Background: nSMase2 is a phospho-protein presenting a novel target in lung injury.
Results: We identified five phosphorylated serines in nSMase2 that control its activity and stability. Both depend on enhanced phosphorylation but could be regulated independently.
Conclusion: The five serines are conserved and consist of interdependent phosphorylation sites.
Significance: Overall, initial regulatory structure/function of nSMase2 is presented.
Keywords: Ceramide, Epithelial Cell, Oxidative Stress, Protein Phosphorylation, Protein Stability, Sphingomyelinase, Activity, Lung Epithelial Cells, nSMase2
Abstract
We previously presented that the neutral sphingomyelinase 2 (nSMase2) is the only SMase activated in human airway epithelial (HAE) cells following exposure to oxidative stress (ox-stress), yielding ceramide accumulation and thereby inducing apoptosis. Furthermore, we reported that nSMase2 is a phospho-protein in which the level of phosphorylation controls nSMase2 activation induced by ox-stress. Here we identify five specific serines that are phosphorylated in nSMase2 and demonstrate that their phosphorylation controls the nSMase2 activity upon ox-stress exposure in an interdependent manner. Furthermore, we show that the nSMase2 protein stability and thus its level of expression is also post-translationally regulated by these five serine phosphorylation sites. This study provides initial structure/function insights regarding nSMase2 phosphorylation sites and offers some new links for future studies aiming to fully elucidate nSMase2 regulatory machinery.
Introduction
The neutral sphingomyelinase 2 (nSMase2,2 from SMPD3 gene) is a member of the extended family of neutral sphingomyelinase enzymes (1, 2), which consists of a specific type of phospholipase C that hydrolyzes the phosphodiester bond of the membrane sphingolipid sphingomyelin (SM) to yield ceramide and phosphocholine (3). Ceramide is a well established second messenger that regulates biological processes such as apoptosis, cell cycle arrest, and senescence, and thus, the modulation of SMase function that controls ceramide generation has been the subject of intense investigation (3–15). Indeed, SM hydrolysis by SMases is considered the major pathway for cell stress-induced ceramide generation and subsequent signaling (4, 8, 10, 16–21).
Consistently, we have recently demonstrated that nSMase2 is a redox-sensitive enzyme that is activated in human airway epithelial (HAE) cells in response to oxidative stress (ox-stress) induced by H2O2 or cigarette smoke (CS) exposure (7, 22). Importantly, we demonstrated that nSMase2 functions as a key target in CS/ox-stress-induced lung injury. Indeed, we not only found that nSMase2 is necessary in elevating ceramide and inducing apoptosis in lungs of rodents exposed to CS, but also that nSMase2 is overexpressed in those rodent lungs and in the lungs of humans (smokers) diagnosed with pulmonary emphysema (19). With such a critical role for nSMase2 in generating ceramide and thereby inducing apoptosis and lung injury, it has became crucial to elucidate the molecular mechanism(s) that regulates its function in response to ox-stress.
We have recently shown that nSMase2 is a phospho-protein in which the level of phosphorylation is modulated by ox-stress, which also controls nSMase2 function (21). We demonstrated that calcineurin (CaN) phosphatase (also known as protein phosphatase 2B) directly binds to nSMase2, dephosphorylates it, and thereby, leads to its deactivation. However, under H2O2-induced ox-stress, when CaN is degraded, nSMase2 is fully phosphorylated and activated, downstream of protein kinase C (PKC) and p38 mitogen-activated protein kinase (MAPK) (21).
In this study, we sought to determine the specific phosphorylation site(s) of nSMase2 and their role in regulating nSMase2 function under exposure of HAE cells to ox-stress. We present herein five serines that are phosphorylated in nSMase2 and demonstrate that the phosphorylation of these serines controls nSMase2 activity in an interdependent manner. Lastly, we found that the nSMase2 protein stability, and thus, its level of expression is determined post-translationally by the phosphorylation of those five serines.
EXPERIMENTAL PROCEDURES
Cell Culture, Treatments, and Reagents
A549 adenocarcinoma (ATCC) and immortalized human bronchial epithelial (HBE1) cells (from Dr. Reen Wu, University of California at Davis (UCD)) were employed in this study. A549 cells were cultured in F12K medium (Invitrogen) supplemented with 10% FBS (Invitrogen) and 1% penicillin/streptomycin (Invitrogen). HBE1 cells were cultured as described previously (23, 24). Cells were grown in serum-free medium supplemented with insulin (Sigma 5 μg/ml), transferrin (Sigma 5 μg/ml), epidermal growth factor (EGF from Upstate Biotech Millipore, 5 ng/ml), dexamethasone (Sigma, 0.1 μm), cholera toxin (Sigma, 20 ng/ml), and bovine hypothalamus extract (Sigma, 15 μg/ml). Within this study, A549 cells and HBE1 cells are collectively referred to as HAE cells. DMEM medium (Invitrogen) was used for the treatments with no FBS supplementation. Hydrogen peroxide (H2O2, from Sigma) was added directly into the treatment medium at a final concentration of 250 μm. Cycloheximide (Sigma) was dissolved in dimethyl sulfoxide (DMSO, from Sigma) and then diluted in the treatment medium at a final concentration of 100 μm. Phosphate-buffered saline (PBS from Invitrogen) or DMSO was added at the appropriate concentration to the control-untreated cells where needed. Cells were collected by scraping either in PBS or directly in the lysis buffer: 1% Nonidet P-40 (IGEPAL, from Sigma), 50 mm Tris, 10% glycerol, 0.02% NaN3, 150 mm NaCl, pH 7.4 (all from Fisher Chemical), containing a mixture of phosphatase and protease inhibitors (Sigma) as well as 1 mm NaF and Na3VO4 (both from Sigma). Lysates were passed five times through a 30-gauge needle prior to centrifugation and further processing of the samples.
Transient transfections of V5-nSMase2 were performed using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer's protocol. The original murine V5-nSMase2 (3′-V5 tag) in the pEF6/V5-His-TOPO vector was kindly provided by Dr. Yusuf Hannun (Medical University of South Carolina), and V5-nSMase2 mutants were prepared by site-directed mutagenesis. Mutagenesis of the plasmid was performed using the Stratagene QuikChange II kit from Agilent Technologies according to the manufacturer's instructions. Briefly, mutagenic primers were designed using the QuikChange Primer Design program on the Agilent Technologies website and were used to amplify nascent plasmid containing the desired mutation(s). Pfu Turbo DNA polymerase enzyme was used in the mutagenetic PCR. Following PCR amplification, the PCR reaction was digested with DpnI restriction enzyme (New England Biolabs) to destroy the parent plasmid, and the remaining mutated plasmid was transformed into chemically competent Escherichia coli (strain DH5). After bacterial colony selection and growth (standard procedures) using lysogeny broth supplemented with 100 μg/ml ampicillin (Roche Applied Science), plasmid DNA was extracted using the maxi prep kit from E&K Scientific (according to the manufacturer's instructions) and resuspended in TE buffer (10 mm Tris base (Fisher Scientific), 1 mm EDTA (from Fisher Scientific), pH 8). As a standard control procedure, all mutant plasmids described in this study were sequenced (commercially available sequencing service). All the other reagents listed below were from Sigma, if not differently indicated.
Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE)
6–12% acrylamide gels were prepared following common procedures (not described) and run via a two-cell system (Bio-Rad) for 1–3 h at 100 V at room temperature.
Immuno-precipitation (IP)
200–400 μg of total protein extracts were incubated overnight with 2–4 μg of antibody (Ab): α-V5 (Invitrogen). 50 μl of 50% protein A-agarose bead complexes (Repligen) were added to the samples and incubated for 90 min. Four washes (by sequential centrifugation and resuspension) with the Nonidet P-40-lysis buffer were done prior to resuspending the immuno-precipitates in 50 μl of 2× loading dye (for SDS-PAGE).
Immuno-blotting (IB)
20–100 μg of total protein extracts or the IP samples were loaded into each well of the SDS-PAGE in the presence of an SDS/dithiothreitol (DTT) reducing loading dye (common concentrations/conditions: samples heated for 5 min at 95 °C). After SDS-PAGE separation, the proteins were transferred to a nitrocellulose membrane and “blocked” with 5% skim milk in Tris-buffered saline with 0.05% Tween 20 (TBST) for 120 min or overnight. Primary Abs were incubated in TBST for 2 h at room temperature. Secondary Abs, either goat α-mouse-horseradish peroxidase (HRP)-conjugated or goat α-rabbit-HRP-conjugated (Jackson ImmunoResearch Laboratories), were incubated for 90 min at room temperature at 1:10,000 dilution in TBST. Bands were visualized by enhanced chemiluminescence (ECL, Pierce). Extensive washes in TBST were done in between each step. Primary Abs used in this study for the immuno-blots were α-V5 (Invitrogen, 1:5000) and α- β-actin (Sigma, 1:10,000).
MALDI-TOF Mass Spectrometry for Phosphorylation Sites
V5-nSMase2 was transfected in HBE1 cells. 24 h after transfection, V5-nSMase2 was immuno-precipitated with the α-V5 Ab and separated by SDS-PAGE. The band relative to V5-nSMase2 was visualized in the gel by staining with Imperial protein stain (Pierce). The piece of gel containing the band was cut out using a scalpel, washed twice with 100 mm ambic, and then dehydrated with 100% acetonitrile followed by speed-vacuum spin (to dry the sample). The gel was rehydrated with 10 mm DTT in 100 mm ambic and heated at 56 °C for 30 min. The DTT-reduced sample was dehydrated with acetonitrile and dried by speed-vacuum spin, then resuspended in 55 mm iodoacetamide in 100 mm ambic to alkylate cysteines and subsequently washed with 100 mm ambic, and then dehydrated and dried with acetonitrile and speed-vacuum spin, respectively. Digestion Buffer was added overnight (50 mm ambic solution containing sequencing grade-modified trypsin from Promega at 13 ng/μl (37 °C)). The reaction was blocked by 60% acetonitrile, the piece of gel was sonicated for 10 min, and the supernatant was collected and dried in the speed-vacuum. 3% trifluoroacetic acid was added and the sample was sonicated again. The sample was then analyzed by MALDI-TOF mass spectrometry in the proteomic core of the Genome and Biomedical Sciences Facility (UCD) by the core staff. Results were analyzed using the program Mascot (Matrix Science).
nSMase Activity Assay
The enzyme activity of nSMase was determined as described (25, 26). Briefly, an enzyme preparation of 10 μg of total protein (from V5-nSMase2-transfected cells) in 20 mm Tris-HCl, pH 7.4, was mixed with [14C]SM (10 nmol/100,000 dpm) in 20 mm Tris-HCl, pH 7.4, containing 0.1% Triton X-100, 10 mm MgCl2, 10 mm DTT, 10 nm phosphatidylserine (Avanti Polar Lipids), and 1 mg/ml BSA. The incubation time was 30 min at 37 °C. The reaction was terminated by the addition of 1 ml of chloroform:methanol (2:1) followed by 0.2 ml of 0.5 m NaCl (Fisher Chemical). After phase separation, the upper phase was collected, and the radioactivity was determined by liquid scintillation counting. The nSMase activity was normalized to the level of transfection of V5-nSMase2, determined by IB with the α-V5 Ab in parallel experiments.
32P in Vivo Labeling for Revealing Phosphorylation of nSMase2
Cells were transfected with various V5-nSMase2 constructs for 24 h and then incubated in a phosphate-free DMEM (MP Biochemicals) for 4 h in the presence of [32P]orthophosphate (MP Biochemicals) and then treated (or not treated) for 30 min with 250 μm H2O2. After cell lysis and IP of V5-nSMase2, radioactivity of nSMase2 was assessed by autoradiography and normalized to the amount of immuno-precipitated V5-nSMase2 measured by IB.
Protein Stability Assay
A549 cells were transiently transfected with various V5-nSMase2 constructs, and 24 h after transfection, they were continuously incubated up until 12 h in 100 μm cycloheximide to block new protein synthesis. At several time points, the cells were collected by scraping them in the Nonidet P-40 lysis buffer. 30 μg of total proteins were discriminated by SDS-PAGE and immuno-blotted for V5-nSMase2 and β-actin protein expression using specific Abs.
Statistical Analysis
Each experiment was repeated at least three times. The plotted data are reported as mean ± S.D. Statistical significance was determined by Student's t test, and p value < 0.05 was considered statistically significant. The program GraphPad Prism 5.01 (GraphPad Software Inc.) was used to calculate the p value between the intercepts on the x axis of linear regressions.
RESULTS
Identification of Five Phospho-serines in nSMase2
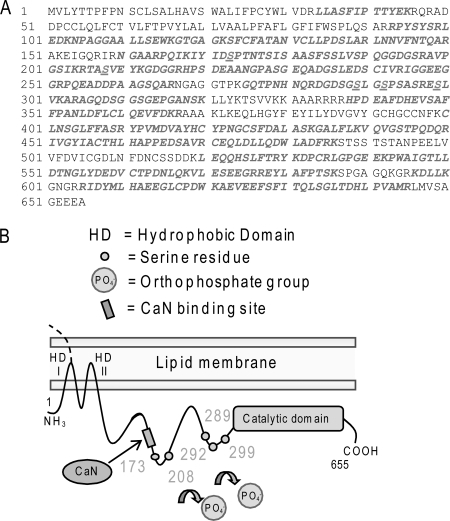
We aimed to elucidate the specific phospho-residues involved in the regulation of nSMase2 function. HBE1 cells were transiently transfected with murine V5-nSMase2. 24 h after transfection, the overexpressed V5-nSMase2 was immuno-precipitated from the total cell lysates by the α-V5 Ab, isolated by SDS-PAGE, eluted, and digested from the gel using trypsin enzyme and then analyzed by MALDI-TOF mass spectrometry for phosphorylated residues, as described under “Experimental Procedures.” Five serines were identified to be phosphorylated in nSMase2: serines 173, 208, 289, 292, and 299 (accession number: NP_067466.1; GI: 10946902). As shown in Fig. 1, two of these phospho-serines (Ser-173 and Ser-208) are close to the CaN binding site that we previously identified (21), whereas the remaining three phospho-serines (Ser-289, Ser-292, and Ser-299) are clustered before the predicted catalytic domain of nSMase2 (16). These serines are all conserved and match with the human nSMase2 serines 173, 209, 291, 294, and 301, respectively (accession number: NP_061137.1; GI: 8923946).
FIGURE 1.
Identification of five phospho-serines in nSMase2. A, the nSMase2 sequence has been screened by MALDI-TOF mass spectrometry, and five phospho-serines have been identified; the screened sequence is in italic/bold font, and the phospho-serines are also underlined. B, topology model of nSMase2 showing the two hydrophobic domains (HD I and II) at the N terminus, the PQIKIY binding site for CaN, the five identified phospho-serines (small, numbered circles along the sequence) and the catalytic domain at the C terminus.
Supporting our findings, out of five serines that we identified in nSMase2, four, Ser-173, Ser-208, Ser-289, and Ser-292, were also reported in the proteome-wide phospho-protein analyses obtained from murine brain tissue or brown fat (27, 28). The proteome-wide analyses also predict four additional putatively phosphorylated serines in nSMase2. Three (Ser-178, Ser-183, and Ser-186) are in the proximity of the Ser-173, and one is in position 294. Of note is that the proteome-wide analysis suggests that a tyrosine residue, Tyr-170, may also be phosphorylated.
Although it is possible that additional serines could be phosphorylated in nSMase2 of HAE cells, the phosphorylation of any Tyr residues of nSMase2 was completely excluded by our studies. This was assessed first by partial acid hydrolysis of the 32P radiolabeled immuno-precipitated nSMase2 followed by thin layer electrophoresis, as reported previously (21), and also by immuno-blot analysis with a specific anti-phospho-tyrosine Ab (α-pY20, from Santa Cruz Biotechnology) of nSMase2 that had been immuno-precipitated from lysates of HAE cells exposed (or not exposed) to ox-stress (not shown). Both methods excluded any Tyr phosphorylation in nSMase2.
A Group of Conserved Phosphorylated Serines Interdependently Controls Neutral Sphingomyelinase 2 Activity in Response to Oxidative Stress
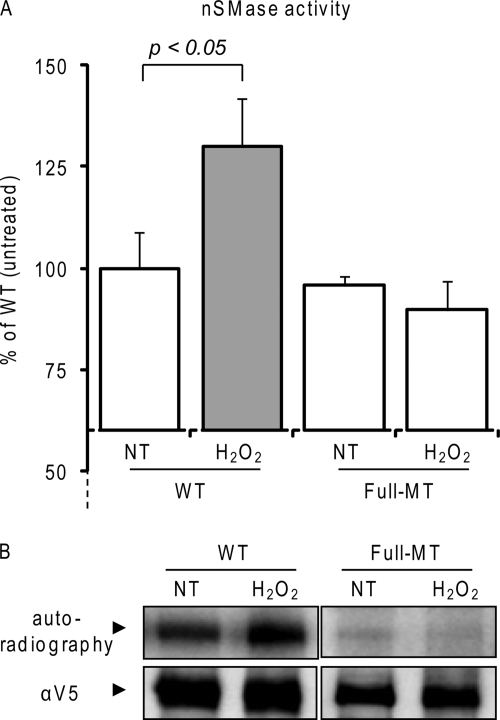
We next investigated whether these five identified phospho-serines play a role in the activation of nSMase2 under ox-stress. We used site-directed mutagenesis to generate a full mutant of nSMase2, where all five serines were replaced with alanines (Full-MT).3 Subsequently, the Full-MT and the wild type (WT) nSMase2 were overexpressed in HBE1 cells at comparable levels, as assessed by quantitative RT-PCR (not shown). 24 h after transfection, the cells were exposed (or not exposed) to a pulse-chase dose of 250 μm H2O2 for 30 min. After cell homogenization and centrifugation, the postnuclear supernatant (PNS) of each sample (10 μg of total proteins) was assayed for nSMase activity as described under “Experimental Procedures.” Fig. 2A demonstrates that the Full-MT nSMase2 conserves a basal in vitro nSMase activity per protein unit that is comparable with that of the WT nSMase2. However, although the WT activity is increased following 30 min of exposure to H2O2, the Full-MT no longer responds to H2O2. Comparable results were obtained in A549 cells (not shown). Next, we investigated the levels of phosphorylation of the Full-MT and WT nSMase2 before and after the H2O2 exposure. Cells were radiolabeled with [32P]orthophosphate and then exposed (or not exposed) to H2O2, as described under “Experimental Procedures.” The Full-MT and the WT nSMase2 were immuno-precipitated from the respective cell lysates, isolated by SDS-PAGE, and transferred to nitrocellulose membrane. Subsequently, autoradiography was used to measure the phosphorylation levels of nSMase2, whereas total immuno-precipitated nSMase2 levels were assessed by IB with the α-V5 Ab. Fig. 2B shows that H2O2 exposure induces an increase of phosphorylation of the WT-nSMase2 (as reported previously by us (21)), but not of the Full-MT nSMase2. This demonstrates that the phosphorylation that regulates nSMase2 activity under ox-stress is dependent upon the five identified serines.
FIGURE 2.
Serine phosphorylation regulates activation of nSMase2 under ox-stress. A, wild type (WT) or Full-MT V5-tagged nSMase2 was transfected in HBE1 cells; 24 h after transfection, the cells were treated (or not treated (NT)) with 250 μm H2O2 for 30 min. Total nSMase activity of 10 μg of PNS cell homogenate was measured as described under “Experimental Procedures.” The data are reported as the percentage of the activity of the untreated WT nSMase2 after normalization to the transfection levels that were assessed by IB equal amounts of cell homogenates with the α-V5 Ab. Standard deviations are indicated. B, V5-tagged nSMase2-transfected cells were labeled in vivo with [32P]orthophosphate for 4 h; cells were treated (or not treated) for the last 30 min of labeling with 250 μm H2O2. V5-nSMase2 was immuno-precipitated from the total cell lysates with the α-V5 Ab, separated by SDS-PAGE, and transferred to nitrocellulose membrane; then, nSMase2 phosphorylation levels and total immuno-precipitated nSMase2 were measured by autoradiography and IB (with α-V5 Ab), respectively.
Ser-173 Phosphorylation Controls nSMase2 Activation following ox-stress Exposure of HAE Cells
Next, we investigated which specific phosphorylation site(s) among the identified five serines was necessary for the ox-stress-inducible activity of nSMase2. Thus, we generated several nSMase2 constructs with various combinations of mutated serines and tested them for basal and H2O2-induced activity, as described below.
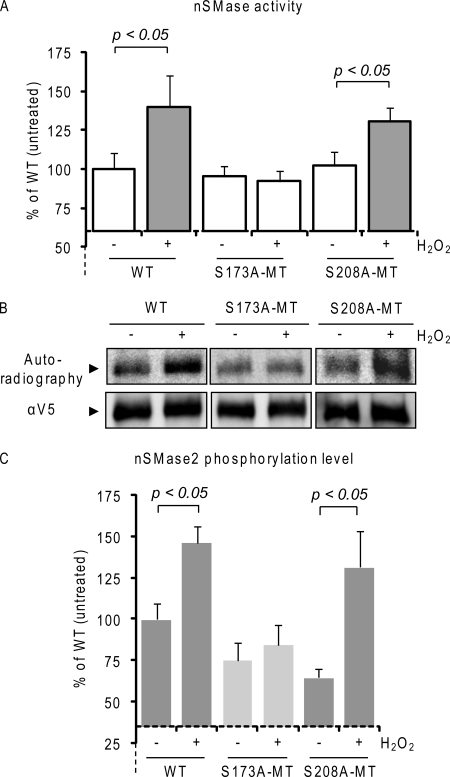
First, we mutated (to alanine) serine 173 (S173A-MT) and serine 208 (S208A-MT), which, out of the five identified serines, are the two closest to the CaN binding site (Fig. 1). 24 h after transfection, A549 cells were transiently transfected with the above nSMase2 constructs at comparable levels and exposed to 250 μm H2O2 for 30 min. As shown in Fig. 3A, by measuring nSMase activity in 10 μg of PNS cell homogenates, we demonstrate that the single point mutant of Ser-173 (S173A-MT nSMase2) loses its ability to be activated upon H2O2-induced ox-stress exposure, whereas the single point S208A-MT could still be activated similarly to the WT nSMase2. In parallel, Fig. 3B demonstrates that following 32P radiolabeling, S173A-MT lost the capacity to be phosphorylated under H2O2 exposure, whereas the S208A-MT kept the H2O2-inducible up-regulation of phosphorylation levels, similarly to that of WT nSMase2 (Fig. 3B). Comparable results were also obtained in HBE1 cells (not shown). These data demonstrate that the inducible activity of nSMase2 observed under exposure to ox-stress is regulated by the phosphorylation of the serine in position 173, whereas the serine 208 does not appear to be essential.
FIGURE 3.
Serine 173 phosphorylation controls nSMase2-inducible activity upon ox-stress. A, wild type (WT) and single point mutants S173A-MT and S208A-MT V5-tagged nSMase2 were transfected in A549 cells; 24 h after transfection, the cells were treated (or not treated) with 250 μm H2O2 for 30 min. Total nSMase activity of 10 μg of PNS cell homogenate was measured as described under “Experimental Procedures.” The data are reported as the percentage of the activity of the untreated WT nSMase2 after normalization to the transfection levels that were assessed by IB equal amounts of cell lysates with the α-V5 Ab. Standard deviations are indicated. B, V5-tagged nSMase2-transfected cells were labeled in vivo with [32P]orthophosphate for 4 h. Cells were treated (or not treated) for the last 30 min of labeling with 250 μm H2O2. V5-nSMase2 was immuno-precipitated from the total cell lysates with the α-V5 Ab; then, nSMase2 phosphorylation levels and total immuno-precipitated nSMase2 were measured by autoradiography and IB (with α-V5 Ab), respectively. C, data represent nSMase2 phosphorylation levels of three independent experiments; standard deviations are indicated.
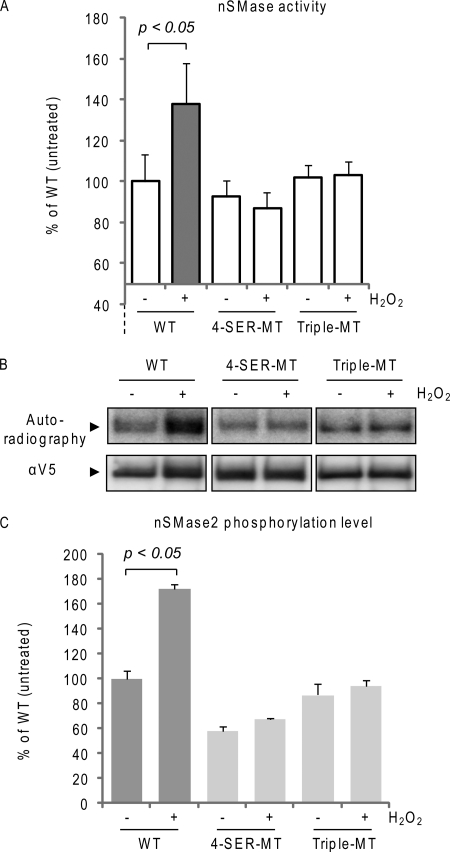
However, when we mutated all five serines, except for the Ser-173, i.e. four serines (4-SER-MT) were mutated, S208A/S289A/S292A/S299A, the ox-stress-dependent activation of nSMase2 was lost for this 4-SER-MT (Fig. 4A). Moreover, the same loss of ox-stress-dependent activation was found for a triple mutant S289A/S292A/S299A (Triple-MT) nSMase2 (Fig. 4A). In parallel, we assessed the phosphorylation levels of the 4-SER- and 3-SER-MTs and found that they no longer respond to ox-stress with increased phosphorylation levels (Fig. 4, B and C). This demonstrates that intact Ser-289, Ser-292, and Ser-299 are also necessary for nSMase2 activation by ox-stress and indicate that there could be interdependent regulation between the phosphorylation of Ser-173 and the other phospho-serines (most probably with the cluster Ser-289-Ser-292-Ser-299 and not with the Ser-208).
FIGURE 4.
Phosphorylation sites in nSMase2 are interdependently regulated. A, wild type (WT) and 4-SER-MT and Triple-MT nSMase2 were transfected in A549 cells; 24 h after transfection, the cells were treated (or not treated) with 250 μm H2O2 for 30 min. Total nSMase activity of 10 μg of PNS cell homogenate was measured as described under “Experimental Procedures.” The data are reported as the percentage of the activity of the untreated WT nSMase2 after normalization to the transfection levels that were assessed by IB equal amounts of cell homogenates with the α-V5 Ab. Standard deviations are indicated. B, V5-tagged nSMase2-transfected cells were labeled in vivo with [32P]orthophosphate for 4 h. Cells were treated (or not treated) for the last 30 min of labeling with 250 μm H2O2. V5-nSMase2 was immuno-precipitated from the total cell lysates with the α-V5 Ab; then, nSMase2 phosphorylation levels and total immuno-precipitated nSMase2 were measured by autoradiography and IB (with α-V5 Ab), respectively. C, data represent nSMase2 phosphorylation levels of three independent experiments; standard deviations are indicated.
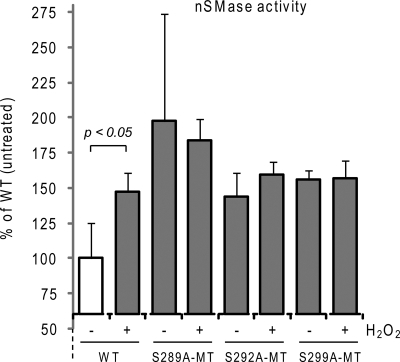
To investigate this possibility, we generated single point MTs of Ser-289, Ser-292, and Ser-299 and tested them for nSMase activity in response to ox-stress. Fig. 5 shows that the activity of these MTs no longer responds to ox-stress. However, in contrast to what we observed for the S173A- and S208A-MTs (Fig. 3), the S289A-, S292A- and S299A-MTs presented a basal nSMase activity per protein unit that was higher than that of the WT nSMase2 (Fig. 5), as if these MTs were already constitutively induced. Thus, we conclude that the phospho-serines Ser-289, Ser-292, and Ser-299 all present an equal role in the maintenance of a proper nSMase2 structure/function.
FIGURE 5.
Phospho-serines Ser-289, Ser-292, and Ser-299 all present an equal role in the maintenance of proper nSMase2 structure/function. Wild type (WT) and single point MTs S289A-MT, S292A-MT, and S299A-MT V5-tagged nSMase2 were transfected in A549 cells; 24 h after transfection, the cells were treated (or not treated) with 250 μm H2O2 for 30 min. Total nSMase activity of 10 μg of PNS cell homogenate was measured as described under “Experimental Procedures.” The data are reported as the percentage of the activity of the untreated WT nSMase2 after normalization to the transfection levels that were assessed by IB equal amounts of cell lysates with the α-V5 Ab. Standard deviations are indicated.
Serine Phosphorylation Regulates nSMase2 Protein Stability
A cluster of conserved phosphorylation sites could affect the conformation of nSMase2, thereby affecting its overall protein stability. Therefore, we used the various nSMase2 constructs to test whether the respective phosphorylated serines affect nSMase2 protein stability.
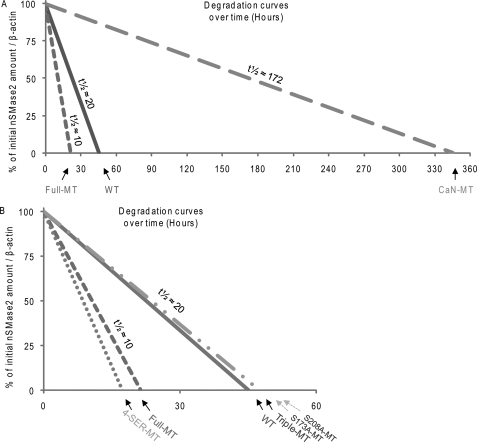
A549 cells were transiently transfected with V5-nSMase2 constructs, and 24 h after transfection, they were incubated with 100 μm cycloheximide for up to 12 h to block new protein synthesis. The cells were harvested and lysed at various time points, and then 30 μg of total proteins were assessed by IB with a specific α-V5 Ab for the levels of V5-nSMase2 expressed. Three of the nSMase2 constructs were tested first: the WT, the Full-MT (described above), and a mutant of nSMase2 that does not bind CaN (CaN-MT), and therefore, is constitutively more phosphorylated than the WT nSMase2, as we reported previously (21). Fig. 6A represents the degradation curves over time of the various V5-nSMase2 constructs, measured at several time points in at least three independent experiments and normalized to the expression levels of β-actin. We found that the Full-MT is less stable than the WT nSMase2 (p < 0.05), which in turn is less stable than the CaN-MT (Fig. 6A, p < 0.05), suggesting that serine phosphorylation regulates not only the constitutive and ox-stress-inducible activity of nSMase2 (Figs. 2 and 3), but may also regulate its expression levels by affecting the protein stability (Fig. 6A). The data also imply that CaN is involved in such regulation of protein expression.
FIGURE 6.
Study of nSMase2 phosphorylation and its protein stability. WT, Full-MT, CaN-MT (21)), 4-SER-MT, Triple-MT, S173A-MT, S208A-MT, and nSMase2 were transfected in A549 cells; 24 h after transfection, the cells were continuously incubated with 100 μm cycloheximide to block new protein synthesis. The cells were collected at several time points between 0 and 12 h after the treatment started, and V5-nSMase2 protein expression in the cell lysates was assessed by IB. A and B show the degradation curves of the various nSMase2 mutants over time, relative to the initial amount (100%), after normalization to the β-actin protein expression levels. A demonstrates results for WT, Full- and CaN-MT, whereas B demonstrates results for WT, Full-, 4-SER-, Triple-, S173A- and S208A-MT. Estimated protein half-lives (t½), calculated from the turnover of each mutant, are also shown.
To further investigate which specific phospho-site(s) out of the five identified serines directly regulates nSMase2 protein stability, we next assessed the stability of the other nSMase2 phospho-serine(s) mutants. Fig. 6B shows that the Triple-MT nSMase2 (cluster of Ser-289, Ser-292, and Ser-299 mutated to alanines) presents a protein stability that is comparable with that of the WT-nSMase2. However, the 4-SER-MT (S208A/S289A/S292A/S299A cluster) is less stable than the WT; the stability of this 4-SER-MT is comparable with that of the Full-MT (Fig. 6B).
Overall, Ser-289, Ser-292, and Ser-299 all mutated to alanines (Triple-MT) do not significantly change the protein stability of nSMase2 in comparison with that of the WT. However, the 4-SER-MT, having Ser-208 mutated in addition to those of the Triple-MT, is less stable than the WT nSMase2 (4-SER-MT as compared with WT nSMase2 in Fig. 6B, p < 0.05). This suggests that the phosphorylation of serine 208 positively regulates nSMase2 stability. However, when only Ser-208 was mutated to alanine (single point S208A-MT), the stability of nSMase2 was not different from that of the WT. Thus, we conclude that the dephosphorylation of Ser-208 down-regulates nSMase2 protein stability, but only upon concurrent dephosphorylation on the cluster Ser-289-Ser-292-Ser-299. Importantly, the observation that the S173A-MT presents protein stability comparable with that of the WT nSMase2 (Fig. 6B) demonstrates that its lack of induced phosphorylation and activation under ox-stress (Fig. 3) is not just an outcome of changes in protein stability/expression, as discussed below.
DISCUSSION
nSMase2 was previously found in the brain (29), and recent studies indicate a role for nSMase2 in aging (17) and in Alzheimer disease (30). At the same time, we reported that ox-stress induced by H2O2 or CS up-regulates ceramide generation by sphingomyelin hydrolysis and causes pathological cell death in HAE cells (4, 7–8, 22, 31). Importantly, airway epithelial cells are the first line of defense for the lung and are thus extensively exposed to reactive oxidants.
This led to the proposal that there must be a specific SMase that is modulated by ox-stress in lung epithelial cells, which was followed by the isolation of the novel nSMase2 from monkey lung tissue and from HAE cells (7). It was then demonstrated (7, 22) that upon siRNA silencing of nSMase2, lung epithelial cells could not undergo cell death in response to CS exposure or H2O2-induced ox-stress, suggesting that nSMase2 could be a critical target not only in the brain pathogenesis but also in ox-stress/CS-induced lung injury in respiratory diseases.
According to current models, sphingomyelin may be constitutively metabolized to ceramide by several SMases, but some SMases may have special patho-physiological significance (32). Levy et al. (7, 22), Filosto et al. (21), and Rutkute et al. (33) have reported that the major SMase enzyme that becomes activated by ox-stress appears to be nSMase2 (SMPD3), which is expressed in the Golgi, in the plasma membrane, and in endosomal compartments (7, 14, 34).
Our recent studies in vivo (19) demonstrated that mice heterozygous for nSMase2 had less ceramide accumulation in the lung in comparison with WT mice when exposed to CS-induced ox-stress; on the other hand, knock-out mice for acidic SMase could accumulate ceramide under CS exposure as much as the WT mice, demonstrating that only nSMase2 and not acidic SMase is modulated by CS (19). Finally, we found that lung tissues from patients with emphysema (smokers) displayed significantly higher levels of nSMase2 expression as compared with lung tissues from control subjects. Together, these data establish the central in vivo role of nSMase2 in ceramide generation, aberrant apoptosis, and lung injury under CS exposure, underscoring its promise as a novel target for the prevention of CS/ox-stress-induced airspace destruction, and thus, the importance of elucidating the molecular mechanism of nSMase2 activation under ox-stress (21).
Here we show that nSMase2 is a phospho-protein exclusively phosphorylated at serine residues. Previously, we reported (21) that the level of nSMase2 phosphorylation can be modulated by treatment with anisomycin or phorbol 12-myristate 13-acetate (12-O-tetradecanoylphorbol-13-acetate), suggesting that p38 MAPK and protein kinases C(s) are upstream of nSMase2 phosphorylation. ox-stress enhances both the activity and the phosphorylation of nSMase2. Importantly, nSMase2 was found to be bound directly by the phosphatase CaN, which acts as an on/off switch for nSMase2 phosphorylation in the presence or absence of ox-stress. Specifically, CaN is being inhibited/degraded and therefore does not bind nSMase2 under ox-stress, and a mutant nSMase2 that lacks the CaN binding site exhibits constitutively elevated phosphorylation and activity relative to WT nSMase2. Importantly, the phosphorylation and activity of the mutant no longer respond to ox-stress, confirming that CaN is a critical link that allows ox-stress to modulate nSMase2 phosphorylation and function. Here, we moved forward to identify the five serines that are phosphorylated in nSMase2 and found that these serines consist of a group of conserved phosphorylation sites: Ser-173 and Ser-208, which are located in proximity of the CaN binding site (21), and Ser-289, Ser-292, and Ser-299, which are adjacent to the catalytic domain of nSMase2.
Acknowledging the complexity of the nSMase2 regulatory mechanism(s), we speculated that these conserved sites may both affect the overall nSMase2 protein stability and modulate its activity. Moreover, we predicted that these phosphorylation sites could be interdependently regulated. Indeed, Dr. Hannun and colleagues (1) speculated that not only is it possible that different phosphorylation site(s) can control different aspects of nSMase2 function (activity, subcellular localization/trafficking, and protein stability), but also that those phosphorylation sites can either positively or negatively control each other.
We first identified Ser-173 to have a significant role in the ability of nSMase2 to be activated under ox-stress. Equally important, we found that Ser-208 has a critical role in maintaining the stability of nSMase2. Interestingly, these residues are located in proximity to the CaN binding site. However, despite their essential roles, these sites of phosphorylation turned out to be somehow dependent on the other cluster of three serines, Ser-289-Ser-292-Ser-299, which are adjacent to the nSMase2 catalytic domain.
We first generated an nSMase2 mutant lacking all five phospho-serines (Full-MT) and showed that in comparison with the WT nSMase2, such a Full-MT loses its receptiveness to ox-stress, being unable to be both phosphorylated and activated under H2O2 exposure (Fig. 2). Subsequently, we mutated only the serine 173 to alanine (S173A-MT) and found that this S173A-MT behaves like the Full-MT in terms of lack of ox-stress-inducible phosphorylation and activation (Fig. 3), demonstrating that the phosphorylation of Ser-173 controls nSMase2 activation in response to ox-stress. However, when we mutated either the serines Ser-289-Ser-292-Ser-299 (Triple-MT) or all the phospho-serines except the serine 173 (4-SER-MT), the ox-stress-dependent activation of nSMase2 was also lost, as it was for the single point mutant S173A-MT. Moreover, the 4-SER-MT and the Triple-MT nSMase2, although still harboring an intact serine 173, did not show increased phosphorylation levels upon ox-stress (Fig. 4, B and C), demonstrating that Ser-173 phosphorylation is not modulated by ox-stress independently of the serines Ser-289, Ser-292, and Ser-299. The three serines 289, 292, and 299 turn out to be all equally important in controlling nSMase2 activity (Fig. 5). However, the single point mutation of either Ser-289, Ser-292, or Ser-299 causes a constitutive increase of nSMase activity (in respect to WT activity) that is not observed for the mutant of Ser-173 (or Ser-208) (Fig. 3A). This indicates that although Ser-289, Ser-292, and Ser-299 have a certain role in nSMase2 structure/function, it is only Ser-173 phosphorylation that controls the ox-stress-inducible nSMase2 activation. In addition, it is possible that the phosphorylation on Ser-289, Ser-292, or Ser-299, respectively, may have a role in maintaining nSMase2 in an autoinhibited protein structure, which would be released by the phosphorylation of serine 173.
We have previously shown that nSMase2 protein levels are increased in lungs of rodents exposed to CS-induced ox-stress, as well as in the lungs of human smokers with pulmonary emphysema (19). Here we demonstrate that a mutant nSMase2 that cannot be bound by CaN (CaN-MT), and thus, is more phosphorylated than the WT nSMase2 (21), maintains a protein half-life of roughly nine times higher than that of the WT nSMase2 (Fig. 6). It is therefore likely that one possible mechanism that may lead to higher nSMase2 expression in the lungs upon ox-stress (19) is dependent on the post-translational stabilization of nSMase2 that cannot bind CaN under ox-stress exposure (see model in Fig. 7). However, the fine-tuning of the molecular signals connecting nSMase2 phosphorylation and protein stability are yet to be better defined. For example, we found that the phosphorylation of serine 173, which controls the activation of nSMase2 upon ox-stress, may not be involved in stabilizing the protein, whereas serine 208 phosphorylation may be necessary for nSMase2 protein stability. At the same time, serine 208 phosphorylation stabilizes nSMase2 protein only in cooperation with the phosphorylation of the cluster Ser-289-Ser-292-Ser-299, further indicating that intact phosphorylation of Ser-289, Ser-292, and Ser-299 is essential for the proper nSMase2 structure/function. Very importantly, one plausible conclusion is that although enhanced stability and increased activity of the nSMase2 protein may both depend on its enhanced phosphorylation, the two may be regulated independently.
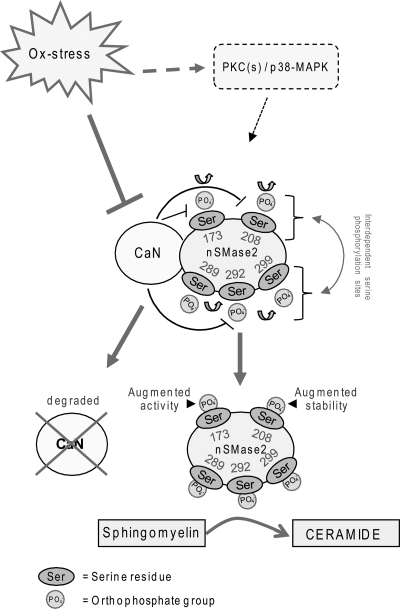
FIGURE 7.
Proposed model of nSMase2 phosphorylation machinery that affects its function. nSMase2 is constitutively phosphorylated on serine residues downstream of p38-MAPK (p38) and PKC(s). CaN phosphatase directly binds to nSMase2, dephosphorylates it, and thereby, deactivates it, as shown previously by us (21). ox-stress causes CaN degradation, leading to nSMase2 phosphorylation. We identified five serines phosphorylated in nSMase2 (see also Fig. 1). Phosphorylation of serine 173 is necessary to impose the activation of nSMase2 following ox-stress exposure of HAE cells, whereas the phosphorylation of serine 208 may be involved in increasing nSMase2 protein stability while interdependent regulation between phosphorylation sites occurs. Overall, increased phosphorylation of nSMase2 leads to its activation and its augmented protein stability, and as a result of these steps, ceramide levels are elevated by sphingomyelin hydrolysis.
In summary, we have identified five specific serines that are phosphorylated in nSMase2 and further demonstrate that phosphorylation of those serines controls both nSMase2 activity and nSMase2 stabilization. We found that intact phospho-serines 173, 289, 292, and 299 are required for nSMase2 activation following ox-stress exposure of HAE cells, whereas phosphorylation of serine 208 seems to play a role in stabilizing the nSMase2 protein. Overall, this study provides initial structure/function insights regarding nSMase2 phosphorylation sites and offers some new links for future studies aiming to fully elucidate nSMase2 regulatory machinery (see model in Fig. 7).
Acknowledgments
We thank Dr. Yusuf Hannun (Medical University of South Carolina) for providing the original V5-nSMase2 construct. We thank Cathleen Becker for critical reading of the manuscript and technical assistance. We thank Dr. David Baston (UCD) for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants HL-71871 and HL-66189 (to T. G.). This work was also supported by Tobacco-related Disease Research Program (TRDRP) Grants 17RT-0131 (to T. G.) and 20FT-0087 (to S. F.).
Throughout this study, Full-MT represents a S173A/S208A/S289A/S292A/S299A nSMase2 mutant; 4-SER-MT represents a S208A/S289A/S292A/S299A nSMase2 mutant; CaN-MT represents a mutant of nSMase2 that does not bind CaN; and Triple-MT represents a S289A/S292A/S299A nSMase2 mutant.
- nSMase
- neutral sphingomyelinase
- SMase
- sphingomyelinase
- SM
- sphingomyelin
- Ab
- antibody
- CaN
- calcineurin
- CS
- cigarette smoke
- DMSO
- dimethyl sulfoxide
- HAE
- human airway epithelial
- HBE
- human bronchial epithelial
- IB
- immuno-blotting
- IP
- immuno-precipitation
- MT
- mutant
- ox-stress
- oxidative stress
- PNS
- postnuclear supernatant
- ambic
- ammonium bicarbonate.
REFERENCES
- 1. Clarke C. J., Wu B. X., Hannun Y. A. (2011) Adv. Enzyme Regul. 51, 51–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Clarke C. J., Snook C. F., Tani M., Matmati N., Marchesini N., Hannun Y. A. (2006) Biochemistry 45, 11247–11256 [DOI] [PubMed] [Google Scholar]
- 3. Bartke N., Hannun Y. A. (2009) J. Lipid Res. 50, (suppl.) S91–S96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goldkorn T., Balaban N., Shannon M., Chea V., Matsukuma K., Gilchrist D., Wang H., Chan C. (1998) J. Cell Sci. 111, 3209–3220 [DOI] [PubMed] [Google Scholar]
- 5. Goldkorn T. (2001) Clin. Appl. Immunol. Rev. 1, 173–179 [Google Scholar]
- 6. Lavrentiadou S. N., Chan C., Kawcak T., Ravid T., Tsaba A., van der Vliet A., Rasooly R., Goldkorn T. (2001) Am. J. Respir. Cell Mol. Biol. 25, 676–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Levy M., Castillo S. S., Goldkorn T. (2006) Biochem. Biophys. Res. Commun. 344, 900–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ravid T., Tsaba A., Gee P., Rasooly R., Medina E. A., Goldkorn T. (2003) Am. J. Physiol. Lung Cell. Mol. Physiol. 284, L1082–L1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barak A., Morse L. S., Goldkorn T. (2001) Invest. Ophthalmol. Vis. Sci. 42, 247–254 [PubMed] [Google Scholar]
- 10. Clarke C. J., Guthrie J. M., Hannun Y. A. (2008) Mol. Pharmacol. 74, 1022–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clarke C. J., Truong T. G., Hannun Y. A. (2007) J. Biol. Chem. 282, 1384–1396 [DOI] [PubMed] [Google Scholar]
- 12. Kolesnick R., Hannun Y. A. (1999) Trends Biochem. Sci. 24, 224–225; author reply 227 [DOI] [PubMed] [Google Scholar]
- 13. Luberto C., Hannun Y. A. (1999) Lipids 34, S5–11 [DOI] [PubMed] [Google Scholar]
- 14. Marchesini N., Osta W., Bielawski J., Luberto C., Obeid L. M., Hannun Y. A. (2004) J. Biol. Chem. 279, 25101–25111 [DOI] [PubMed] [Google Scholar]
- 15. Marchesini N., Hannun Y. A. (2004) Biochem. Cell Biol. 82, 27–44 [DOI] [PubMed] [Google Scholar]
- 16. Wu B. X., Clarke C. J., Hannun Y. A. (2010) Neuromolecular Med. 12, 320–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nikolova-Karakashian M., Karakashian A., Rutkute K. (2008) Subcell. Biochem. 49, 469–486 [DOI] [PubMed] [Google Scholar]
- 18. Rutkute K., Karakashian A. A., Giltiay N. V., Dobierzewska A., Nikolova-Karakashian M. N. (2007) Hepatology 46, 1166–1176 [DOI] [PubMed] [Google Scholar]
- 19. Filosto S., Castillo S., Danielson A., Franzi L., Khan E., Kenyon N., Last J., Pinkerton K., Tuder R., Goldkorn T. (2011) Am. J. Respir. Cell Mol. Biol. 44, 350–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goldkorn T., Filosto S. (2010) Am. J. Respir. Cell Mol. Biol. 43, 259–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Filosto S., Fry W., Knowlton A. A., Goldkorn T. (2010) J. Biol. Chem. 285, 10213–10222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Levy M., Khan E., Careaga M., Goldkorn T. (2009) Am. J. Physiol. Lung Cell. Mol. Physiol. 297, L125–L133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen Y., Zhao Y. H., Wu R. (2001) Am. J. Respir. Cell Mol. Biol. 25, 409–417 [DOI] [PubMed] [Google Scholar]
- 24. Robinson C. B., Wu R. (1991) Methods Cell Sci. 13, 95–102 [Google Scholar]
- 25. Okazaki T., Bielawska A., Domae N., Bell R. M., Hannun Y. A. (1994) J. Biol. Chem. 269, 4070–4077 [PubMed] [Google Scholar]
- 26. Lawler J. F., Jr., Yin M., Diehl A. M., Roberts E., Chatterjee S. (1998) J. Biol. Chem. 273, 5053–5059 [DOI] [PubMed] [Google Scholar]
- 27. Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villén J., Haas W., Sowa M. E., Gygi S. P. (2010) Cell 143, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hsu P. P., Kang S. A., Rameseder J., Zhang Y., Ottina K. A., Lim D., Peterson T. R., Choi Y., Gray N. S., Yaffe M. B., Marto J. A., Sabatini D. M. (2011) Science 332, 1317–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hofmann K., Tomiuk S., Wolff G., Stoffel W. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5895–5900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Satoi H., Tomimoto H., Ohtani R., Kitano T., Kondo T., Watanabe M., Oka N., Akiguchi I., Furuya S., Hirabayashi Y., Okazaki T. (2005) Neuroscience 130, 657–666 [DOI] [PubMed] [Google Scholar]
- 31. Chan C., Goldkorn T. (2000) Am. J. Respir. Cell Mol. Biol. 22, 460–468 [DOI] [PubMed] [Google Scholar]
- 32. Uhlig S., Gulbins E. (2008) Am. J. Respir. Crit. Care Med. 178, 1100–1114 [DOI] [PubMed] [Google Scholar]
- 33. Rutkute K., Asmis R. H., Nikolova-Karakashian M. N. (2007) J. Lipid Res. 48, 2443–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Milhas D., Clarke C. J., Idkowiak-Baldys J., Canals D., Hannun Y. A. (2010) Biochim. Biophys. Acta 1801, 1361–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]