Background: The substrates and regulation of DHHC protein palmitoyl acyltransferases (PATs) are largely unknown.
Results: Flotillin-2 palmitoylation is abolished in DHHC5 gene-targeted neural stem cells, and neuronal differentiation induces DHHC5 turnover.
Conclusion: Flotillin-2 is a substrate for DHHC5, which is regulated at the protein level.
Significance: The paper describes an approach to PAT substrate identification and a new PAT regulation mechanism.
Keywords: Chemical Biology, Lipid Raft, Membrane Enzymes, Neurodifferentiation, Neuroprogenitor Cell, Protein Acylation, Protein Palmitoylation, Protein Turnover, Proteomics
Abstract
Post-translational palmitoylation of intracellular proteins is mediated by protein palmitoyltransferases belonging to the DHHC family, which share a common catalytic Asp-His-His-Cys (DHHC) motif. Several members have been implicated in neuronal development, neurotransmission, and synaptic plasticity. We previously observed that mice homozygous for a hypomorphic allele of the ZDHHC5 gene are impaired in context-dependent learning and memory. To identify potentially relevant protein substrates of DHHC5, we performed a quantitative proteomic analysis of stable isotope-labeled neuronal stem cell cultures from forebrains of normal and DHHC5-GT (gene-trapped) mice using the bioorthogonal palmitate analog 17-octadecynoic acid. We identified ∼300 17-octadecynoic acid-modified and hydroxylamine-sensitive proteins, of which a subset was decreased in abundance in DHHC5-GT cells. Palmitoylation and oligomerization of one of these proteins (flotillin-2) was abolished in DHHC5-GT neuronal stem cells. In COS-1 cells, overexpression of DHHC5 markedly stimulated the palmitoylation of flotillin-2, strongly suggesting a direct enzyme-substrate relationship. Serendipitously, we found that down-regulation of DHHC5 was triggered within minutes following growth factor withdrawal from normal neural stem cells, a maneuver that is used to induce neural differentiation in culture. The effect was reversible for up to 4 h, and degradation was partially prevented by inhibitors of ubiquitin-mediated proteolysis. These findings suggest that protein palmitoylation can be regulated through changes in DHHC PAT levels in response to differentiation signals.
Introduction
Palmitoylation is a reversible post-translational modification important for trafficking, stability, and activity of numerous intracellular proteins in eukaryotic cells. Many proteins involved in neural development, neurotransmission, and synaptic plasticity are known to be palmitoylated, including signaling proteins, ion channels, G protein-coupled receptors, cell adhesion molecules, transporters, adaptors, and scaffolding proteins (1–4). Protein palmitoylation has a prominent role in targeting proteins to specialized membrane compartments, which is central to axon and dendrite outgrowth and the formation of synapses. Therefore, understanding the underlying mechanisms and regulation of protein palmitoylation is essential for understanding the molecular mechanisms controlling neuronal function.
Protein palmitoylation is carried out by a family of 23 mammalian palmitoyl acyltransferases (PATs),3 referred to as the DHHC family, that catalyze palmitoylation of intracellular proteins on cysteine residues (5–9). The cellular and physiological roles of the DHHC PATs are just beginning to be explored. A particular challenge has been the identification of physiologically relevant enzyme-substrate pairs. Genetic approaches have proven powerful, especially in lower organisms (see, for example, Refs. 10 and 11) and, more recently, in mammalian systems. These approaches, coupled with powerful new methods to profile palmitoylated proteins in cells and tissues, are providing opportunities for delineating pathways controlled by protein palmitoylation (reviewed in Refs. 4 and 9).
A second challenge has been to understand how protein palmitoylation is regulated. The mechanisms are incompletely understood and limited to only a few examples, in particular, the palmitoylation of G protein α subunit Gαs (12) and the neuronal scaffolding protein PSD-95 (13). In both instances, palmitate turnover is accelerated in response to extracellular signals by receptor activation, contributing to down-regulation of these signaling pathways. Positive regulation of palmitoylation by DHHC PAT recruitment to cellular compartments has more recently been proposed as a mechanism to increase palmitoylation. For example, DHHC2 translocates to postsynaptic membranes upon suppression of neuronal activity, thereby increasing PSD-95 palmitoylation and promoting its synaptic accumulation (14). The effect is mediated by the carboxyl-terminal domain of DHHC2 (15). However, the regulation of overall DHHC protein levels in cells has not been previously described.
We have previously shown that mice carrying a hypomorphic allele of DHHC5 (designated DHHC5-GT, for gene-targeted) have a defect in fear learning and memory. In the brain, DHHC5 localizes to the post-synaptic density and interacts with PSD95 but does not palmitoylate it (16). In the current study, we identify the abundant lipid raft protein flotillin-2 as a substrate for DHHC5 and show that it is under-palmitoylated in DHHC5-GT neural stem cells and brain. Furthermore, we find that DHHC5 declines rapidly (within minutes) on induction of neural differentiation, demonstrating a previously unsuspected mode of DHHC PAT regulation.
EXPERIMENTAL PROCEDURES
Materials
FBS was purchased from Gemini. MG-132 was purchased from Boston Biochem (Cambridge, MA). N-Acetyl-l-leucyl-l-leucinylnorleucinal (ALLN) was from Calbiochem. Forskolin was purchased from Sigma-Aldrich. Poly-d-lysine-coated 60-mm culture dishes were purchased from BD Biosciences. Protease inhibitor mixture was from Roche Applied Science. Disuccinimidyl suberate was purchased from Thermo Scientific. Other materials are listed in the supplemental “Experimental Procedures.”
Plasmids
pEF-BOS-HA-DHHC5 (17) was a gift of Dr. Fusaka Masaki (National Institute for Physiological Sciences, Aichi, Japan). This plasmid directs transcription of a full-length mouse DHHC5 cDNA corresponding to NCBI RefSeq NM_144887. pEF-BOS-HA-mDHHC5 (CΔS) was generated by site-directed mutagenesis (QuikChange II XL; Stratagene), resulting in a single amino acid change C134S within the DHHC motif. A full-length human flotillin-2 cDNA clone (Open Biosystems, MHS1010–57825, corresponding to the human flotillin-2 cDNA in GenBankTM BC_003683) was cloned into pcDNA3.1/Myc-His(−) (Invitrogen) using XhoI and BamHI sites and is designated pcDNA/Myc-His-FLOT2. Site-directed mutagenesis of pcDNA/Myc-His-FLOT2 to create cysteine to serine (palmitoylation defective) substitutions at Cys-4, -19, and -20 was also performed as above.
Quantitative Profiling of Protein Palmitoylation in Mutant Neuronal Stem Cells
NSCs derived from DHHC5-GT mice and littermate controls were profiled according to Ref. 18. Full experimental procedures are available in the supplemental materials.
Acyl-Biotin Exchange (ABE) Assay
NSCs (60 × 106 cells) growing in suspension culture in NSC growth medium (DMEM/F-12/N2 containing 20 ng/ml EGF, 20 ng/ml FGF2, and 5 μg/ml heparin) were harvested by centrifugation, washed three times in PBS, and lysed in 1 ml of radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 7.2, 1% Triton X-100, 150 mm NaCl, 1% sodium deoxycholate, 1 mm EDTA, 0.1% SDS, and protease inhibitor mixture (“lysis buffer”) containing 10 μl of freshly added 1 m N-ethylmaleimide. ABE assays were performed according to (10). Briefly, proteins were precipitated using chloroform-methanol, treated exhaustively with N-ethylmaleimide to block free thiol groups, which was removed by sequential chloroform-methanol precipitation and then treated with hydroxylamine and biotin-HPDP to exchange thiol-bound fatty acids for biotin; proteins were precipitated to remove free biotin and recover the biotinylated proteins. Equal amounts of solubilized protein were incubated with neutravidin beads, and eluted with 1% β-mercaptoethanol. Proteins eluted from the beads were concentrated and resuspended in a final volume of 50 μl, and one-fifth of the entire sample was loaded onto an SDS-PAGE gel for subsequent transfer to nitrocellulose membranes and immunoblotting.
Immunoblotting
Whole cell lysates or eluates from neutravidin beads were subjected to SDS-PAGE and transferred to nitrocellulose filters, and immunoblots were performed using the following primary antibodies: mouse monoclonal anti-flotillin-2 (1:5000; BD Transduction Laboratories, catalogue number 610383), rabbit polyclonal anti-Rap2b (1:1000, ProteinTech, 16266–1-AP), rabbit polyclonal anti-calnexin antibody (1:1000, Abcam), anti-DHHC5 polyclonal antibody (1:1000; Sigma-Aldrich HPA014670), mouse monoclonal α-adaptin antibody (1:1000, Abcam), mouse anti-β-actin monoclonal antibody (1:5000, Sigma-Aldrich), and mouse monoclonal anti-GAP-43 (Santa-Cruz, SC-17790). Bound antibodies were visualized by chemiluminescence using either or sheep anti-mouse IgG, peroxidase-linked species specific F(ab′)2 fragment or HRP-linked donkey anti-rabbit IgG (both from GE Healthcare). The filters were exposed to Kodak X-Omat Blue XB-1 film at room temperature for times varying between 5 s and 15 min.
Oligomerization of Flotillins
Oligomerization of flotillins in NSCs was assessed according to Ref. 19. Disuccinimidyl suberate was prepared in Me2SO (25 mm). NSCs were washed three times with ice-cold PBS (pH 8.0), and the cells were suspended at a density of ∼25 × 106 cells/ml in PBS (pH 8.0). Disuccinimidyl suberate (or vehicle alone) was added to the cell suspension to a final concentration of 1 mm. After 30 min of incubation at room temperature, 1 m Tris-HCl (pH 7.5) was added to a final concentration of 20 mm. The cells were harvested by centrifugation, and cell lysates were prepared as described above and analyzed by immunoblotting using an anti-flotillin-2 monoclonal antibody.
NSC Differentiation and Immunofluorescence
DHHC5-WT or -GT NSCs (∼50,000) were seeded in wells of a 24-well plate on polylysine-coated coverslips in 500 μl of NSC differentiation medium. After 2 days, one-half of the spent medium was replaced with 250 μl of fresh NSC differentiation medium. Three days after seeding, NSCs were fixed with 4% paraformaldehyde for 15 min at room temperature and permeabilized with 0.1% Triton X-100 on ice for 5 min. The cells were then incubated with rabbit anti-Tuj 1 polyclonal antibody (1:2000, Covance) and mouse anti-GFAP monoclonal antibody (1:300, Cell Signaling) at 37 °C for 30 mins and then with Alexa Fluor 488 goat anti-rabbit IgG secondary antibody (1:200, Invitrogen) and Alexa Fluor 594 goat anti-mouse IgG secondary antibody (1:200, Invitrogen) at 37 °C for 30 min. The cover glasses were then mounted using Prolong Gold antifade reagent with DAPI (Invitrogen).
RESULTS
Quantitative Profiling of Protein Palmitoylation in Mutant Neuronal Stem Cells
To study the cellular role of DHHC5, we dissected the forebrains of post-natal day 1 DHHC5-GT mice and littermate controls and cultured them under conditions that select for cells maintaining neural stem cell pluripotency (20). Virtually 100% strongly expressed Sox2, a marker of NSCs (supplemental Fig. S1). These cultures have been continually passaged for 1 year and maintain elevated Sox2 expression with greatly reduced levels of DHHC5 (∼5% of normal, the residual caused by splicing around the gene trap cassette) (16).
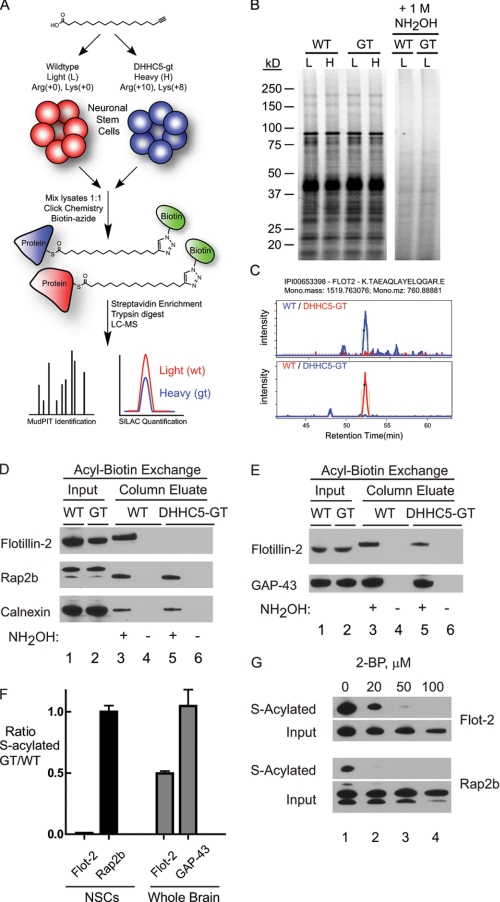
To identify potential substrates of DHHC5, we utilized stable isotope labeling with amino acids in cell culture (SILAC) methods, culturing both wild type and DHHC5-GT cells in normal or isotopic arginine (+8) and lysine (+10) for several passages and then incubating the cells with the fatty acid analog 17-octadecynoic acid (17-ODYA) to metabolically label endogenous sites of palmitoylation (18) as illustrated in Fig. 1A. Membrane lysates were reacted with rhodamine-azide by copper(I)-catalyzed azide-alkyne cycloaddition (click) chemistry for gel-based fluorescent analysis (21). Labeling was shown to be hydroxylamine-sensitive, yet no significant changes in 17-ODYA labeling were visibly different between wild type and DHHC5-GT cells (Fig. 1B).
FIGURE 1.
Identification of flotillin-2 as a substrate for DHHC5 in neuronal stem cells. A, schematic illustrating quantitative chemo-proteomic profiling of DHHC5-regulated proteins in neuronal stem cells. Isotopically labeled neuronal stem cells were labeled with 17-ODYA, then lysed, and mixed for click chemistry ligation with biotin-azide. Streptavidin-enriched proteins are digested for high resolution proteomic analysis and quantification. B, membrane lysates from 17-ODYA-labeled cells were reacted with rhodamine-azide for SDS-PAGE separation and in-gel fluorescence analysis. Treatment with 1 m hydroxylamine (NH2OH) releases 17-ODYA from essentially all labeled proteins. Representative MS1 spectra of flotillin-2 from 17-ODYA-enriched membrane proteomes demonstrate specific depletion of palmitoylated flotillins in DHHC5-GT neuronal stem cells. L, light; H, heavy isotope labeling. C, extracted ion chromatographs for FLOT2. D, acyl-biotin exchange analysis of flotillin-2 in NSCs from DHHC5-WT and DHHC5-GT mice. Methods are detailed under “Experimental Procedures.” Lanes labeled Input consisted of 15 μg of whole cell lysate (NSCs) or 25 μg of total brain extract. One-fifth of the total column eluates were loaded. No proteins eluted in the absence of neutral hydroxylamine (NH2OH), demonstrating the presence of hydroxylamine-sensitive linkages. Rap2b migration differences in input (lanes 1 and 2) versus column eluate (lanes 3 and 5) shown on this gel reflect buffer composition differences in the samples loaded, because the relative positions varied from experiment to experiment and were usually less than what is shown here. E, acyl-biotin exchange analysis of flotillin-2 using whole brain from DHHC5-WT and DHHC5-GT mice. Wild type and DHHC-GT mice were sacrificed by CO2 narcosis, and the brains were removed, homogenized in lysis buffer, and subjected to acyl-biotin exchange assay as described above. F, quantitation of acyl-biotin immunoblots. Immunoblots from ABE experiments using WT and DHHC5-GT NSCs and mouse brains were quantified using ImageQuantTM TL software. The values are expressed as the means ± S.D. of the ratio of DHHC5-GT to WT (n = 3, independent experiments). G, 2-bromopalmitate (2-BP) inhibition of S-acylation of flotillin-2 in NSCs. NSCs were pretreated with 2-bromopalmitate at 37 °C for 11 h and then subjected to the acyl-biotin exchange assay. Aliquots (15 μg) of cell lysate (Input) and one-third of total column eluates were subjected to SDS-PAGE and immunoblotting. The results shown were from one of two independent experiments yielding similar results.
To profile the substrates of DHHC5, we first defined a list of palmitoylated proteins in neuronal stem cells. Heavy and light wild type cells were treated overnight with 17-ODYA or palmitic acid (PA) as a negative control. (PA is expected to be incorporated into proteins but lacks the functional alkyne group required for protein capture.) An additional set of control experiments was performed where heavy and light cells were both treated with 17-ODYA, and then one of the samples was treated with hydroxylamine. After precipitation, equal amounts of membrane lysate from each of the light and heavy pairs were combined, mixing 17-ODYA and PA (or hydroxylamine)-treated lysates for reaction with biotin-azide and enrichment of bioorthogonally labeled palmitoylated proteins. Tryptic peptides were separated in-line by multidimensional protein identification technology (MudPIT) and analyzed using a high resolution mass spectrometer. Quantitative proteomic analysis of these samples revealed ∼400 proteins that were enriched more than 5-fold and were present in reciprocally treated sample pairs from the PA control experiments (supplemental Table S1) and several hundred more proteins in the hydroxylamine control experiments (supplemental Table S2). In total, ∼300 proteins were identified in both approaches, defining a list of high confidence palmitoylated proteins. This list includes many previously known neuronal palmitoylated proteins, including N-Ras (22), RhoB (23), glutamate receptor GluR2 (24), nicastrin, (25), SNAP25 (26), and GAP43 (27).
Next, isotopically labeled wild type and DHHC-GT cells were labeled overnight with 17-ODYA, reacted with biotin-azide, selectively enriched on streptavidin beads, and digested for comparative proteomic analysis. As expected, DHHC5 showed the greatest WT/GT ratio, as a consequence of gene-targeting. (DHHC5 is S-acylated at three sites downstream of the catalytic DHHC motif as shown in a prior proteomic study that used the acyl-biotin exchange method (28).) Of the high confidence 17-ODYA-enriched proteins identified above, ∼20 proteins demonstrated elevated SILAC ratio changes (WT/GT) of 2-fold or greater, suggestive of DHHC5 substrates (supplemental Table S3). However, unenriched tryptic digests of combined heavy and light membrane lysates from pairs of wild type and DHHC5-GT NSCs were analyzed, allowing comparison of the absolute abundance to 17-ODYA enrichment (supplemental Table S4) for the most abundant of these proteins. Athough this analysis only measured the abundance of ∼50% of the putative DHHC5 substrates, in each case, the WT/GT ratio difference in the 17-ODYA-treated samples reflected an underlying difference in overall membrane protein abundance (supplemental Table S5). Therefore, we did not find strong evidence for a steady-state pool of unacylated proteins in the DHHC5-GT cells.
Flotillin-2 Palmitoylation Is Abolished in DHHC5 Neural Stem Cells
Flotillins are palmitoylated proteins implicated in lipid raft assembly and function (29, 30). We observed a SILAC ratio (WT/GT) for flotillin-2 of 3.06 ± 0.12 (S.E.), making it a candidate DHHC5 substrate (Fig. 1C). To evaluate flotillin-2 palmitoylation by an independent method, we used an orthogonal palmitoylation assay (acyl-biotin exchange) to assess flotillin-2 palmitoylation. In this approach, hydroxylamine-sensitive, palmitoylated cysteines are exchanged for a disulfide biotin for capture on a neutravidin column (31, 32). As shown in Fig. 1D, the total amount of flotillin-2 was decreased ∼50% in the DHHC5-GT as compared with wild type lysates; however, the amount of flotillin-2 captured on the biotin column was nearly undetectable. Other known palmitoylated proteins, such as Rap2b and calnexin (31, 33) (Fig. 1D, lower two panels, lanes 3 and 5) showed no change in palmitoylation, similar to the calculated 17-ODYA SILAC ratios of 0.91 ± 0.14 and 1.27 ± 0.23 (means ± S.D.) respectively. In whole mouse brain lysates (Fig. 1E), absolute levels of flotillin-2 were unchanged, but palmitoylated flotillin-2 was reduced in the DHHC5-GT mice as compared with wild type (Fig. 1E), further supporting the observation that flotillin-2 palmitoylation is dependent upon DHHC5. Quantitative immunoblots from three independent experiments confirmed a greater than 10-fold decrease in the S-acylated fraction in the DHHC5-GT cells (Fig. 1F). Furthermore, incubation with increasing concentrations of the palmitoylation inhibitor 2-bromopalmitate abolished the S-acylated fraction of flotillin-2 in the neural stem cells, confirming a fatty acid as the hydroxylamine-sensitive modification (Fig. 1G).
DHHC5 Stimulates Flotillin-2 Palmitoylation in Heterologous Cells
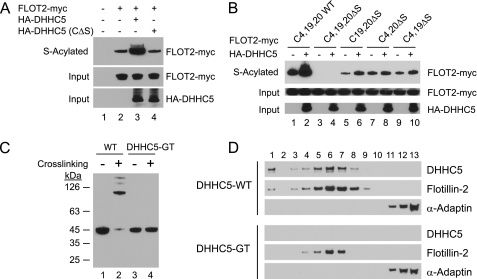
We next sought to determine whether DHHC5 would direct the palmitoylation of flotillin-2 in a heterologous cell assay system. COS-1 cells transfected with an epitope-tagged flotillin-2 and DHHC5, but not the catalytic mutant DHHC5 (C134S), showed greatly enhanced enrichment of flotillin-2 by acyl biotin exchange, demonstrating the ability of DHHC5 to palmitoylate flotillin-2 in heterologous cells (Fig. 2A). These data, together with the data presented in Fig. 1, provide strong evidence for a direct enzyme-substrate interaction for DHHC5-flotillin-2. Flotilin-2 is myristoylated at Gly-2 and palmitoylated at three cysteine residues located near the amino terminus (Cys-4, -19, and -20), with Cys-4 showing the most robust palmitate incorporation in HeLa cells (34). Mutation of all three cysteine residues blocked palmitoylation of flotillin-2 by endogenous COS cells enzymes (Fig. 2B, lane 3) and by DHHC5 (compare lanes 2 and 4). Mutation of cysteine residues in various combinations (Fig. 2B, lanes 5–10) reveals that DHHC5 stimulated palmitoylation most prominently at Cys-4 but also at Cys-20.
FIGURE 2.
DHHC5-mediated flotillin-2 palmitoylation in COS cells and flotillin-2 cross-linking in DHHC5-GT neural stem cells. A, DHHC5-mediated stimulation of flotillin-2 palmitoylation. COS-1 cells (∼60 × 106/plate) were transfected on day 0 with pEF-BOS-HA/pcDNA3.1-Myc-His(−), pEF-BOS-HA/Flot2-Myc-His, HA-DHHC5/Flot2-Myc-His, or HA-DHHC5 (CΔS)/Flot2-Myc-His using FuGENE 6 transfection reagent (Roche Applied Science). On day 2, the cells were harvested, washed three times with PBS, lysed in 1 ml of lysis buffer, and processed for ABE as described above. The cell lysates (10 μg, Input) and eluates from the neutravidin beads were analyzed by immunoblotting as described under “Experimental Procedures.” B, flotillin-2 palmitoylation sites. COS-1 cells (∼60 × 106/plate) were transfected on day 0 with Flot2-Myc-His or mutants as shown together with pEF-BOS-HA (empty vector) or HA-DHHC5. On day 2, the cells were harvested and processed for ABE as described above. C, flotillin-2 cross-linking in WT and DHHC5-GT NSCs. Oligomerization of flotillins in NSCs was assessed according to Ref. 19 using disuccinimidyl suberate cross-linking and immunoblotting as described under “Experimental Procedures.” D, sucrose density gradient fractionation of detergent resistant cell lysates from WT and DHHC5-GT NSCs. The isolation of detergent-resistant membranes was performed according to a previously published protocol (55). NSCs growing in growth medium (∼300 μl of packed cells) were harvested and washed three times with ice-cold PBS. The cells were gently resuspended by pipetting up and down in 700 μl of 1% Triton X-100 in HEPES buffer (25 mm HEPES-HCl, pH 6.5, 150 mm NaCl, 1 mm EDTA, and protease inhibitor mixture), homogenized using a Teflon-coated Dounce homogenizer (30 strokes), then incubated on ice for 30 min, and fractionated on a sucrose density gradient. The fractions were collected from top (fraction 1) to bottom (fraction 13) and analyzed by immunoblotting. The results shown were from one of two independent experiments giving similar results. α-Adaptin is a marker for clathrin-coated vesicles.
Lack of Flotillin-2 Oligomerization in DHHC5-GT Cells
Flotillin-1 and flotillin-2 function as homo-and hetero-oligomers in cells through association of their carboxyl-terminal domains (19). To assess oligomerization of flotillin-2 in the NSCs, we performed a previously described analysis that involves cross-linking of adjacent lysine residues in proteins using a chemical reagent, disuccinimidyl suberate, and assessment of flotillin-2 migration by SDS-PAGE and immunoblotting (19). In wild type NSCs (Fig. 2C, lanes 1 and 2), cross-linking of cell lysates reveals that flotillin-2, which normally exists as a monomer at 45 kDa, shifts almost entirely to a high molecular weight (presumably the tetramer as previously reported (19)). Interestingly, no high molecular weight flotillin-2 complexes were observed in the DHHC5-GT NSCs (Fig. 2C, lanes 3 and 4). These results are consistent with the known requirement of flotillin-2 palmitoylation for formation of higher order complexes (35, 36). Somewhat surprisingly, the buoyant density of flotillin-2 as assessed by sucrose density centrifugation was similar between WT and DHHC5-GT cells (Fig. 2D). Under these conditions, α-adaptin, a marker for clathrin coated vesicles, was found near the bottom of the gradient (Fig. 2D, lanes 11–13), whereas flotillin-2 was found in fractions 5–8, peaking in fraction 6 for both cell types. Although a trace of flotillin-2 could be detected at the top of the gradient only in WT extracts (in two experiments), whether this was a true difference remains to be determined, because flotillin-2 levels were reduced in the knockout, and flotillin-2 in fraction 1 in WT cells was at the limits of detection in the assay. However, it is clear that the sedimentation of the vast majority of flotillin-2 was unaffected by the lack of DHHC5.
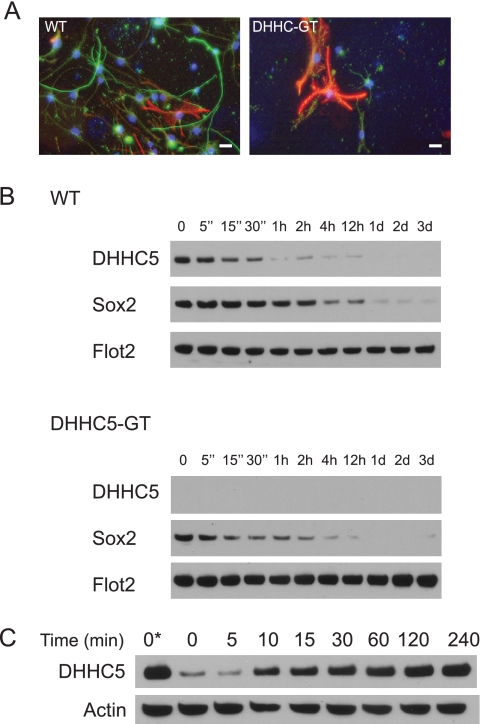
Rapid Loss of DHHC5 upon Induction of Neural Stem Cell Differentiation
Flotillin-2 palmitoylation has been shown to be required for neurite outgrowth (35, 36), a process that can be induced in cell culture by changes in the culture medium that include growth factor withdrawal (20). Consistent with this observation, we observed that DHHC5-GT NSCs did not extend neurites and showed little evidence of neuronal differentiation, at least as evidenced by expression of the neuronal marker, β-tubulin III (Tuj 1) (Fig. 3A). Interestingly, in the course of these experiments, we tracked normal DHHC5 protein expression during induction of neuronal differentiation in WT cells and discovered that within 5 min of shifting to the differentiation media, DHHC5 levels begin to decrease rapidly (Fig. 3B, top panel), reaching a level of 10% of previous levels after 1 h. The decline closely paralleled the disappearance of the stem cell marker, Sox2 (middle panel). Flotillin-2 levels were unaffected (bottom panel). This suggests that DHHC5 is under the control of upstream signals that either promote neurogenesis or maintain stem cell pluripotency. Of note, there was no difference in the decline of Sox2 between wild type and DHHC5-GT cells (Fig. 3B, middle panels), indicating that the loss of DHHC5 does not affect signaling to Sox2. Additionally, adding back growth factors to the cells after a period of growth factor withdrawal (4 h) was sufficient to rapidly induce DHHC5, with an increase that was half-maximal at 10 min and complete within 1–2 h (Fig. 3C), indicating that the changes are rapid and reversible.
FIGURE 3.
Induction of neuronal differentiation. A, fluorescence imaging of DHHC5-WT and -GT neural stem cells upon shift to differentiation medium (green, anti-Tuji 1, neuronal marker; red, anti-GFAP, astrocyte marker; blue, DAPI, nuclei). The results shown were from one of two independent experiments giving similar results. Bars, 20 μm. B, rapid changes in DHHC5 immunoreactivity following growth factor withdrawal. NSCs maintained in NSC growth medium were washed three times with NSC differentiation medium (DMEM/F-12 (1:1) with 1× N2 supplement containing 5 μm forskolin, 0.5% FBS, and 1× antibiotic-antimycotic) in the absence of added EGF, FGF, and heparin and then seeded onto the poly-d-lysine-coated 60-mm plates at a density of 2 × 106/plate in NSC differentiation medium. The entire washing procedure was carried out in under 5 min. The cells were then harvested at the times indicated, and whole cell lysates were analyzed by immunoblotting. C, time course of restoration of DHHC5 levels upon adding back EGF, FGF2, and heparin. The cells were grown in growth medium containing EGF, FGF2, and heparin (lane 1) or washed and incubated for 4 h in DMEM/F-12/N2 containing no additives (lanes 2–9) to down-regulate DHHC5, after which EGF, FGF2, and heparin were added to the medium (at concentrations above) for the time indicated, and whole cell lysates were analyzed by immunoblotting.
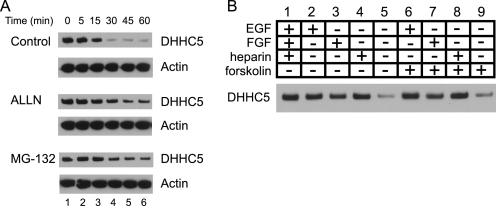
The loss of DHHC5 suggested that DHHC5 is subject to rapid degradation in response to the shift to the differentiation medium. We therefore tested the effect of inhibitors of proteosome-mediated degradation (ALLN (37) and MG-132 (38)) on the time course of DHHC5 down-regulation. A short preincubation with ALLN or MG-132 largely prevented the loss of DHHC5 in response to medium shift (Fig. 4A).
FIGURE 4.
Time course of DHHC5 down-regulation in presence and absence of inhibitors of proteolysis and growth factor requirements for DHHC5 stabilization. A, cells were preincubated for 2 h at 37 °C in NSC growth medium containing 25 μg/ml ALLN (25 μg/ml), MG-132 (10 μm), or Me2SO (vehicle); washed three times in the same solution; and then seeded at a density of 2 × 106 onto poly-d-lysine-coated 60-mm plates. The cells were harvested at the times indicated, and whole cell lysates were analyzed by immunoblotting. Shown is one of three experiments giving similar results. B, cells growing in NSC growth medium (DMEM/F-12/N2 containing EGF (20 ng/ml), FGF (20 ng/ml), and heparin (5 μg/ml) were washed and incubated for 45 min in DMEM/F-12/N2 with the individual additives shown. The whole cell lysates were analyzed by immunoblotting as described under “Experimental Procedures.”
To begin to understand the growth factor requirements for DHHC5 stability in NSCs, we first cultured cells in growth medium, shifted the cells to medium containing various combinations of medium supplements, and assayed for DHHC5 immunoreactivity 45 min later (Fig. 4B). NSCs are maintained in an undifferentiated state in serum-free medium containing EGF, FGF2, and heparin and induced to differentiate by the addition of forskolin and FBS (0.5%) (20). As shown in Fig. 4B, rapid degradation of DHHC5 occurs in medium lacking growth factors regardless of the presence or absence of 0.5% FBS or forskolin (Fig. 4B, compare lanes 5 and 9). Supplementation with EGF, FGF2, or heparin (which is understood to act via stabilization of endogenous FGF signaling complexes (39)) was sufficient to stabilize DHHC5. Therefore, we conclude that continued signaling through EGF or FGF2 is sufficient to prevent DHHC5 turnover.
DISCUSSION
Here we identify flotillin-2 as an endogenous substrate for DHHC5 and demonstrate down-regulation of DHHC5 in neural stem cells in response to growth factor withdrawal. Flotillins-1 and -2 (also known as reggie-2 and reggie-1, respectively) were originally described as abundant components of detergent-insoluble cell membranes up-regulated after sectioning of the goldfish optic nerve (40, 41). More recently, flotillins are understood as key components of a clathrin-independent endocytic pathway (42, 43) involved in various cellular processes such as insulin and epidermal growth factor receptor signaling, T cell activation, phagocytosis, cell motility, and transformation (reviewed in Ref. 44). Flotillins are organized as homo- and hetero-oligomers at the plasma membrane via contact between their carboxyl-terminal coiled-coil domains (45, 46) and are required for the extension of hippocampal neurites in cell culture (34) in a process that requires flotillin-2 palmitoylation. In the current paper, we independently confirm that a loss of flotillin-2 palmitoylation is associated with a loss of higher order flotillin-2 complexes in DHHC5-GT NSCs and failure of the cells to extend neurites.
Whether the failure of neurite extension is directly linked to flotillin-2 palmitoylation or results from a more global failure of differentiation remains to be determined. One way to interpret our findings is that growth factor withdrawal, by inducing DHHC5 down-regulation, unmasks secondary events that become rate-limiting in the DHHC-GT cells, which are already quite depleted of DHHC5. The relevance of this finding to normal brain function, if any, also remains to be determined. We did not observe gross abnormalities in brain development in the DHHC5 mice, despite their measureable learning and memory defects (16). Of course, more subtle derangements in neuronal development or synapse formation may be responsible for the observed deficits. Flotillins have also been described as important components of a discrete set of vesicles involved in a rapid, caveolin-independent (but dynamin-dependent) endocytic pathway (42, 47). In non-neuronal cells, overexpression of flotillins in stoichiometric proportions induces caveolar-like structures (48). These results predict the interesting possibility that DHHC5-GT NSCs may lack a flotillin-dependent endocytic pathway. In the nervous system, neurotransmitter transporters for dopamine and glutamate are cargo in a flotillin-mediated endocytic pathway (49). It will be of interest to investigate these pathways in the DHHC5-GT NSCs and mice, especially in light of the learning phenotype.
We unexpectedly observed that DHHC5 is rapidly degraded in response to growth factor withdrawal and restored in response to growth factor repletion. These observations suggest that DHHC5 is subject to constant rapid turnover, and the levels of DHHC5 are tightly controlled by extracellular signals, which may in turn influence the palmitoylation of other proteins. It will be of interest to see whether other DHHC family members are subject to this mode of regulation, because the mechanisms whereby protein palmitoylation is regulated in response to extracellular signals in a broader context are currently largely unknown. For example, NCAM1 palmitoylation is stimulated by FGF receptor activation through a candidate PAT, DHHC7 by an unknown mechanism (50); stabilization of PAT turnover as a potential mechanism should be examined.
Finally, in the course of this study, we identified hundreds of proteins in neural stem cells metabolically labeled by a fatty acid analog, 17-ODYA, through hydroxylamine-sensitive linkages. Many of these (if not most) will represent palmitoylated proteins. A number of these 17-ODYA-tagged proteins were decreased in abundance in the DHHC5-GT samples, representing potential substrates of DHHC5. Two previously reported DHHC5 substrates, STREX, a splice variant of the large conductance calcium-activated potassium (BK) channel (51), and SSTR5, somatostatin receptor Type 5 (52), were not identified in this screen. This may be due to low abundance of the substrates in neuronal tissues, a weak contribution of DHHC5 to the overall palmitoylation of these substrates (for example, STREX is palmitoylated by multiple DHHC enzymes), or other factors that remain to be determined. (Interestingly, there are no obvious similarities in the substrates that would suggest a common DHHC5 recognition site).
It was somewhat puzzling, but potentially important, that in all instances for which data were available, the decrease in palmitoylation was accompanied by a decrease in overall protein abundance. Several explanations are possible. One potential explanation is that because only membrane proteins were profiled, unacylated proteins were lost from the membrane fraction, decreasing overall abundance. However, we did not detect a shift of flotillin-2 (or a small sampling of other proteins) from the membrane fractions of DHHC5-GT cells. A second more compelling explanation is that if protein palmitoylation were tightly linked to protein stability, decreased palmitoylation would also be reflected in protein abundance. An interplay between protein palmitoylation and protein stability has been increasingly appreciated, as discussed in a recent review (3). This explanation poses a serious limitation for the 17-ODYA enrichment method, because changes in protein abundance will increase the number of false positive candidates. Further work will be required to determine whether other candidates whose abundance was reduced in DHHC5-GT cells in addition to flotillin-2 are substrates for DHHC5. Recent improvements in the methods for examining the palmitoyation status of individual proteins (for example, as in Refs. 53 and 54) will facilitate a more comprehensive examination of additional DHHC5 candidate substrates in future experiments.
Supplementary Material
Acknowledgments
We thank Dr. Chun-Lu Zhang for advice regarding deriving neuronal stem cells from mice and Drs. Russell DeBose-Boyd, Abigail Soyombo, and Jui-Yun Lu and members of the Cravatt lab for helpful advice. We also thank Chu Wang, Melissa Dix, and Anna Spears for mass spectrometry and informatics support.
This work was supported, in whole or in part, by National Institutes of Health Grants R37 NS36867 (to S. L. H.) and K99CA151460 (B. R. M.). This work was also supported by Robert A. Welch Foundation Grant I-1187 (to S. L. H.).

This article contains supplemental text, Tables S1–S5, and Fig. S1.
- PAT
- palmitoyl acyltransferase
- ALLN
- N-acetyl-l-leucyl-l-leucinylnorleucinal
- PA
- palmitic acid
- NSC
- neural stem cell
- GT
- gene-targeted
- 17-ODYA
- 17-octadecynoic acid
- ABE
- acyl-biotin exchange
- SILAC
- stable isotope labeling with amino acids in cell culture.
REFERENCES
- 1. Huang K., El-Husseini A. (2005) Curr. Opin. Neurobiol. 15, 527–535 [DOI] [PubMed] [Google Scholar]
- 2. Resh M. D. (2006) Sci. STKE 2006, re14. [DOI] [PubMed] [Google Scholar]
- 3. Linder M. E., Deschenes R. J. (2007) Nat. Rev. Mol. Cell Biol. 8, 74–84 [DOI] [PubMed] [Google Scholar]
- 4. Fukata Y., Fukata M. (2010) Nat. Rev. Neurosci. 11, 161–175 [DOI] [PubMed] [Google Scholar]
- 5. Bartels D. J., Mitchell D. A., Dong X., Deschenes R. J. (1999) Mol. Cell. Biol. 19, 6775–6787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lobo S., Greentree W. K., Linder M. E., Deschenes R. J. (2002) J. Biol. Chem. 277, 41268–41273 [DOI] [PubMed] [Google Scholar]
- 7. Roth A. F., Feng Y., Chen L., Davis N. G. (2002) J. Cell Biol. 159, 23–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mitchell D. A., Vasudevan A., Linder M. E., Deschenes R. J. (2006) J. Lipid Res. 47, 1118–1127 [DOI] [PubMed] [Google Scholar]
- 9. Greaves J., Chamberlain L. H. (2011) Trends Biochem. Sci 36, 245–253 [DOI] [PubMed] [Google Scholar]
- 10. Roth A. F., Wan J., Bailey A. O., Sun B., Kuchar J. A., Green W. N., Phinney B. S., Yates J. R., 3rd, Davis N. G. (2006) Cell 125, 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Montoro A. G., Ramirez S. C., Quiroga R., Taubas J. V. (2011) PLoS One 6, e16969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wedegaertner P. B., Bourne H. R. (1994) Cell 77, 1063–1070 [DOI] [PubMed] [Google Scholar]
- 13. El-Husseini Ael-D., Schnell E., Dakoji S., Sweeney N., Zhou Q., Prange O., Gauthier-Campbell C., Aguilera-Moreno A., Nicoll R. A., Bredt D. S. (2002) Cell 108, 849–863 [DOI] [PubMed] [Google Scholar]
- 14. Noritake J., Fukata Y., Iwanaga T., Hosomi N., Tsutsumi R., Matsuda N., Tani H., Iwanari H., Mochizuki Y., Kodama T., Matsuura Y., Bredt D. S., Hamakubo T., Fukata M. (2009) J. Cell Biol. 186, 147–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Greaves J., Carmichael J. A., Chamberlain L. H. (2011) Mol. Biol. Cell 22, 1887–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li Y., Hu J., Höfer K., Wong A. M., Cooper J. D., Birnbaum S. G., Hammer R. E., Hofmann S. L. (2010) J. Biol. Chem. 285, 13022–13031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fukata M., Fukata Y., Adesnik H., Nicoll R. A., Bredt D. S. (2004) Neuron 44, 987–996 [DOI] [PubMed] [Google Scholar]
- 18. Martin B. R., Wang C., Adibekian A., Tully S. E., Cravatt B. F. (2011) Nat. Methods, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Solis G. P., Hoegg M., Munderloh C., Schrock Y., Malaga-Trillo E., Rivera-Milla E., Stuermer C. A. (2007) Biochem. J. 403, 313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang C. L., Zou Y., He W., Gage F. H., Evans R. M. (2008) Nature 451, 1004–1007 [DOI] [PubMed] [Google Scholar]
- 21. Martin B. R., Cravatt B. F. (2009) Nat. Methods 6, 135–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hancock J. F., Magee A. I., Childs J. E., Marshall C. J. (1989) Cell 57, 1167–1177 [DOI] [PubMed] [Google Scholar]
- 23. Adamson P., Marshall C. J., Hall A., Tilbrook P. A. (1992) J. Biol. Chem. 267, 20033–20038 [PubMed] [Google Scholar]
- 24. Hayashi T., Rumbaugh G., Huganir R. L. (2005) Neuron 47, 709–723 [DOI] [PubMed] [Google Scholar]
- 25. Cheng H., Vetrivel K. S., Drisdel R. C., Meckler X., Gong P., Leem J. Y., Li T., Carter M., Chen Y., Nguyen P., Iwatsubo T., Tomita T., Wong P. C., Green W. N., Kounnas M. Z., Thinakaran G. (2009) J. Biol. Chem. 284, 1373–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hess D. T., Slater T. M., Wilson M. C., Skene J. H. (1992) J. Neurosci. 12, 4634–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Skene J. H., Virág I. (1989) J. Cell Biol. 108, 613–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang W., Di Vizio D., Kirchner M., Steen H., Freeman M. R. (2010) Mol. Cell. Proteomics 9, 54–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Morrow I. C., Parton R. G. (2005) Traffic 6, 725–740 [DOI] [PubMed] [Google Scholar]
- 30. Stuermer C. A. (2011) J. Neurochem. 116, 708–713 [DOI] [PubMed] [Google Scholar]
- 31. Kang R., Wan J., Arstikaitis P., Takahashi H., Huang K., Bailey A. O., Thompson J. X., Roth A. F., Drisdel R. C., Mastro R., Green W. N., Yates J. R., 3rd, Davis N. G., El-Husseini A. (2008) Nature 456, 904–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wan J., Roth A. F., Bailey A. O., Davis N. G. (2007) Nat. Protoc. 2, 1573–1584 [DOI] [PubMed] [Google Scholar]
- 33. Uechi Y., Bayarjargal M., Umikawa M., Oshiro M., Takei K., Yamashiro Y., Asato T., Endo S., Misaki R., Taguchi T., Kariya K. (2009) Biochem. Biophys. Res. Commun. 378, 732–737 [DOI] [PubMed] [Google Scholar]
- 34. Neumann-Giesen C., Falkenbach B., Beicht P., Claasen S., Lüers G., Stuermer C. A., Herzog V., Tikkanen R. (2004) Biochem. J. 378, 509–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Munderloh C., Solis G. P., Bodrikov V., Jaeger F. A., Wiechers M., Málaga-Trillo E., Stuermer C. A. (2009) J. Neurosci. 29, 6607–6615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Langhorst M. F., Reuter A., Jaeger F. A., Wippich F. M., Luxenhofer G., Plattner H., Stuermer C. A. (2008) Eur. J. Cell Biol. 87, 211–226 [DOI] [PubMed] [Google Scholar]
- 37. Laing J. G., Beyer E. C. (1995) J. Biol. Chem. 270, 26399–26403 [DOI] [PubMed] [Google Scholar]
- 38. Hartman I. Z., Liu P., Zehmer J. K., Luby-Phelps K., Jo Y., Anderson R. G., DeBose-Boyd R. A. (2010) J. Biol. Chem. 285, 19288–19298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Caldwell M. A., Garcion E., terBorg M. G., He X., Svendsen C. N. (2004) Exp. Neurol. 188, 408–420 [DOI] [PubMed] [Google Scholar]
- 40. Schulte T., Paschke K. A., Laessing U., Lottspeich F., Stuermer C. A. (1997) Development 124, 577–587 [DOI] [PubMed] [Google Scholar]
- 41. Bickel P. E., Scherer P. E., Schnitzer J. E., Oh P., Lisanti M. P., Lodish H. F. (1997) J. Biol. Chem. 272, 13793–13802 [DOI] [PubMed] [Google Scholar]
- 42. Glebov O. O., Bright N. A., Nichols B. J. (2006) Nat. Cell Biol. 8, 46–54 [DOI] [PubMed] [Google Scholar]
- 43. Saslowsky D. E., Cho J. A., Chinnapen H., Massol R. H., Chinnapen D. J., Wagner J. S., De Luca H. E., Kam W., Paw B. H., Lencer W. I. (2010) J. Clin. Invest. 120, 4399–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Babuke T., Tikkanen R. (2007) Eur J. Cell Biol. 86, 525–532 [DOI] [PubMed] [Google Scholar]
- 45. Riento K., Frick M., Schafer I., Nichols B. J. (2009) J. Cell Sci. 122, 912–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Babuke T., Ruonala M., Meister M., Amaddii M., Genzler C., Esposito A., Tikkanen R. (2009) Cell Signal. 21, 1287–1297 [DOI] [PubMed] [Google Scholar]
- 47. Damm E. M., Pelkmans L., Kartenbeck J., Mezzacasa A., Kurzchalia T., Helenius A. (2005) J. Cell Biol. 168, 477–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Frick M., Bright N. A., Riento K., Bray A., Merrified C., Nichols B. J. (2007) Curr. Biol. 17, 1151–1156 [DOI] [PubMed] [Google Scholar]
- 49. Cremona M. L., Matthies H. J., Pau K., Bowton E., Speed N., Lute B. J., Anderson M., Sen N., Robertson S. D., Vaughan R. A., Rothman J. E., Galli A., Javitch J. A., Yamamoto A. (2011) Nat. Neurosci. 14, 469–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ponimaskin E., Dityateva G., Ruonala M. O., Fukata M., Fukata Y., Kobe F., Wouters F. S., Delling M., Bredt D. S., Schachner M., Dityatev A. (2008) J. Neurosci. 28, 8897–8907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tian L., McClafferty H., Jeffries O., Shipston M. J. (2010) J. Biol. Chem. 285, 23954–23962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kokkola T., Kruse C., Roy-Pogodzik E. M., Pekkinen J., Bauch C., Hönck H. H., Hennemann H., Kreienkamp H. J. (2011) FEBS Lett. 585, 2665–2670 [DOI] [PubMed] [Google Scholar]
- 53. Zhang M. M., Tsou L. K., Charron G., Raghavan A. S., Hang H. C. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 8627–8632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Forrester M. T., Hess D. T., Thompson J. W., Hultman R., Moseley M. A., Stamler J. S., Casey P. J. (2011) J. Lipid Res. 52, 393–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim K. B., Lee J. S., Ko Y. G. (2008) Methods Mol. Biol. 424, 413–422 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.