Background: Humanized PET reporter gene (PRG) systems are needed to replace immunogenic, viral-derived systems.
Results: Employing a structure-guided approach, we developed a highly sensitive humanized PRG characterized by reduced activity for its natural substrates.
Conclusion: Sensitivity of PRGs can be improved by reducing their endogenous activities.
Significance: Our method can be employed to rapidly develop highly sensitive humanized PRGs.
Keywords: Enzyme Kinetics, Enzymes, In Vivo Imaging, Molecular Modeling, Mutagenesis, L-18F-FMAU, PET Reporter Gene, Non-invasive Imaging, Nucleoside Analog PET Probes, Thymidine Kinase 2
Abstract
Positron emission tomography (PET) reporter gene imaging can be used to non-invasively monitor cell-based therapies. Therapeutic cells engineered to express a PET reporter gene (PRG) specifically accumulate a PET reporter probe (PRP) and can be detected by PET imaging. Expanding the utility of this technology requires the development of new non-immunogenic PRGs. Here we describe a new PRG-PRP system that employs, as the PRG, a mutated form of human thymidine kinase 2 (TK2) and 2′-deoxy-2′-18F-5-methyl-1-β-l-arabinofuranosyluracil (l-18F-FMAU) as the PRP. We identified l-18F-FMAU as a candidate PRP and determined its biodistribution in mice and humans. Using structure-guided enzyme engineering, we generated a TK2 double mutant (TK2-N93D/L109F) that efficiently phosphorylates l-18F-FMAU. The N93D/L109F TK2 mutant has lower activity for the endogenous nucleosides thymidine and deoxycytidine than wild type TK2, and its ectopic expression in therapeutic cells is not expected to alter nucleotide metabolism. Imaging studies in mice indicate that the sensitivity of the new human TK2-N93D/L109F PRG is comparable with that of a widely used PRG based on the herpes simplex virus 1 thymidine kinase. These findings suggest that the TK2-N93D/L109F/l-18F-FMAU PRG-PRP system warrants further evaluation in preclinical and clinical applications of cell-based therapies.
Introduction
The inability to routinely monitor the tissue pharmacokinetics of therapeutic genes and cells and correlate this information with therapeutic outcomes represents a significant roadblock in the clinical adoption of these emerging therapies. Most cell/gene therapy trials use invasive biopsy techniques to localize therapeutic genes or therapeutic cells at target sites. However, invasive techniques are prone to sampling errors and carry risks for the patients. There is an unmet need for techniques to monitor the whole-body tissue distribution of therapeutic cells and therapeutic genes, to quantify therapeutic cells, and to measure therapeutic gene expression at all locations non-invasively and sequentially after treatment. This unmet need can be addressed by PET3 reporter gene (PRG) imaging (1). A PRG encodes a protein that mediates the specific accumulation of a PET reporter probe (PRP) labeled with a positron-emitting isotope (2). Such non-invasive PET measurements may predict and/or evaluate treatment efficacy and the risk of side effects; they can provide information that complements data obtained using invasive techniques, such as serial biopsies (2). PRGs developed to date encode proteins with various activities, including enzymes, transporters, and receptors (for review, see Ref. 3). In theory, enzyme-encoding PRGs should have the highest sensitivity among different classes of PRGs as a result of signal amplification by the catalytic turnover of the enzymatic reaction that traps the probe. The most commonly used PRGs are based on herpes simplex virus type 1 thymidine kinase (HSV1-tk) (4) and its optimized mutant, sr39tk (5). Both wild type (WT) HSV1-tk and sr39tk have been used to study the kinetics of therapeutic cells in preclinical settings (6–9). Several PRPs can be used to image cells engineered to express HSV1-tk-based PRGs: 9-[4-18F-3-(hydroxymethyl)butyl]guanine (18F-FHBG) (10–12), 2′-deoxy-2′-18F-5-ethyl-1-β-d-arabinofuranosyluracil (18F-FEAU) (13–15), and 2′-deoxy-2′-18F-5-iodo-1-β-d-arabinofuranosyluracil (18F-FIAU) (15). To date, HSV1-tk is the only PRG that has been used to image therapeutic cells in patients (10).
The main disadvantage of HSV1-tk as a PRG is its immunogenicity, which can lead to immune-mediated elimination of therapeutic cells. This phenomenon has been documented in clinical trials (16, 17). The immunogenicity problem may be solved by replacing the viral kinase with a human orthologue (18). Two potentially non-immunogenic candidate PRGs based on human nucleoside kinases have been developed; that is, a double mutant of deoxycytidine kinase (dCK) (19) and a truncated form of mitochondrial thymidine kinase 2 (TK2) (20). These PRGs phosphorylate and trap the PRP 18F-FEAU. The sensitivity of the dCK-double mutant/18F-FEAU PRG-PRP system was comparable with that of HSV1-tk/18F-FEAU, whereas TK2/18F-FEAU had lower sensitivity. In non-human primates 18F-FEAU has a favorable biodistribution as a candidate PRP, with tracer accumulation in the liver, small intestine, kidneys, and urinary bladder (21) but not in other organs and tissues. Human biodistribution data for this candidate PRP are not available.
The utility of a PRG-PRP system is dependent on its sensitivity (the ability to detect few therapeutic cells at various anatomical locations) and specificity (the probe should accumulate only in cells engineered to express the PRG). Another equally important parameter is the requirement that a PRG should be biologically inert. In other words its ectopic expression in therapeutic cells should not alter the metabolism or normal function of these cells. This requirement is especially important in the case of nucleoside kinase PRGs. Ectopic expression of a nucleoside kinase could perturb the normal regulation of nucleotide metabolism through excess phosphorylation of endogenous nucleosides. Such metabolic alterations can lead to imbalanced nucleotide pools and increased risk of genotoxicity (22–25). In this context the dCK-double mutant has significantly higher activity than WT dCK toward endogenous nucleosides such as deoxycytidine and thymidine (26). Truncated TK2 also retains normal activity with natural substrates. Whether these new PRGs fulfill the critical requirement of being biologically inert remains to be determined.
Here we describe the development of a new PRG-PRP system that meets the specifications mentioned above. We determined the biodistribution of l-18F-FMAU, the candidate PRP, in mice and humans. We used enzyme engineering to develop a mutant PRG enzyme that is orthogonal to the wild type enzyme regarding its ability to phosphorylate endogenous nucleosides. The resulting PRG-PRP system, TK2-N93D/L109F as PRG and l-18F-FMAU as PRP, should find utility in various preclinical and clinical therapeutic cell tracking applications. The approach used to develop this system should be generalizable to the identification and evaluation of other pairs of nucleoside analogs and nucleoside kinases for PET reporter gene imaging applications.
EXPERIMENTAL PROCEDURES
Radiochemical Synthesis of 18F-Labeled PET Probes
18F-FHBG was synthesized as previously described (12). The radiochemical synthesis of l-18F-FMAU is described in the supplemental material.
Molecular Modeling of Human TK2
We generated a homology model of TK2 using the SWISS-MODEL server (27). The solved structures of human dCK (35% identity, 50% homology to TK2) in both its closed (PDB ID 1P5Z) and open conformation (PDB ID 3QEO) (28, 29) served as templates.
Generation of TK2 Mutants
We used the Δ50N truncation variant of TK2, and we are referring to this truncated form (which lacks the mitochondrial sorting signal) as the WT enzyme. Numbering of residues is based on the full-length sequence of human TK2 (Uniprot ID O00142). Cloning of human TK2 has been described previously (30). Mutants were produced on the WT TK2 sequence that was present in both the pMSCV vector for retroviral transduction and a modified pET14b expression vector for production of recombinant protein.
Expression and Purification of Recombinant TK2 Proteins
Expression and purification of TK2 have been described previously (30). In short, Escherichia coli BL21 (DE3) C41 harboring the modified pET14b vector (to include a SUMO tag between the hexahistidine sequence and TK2) were grown at 37 °C until an optical density of ∼0.8 was reached. At that point the temperature was reduced to 18 °C; the culture was induced with 0.5 mm isopropyl β-d-1-thiogalactopyranoside and left to shake overnight. Cells were harvested by centrifugation, washed, and stored at −80 °C until use. Purification involved two steps. The first step used a metal affinity column (HisTRAP HP column, GE Healthcare); after elution of the His-SUMO-TK2 fusion protein, the SUMO protease was added. The cleaved protein was reapplied onto the nickel column to separate TK2 from the His-SUMO tag. The second step involved a gel filtration column (S200, GE Healthcare) equilibrated with 25 mm Tris, pH 7.5, 200 mm NaCl, and 3 mm DTT. Pure TK2 was pooled, concentrated to ∼10 mg/ml, separated into aliquots, flash-frozen in liquid nitrogen, and stored at −80 °C until use.
Kinetic Analyses of TK2-based Candidate PRGs
We used an NADH-dependent enzyme coupled assay (31). Using a Cary UV spectrophotometer, measurements were made in triplicate at 37 °C in a buffer containing 100 mm Tris, pH 7.5, 100 mm KCl, 5 mm MgCl2, and 1 mm ATP. For data in which kobs is given, a single nucleoside concentration of 200 μm was used. For data in which both Km and kcat are given, the nucleoside concentration was varied between 15 and 500 μm. TK2 concentration in the cuvette was 400 nm. Data were fit to the Michaelis-Menten equation using SigmaPlot. Of note, in some previous reports, negative cooperativity was observed with thymidine but not with deoxycytidine (32, 33). When we fit our data for WT TK2 using the Hill equation, we also see the same magnitude of negative cooperativity as reported by others (n = ∼0.7) with thymidine and the analogs tested. However, the quality of the fit of the data is only marginally improved compared with that using the simple Michaelis-Menten equation. When the data of the TK2 mutants are fit using the Hill equation, a more complicated behavior is observed, with some conditions having a Hill coefficient below 1, some above 1, and some nearly one. Here again, the quality of the fit is not dramatically improved by adding the extra parameter of the Hill coefficient. Therefore, we present all of the kinetic data using the Michaelis-Menten equation without the Hill coefficient.
Cell Lines
The L1210 cell line (34) was a gift from Charles Dumontet (Université Claude Bernard Lyon I, Lyon, France). Cells were cultured at 5% v/v CO2 and 37 °C in RPMI supplemented with 5% v/v FCS. Murine stem cell virus (pMSCV)-based helper-free retroviruses encoding the TK2 mutants (or sr39tk), an internal ribosomal entry site, and the yellow fluorescent protein (YFP) were produced by transient co-transfection of the amphotrophic retrovirus packaging cell line Phoenix (American Type Culture Collection, SD 3443) (35). L1210 cells underwent spinfection with the pMSCV-TK2 mutants- internal ribosomal entry site-YFP retrovirus with 2 μg/ml Polybrene (1000 × g, 120 min, 37 °C). L1210 cells expressing various PRGs, (L1210-PRG) were FACS-sorted to ensure that each population had equivalent levels of PRG expression.
Probe Uptake Assays Using Transduced L1210 Cell Lines
L1210 cells transduced with the indicated PET reporter genes (L1210-PRG) were seeded at a density of 500,000 cells/well in 24-well plates. 5 μCi of l-18F-FMAU were added to the L1210-PRG cells simultaneously with the indicated amounts of d-thymidine (d-dT) at a final volume of 1 ml/well. After 1 h at 37 °C, cells were harvested and washed four times with ice-cold PBS. Radioactivity was measured using a gamma counter.
MicroPET/CT Imaging Studies in Mice
Animal studies were approved by the UCLA Animal Research Committee and were carried out according to the guidelines of the Department of Laboratory Animal Medicine at UCLA. C57/BL6 mice were injected with the indicated probe and underwent microPET/CT analyses at 1- and 3-h post probe injection (Inveon, Siemens Medical Solutions USA Inc.; microCAT; Imtek Inc.). For tumor imaging studies, SCID mice were injected subcutaneously on day −7 in the right and left flanks with 1 × 106 L1210-PRG-expressing cells in 50% v/v phosphate-buffered saline and 50% v/v MatrigelTM (BD Biosciences). For imaging experiments, mice were kept warm and under gas anesthesia (2% v/v isoflurane) and were injected intravenously with 200 μCi of 18F-labeled probes. A 3-h interval was allowed between probe administration and microPET/CT scanning. Static microPET images were acquired for 600 s. Image data were evaluated in three-dimensional histograms and reconstructed with a zoom factor of 2.1 using three-dimensional ordered set expectation maximization (OSEM) with 2 iterations followed by MAP (maximum a posteriori) reconstruction with 18 iterations (beta = 0.1). Images were analyzed using OsiriX Imaging Software Version 3.8.
Human PET/CT Studies
All studies involving human volunteers were approved by the UCLA Medical Internal Review Board. l-18F-FMAU was approved by the UCLA Radioactive Drug Research Committee. l-18F-FHBG has an FDA investigational new drug approval (IND #61,880) and was also approved by the UCLA Radioactive Drug Research Committee. A 53-year-old healthy male and a 44-year-old healthy female volunteer were recruited for the l-18F-FMAU biodistribution study. Each volunteer received a bolus intravenous injection of ∼56 MBq (1.5 mCi) sterile l-18F-FMAU and had four consecutive whole-body (starting from just above the head to above the knees, 6 bed positions, 5-min scan at each bed position) PET scans (Biograph 64, Siemens), with the first scan starting shortly after intravenous injection of l-18F-FMAU. A low dose CT scan was also obtained for attenuation correction. Volunteers urinated after all scans had been performed. The region of interest analysis was performed to measure mean standard uptake values of l-18F-FMAU in major organs/tissues. To illustrate the biodistribution of 18F-FHBG, we used an unpublished scan from a previous study (10).
Statistical Analysis
Data are presented as the means ± S.E. All p values are two-tailed, and p values of <0.05 are considered to be statistically significant. Graphs were generated and analyzed using the Prism 5 software (GraphPad).
RESULTS
Comparison of Biodistribution of l-18F-FMAU and 18F-FHBG in Mice
Nucleoside analogs are being increasingly used as PET probes for assaying nucleotide metabolism, cell proliferation, and mitochondrial function (36–40). Nucleosides can adopt one of two enantiomeric configurations. Naturally occurring nucleosides are in the d configuration (41). Recently there has been increasing interest in using nucleoside analogs with the non-natural l configuration as PET probes to image the activity of endogenous nucleoside kinases (42–45). To date, l nucleosides have not been evaluated as PRPs. To start determining the potential value of l nucleosides as PRPs, we focused on l-18F-FMAU, the non-natural counterpart of d-18F-FMAU, one of the pyrimidine analogs that has been previously evaluated as a candidate PRP for the HSV1-tk PRG (46).
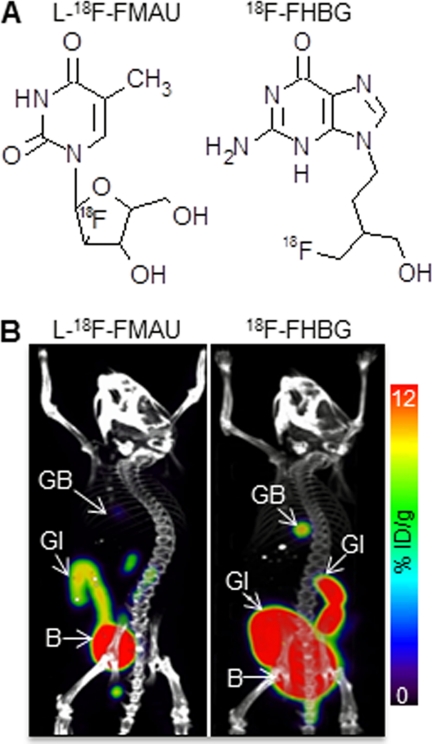
We compared the biodistribution of l-18F-FMAU in mice with that of 18F-FHBG, a well characterized and frequently used PRP (12, 47, 48). To achieve optimal signal to noise ratios, PRPs should not accumulate in cells and tissues that do not express the corresponding PRG. For instance, the accumulation of the candidate PRP should be minimal or undetectable in all tissues, except in those involved in probe clearance from the body. C57/BL6 mice were scanned 3 h after administration of either l-18F-FMAU or 18F-FHBG (Fig. 1A). Three-dimensional reconstructions of the whole body microPET/CT images are shown in Fig. 1B. Quantification of the signals is presented in supplemental Table 1. Both l-18F-FMAU and 18F-FHBG had very low retention in the thoracic cavity. At the 3-h time point neither probe showed any accumulation in the liver. Accumulation in the gallbladder was 4 times higher for 18F-FHBG (7.45 ± 5.31% injected dose/g) than for l-18F-FMAU (1.68 ± 0.46% injected dose/g). Retention in the abdominal cavity was three times higher for 18F-FHBG than for l-18F-FMAU. This was likely due to higher biliary excretion of 18F-FHBG. Elevated 18F-FHBG accumulation was detected throughout the GI tract. In contrast, in mice injected with l-18F-FMAU signals were only detected in the lower GI tract. Thus, the biodistribution of l-18F-FMAU in mice was at least comparable with, if not better than that of 18F-FHBG.
FIGURE 1.
Biodistribution of l-18F-FMAU and 18F-FHBG in mice. A, chemical structures of l-18F-FMAU and 18F-FHBG are shown. B, MicroPET/CT scans of C57/BL6 mice 3 h after injection of l-18F-FMAU (left) and 18F-FHBG (right) are shown. Images are co-registered displays of the separate microPET and microCT scans. Quantifications of the PET signals are listed in supplemental Table 1. B, bladder; GB, gallbladder. %ID/g, % injected dose/g.
Development of New PRG to Be Used in Conjunction with l-18F-FMAU Candidate PRP
l-FMAU has been shown to be a substrate for human TK2, a nucleoside kinase that due to its lack of enantiomeric specificity can phosphorylate both d and l nucleosides (49). Ideally, modifications to the TK2 sequence should achieve two objectives; (i) increase sensitivity by reducing the negative feedback regulation of the enzyme and by increasing the phosphorylation rate of the l-FMAU PRP; (ii) reduce the activity of the PRG kinase for the endogenous substrates thymidine and deoxycytidine (to avoid competition between l-FMAU and endogenous nucleosides and potentially genotoxic perturbations of endogenous nucleotide pools).
The enzymatic activity of TK2 is regulated by thymidine triphosphate (dTTP) through negative feedback inhibition (50). dTTP is produced by de novo synthesis and through the salvage of thymidine (via the cytosolic nucleoside kinase TK1). dTTP levels fluctuate throughout the cell cycle and are highest during the S phase, when they increase by as much as 2.5–20-fold compared with the G1 phase (51, 52). It is possible that fluctuations in dTTP levels during the cell cycle will reduce sensitivity and result in difficult to interpret changes in PET signals.
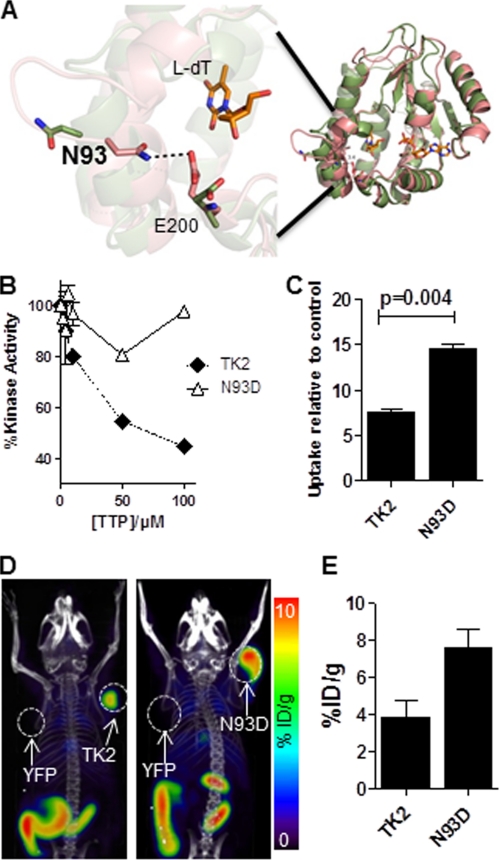
To reduce the susceptibility of TK2 to dTTP-mediated feedback inhibition, we took advantage of the 40% sequence identity between human TK2 and Drosophila melanogaster deoxyribonucleoside kinase (Dm-DNK) (53) and of the identification of a point mutation (N64D) in Dm-DNK that has been shown to reduce the effect of dTTP feedback inhibition (54). The residue in TK2 corresponding to Asn-64 in D. melanogaster deoxyribonucleoside kinase is Asn-93; the corresponding mutation in TK2 is N93D. To predict the effects of the N93D mutation on the structure of TK2, we used molecular modeling. We took advantage of the fact that dCK belongs to the same family of nucleoside kinases as TK2. The sequence identity and homology between dCK and TK2 are 35 and 50%, respectively. Based on our previous work with dCK (28, 29), we obtained a homology model of TK2 (Fig. 2A). We hypothesized that, similar to dCK, TK2 will also adopt an open or a closed conformation. The enzyme is expected to be active in the closed conformation and inactive in the open conformation. In our model, when TK2 is in the closed conformation, Asn-93 is involved in hydrogen bonding with the glutamine at position 200 (E200, Fig. 2A). When the enzyme is in the open conformation, the residues are too far apart to interact. Thus, the N93D mutation would be expected to disfavor the closed conformation due to disruption of the interaction between Asn-93 and Glu-200 (Fig. 2A). dTTP should be able to exert its negative feedback inhibition on TK2 only if the enzyme is in the closed conformation. Because the N93D mutation favors the open conformation of the enzyme, we predicted there would be a reduced probability for dTTP to bind and exert its inhibitory effect.
FIGURE 2.
Evaluation of the TK2-N93D mutant. A, model of WT TK2 bound with l-dT in both the closed (green) and open (pink) conformation of the enzyme. ADP is bound in the phosphate donor pocket shown in this model. The enzyme is active in the closed conformation, which is stabilized by bonds between residues Asn-93 and Glu-200. When asparagine 93 is mutated to a glutamine, the bonds are disrupted, and the enzyme is predicted to switch to an open (inactive) conformation. B, l-FMAU kinase assay using recombinant WT TK2 and TK2-N93D in the presence of increasing concentrations of dTTP is shown. C, l-18F-FMAU uptake assay using WT TK2- or TK2-N93D-expressing L1210 cells is shown. Probe uptake values are reported relative to a control L1210 cell line that expresses YFP. Results are for a representative experiment or n = 2 experiments. D, l-18F-FMAU microPET/CT scans of mice bearing L1210 tumors engineered to express various PRGs (TK2, L1210-TK2; N93D, L1210-TK2-N93D; YFP, L1210-YFP). E, shown is quantification of PET scans from panel D. %ID/g, % injected dose/g.
To test this hypothesis we performed kinase assays using l-FMAU and recombinant WT TK2 and TK2-N93D in the presence of varying amounts of dTTP (Fig. 2B). WT TK2 activity decreased by 20% in the presence of 10 μm dTTP. In contrast, the activity of the N93D mutant decreased by only 4%. When the dTTP concentration was increased to 100 μm, the activity of WT TK2 decreased by 55%, whereas that of TK2-N93D decreased by less than 5%.
We then used cell-based uptake assays to determine whether the decreased susceptibility to feedback inhibition conferred by the N93D mutation increases l-18F-FMAU uptake. As shown in Fig. 2C, we observed a 1.5-fold increase in l-18F-FMAU uptake by the N93D TK2 expressing L1210 cells relative to cells expressing similar levels of WT TK2.
To confirm that the increase in signal can also be detected in vivo, we used mice implanted with L1210 cells transduced with the WT TK2 and mutant TK2-N93D PRGs (Fig. 2D). In vivo, l-18F-FMAU uptake by TK2-N93D PRG-expressing cells was nearly double of that observed with the WT TK2-expressing cells (Figs. 2, D and E). Thus, by engineering a TK2 mutant that is less sensitive to feedback inhibition, we were able to improve the sensitivity of this candidate PRG for l-18F-FMAU.
Further Improvements of Selectivity and Affinity of TK2-derived PRG for l-18F-FMAU
For enzymatic PRGs, the higher the catalytic turnover (kcat) of the enzyme, the more the PRP will accumulate per unit time, leading to a higher PET signal. We determined the kcat of mutated TK2 PRG for l analogs compared with the endogenous substrate, d-dT. Relative to WT TK2, the N93D mutation reduced the kcat of the enzyme toward d-dT, l-dT, and l-FMAU (Table 1). However, the activity toward d-dT decreased by 77%, whereas that for l-dT and l-FMAU decreased only 48 and 32%, respectively. The kcat (l-FMAU)/kcat (d-dT) ratio for N93D nearly triples when compared with wild type. kcat/Km gives a measure of the substrate preference of an enzyme. Compared with WT TK2, the kcat/Km of N93D for l-FMAU increased by 77%, whereas the kcat/Km for d-dT decreased by 60%. Thus, the N93D mutation also achieved the goal of increasing the preference of the enzyme for l-FMAU over the natural substrate.
TABLE 1.
Kinetic analyses of recombinant TK2 mutants with d-dT, l-dT, and l-FMAU
Values were measured at 37 °C using 1 mm ATP as phosphoryl donor. kcat is in s−1, Km is in μm, and kcat/ Km is in m−1 × s−1.
| Nucleoside | WT | N93D | L109F | N93D/L109F | |
|---|---|---|---|---|---|
| d-dT | kcat | 0.123 ± 0.002 | 0.029 ± 0.001 | 0.459 ± 0.005 | 0.063 ± 0.001 |
| Km | 14.6 ± 0.9 | 8.9 ± 1.3 | 8.2 ± 0.6 | 9.8 ± 0.7 | |
| kcat/Km | 8.42 10−3 | 3.26 × 10−3 | 55.98 × 10−3 | 6.43 × 10−3 | |
| l-dT | kcat | 0.370 ± 0.012 | 0.195 ± 0.003 | 1.176 ± 0.033 | 0.572 ± 0.012 |
| Km | 66.9 ± 7.5 | 18.2 ± 1.4 | 34.1 ± 3.9 | 17.4 ± 1.8 | |
| kcat/Km | 5.53 × 10−3 | 10.71 × 10−3 | 34.49 × 10−3 | 32.87 × 10−3 | |
| l-FMAU | kcat | 0.461 ± 0.016 | 0.316 ± 0.009 | 1.079 ± 0.026 | 0.940 ± 0.026 |
| Km | 37.6 ± 5.2 | 14.5 ± 2.1 | 36.7 ± 3.8 | 66.0 ± 6.3 | |
| kcat/Km | 12.26 × 10−3 | 21.79 × 10−3 | 29.40 × 10−3 | 14.24 × 10−3 | |
| Ratio kcat (l-FMAU/d-dT) | 3.7 | 10.9 | 2.4 | 14.9 |
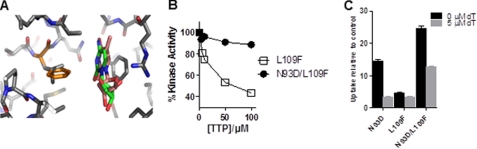
To identify additional mutations that may further improve the selectivity of the TK2 PRG for l analogs, high resolution structures of dCK in complex with l and d substrates (55) were used to generate a homology model of TK2 with bound l-dT and d-dT (Fig. 3A). We then used this model to identify residues that, when mutated, would result in an enzyme with increased affinity for l-dT and decreased affinity for d-dT. This approach led to the identification of residue Leu-109 (Fig. 3A). According to our homology model, this residue interacts with the pyrimidine base. We surmised that if Leu-109 were mutated to an amino acid with a bulkier side chain (e.g. phenylalanine), this would induce a steric clash with d nucleosides but less so with l nucleosides. In turn, this would lead to preferential binding of l versus d nucleosides. Contrary to our expectations, the L109F mutation led to a decrease in the Km for both the d and l forms of dT (Table 1). Notably, the L109F mutation made the enzyme faster at phosphorylating all of the substrates tested, with a bigger effect on d-dT. Thus, for WT TK2, the kcat (l-FMAU)/kcat (d-dT) ratio is 3.7, whereas for TK2-L109F this is 2.4 (Table 1). Compared with WT TK2, the kcat/Km of L109F for l-FMAU increased 2.4 times, whereas the kcat/Km for d-dT increased 6.7 times. Thus, contrary to the prediction, the L109F mutation increased the preference of the enzyme for d-dT compared with l-FMAU. This demonstrates that although a homology model can be sufficient to identify “hot spots” for mutagenesis (in this case, position 109), such a model may lack accuracy that can only be attained by an experimentally derived model. Nevertheless, although the L109F did not provide the desired increase in selectivity toward l nucleosides, it is important to note that the L109F mutation did increase the overall speed of the enzyme for all tested substrates.
FIGURE 3.
Evaluation of L109F and N93D/L109F TK2 mutants. A, a homology model of TK2 bound with l-dT (pink) and dT (green) is shown. The TK2 model (solid residues) is overlaid on a crystal structure of dCK (light colored residues) with bound substrates. The TK2 residue Leu-109 is highlighted in gold. B, a l-FMAU kinase assay using recombinant TK2-L109F and TK2-N93D/L109F in the presence of increasing dTTP concentrations is shown. C, shown is an in vitro l-18F-FMAU uptake assay using L1210 cells expressing either TK2-N93D, TK2-L109F, or TK2-N93D/L109F. The assay was done in either the presence or absence of 5 μm d-dT. Probe uptake values are reported relative to a control L1210 cell line that expresses YFP. Results are for a representative experiment or n = 2 experiments. p = 0.005 between N93D and N93D/L109F in the presence of 0 μm dT, and p = 0.0008 between N93D and N93D/L109F in the presence of 5 μm dT.
Based on these observations, we generated TK2-N93D/L109F with the expectation that this double mutant will combine the enzymatic properties of the two single mutants. As shown in Table 1, this was indeed the case. Compared with TK2, the N93D/L109F double mutant had decreased kcat with d-dT (down 49%) but increased kcat with l-dT (up 54%) and l-FMAU (up 100%). The kcat (l-FMAU)/kcat (d-dT) ratio for the TK2-N93D/L109F mutant is 14.9, 4-fold higher than that for TK2 and nearly 40% higher than that for TK2-N93D. Importantly, the TK2-N93D/L109F mutant still retained resistance to inhibition by dTTP (Fig. 3B). In the presence of 10 μm dTTP, the kinase activity of recombinant TK2-N93D/L109F decreased by only 4%, whereas that of TK2-L109F decreased by 25%. At 100 μm dTTP, TK2-N93D/L109F decreased by only 11%, whereasTK2-L109F decreased by 56%.
To determine the preference of the TK2 mutants for l-18F-FMAU over d-dT, we performed uptake assays using L1210 cells in the presence or absence of 5 μm d-dT (Fig. 3C). l-18F-FMAU uptake by TK2-N93D/L109F-expressing L1210 cells in the absence of d-dT was >1.5 times higher than that of TK2-N93D cells and nearly 4 times higher than that of TK2-L109F cells. In the presence of 5 μm d-dT, l-18F-FMAU uptake by TK2-N93D/L109F cells decreased by 47%, whereas that of TK2-N93D cells decreased by 75%. Although the l-18F-FMAU uptake of TK2-L109F in the presence of 5 μm d-dT decreased by 37%, it was still only 31% of the corresponding uptake for TK2-N93D/L109F.
Next, we investigated whether TK2-N93D/L109F had low activity toward deoxycytidine, the other endogenous nucleoside that is phosphorylated by WT TK2. TK2-N93D/L109F has a kobs (dC) that is 62% that of TK2 (supplemental Table 2). These data indicate that TK2-N93D/L109F is orthogonal to wild type TK2, with increased activity toward the l-18F-FMAU PRP and decreased activity toward the endogenous nucleosides thymidine and deoxycytidine.
In Vivo Comparison between TK2-N93D/L109F/l-18F-FMAU and HSV1-sr39tk/18F-FHBG PRG-PRP Systems
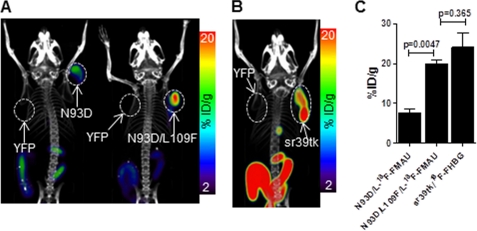
Mice implanted with L1210 cells expressing TK2-based PRGs were scanned by microPET/CT using l-18F-FMAU (Fig. 4A). For comparison, mice implanted with L1210 cells expressing HSV1-sr39tk were scanned by microPET/CT using 18F-FHBG (Fig. 4B). l-18F-FMAU uptake by the TK2-N93D/L109F-expressing L1210 cells was 2.6-fold higher than that of TK2-N93D-expressing cells (Fig. 4C). 18F-FHBG accumulation into sr39tk-expressing L1210 cells was comparable with that of l-18F-FMAU into L1210 cells expressing TK2-N93D/L109F (24.1 ± 6.2 versus 19.9 ± 1.5% injected dose/g; p = 0.37). Taken together, these findings demonstrate that the sensitivity of the TK2 N93D/L109F PRG is higher than that of the TK2-N93D PRG and is not significantly different from that of the sr39tk/18F-FHBG pair.
FIGURE 4.
Comparison of ΔTK2/l-18F-FMAU and sr39tk/18F-FHBG PET reporter gene systems. A, shown are l-18F-FMAU microPET/CT scans of mice bearing L1210 tumors engineered to express various TK2-based PRGs. B, shown are 18F-FHBG microPET/CT scans of mice bearing L1210 tumors engineered to express sr39tk. C, shown is quantification of probe uptake in L1210 tumors from A and B. %ID/g, % injected dose/g.
l-18F-FMAU Biodistribution in Humans
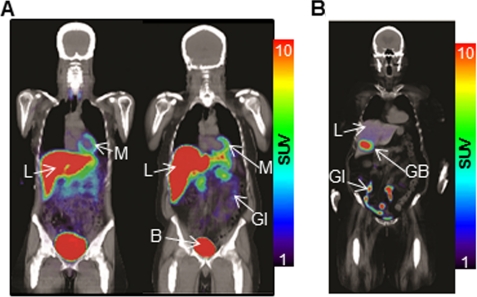
As the first step toward clinical translation of the newly developed PRG-PRP system, we determined the biodistribution of l-18F-FMAU in humans. Fig. 5 illustrates the biodistribution of l-18F-FMAU in two healthy volunteers and the biodistribution of 18F-FHBG in a female volunteer 2 h post-administration of the PRPs. Mean standard uptake values of the probes in different tissues for 18F-FHBG and l-18F-FMAU are listed in supplemental Table 3. For both probes, relatively high signals were observed in liver, kidneys, gall bladder, bladder, and the GI tract. l-18F-FMAU accumulation was also observed in the myocardium. At 2 h, more intense activity was observed in the liver after l-18F-FMAU injections than after 18F-FHBG administration. However, l-18F-FMAU activity was lower than that of 18F-FHBG within the GI tract region.
FIGURE 5.
Biodistribution of l-18F-FMAU and 18F-FHBG in humans. PET/CT scans of a healthy female (left) and a healthy male (right) volunteer 2 h after injection of l-18F-FMAU (A) and pretreatment glioma patient 2 h after injection of 18F-FHBG (B). B, bladder; GB, gallbladder; L, liver; M, myocardium. SUV, standard uptake value.
DISCUSSION
To develop a PRG that can be used in conjunction with l-18F-FMAU, a thymidine analog with the unnatural l-conformation, we removed the mitochondrial sorting sequence in human TK2. As shown previously, the truncated protein is expected to localize in the cytosol rather than in the mitochondria (20). Rational design was then used to improve the sensitivity and selectivity of the TK2 PRG. This led to the development of TK2-N93D/L109F, a double mutant TK2 kinase characterized by reduced affinity for the natural substrates d-thymidine and d-deoxycytidine and increased affinity for l-FMAU. Studies in mice indicated that the TK2-N93D/L109F PRG has comparable sensitivity to that of the widely used HSV1-sr39tk/18F-FHBG system. We have also determined the biodistribution of l-FMAU in humans.
Advantages of TK2-N93D/L109F/l-18F-FMAU PRG System
In mice, l-18F-FMAU accumulates in the liver 1-h post injection (data not shown). The progression of the signal from the liver to the gallbladder and then to the GI indicates that l-18F-FMAU is excreted via a hepato-biliary mechanism, similar to that observed for 18F-FHBG (12). However, the GI activity in l-18F-FMAU-injected mice is significantly less intense than that observed in mice injected with 18F-FHBG. The intense signal in the GI of mice injected with 18F-FHBG leads to spillover in other organs in the lower abdomen, limiting the utility of 18F-FHBG for cell tracking applications in mice if these cells localize in the abdominal cavity.
In addition to its human origin (which is expected to reduce immunogenicity compared with the viral PRGs), the TK2-N93D/L109F PRG also has the advantage of reduced activity toward the endogenous nucleosides, d-thymidine and d-deoxycytidine. PRGs are typically overexpressed in therapeutic cells. In this context, if the mutant PRG retains the ability to efficiently phosphorylate thymidine and/or deoxycytidine, then this may alter cellular metabolism due to overproduction of dTTP and/or dCTP. Such effects would be of particular concern in preclinical settings as serum levels of thymidine in mice and rats are 9–15 times higher than those in humans (56). Any changes in nucleotide metabolism and dNTP pools in therapeutic cells may have genotoxic consequences, especially when prolonged persistence in vivo of these cells is anticipated (for example in the case of stem cells). In contrast to previously reported PRG such as dCK-double mutant, TK2-N93D/L109F is less likely to perturb cellular nucleotide metabolism and genomic integrity due to the decreased activity of the double mutant enzyme toward natural substrates.
Disadvantages of New TK2-N93D/L109F/l-18F-FMAU PRG System and Potential Improvements
The observed differences between the biodistribution of l-18F-FMAU biodistribution in mice and humans underscore the importance of performing such comparative studies at an early stage of the development process for new PRG-PRP systems. In contrast to the excellent biodistribution in mice, in humans l-18F-FMAU accumulates in the myocardium and liver. Regarding l-18F-FMAU accumulation in the heart, the myocardium has a very high density of mitochondria (57). Moreover, the reported activity of the WT mitochondrial TK2 enzyme from human heart tissue is nearly 10 times higher than that of the enzyme from mouse heart tissue (58, 59). This difference may explain the observed differences in l-18F-FMAU myocardial accumulation between mice and humans. l-18F-FMAU is taken up by the liver of both mice and humans. However, l-18F-FMAU eventually clears the murine liver but is retained in the human liver. One reason for this difference may be that, similar to 18F-FLT (3′-deoxy-3′-18fluorothymidine) (60), l-18F-FMAU may also undergo glucuronidation in human liver tissue. Glucuronidation of thymidine analogs is significantly less extensive in mice than in humans (61). Given the accumulation of l-18F-FMAU in human liver and myocardium, this probe may not be useful for PET imaging of therapeutic cells at these sites. Myocardial accumulation may be reduced if l-18F-FMAU is modified to decrease its phosphorylation by WT TK2. Replacing the 5-methyl group with a larger substituent such as ethyl or propyl may achieve this objective.
Conclusion
New PET reporter gene/probe systems are needed to assist the development and clinical translation of cell-based therapies. In the current study we used a structure-guided approach to develop a human nucleoside kinase-based PRG characterized by high specificity and selectivity for l-18F-FMAU, a non-natural nucleoside analog PRP. The initial findings in mice and the observed biodistribution of l-18F-FMAU in mice and humans warrant additional studies in both species and suggest potential strategies to further improve the sensitivity and specificity of the new human TK2-based PET reporter gene assay.
Supplementary Material
Acknowledgments
We thank Lisa Ta for excellent technical assistance. We thank Liu Wei and Larry Pang for assistance with microPET/CT imaging and the cyclotron group for producing the PET probes used in this study.
This work was supported, in whole or in part, by National Institutes of Health Grants P50 CA86306 (In Vivo Cellular and Molecular Imaging Centers Developmental Project) 1R01EB13685 and 1R01CA16077001 (National Institute of Biomedical Imaging and Bioengineering). This work was also funded by California Institute for Regenerative Medicine Grant RT1-01126-1 and United States Department of Energy Contract DE-FG02-06ER64249. Conflict of interest statement: Dean Campbell, Caius Radu, Arnon Lavie, Nagichettiar Satyamurthy, Johannes Czernin, and Shahriar Yaghoubi are among the inventors of the national and Patent Cooperation Treaty (PCT) patent applications for the PRG technology referred to in this article. Shahriar Yaghoubi is also a co-founder and the Chief Scientific Officer of CellSight Technologies, Inc., a molecular imaging company that specializes in imaging tools for cell and gene therapies that has licensed this intellectual property.

This article contains supplemental Tables 1–3.
- PET
- positron emission tomography
- PRG
- PET reporter gene
- PRP
- PET reporter probe
- TK2
- thymidine kinase 2
- l-18F-FMAU
- 2′-deoxy-2′-18F-5-methyl-1-β-L-arabinofuranosyluracil
- HSV1-tk
- herpes simplex virus type 1 thymidine kinase
- 18F-FHBG
- 9-[4-18F-3-(hydroxymethyl)butyl]guanine
- CT
- computed tomography
- GI
- gastrointestinal
- d-dT
- d-thymdine
- l-dT
- l-thymidine
- SUMO
- small ubiquitin-like modifier
- dCK
- deoxycytidine kinase.
REFERENCES
- 1. Herschman H. R. (2004) Crit. Rev. Oncol. Hematol. 51, 191–204 [DOI] [PubMed] [Google Scholar]
- 2. Gambhir S. S., Yaghoubi S. S. (eds) (2010) Molecular Imaging With Reporter Genes, pp. 258–274, Cambridge University Press, Cambridge, UK [Google Scholar]
- 3. Nair-Gill E. D., Shu C. J., Hildebrandt I. J., Campbell D. O., Witte O. N., Radu C. G. (2010) in Molecular Imaging with Reporter Genes (Gambhir S. S., Yaghoubi S. S., eds) pp. 258–274 Cambridge University Press, Cambridge, UK [Google Scholar]
- 4. Tjuvajev J. G., Finn R., Watanabe K., Joshi R., Oku T., Kennedy J., Beattie B., Koutcher J., Larson S., Blasberg R. G. (1996) Cancer Res. 56, 4087–4095 [PubMed] [Google Scholar]
- 5. Gambhir S. S., Bauer E., Black M. E., Liang Q., Kokoris M. S., Barrio J. R., Iyer M., Namavari M., Phelps M. E., Herschman H. R. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 2785–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shu C. J., Guo S., Kim Y. J., Shelly S. M., Nijagal A., Ray P., Gambhir S. S., Radu C. G., Witte O. N. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17412–17417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yaghoubi S. S., Creusot R. J., Ray P., Fathman C. G., Gambhir S. S. (2007) J. Biomed. Opt. 12, 064025. [DOI] [PubMed] [Google Scholar]
- 8. Wu J. C., Chen I. Y., Sundaresan G., Min J. J., De A., Qiao J. H., Fishbein M. C., Gambhir S. S. (2003) Circulation 108, 1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hung S. C., Deng W. P., Yang W. K., Liu R. S., Lee C. C., Su T. C., Lin R. J., Yang D. M., Chang C. W., Chen W. H., Wei H. J., Gelovani J. G. (2005) Clin. Cancer Res. 11, 7749–7756 [DOI] [PubMed] [Google Scholar]
- 10. Yaghoubi S. S., Jensen M. C., Satyamurthy N., Budhiraja S., Paik D., Czernin J., Gambhir S. S. (2009) Nat. Clin. Pract. Oncol. 6, 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peñuelas I., Mazzolini G., Boán J. F., Sangro B., Martí-Climent J., Ruiz M., Ruiz J., Satyamurthy N., Qian C., Barrio J. R., Phelps M. E., Richter J. A., Gambhir S. S., Prieto J. (2005) Gastroenterology 128, 1787–1795 [DOI] [PubMed] [Google Scholar]
- 12. Yaghoubi S., Barrio J. R., Dahlbom M., Iyer M., Namavari M., Satyamurthy N., Goldman R., Herschman H. R., Phelps M. E., Gambhir S. S. (2001) J. Nucl. Med. 42, 1225–1234 [PubMed] [Google Scholar]
- 13. Chin F. T., Namavari M., Levi J., Subbarayan M., Ray P., Chen X., Gambhir S. S. (2008) Mol. Imaging Biol. 10, 82–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miyagawa T., Gogiberidze G., Serganova I., Cai S., Balatoni J. A., Thaler H. T., Ageyeva L., Pillarsetty N., Finn R. D., Blasberg R. G. (2008) J. Nucl. Med. 49, 637–648 [DOI] [PubMed] [Google Scholar]
- 15. Alauddin M. M., Shahinian A., Park R., Tohme M., Fissekis J. D., Conti P. S. (2007) Eur. J. Nucl. Med. Mol. Imaging 34, 822–829 [DOI] [PubMed] [Google Scholar]
- 16. Traversari C., Marktel S., Magnani Z., Mangia P., Russo V., Ciceri F., Bonini C., Bordignon C. (2007) Blood 109, 4708–4715 [DOI] [PubMed] [Google Scholar]
- 17. Berger C., Flowers M. E., Warren E. H., Riddell S. R. (2006) Blood 107, 2294–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Arnér E. S., Eriksson S. (1995) Pharmacol. Ther. 67, 155–186 [DOI] [PubMed] [Google Scholar]
- 19. Likar Y., Zurita J., Dobrenkov K., Shenker L., Cai S., Neschadim A., Medin J. A., Sadelain M., Hricak H., Ponomarev V. (2010) J. Nucl. Med. 51, 1395–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ponomarev V., Doubrovin M., Shavrin A., Serganova I., Beresten T., Ageyeva L., Cai C., Balatoni J., Alauddin M., Gelovani J. (2007) J. Nucl. Med. 48, 819–826 [DOI] [PubMed] [Google Scholar]
- 21. Dotti G., Tian M., Savoldo B., Najjar A., Cooper L. J., Jackson J., Smith A., Mawlawi O., Uthamanthil R., Borne A., Brammer D., Paolillo V., Alauddin M., Gonzalez C., Steiner D., Decker W. K., Marini F., Kornblau S., Bollard C. M., Shpall E. J., Gelovani J. G. (2009) Mol. Imaging 8, 230–237 [PMC free article] [PubMed] [Google Scholar]
- 22. Kumar D., Abdulovic A. L., Viberg J., Nilsson A. K., Kunkel T. A., Chabes A. (2011) Nucleic Acids Res. 39, 1360–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Song S., Wheeler L. J., Mathews C. K. (2003) J. Biol. Chem. 278, 43893–43896 [DOI] [PubMed] [Google Scholar]
- 24. Sargent R. G., Mathews C. K. (1987) J. Biol. Chem. 262, 5546–5553 [PubMed] [Google Scholar]
- 25. Kumar D., Viberg J., Nilsson A. K., Chabes A. (2010) Nucleic Acids Res. 38, 3975–3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hazra S., Sabini E., Ort S., Konrad M., Lavie A. (2009) Biochemistry 48, 1256–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 28. Sabini E., Ort S., Monnerjahn C., Konrad M., Lavie A. (2003) Nat. Struct. Biol. 10, 513–519 [DOI] [PubMed] [Google Scholar]
- 29. Hazra S., Szewczak A., Ort S., Konrad M., Lavie A. (2011) Biochemistry 50, 2870–2880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hazra S., Ort S., Konrad M., Lavie A. (2010) Biochemistry 49, 6784–6790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Agarwal K. C., Miech R. P., Parks R. E., Jr. (1978) Methods Enzymol. 51, 483–490 [DOI] [PubMed] [Google Scholar]
- 32. Barroso J. F., Carvalho R. N., Flatmark T. (2005) Biochemistry 44, 4886–4896 [DOI] [PubMed] [Google Scholar]
- 33. Wang L., Saada A., Eriksson S. (2003) J. Biol. Chem. 278, 6963–6968 [DOI] [PubMed] [Google Scholar]
- 34. Jordheim L. P., Cros E., Gouy M. H., Galmarini C. M., Peyrottes S., Mackey J., Perigaud C., Dumontet C. (2004) Clin. Cancer Res. 10, 5614–5621 [DOI] [PubMed] [Google Scholar]
- 35. Hawley R. G., Lieu F. H., Fong A. Z., Hawley T. S. (1994) Gene. Ther. 1, 136–138 [PubMed] [Google Scholar]
- 36. Radu C. G., Shu C. J., Nair-Gill E., Shelly S. M., Barrio J. R., Satyamurthy N., Phelps M. E., Witte O. N. (2008) Nat. Med. 14, 783–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shields A. F. (2003) J. Nucl. Med. 44, 1432–1434 [PubMed] [Google Scholar]
- 38. Sun H., Mangner T. J., Collins J. M., Muzik O., Douglas K., Shields A. F. (2005) J. Nucl. Med. 46, 292–296 [PubMed] [Google Scholar]
- 39. Mangner T. J., Klecker R. W., Anderson L., Shields A. F. (2003) Nucl. Med. Biol. 30, 215–224 [DOI] [PubMed] [Google Scholar]
- 40. Namavari M., Chang Y. F., Kusler B., Yaghoubi S., Mitchell B. S., Gambhir S. S. (2011) Mol. Imaging Biol. 13, 812–818 [DOI] [PubMed] [Google Scholar]
- 41. de Leder Kremer R. M., Gallo-Rodriguez C. (2004) Adv. Carbohydr. Chem. Biochem. 59, 9–67 [DOI] [PubMed] [Google Scholar]
- 42. Shu C. J., Campbell D. O., Lee J. T., Tran A. Q., Wengrod J. C., Witte O. N., Phelps M. E., Satyamurthy N., Czernin J., Radu C. G. (2010) J. Nucl. Med. 51, 1092–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nishii R., Volgin A. Y., Mawlawi O., Mukhopadhyay U., Pal A., Bornmann W., Gelovani J. G., Alauddin M. M. (2008) Eur. J. Nucl. Med. Mol. Imaging 35, 990–998 [DOI] [PubMed] [Google Scholar]
- 44. Mukhopadhyay U., Pal A., Gelovani J. G., Bornmann W., Alauddin M. M. (2007) Appl. Radiat. Isot. 65, 941–946 [DOI] [PubMed] [Google Scholar]
- 45. Schwarzenberg J., Radu C. G., Benz M., Fueger B., Tran A. Q., Phelps M. E., Witte O. N., Satyamurthy N., Czernin J., Schiepers C. (2011) Eur. J. Nucl. Med. Mol. Imaging 38, 711–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Alauddin M. M., Shahinian A., Gordon E. M., Conti P. S. (2004) Mol. Imaging 3, 76–84 [DOI] [PubMed] [Google Scholar]
- 47. Alauddin M. M., Conti P. S. (1998) Nucl. Med. Biol. 25, 175–180 [DOI] [PubMed] [Google Scholar]
- 48. Alauddin M. M., Shahinian A., Gordon E. M., Bading J. R., Conti P. S. (2001) J. Nucl. Med. 42, 1682–1690 [PubMed] [Google Scholar]
- 49. Wang J., Choudhury D., Chattopadhyaya J., Eriksson S. (1999) Biochemistry 38, 16993–16999 [DOI] [PubMed] [Google Scholar]
- 50. Radivoyevitch T., Munch-Petersen B., Wang L., Eriksson S. (2011) Nucleosides Nucleotides Nucleic Acids 30, 203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bianchi V., Borella S., Rampazzo C., Ferraro P., Calderazzo F., Bianchi L. C., Skog S., Reichard P. (1997) J. Biol. Chem. 272, 16118–16124 [DOI] [PubMed] [Google Scholar]
- 52. Spyrou G., Reichard P. (1988) Mutat. Res. 200, 37–43 [DOI] [PubMed] [Google Scholar]
- 53. Eriksson S., Munch-Petersen B., Johansson K., Eklund H. (2002) Cell. Mol. Life Sci. 59, 1327–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Welin M., Skovgaard T., Knecht W., Zhu C., Berenstein D., Munch-Petersen B., Piskur J., Eklund H. (2005) FEBS J. 272, 3733–3742 [DOI] [PubMed] [Google Scholar]
- 55. Sabini E., Hazra S., Konrad M., Lavie A. (2007) J. Med. Chem. 50, 3004–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nottebrock H., Then R. (1977) Biochem. Pharmacol. 26, 2175–2179 [DOI] [PubMed] [Google Scholar]
- 57. Schaper J., Meiser E., Stämmler G. (1985) Circ. Res. 56, 377–391 [DOI] [PubMed] [Google Scholar]
- 58. Saada A., Shaag A., Elpeleg O. (2003) Mol. Genet. Metab. 79, 1–5 [DOI] [PubMed] [Google Scholar]
- 59. Wang L., Eriksson S. (2000) Biochem. J. 351, 469–476 [PMC free article] [PubMed] [Google Scholar]
- 60. Shields A. F., Grierson J. R., Dohmen B. M., Machulla H. J., Stayanoff J. C., Lawhorn-Crews J. M., Obradovich J. E., Muzik O., Mangner T. J. (1998) Nat. Med. 4, 1334–1336 [DOI] [PubMed] [Google Scholar]
- 61. Barthel H., Cleij M. C., Collingridge D. R., Hutchinson O. C., Osman S., He Q., Luthra S. K., Brady F., Price P. M., Aboagye E. O. (2003) Cancer Res. 63, 3791–3798 . [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.