ahmed M., Forsberg J., Bergsten P. (2005) Protein profiling of human pancreatic islets by two-dimensional gel electrophoresis and mass spectrometry. J. Proteome Res. 4, 931–940 [DOI] [PubMed] [Google Scholar]
- 6. Karala A. R., Lappi A. K., Ruddock L. W. (2010) Modulation of an active-site cysteine pKa allows PDI to act as a catalyst of both disulfide bond formation and isomerization. J. Mol. Biol. 396, 883–892 [DOI] [PubMed] [Google Scholar]
- 7. Wang C. C. (1998) Isomerase and chaperone activities of protein disulfide isomerase are both required for its function as a foldase. Biochemistry 63, 407–412 [PubMed] [Google Scholar]
- 8. Weiss M. A. (2009) Proinsulin and the genetics of diabetes mellitus. J. Biol. Chem. 284, 19159–19163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu M., Hodish I., Haataja L., Lara-Lemus A. R., Rajpal G., Wright J., Arvan P. (2010) Proinsulin misfolding and diabetes: Mutant INS gene-induced diabetes of Youth. Trends Endocrinol. Metab. 21, 652–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Winter J., Klappa P., Freedman R. B., Lilie H., Rudolph R. (2002) Catalytic activity and chaperone function of human protein-disulfide isomerase are required for the efficient refolding of proinsulin. J. Biol. Chem. 277, 310–317 [DOI] [PubMed] [Google Scholar]
- 11. Tang J. G., Wang C. C., Tsou C. L. (1988) Formation of native insulin from the scrambled molecule by protein disulphide-isomerase. Biochem. J. 255, 451–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kitiphongspattana K., Khan T. A., Ishii-Schrade K., Roe M. W., Philipson L. H., Gaskins H. R. (2007) Protective role for nitric oxide during the endoplasmic reticulum stress response in pancreatic beta-cells. Am. J. Physiol. Endocrinol. Metab. 292, E1543–1554 [DOI] [PubMed] [Google Scholar]
- 13. Zito E., Chin K. T., Blais J., Harding H. P., Ron D. (2010) ERO1-beta, a pancreas-specific disulfide oxidase, promotes insulin biogenesis and glucose homeostasis. J. Cell Biol. 188, 821–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khoo C., Yang J., Rajpal G., Wang Y., Liu J., Arvan P., Stoffers D. A. (2011) Endoplasmic reticulum oxidoreductin-1-like β (ERO1lβ) regulates susceptibility to endoplasmic reticulum stress and is induced by insulin flux in β-cells. Endocrinology 152, 2599–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Puig A., Lyles M. M., Noiva R., Gilbert H. F. (1994) The role of the thiol/disulfide centers and peptide binding site in the chaperone and anti-chaperone activities of protein disulfide isomerase. J. Biol. Chem. 269, 19128–19135 [PubMed] [Google Scholar]
- 16. Tsai B., Rodighiero C., Lencer W. I., Rapoport T. A. (2001) Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell 104, 937–948 [DOI] [PubMed] [Google Scholar]
- 17. Guo Z. Y., Qiao Z. S., Feng Y. M. (2008) The in vitro oxidative folding of the insulin superfamily. Antioxid. Redox Signal. 10, 127–139 [DOI] [PubMed] [Google Scholar]
- 18. Lundström J., Holmgren A. (1990) Protein disulfide-isomerase is a substrate for thioredoxin reductase and has thioredoxin-like activity. J. Biol. Chem. 265, 9114–9120 [PubMed] [Google Scholar]
- 19. Zhang L., Lai E., Teodoro T., Volchuk A. (2009) GRP78, but Not Protein-disulfide Isomerase, Partially Reverses Hyperglycemia-induced Inhibition of Insulin Synthesis and Secretion in Pancreatic {beta}-Cells. J. Biol. Chem. 284, 5289–5298 [DOI] [PubMed] [Google Scholar]
- 20. Schulman S., Wang B., Li W., Rapoport T. A. (2010) Vitamin K epoxide reductase prefers ER membrane-anchored thioredoxin-like redox partners. Proc. Natl. Acad. Sci. U.S.A. 107, 15027–15032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rutkevich L. A., Cohen-Doyle M. F., Brockmeier U., Williams D. B. (2010) Functional relationship between protein disulfide isomerase family members during the oxidative folding of human secretory proteins. Mol. Biol. Cell 21, 3093–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu M., Haataja L., Wright J., Wickramasinghe N. P., Hua Q. X., Phillips N. F., Barbetti F., Weiss M. A., Arvan P. (2010) Mutant INS-gene induced diabetes of youth: proinsulin cysteine residues impose dominant-negative inhibition on wild-type proinsulin transport. PLoS One 5, e13333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Forster M. L., Sivick K., Park Y. N., Arvan P., Lencer W. I., Tsai B. (2006) Protein disulfide isomerase-like proteins play opposing roles during retrotranslocation. J. Cell Biol. 173, 853–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Honjo Y., Ito H., Horibe T., Takahashi R., Kawakami K. (2010) Protein disulfide isomerase-immunopositive inclusions in patients with Alzheimer disease. Brain Res. 1349, 90–96 [DOI] [PubMed] [Google Scholar]