Background: TFIIB and TFIIH are required for transcription initiation by RNA polymerase II.
Results: TFIIB and the Ssl2 helicase of TFIIH functionally interact at both promoter and terminator.
Conclusion: Ssl2 affects TFIIB-mediated gene looping and start site selection.
Significance: These results underscore the intriguing role of promoter-terminator interactions during the transcription cycle.
Keywords: Gene Regulation, Transcription, Transcription Initiation Factors, Transcription Termination, Yeast Genetics, DNA Helicase, Ssl2, TFIIB, TFIIH, XPB
Abstract
TFIIB is essential for transcription initiation by RNA polymerase II. TFIIB also cross-links to terminator regions and is required for gene loops that juxtapose promoter-terminator elements in a transcription-dependent manner. The Saccharomyces cerevisiae sua7-1 mutation encodes an altered form of TFIIB (E62K) that is defective for both start site selection and gene looping. Here we report the isolation of an ssl2 mutant, encoding an altered form of TFIIH, as a suppressor of the cold-sensitive growth defect of the sua7-1 mutation. Ssl2 (Rad25) is orthologous to human XPB and is a member of the SF2 family of ATP-dependent DNA helicases. The ssl2 suppressor allele encodes an arginine replacement of the conserved histidine residue (H508R) located within the DEVH-containing helicase domain. In addition to suppressing the TFIIB E62K growth defect, Ssl2 H508R partially restores both normal start site selection and gene looping. Moreover, Ssl2, like TFIIB, associates with promoter and terminator regions, and the diminished association of TFIIB E62K with the PMA1 terminator is restored by the Ssl2 H508R suppressor. These results define a novel, functional interaction between TFIIB and Ssl2 that affects start site selection and gene looping.
Introduction
Promoter recognition and transcription initiation by RNA polymerase II (Pol II)3 require five general transcription factors: TFIIB, TFIID, TFIIE, TFIIF, and TFIIH (1–3). Promoters containing a TATA box require the TATA-binding protein (TBP) subunit of TFIID for promoter recognition. TFIIB binds the DNA-TBP complex, contacting promoter DNA both upstream and downstream of TATA. Pol II, in association with TFIIF, binds the DNA-TBP-TFIIB ternary complex, forming a closed promoter complex. TFIIE and TFIIH complete formation of the preinitiation complex (PIC). TFIIE and TFIIH are essential for transcription initiation in vivo but are dispensable for Pol II transcription from negatively supercoiled DNA in vitro, indicating that these factors play an essential role in promoter melting (4). Biochemical probes of DNA-protein interactions mapped the path of promoter DNA within human and yeast PICs (5–13). The topology of the PIC deduced from these in vitro experiments agrees remarkably well with the consensus location of the general transcription factors revealed by a genome-wide analysis of their locations in yeast (14).
This paper focuses on TFIIB and TFIIH. TFIIB is a single subunit protein consisting of an N-terminal zinc ribbon (“B-ribbon”) followed by a ∼60-amino acid sequence that links the B-ribbon to two cyclin repeats (“B-core”) that form the C-terminal two-thirds of the protein. The yeast gene encoding TFIIB (SUA7) was initially identified based on mutations that shift start site selection downstream of normal (15). The sua7-1 mutation encodes an E62K replacement between the B-ribbon and B-core; other replacements spanning residues Glu-62 to Val-79 were also found to affect start site selection (16–22). X-ray structures of yeast Pol II-TFIIB complexes revealed two alternative structures for this region, denoted the “B-finger” and “B-reader” (23–25). The B-reader has been proposed to “scan” template DNA for acceptable start sites (24). A conformational switch between the B-reader and B-finger subsequent to initiation facilitates the transition from abortive initiation to promoter escape (25, 26).
Genetic suppressors of the sua7-1 mutation identified other components of the PIC that affect start site selection. Two tfg1 alleles (ssu71-1 and ssu71-2), encoding single amino acid replacements in the Tfg1 subunit of TFIIF, shift initiation upstream of normal and partially restore the normal initiation pattern in the sua7-1 mutant (27, 28). Other amino acid replacements in Tfg1 or Tfg2 also affect start site selection, in each case shifting initiation upstream (29). Mutations in the RPB2 and RPB9 genes, which encode two subunits of Pol II, were also identified as suppressors of either sua7-1 or the related sua7-3 mutation (TFIIB R78C) (30, 31). The architecture of the PIC suggests that Rpb2, Rbp9, and TFIIF affect start site selection allosterically, via a protein interaction network that extends from Rpb9 at the leading edge of the Pol II through TFIIF to TFIIB (9).
TFIIH is an 11-subunit complex that includes two DNA helicases and a kinase-cyclin subcomplex that targets the C-terminal repeat domain of the Rpb1 subunit of Pol II (32). XPB and XPD are the helicase subunits of human TFIIH and are orthologs of yeast Ssl2 (also known as Rad25) and Rad3, respectively. XPB/Ssl2 is the only TFIIH subunit in immediate proximity to promoter DNA and is located at the leading edge of Pol II (5, 7). XPB is required for open complex formation (33), although not as a conventional helicase but apparently by acting as a “molecular wrench” that rotates DNA downstream of the promoter relative to a fixed upstream position (5). The location of Ssl2 within the yeast PIC is consistent with this model for promoter opening (7). The location and function of Ssl2/XPB suggest that it might affect start site selection, perhaps by feeding template DNA into the active center (7), although no direct evidence for TFIIH involvement in start site selection has been reported.
In addition to the role of TFIIB in start site selection, we have discovered a function for TFIIB in the formation of gene loops that juxtapose promoter and terminator regions (34). Furthermore, TFIIB occupies the terminator region of Pol II-transcribed genes and does so in a manner dependent upon transcription. The sua7-1 mutation adversely affects gene looping and TFIIB-terminator association without affecting PIC assembly. The physiological role of gene loops remains to be established, although mutations that block gene looping, including sua7-1, adversely affect “transcriptional memory” of GAL genes (35, 36). In an effort to learn more about the role of TFIIB in transcription and to further investigate the significance of gene loops, we have isolated additional suppressors of sua7-1. Here we report the identification of ssl2-508, encoding an altered form of the Ssl2 subunit of TFIIH, as a suppressor of TFIIB E62K. Our results define a novel interaction between TFIIB and Ssl2 at promoter and terminator regions that affect both start site selection and gene looping.
EXPERIMENTAL PROCEDURES
Yeast Strains
The Saccharomyces cerevisiae strains used in this study are listed in Table 1. Strains YMH14 (T16), YMH124 (YDW546), and YMH893 (YMH71-9C) were described previously using the names indicated in parentheses. The ssl2 plasmid shuffle strain YMH1142 (ssl2::kanMX [pN863: SSL2-TRP1]) was constructed as follows. W303-1B was transformed with plasmid pN863 (SSL2-TRP1), followed by disruption of the chromosomal SSL2 locus using kanMX as the selectable marker (37). 5′-SSL2-kanMX-3′ oligonucleotides were designed such that the forward and reverse PCR primers included 45 nucleotides of SSL2 sequence immediately upstream of the start codon and immediately downstream of the stop codon. The PCR was performed using plasmid pFA6a-kanMX as template DNA. The PCR product was analyzed by agarose gel electrophoresis and introduced into strain W303-1B [pN863: SSL2-TRP1], selecting for transformants on YPD-G418 medium at 30 °C. A resulting transformant (YMH1142) failed to grow on 5-FAA medium, consistent with disruption of the essential SSL2 gene and confirmed by PCR analysis of the ssl2::kanMX locus. Strains YMH1141 and YMH1143 were created by introduction of plasmids pN861 (SSL2-URA3) and pM1930 (ssl2-508-URA3), respectively, into YMH1142, followed by counterselection of pN863 (SSL2-TRP1) on 5-FAA medium.
TABLE 1.
List of yeast strains
| Straina | Genotype | Source |
|---|---|---|
| W303-1B | MATα leu2-3,112 his3-11,15 trp1-1 ade2-1 can1-100 ura3-1 | Ref. 44 |
| YMH14 | MATα cyc1-5000 cyc7-67 ura3-52 leu2–3,112 | Ref. 16 |
| YMH124 | MATα cyc1-5000 cyc7-67 ura3-52 leu2-3,112 sua7-1 | Ref. 16 |
| YMH893 | MATacyc1-5000 cyc7-67 ura3-52 trp5-48 his5-2 sua7-1 | Ref. 16 |
| YHM1138 | MATa cyc1-5000 cyc7-67 trp5-48 his5-2 ura3-52 sua7-1 ssl2-508 | This study |
| YMH1141 | MATα leu2-3,112 his3-11, trp1-1 ade2-1 can1-100, ura3–1 ssl2::KanMX [pN861: SSL2-URA3] | This study |
| YMH1142 | MATα leu2-3 112 his3-11 trp1-1 ade2-1 can1-100 ura3-1 ssl2::KanMX [pN863: SSL2-TRP1] | This study |
| YMH1143 | MATα leu2-3,112 his3-11, trp1-1 ade2-1 can1-100, ura3-1 ssl2::KanMX [pM1930: ssl2–508-URA3] | This study |
| YMH1165 | MATα cyc1-5000 cyc7-67 ura3-52 leu2-3,112 SSL2-TAP | This study |
| YMH1166 | MATacyc1-5000 cyc7-67 ura3-52 trp5-48 his5-2 sua7-1 SSL2-TAP | This study |
| YMH1167 | MATa cyc1-5000 cyc7-67 trp5-48 his5-2 ura3-52 sua7-1 ssl2-508-TAP | This study |
| DG1657 | MATaura3-167 his3-Δ200 trp1-hisG leu2-hisG Ty1–270 his3-AI Ty1–588neo Ty1–146 [tyb1::lacZ] | Ref. 56 |
| DG1772 | DG1657 ssl2::TRP1 (pRS416: URA3-CEN SSL2) | Ref. 56 |
| DG1773 | DG1657 ssl2::TRP1 [BDG853: LEU2-CEN-SSL2] [pC59: URA3-CEN-SSL2] | Ref. 56 |
| DG1774 | DG1657 ssl2::TRP1 [BDG853: LEU2-CEN-SSL2] | Ref. 56 |
| DG1775 | DG1657 ssl2::TRP1 [pRS416: URA3-CEN-ssl2-rtt] | Ref. 56 |
| DG1776 | DG1657 ssl2::TRP1 [pRS416: URA3-CEN-ssl2-DEAD] | Ref. 56 |
| DG1777 | DG1657 ssl2::TRP1 [URA3-CEN-ssl2-XP] | Ref. 56 |
| DG1778 | DG1657 ssl2::TRP1 [LEU2-CEN-ssl2-1] | Ref. 56 |
a Strains YMH14 (T16), YMH124 (YDW546), and YMH893 (YMH71-9C) were described previously using the names indicated in parentheses (16).
Growth Media and Nomenclature
Rich (YPD), minimal (SD), synthetic complete (SC), and omission (−Ura, −Trp) media were prepared according to standard recipes (38). YPG medium contains 2% galactose supplemented with antimycin A to inhibit respiration. YPD-G418 medium includes 200 μg/ml G418 (37). 5-fluoroorotic acid medium (39) and 5-FAA medium (40), used to select against strains harboring plasmid-borne URA3 and TRP1 genes, respectively, were prepared as described. 6-Azauracil was added to −Ura medium at a final concentration of 50 μg/ml. Yeast X-gal indicator medium was prepared as described (41). For growth assays, strains were grown to saturation at 30 °C in liquid YPD medium, harvested, and diluted in sterile water to ∼1 × 108 cells/ml, followed by spotting of 10-fold serial dilutions onto the indicated medium. Csm− (cold-sensitive) and Tsm− (heat-sensitive) phenotypes refer to distinctly impaired growth at 12 °C (Csm−) and 39 °C (Tsm−), respectively.
Isolation and Genetic Analysis of sua7-1 Suppressors
Strain YMH893 (sua7-1) was streaked on YPD medium and incubated at 30 °C. Single colonies were picked and inoculated into separate 5-ml YPD liquid culture tubes and grown to stationary phase at 30 °C. Cultures were harvested, washed with sterile water, and diluted to 1 × 107 cells/ml. One hundred μl of cells were spread onto YPD plates and incubated at 12 °C for at least 7 days. Single colony Csm+ revertants (a maximum of one colony per plate) were picked, subcloned on YPD medium, and incubated at 12 °C. Dominance/recessiveness of suppressor mutations and linkage of the Csm+ suppressor and Tsm− pleiotropic phenotypes were determined by standard yeast genetic methods involving backcrosses, diploid selection, sporulation, and tetrad dissection, as described (38).
Isolation of SSL2 and ssl2-508 Allele
The SSL2 gene was isolated from a YCp50 genomic library (42) by complementation of the Tsm− phenotype conferred by the ssl2-508 mutation using strain YMH1138 as the host. The complementing DNA was delimited to the SSL2 gene using plasmid pN861 (pEP23), which carries the SSL2 (RAD25) gene (43). The ssl2-508 allele was recovered from genomic DNA by gap repair (44). Briefly, pN861 was digested to completion with restriction enzymes PvuII and SwaI, thereby deleting most of the SSL2 open reading frame. Linear DNA was purified by agarose gel electrophoresis and introduced into strain YMH1138 (ssl2-508 ura3). Ura+ colonies were selected and screened for retention of the Csm+ and Tsm− suppressor phenotypes. Plasmid DNA was recovered, amplified in Escherichia coli, and analyzed by restriction digestion to confirm the presence of the ssl2 ORF. The resulting plasmid (pM1930) failed to complement the Csm+ and Tsm− phenotypes when reintroduced into strain YMH1138, thereby confirming recovery of the ssl2-508 allele. The DNA sequence of the entire ssl2-508 ORF was determined using SSL2-specific primers that span the entire ORF at ∼500-bp intervals. The suppressor mutation was identified by aligning the ssl2-508 sequence with SSL2 sequence obtained from the Saccharomyces Genome Database Web site.
Determination of Transcription Start Sites
Reporter plasmids pM50 and pM107 were used to assess transcription start site changes at the CYC1 promoter, as described previously (15, 45). Primer extension experiments were performed using the ADH1-specific primer oIP87; primer extension products were resolved in an 8% polyacrylamide gel and visualized by autoradiography (15).
Chromatin Immunoprecipitation
Cross-linking and isolation of chromatin were performed as described previously (34) using antibodies directed against either TFIIB or Protein A (Ssl2-TAP) (IgG-agarose; Sigma). Conditions for PCRs to quantify PMA1 DNA were as described previously (46) using [α-32P]dATP in 25-μl reactions containing 1× Standard Taq Buffer (New England Biolabs). PCR products were resolved in 6% polyacrylamide, 1× TBE gels and quantified by a PhosphorImager (Amersham Biosciences). Conditions for PCRs to detect PYK1 DNA were performed as described previously (47), except that PCR products were fractionated in 1.5% agarose gels and visualized by ethidium bromide staining using an AlphaImager 2000 (34). The immunoprecipitate/input ratio of each gene-specific product was normalized to the immunoprecipitate/input ratio of an intergenic region of chromosome V (for PMA1 ChIP) or to the non-transcribed HMR gene (for PYK1 ChIP). Accordingly, the numbers on the y axis depict the -fold enrichment of the ChIP signal over the background signal such that y = 1 represents background. All PCR primer pairs are identical to those described previously (46) except for the HMR primers (48).
Chromosome Conformation Capture
DNA loops were analyzed by a modified version of 3C (49, 50), as described previously (51). Chromatin was extracted and digested overnight at 37 °C with gentle shaking in the presence of the restriction enzyme HindIII (New England Biolabs). Juxtaposition of the SEN1, BLM10, and HEM3 promoter-terminator regions were detected as P1-T1 PCR products (34). Control reactions were performed to establish that P1-T1 PCR products are dependent upon formaldehyde cross-linking, HindIII restriction digestion, and subsequent ligation (data not shown) (51). Control PCRs were carried out using the same intergenic chromosome V convergent primer pair as in ChIP. PCR products were fractionated on a 1.5% agarose gel and visualized by ethidium bromide staining using an AlphaImager 2000.
RT-PCR Analysis
Total RNA was isolated using the RNeasy Mini RNA isolation procedure (Qiagen). RT-PCR was performed using 25 ng of total RNA and SEN1-specific forward and reverse primer pairs according to the One-step RT-PCR system (Qiagen). Samples without reverse transcriptase were used to control for DNA contamination. The PCR products were analyzed as described for the 3C assay.
RESULTS
Isolation of sua7-1 Suppressors
The sua7-1 allele encodes a lysine replacement of glutamic acid at position 62 (E62K) and confers a cold-sensitive growth defect (16). Previous work from our laboratory identified an allele of RPB9, encoding an altered form of Pol II, and two alleles of TFG1, encoding altered forms of the largest subunit of TFIIF, as suppressors of the sua7-1 Csm phenotype; an allele of the RPB2 was also isolated as a suppressor of the related sua7-3 mutation (R78C). In light of new information about TFIIB, including its association with terminator regions and its role in gene looping, we have isolated new suppressors of sua7-1 in an effort to more fully understand the role of TFIIB in the transcription cycle.
Strain YMH1138 (Table 1) was isolated as a spontaneous Csm+ revertant of YMH893 (MATa sua7-1) at 12 °C; this strain also acquired a distinct Tsm− phenotype at 39 °C (Fig. 1). When back-crossed to strain YMH124 (MATα sua7-1), the resulting diploid strain reacquired the Csm− and Tsm+ phenotypes of the primary mutant, indicating that both phenotypes are the result of a recessive mutation(s). The YMH1138 × YMH124 diploid strain was sporulated and dissected, and segregants were scored for growth at 12, 30, and 39 °C. The Csm+ and Tsm− co-segregated, thereby defining Tsm− as a pleiotropic marker of the sua7-1 suppressor.
FIGURE 1.
Phenotypes associated with sua7-1 mutant and ssl2-508 suppressor. 10-Fold serial dilutions of SUA7 (YMH14), sua7-1 (YMH893), and sua7-1 ssl2-508 (YMH1138) haploid strains and diploid strain sua7-1/sua7-1 SSL2/ssl2-508 (YMH1138 × YMH124) were spotted onto YPD medium and photographed after incubation at the indicated temperature for 2 days (30 °C), 5 days (12 °C), or 3 days (39 °C).
To identify the suppressor gene in strain YMH1138, a YCp50 genomic library (URA3) was introduced into YMH1138, and Ura+ transformants were selected at 39 °C. Two Tsm+ transformants were isolated that were also Csm−. When cured of plasmid DNA, the Csm+ and Tsm− phenotypes were restored, indicating that complementation was due to plasmid DNA rather than strain reversion. Plasmid DNA from both strains included identical 13-kb fragments encompassing five ORFs from chromosome IX. Retransformation of YMH1138 with plasmid pN861 (SSL2-URA3-CEN) established that complementation was due solely to the SSL2 (RAD25) gene.
Suppressor of sua7-1 Encodes Ssl2 H508R Replacement
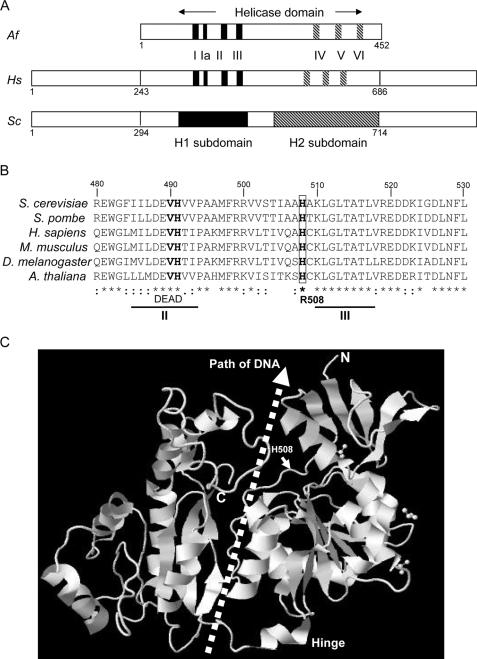
Ssl2 is a subunit of TFIIH and member of the SF2 family of DNA helicases. SF2 proteins are defined by seven helicase motifs, denoted I, Ia, II, III, IV, V, and VI, that form two distinct subdomains, designated H1 and H2 (Fig. 2A). To identify the Ssl2 defect that suppresses TFIIB E62K, we cloned the ssl2 suppressor allele from strain YMH1138 by gap repair (see “Experimental Procedures”). DNA sequence analysis of the entire ssl2 open reading frame revealed a single base pair substitution encoding a histidine to arginine replacement at position 508 (H508R) (Fig. 2B). Accordingly, we designated the suppressor allele ssl2-508.
FIGURE 2.
The ssl2-508 allele encodes an Ssl2 H508R replacement. A, schematic representation of the Ssl2/XPB orthologous proteins from A. fulgidus (Af), human (Hs), and S. cerevisiae (Sc). Ssl2/XPB is a member of the SF2 family of DNA helicases, defined by seven helicase motifs, denoted I, Ia, II, III, IV, V, and VI, that form two distinct subdomains, designated H1 and H2. B, partial sequence alignment of Ssl2/XPB proteins from the indicated organisms, spanning amino acids 480–530 of S. cerevisiae Ssl2. The ssl2-508 suppressor encodes replacement of the phylogenetically invariant histidine at position 508 by arginine (H508R), located between helicase motifs II and III. The alignment was created by the software MultAlin (available on the World Wide Web). C, position of yeast Ssl2 H508 mapped onto the x-ray structure of the A. fulgidus AfXPB protein. His-508 is within the proposed path of DNA between the two helicase subdomains. This structure was generated using Protein Data Bank entry 2FWR in the JMol PDB viewer.
The His-508 residue lies between the DEVH nucleotide-binding motif II and motif III and is phylogenetically invariant. When modeled onto the x-ray structure of the Archaeoglobus fulgidus XPB homolog (AfXPB) (52), His-508 is located near the central groove, between the H1 and H2 helicase subdomains (Fig. 2C). The location of DNA within helicases of known structure, combined with hydroxyradical cleavage of yeast Ssl2, suggests that promoter DNA immediately downstream of the transcription bubble lies in the central groove between the two helicase subdomains (10). From these data, it appears likely that the functional interaction between TFIIB and TFIIH involves the helicase activity of Ssl2, affecting the structure of the PIC downstream of the Pol II active center.
Phenotypes Associated with ssl2 Mutants
We next asked whether the ssl2-508 mutation confers cell growth phenotypes independent of the sua7-1 primary mutation. We constructed an isogenic SSL2 (YMH1141) and ssl2-508 (YMH1143) strain pair in a SUA7 wild type background by plasmid shuffle (see “Experimental Procedures”). The ssl2-508 mutant is viable and, in contrast to the sua7-1 ssl2-508 suppressor strain, does not exhibit a Tsm− phenotype (Fig. 3A). Thus, the Tsm− phenotype of the sua7-1 ssl2-508 strain is synthetic, dependent upon both mutations, a result that underscores the functional interaction between TFIIB and Ssl2. This interaction does not necessarily reflect a direct TFIIB-Ssl2 physical interaction because these two components of the PIC interact with distinct regions of promoter DNA and Pol II (10).
FIGURE 3.
Growth phenotypes of ssl2 mutants. A, 10-fold serial dilutions of W303–1B (WT) and YMH1143, either before (ssl2-508 [SSL2]) or after (ssl2-508) counterselection of plasmid pN863 (SSL2-TRP) on 5-FAA medium, were spotted onto the indicated medium and photographed after incubation for either 2 days (YPD, 30 °C; YPG, 30 °C; 6-azauracil (6-AU), 30 °C) or 3 days (YPD, 37 °C). B, 10-fold serial dilutions of isogenic URA3 plasmid-containing strains SSL2 (DG1772), ssl2-rtt (DG1775), ssl2-DEAD (DG1776), and ssl2-XP (DG1777) or LEU2 plasmid-containing strains SSL2 (DG1774) and ssl2–1 (DG1778) were spotted onto either YPD or YPG medium and incubated at the indicated temperature for 2 days (30 °C) or 3 days (34 and 37 °C).
We also assayed growth of the ssl2-508 mutant on medium containing galactose as the sole carbon source and on medium containing 6-azauracil. A Gal− phenotype is often associated with defects in Pol II transcription initiation or termination (53), whereas sensitivity to 6-azauracil is associated with defects in elongation (54, 55). The ssl2-508 mutant exhibited no discernable sensitivity to 6-azauracil but displayed a distinct Gal− phenotype (Fig. 3A). The previously isolated ssl2-rtt (E556K), ssl2-DEAD (V490A/H491D), and ssl2–1 (W427L) mutants, encoding the indicated (in parentheses) amino acid replacements within the Ssl2 helicase domain (56, 57), also display Gal− phenotypes. We also note, however, that the ssl2-XP mutant, which encodes a C-terminal truncation of Ssl2 that was designed to mimic the truncated form of XPB from patients suffering from xeroderma pigmentosum (58), displays a mild Gal− phenotype (Fig. 3B).
The ssl2-508 Suppressor Affects Transcription Start Site Selection
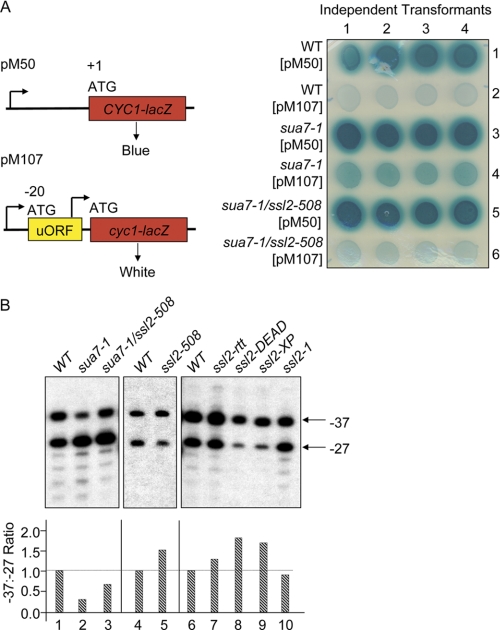
TFIIB Glu-62 and Arg-78 lie on opposite sides of the B-finger, forming a salt bridge at the base of the finger (16, 23). In addition to conferring Csm− phenotypes, the sua7-1 (E62K) and sua7–3 (R78C) mutations shift start site selection downstream of normal at the ADH1 and CYC1 genes (16). The previously defined ssu71 (Tfg1) and ssu73 (Rpb9) suppressors of sua7-1 and sua7-3 shift initiation upstream of normal, partially restoring the normal initiation patterns at these genes (28, 30, 31). To determine whether the ssl2-508 suppressor exerts a similar effect on initiation, we assessed start site selection using a CYC1-lacZ reporter plasmid (Fig. 4A) (15, 45). Plasmid pM50 contains the normal CYC1 promoter and leader region fused in frame to lacZ just downstream of and in-frame with the CYC1 ATG start codon. Plasmid pM107 is identical to pM50 except for a point mutation in the CYC1 leader that creates an aberrant ATG codon, upstream and out-of-frame (uORF) with CYC1-lacZ. pM50 and pM107 were introduced into the wild type (YMH14), primary mutant (YMH893), and suppressor strains (YMH1138). As expected, all three strains harboring pM50 turned dark blue on X-gal indicator medium, whereas the wild type strain harboring pM107 was very pale blue, a consequence of translation initiation at the uORF (Fig. 4A). The sua7-1 mutant harboring pM107 exhibited enhanced blue color (cf. rows 2 and 4), corresponding to a downstream transcription start site shift (−46 to −14), whereas the sua7-1 ssl2-508 suppressor strain was restored to pale blue (row 6), due to transcription start site selection upstream of the uORF.
FIGURE 4.
Effects of Ssl2 defects on transcription start site selection. A, plasmid pM50 or pM107 (URA3 marker) was introduced into the wild type strain (YMH14), sua7-1 mutant strain (YMH893), or sua7-1 ssl2-508 suppressor strain (YMH1138). Four independent transformants of each strain were spotted onto X-gal indicator medium, incubated at 30 °C, and photographed. The E. coli LacZ gene is fused in-frame with the normal CYC1 ATG start codon. pM107 is identical to pM50, except for a single base pair substitution (position −20 relative to the ATG) that creates a short ORF, upstream and out-of-frame with the normal CYC1 gene. The normal CYC1 transcription start site at −46 is indicated by the arrow; the downstream start site associated with the sua7-1 mutation is indicated at position −14 of pM107 (15). B, primer extension analysis of ADH1 transcription start sites. Strains (Table 1) are identical to those described in Fig. 3 plus YMH1141 (WT, lane 4) and YMH1143 (ssl2-508, lane 5). In the wild type strains (lanes 1, 4, and 6), transcription initiates equally at two sites, −37 and −27 (A of ATG is denoted +1). Transcripts initiating at −37 and −27 were quantified by determining the ratio of −37 to −27 and normalized to a ratio of 1.0 in the wild type strains.
To confirm the effect of the ssl2-508 mutation on start site selection, we mapped initiation at the ADH1 gene by primer extension. Results are shown in Fig. 4B. The wild type strain (YMH14) displayed the normal pattern of ADH1 initiation, defined by a 1:1 ratio of start sites at positions −37 and −27 (lane 1). As observed previously, the sua7-1 primary mutant showed diminished initiation at −37 and enhanced initiation downstream of −27 (lane 2). Consistent with the lacZ reporter assay (Fig. 4A), the ssl2-508 allele resulted in a modest upstream shift in the sua7-1 background (lane 3). Furthermore, the ssl2-508 allele alone, as well as the ssl2-rtt, ssl2-DEAD, and ssl2-XP mutants, exhibited upstream shifts in the SUA7 wild type background, in each case increasing the −37/−27 start site ratio from 1.0 to 1.3–2.0 (lanes 4–9). The ssl2-1 mutant, on the other hand, exhibited little or no effect on initiation (lane 10). Taken together, these results demonstrate that the Ssl2 subunit of TFIIH is an effector of start site selection, presumably in a manner dependent upon its helicase activity and via functional interaction with TFIIB.
Ssl2 Cross-links to Promoter and Terminator Regions
In addition to occupying the promoter, TFIIB also localizes to the 3′-ends of genes near the poly(A) site in yeast (34–36, 59) and in mammalian cells (60). To determine whether Ssl2 also occupies terminator regions, we performed ChIP of Ssl2 using a TAP-tagged Ssl2 strain (YMH1165). As expected, Ssl2 cross-links to the promoter of the PMA1 gene but also cross-links to regions 7 and 8, just downstream of the two poly(A) sites (Fig. 5). These results do not reflect the presence of a cryptic promoter because TBP does not cross-link to these regions (34). Ssl2-terminator association is not unique to PMA1 because Ssl2 also cross-links to the terminator region of the PYK1 gene (supplemental Fig. S1). To our knowledge, this is the first demonstration of Ssl2 or XPB occupancy of a terminator region, although earlier ChIP data revealed terminator occupancy of GAL-FMP27 by the Kin28 subunit of TFIIH (61).
FIGURE 5.
Ssl2 cross-links to the terminator region as well as the promoter of PMA1. A, schematic depiction of the PMA1 gene showing the positions of the promoter (TATA box) and the two poly(A) sites. The regions probed by ChIP are depicted by black bars numbered 1, 2, 4, 5, 6, 7, 8, and 9 and correspond to primer pair numbers denoted at the top of each lane in B. B, ChIP analysis of Ssl2-TAP cross-linking to PMA1 using protein A antibody. Strains YMH14 (WT), YMH893 (sua7-1), and YMH1138 (sua7-1 ssl2-508) are described in Table 1. The lower band marked by the asterisk is the PCR product for a non-transcribed region of chromosome V and is included as a control. C, quantification of the ChIP data. Signals are expressed as x-fold over the background, calculated as described under “Experimental Procedures.” Values are the mean of three independent experiments, including the data presented in B, with S.D. (error bars).
We also asked whether Ssl2-terminator occupancy is affected by the sua7-1 mutation and by the ssl2-508 suppressor. Remarkably, sua7-1 significantly enhanced Ssl2 cross-linking to region 7 yet had no apparent effect on cross-linking to either region 1 or region 8 (Fig. 5, middle). Furthermore, the ssl2-508 suppressor restored Ssl2 cross-linking to its normal level at region 7 in the sua7-1 background (Fig. 5, right). The ssl2-508 suppressor appeared to adversely affect Ssl2 cross-linking to the promoter (region 1), although we do not know whether this effect requires the sua7-1 allele.
Ssl2 H508R Affects TFIIB-Terminator Occupancy
We next asked whether the ssl2-508 suppressor affects TFIIB occupancy of the PMA1 gene. Results are shown in Fig. 6. As noted above and consistent with previous results (34), TFIIB cross-links to the promoter and terminator regions in the wild type strain (Fig. 6, left); furthermore, occupancy of the terminator, but not the promoter, is diminished in the sua7-1 mutant (middle). The ssl2-508 mutation suppressed this effect, restoring TFIIB-terminator occupancy (Fig. 6, right). Thus, the sua7-1 primary mutation enhances Ssl2-terminator occupancy (Fig. 5) while diminishing TFIIB-terminator occupancy (Fig. 6), whereas the ssl2-508 suppressor reverses both effects, diminishing Ssl2 occupancy while enhancing TFIIB occupancy. These results demonstrate that Ssl2, presumably as a component of TFIIH, occupies not only the promoter but also the terminator of a Pol II-transcribed gene and does so in a manner that involves functional interaction with TFIIB.
FIGURE 6.
Effects of sua7-1 and ssl2-508 on TFIIB cross-linking to the promoter and terminator regions of PMA1. A, ChIP analysis of TFIIB cross-linking to PMA1 using polyclonal α-TFIIB antibody. ChIP was performed, and results were quantified as described in the legend to Fig. 5 using the same strains. B, quantification of the ChIP data, as in Fig. 5.
Ssl2 Affects Gene Loops
In addition to the sua7-1 effects on start site selection and TFIIB-terminator occupancy, TFIIB E62K adversely affects gene loops that juxtapose promoter and terminator regions (34). To determine whether Ssl2 also affects looping, we assayed gene loops by 3C at the BLM10, SEN1, and HEM3 genes, each of which was shown previously to form transcription-dependent promoter-terminator loops (34, 62). Following cell growth in YPD medium to midlog phase, the wild type (YMH14), sua7-1 (YMH893), and sua7-1 ssl2-508 (YMH1138) strains were assayed for gene looping by 3C using P1 and T1 primer pairs that are specific to promoter and terminator regions of these genes (Fig. 7A). P1-T1 PCR products were quantified by dividing P1-T1 PCR signals by control PCR signals representing an intergenic region of chromosome V (51). Consistent with earlier results, we observed a significant reduction in the P1-T1 PCR signals for all three genes in the sua7-1 mutant (Fig. 7B). At each gene, however, the looping defect is suppressed by the ssl2-508 mutation, restoring looping signals to near wild type levels (Fig. 7, B and C). Thus, Ssl2 H508R suppresses all defects associated with the TFIIB E62K replacement, including cell growth rates, changes in transcription start site selection, diminished TFIIB-terminator association, and impaired gene looping. These results define a novel role for the Ssl2 helicase subunit of TFIIH in start site selection and underscore the intriguing, albeit ill defined, role of promoter-terminator interaction during the transcription cycle.
FIGURE 7.
Effects of ssl2-508 on gene looping. A, schematic depiction of SEN1, BLM10, and HEM3 genes, including the positions of the HindIII sites and the P1 and T1 primer pairs. Approximate length of each ORF is indicated as kb. For description of the 3C assay, see Ref. 51. B, gene looping results detected as P1-T1 PCR products for isogenic strains YMH14 (SUA7), YMH893 (sua7-1), and YMH1138 (sua7-1 ssl2-508). Control PCR represents an intergenic region of chromosome V (51). C, quantification of the 3C data. All PCR products were determined to be in the linear range (51). The P1-T1 PCR products were quantified by dividing the P1-T1 PCR signals by the Chr V PCR signals for each sample. These ratios were then normalized to the sample/control ratio in the wild type. The values plotted for SEN1 and HEM3 are the mean of three independent experiments with S.D. (error bars).
DISCUSSION
The results presented in this paper define a novel, functional interaction between TFIIB and the Ssl2 subunit of TIFIH. First, the ssl2-508 allele suppresses the cold-sensitive growth phenotype of the sua7-1 mutant at 12 °C. Second, the sua7-1 ssl2-508 double mutant displays a synthetic heat-sensitive growth defect at 37 °C, a phenotype dependent upon both mutations. Third, the ssl2-508 mutation affects start site selection by compensating for the downstream shift conferred by sua7-1. Fourth, TFIIB and Ssl2 occupy both the promoter and terminator regions of the PMA1 and PYK1 genes. Fifth, the sua7-1 mutation exerts reciprocal effects on TFIIB- and Ssl2-terminator occupancy, and these effects are reversed by the ssl2-508 suppressor. Finally, ssl2-508 restores gene looping in the sua7-1 mutant, an effect that could position the terminator-TFIIB-Ssl2 complex proximal to the promoter to facilitate transcription reinitiation.
Ssl2 at Promoter
Helicases perform critical roles in essentially all aspects of DNA (replication, repair, and recombination) and RNA (transcription, splicing, and translation) metabolism (63). More than 80 genes in the S. cerevisiae genome include conserved helicase motifs and affect a range of RNA and DNA metabolic activities. The activities of these proteins are not limited to classical helicase activity, defined as NTP-driven duplex unwinding. Instead, many SF2 helicases are processive translocases that catalyze directional movement along either single- or double-stranded nucleic acids. Although yeast Ssl2 and human XPB exhibit classical 3′–5′ DNA helicase activities, their principal roles in transcription initiation are likely to include translocase activities (64).
How does Ssl2 affect start site selection? Based on protein-DNA cross-linking data, human XPB was proposed to act as a “ratchet wrench” that rotates DNA at the leading edge of the PIC relative to a fixed upstream site (5). The ensuing torsional stress melts the promoter, enabling single-stranded template DNA to descend into the Pol II active site cleft (65, 66). Hydroxyradical cleavage data supports this model by showing that DNA at the leading edge of the promoter appears to be positioned within the groove between the two helicase subdomains of Ssl2 (10). Regions of TFIIE and TFIIF are also positioned in proximity to DNA, where they are likely to facilitate and stabilize DNA strand separation (9). X-ray structures of Pol II-TFIIB suggest that the B-reader of TFIIB, located within the active site cleft, then scans single-stranded template DNA for acceptable start sites, a conclusion consistent with the positions of TFIIB amino acid replacements that alter start site selection.
If the position of Ssl2 is “fixed” at the leading edge of the promoter, then its 3′–5′ translocase activity could generate the rotational torque required to melt the promoter and continue to feed template DNA into the active center of Pol II as TFIIB scans for start sites (Fig. 8). As suggested previously (10), DNA might be fed into the active center by a “scrunching” mechanism (67) that could generate long, single-stranded DNA loops emerging from the Pol II central cleft prior to its closure upon initiation. Whereas the TFIIB E62K replacement shifts initiation downstream of normal by scanning past the normal initiator sequence (20, 68), the Ssl2 H508R replacement could suppress this defect by impeding the translocase activity of Ssl2, compensating for the downstream shift by slowing the rate at which template DNA is fed into the active site cleft, past the TFIIB “scanner.”
FIGURE 8.
Model for Ssl2-mediated promoter melting and start site selection. The model proposes that the Ssl2 helicase subunit of TFIIH, located at the leading edge of Pol II, rotates promoter DNA relative to a fixed upstream site, thereby inducing torsional stress that melts the promoter (5). Single-stranded template DNA then descends into the active site cleft, where TFIIB “scans” for acceptable start sites. The translocase activity of Ssl2 might feed template DNA through the active center by a “scrunching” mechanism (67). Amino acid replacements in the B-reader would allow template DNA to be fed past the active center, resulting in a downstream start site shift. Amino acid replacements in Ssl2 that impair translocase activity would slow the rate at which template DNA is fed into the active center, resulting in B-reader recognition of acceptable upstream start sites. Amino acid replacements in the Rpb2 lobe domain or deletion of the Rpb9 subunit of Pol II, located near the leading edge of Pol II and proximal to Ssl2, might affect start site selection either by affecting Ssl2 translocase activity or by transducing their effects via TFIIF to the TFIIB-Pol II catalytic center.
Ssl2 at Terminator
TFIIB cross-links to the terminator as well as the promoter of Pol II-transcribed genes (34–36, 59, 60). The TFIIB E62K replacement, although without affect on PIC assembly (8), impairs TFIIB-terminator cross-linking as well as promoter-terminator looping (34). Remarkably, Ssl2 exhibits a pattern of cross-linking similar to that of TFIIB, occupying DNA downstream of the two poly(A) sites of PMA1 and the terminator region of PYK1 (Fig. 5 and supplemental Fig. S1). Moreover, Ssl2 H508R restores TFIIB E62K occupancy over the PMA1 terminator (Fig. 6) and also restores looping at the SEN1, BLM10, and HEM3 genes (Fig. 7). Thus, Ssl2 functionally interacts with TFIIB at the promoter, affecting transcription start site selection, and with TFIIB at the terminator, affecting juxtaposition of the terminator to the promoter to form gene loops.
Although we do not yet understand the molecular nature of the promoter-terminator interaction or how the 3′-end of a gene can affect start site selection, these effects are not entirely surprising or without precedent. Of particular interest, Jensen and colleagues (60) recently showed that 3′-end processing and termination of the β-globin gene stimulates transcription initiation in a manner involving recycling of Pol II and TFIIB. Although they did not assay looping at the β-globin locus, they speculate that physical interaction between the promoter and terminator regions could allow recycling by direct hand-off of Pol II and TFIIB from the terminator to the promoter (69).
There is also precedent for helicase function at Pol II terminators. Yeast Sen1 is a member of the SF1 family of RNA/DNA helicases and, like Ssl2, exhibits 3′–5′ unwinding activity. Sen1 is required for termination of small nucleolar RNAs and other short transcripts as a member of the Sen1-Nrd1-Nab3 complex and also functionally interacts with the Rat1 exonuclease to promote termination of polyadenylated Pol II transcripts (70–73). Sen1 resolves R-loops (DNA-RNA hybrids) that form in the wake of Pol II, thereby enabling the Rat1 exonuclease to degrade RNA and promote termination (74). A similar function has been attributed to senataxin, the mammalian Sen1 ortholog (75). Furthermore, senataxin depletion reduces Pol II occupancy over the β-actin promoter (75), a result that resonates with cross-talk between terminator and promoter of the β-globin gene (60). Whether the Ssl2 helicase might also be involved in resolution of R-loops and how this might be related to formation or maintenance of gene loops are subjects for future investigations.
Supplementary Material
Acknowledgments
We thank Tom Donahue, David Garfinkel, and Louise Prakash for strains and plasmids.
This work was supported, in whole or in part, by National Institutes of Health Grants GM39484 (to M. H.) and GM068887 (to Claire Moore (Tufts Medical School) and M. H.).

This article contains supplemental Fig. S1.
- Pol II
- RNA polymerase II
- TBP
- TATA-binding protein
- PIC
- preinitiation complex
- 3C
- chromosome conformation capture
- 5-FAA
- 5-fluoroanthranilic acid
- X-gal
- 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside
- uORF
- upstream ORF.
REFERENCES
- 1. Orphanides G., Lagrange T., Reinberg D. (1996) Genes Dev. 10, 2657–2683 [DOI] [PubMed] [Google Scholar]
- 2. Hahn S. (2004) Nat. Struct. Mol. Biol. 11, 394–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Woychik N. A., Hampsey M. (2002) Cell 108, 453–463 [DOI] [PubMed] [Google Scholar]
- 4. Parvin J. D., Sharp P. A. (1993) Cell 73, 533–540 [DOI] [PubMed] [Google Scholar]
- 5. Kim T. K., Ebright R. H., Reinberg D. (2000) Science 288, 1418–1422 [DOI] [PubMed] [Google Scholar]
- 6. Kim T. K., Lagrange T., Wang Y. H., Griffith J. D., Reinberg D., Ebright R. H. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 12268–12273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen H. T., Hahn S. (2004) Cell 119, 169–180 [DOI] [PubMed] [Google Scholar]
- 8. Chen B. S., Mandal S. S., Hampsey M. (2004) Biochemistry 43, 12741–12749 [DOI] [PubMed] [Google Scholar]
- 9. Chen H. T., Warfield L., Hahn S. (2007) Nat. Struct. Mol. Biol. 14, 696–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miller G., Hahn S. (2006) Nat. Struct. Mol. Biol. 13, 603–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Forget D., Langelier M. F., Thérien C., Trinh V., Coulombe B. (2004) Mol. Cell Biol. 24, 1122–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Robert F., Douziech M., Forget D., Egly J. M., Greenblatt J., Burton Z. F., Coulombe B. (1998) Mol. Cell 2, 341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Douziech M., Coin F., Chipoulet J. M., Arai Y., Ohkuma Y., Egly J. M., Coulombe B. (2000) Mol. Cell Biol. 20, 8168–8177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Venters B. J., Wachi S., Mavrich T. N., Andersen B. E., Jena P., Sinnamon A. J., Jain P., Rolleri N. S., Jiang C., Hemeryck-Walsh C., Pugh B. F. (2011) Mol. Cell 41, 480–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pinto I., Ware D. E., Hampsey M. (1992) Cell 68, 977–988 [DOI] [PubMed] [Google Scholar]
- 16. Pinto I., Wu W. H., Na J. G., Hampsey M. (1994) J. Biol. Chem. 269, 30569–30573 [PubMed] [Google Scholar]
- 17. Bangur C. S., Pardee T. S., Ponticelli A. S. (1997) Mol. Cell Biol. 17, 6784–6793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cho E. J., Buratowski S. (1999) J. Biol. Chem. 274, 25807–25813 [DOI] [PubMed] [Google Scholar]
- 19. Pardee T. S., Bangur C. S., Ponticelli A. S. (1998) J. Biol. Chem. 273, 17859–17864 [DOI] [PubMed] [Google Scholar]
- 20. Faitar S. L., Brodie S. A., Ponticelli A. S. (2001) Mol. Cell Biol. 21, 4427–4440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu W. H., Pinto I., Chen B. S., Hampsey M. (1999) Genetics 153, 643–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang D. Y., Carson D. J., Ma J. (2002) Nucleic Acids Res. 30, 3078–3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bushnell D. A., Westover K. D., Davis R. E., Kornberg R. D. (2004) Science 303, 983–988 [DOI] [PubMed] [Google Scholar]
- 24. Kostrewa D., Zeller M. E., Armache K. J., Seizl M., Leike K., Thomm M., Cramer P. (2009) Nature 462, 323–330 [DOI] [PubMed] [Google Scholar]
- 25. Liu X., Bushnell D. A., Wang D., Calero G., Kornberg R. D. (2010) Science 327, 206–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu X., Bushnell D. A., Silva D. A., Huang X., Kornberg R. D. (2011) Science 333, 633–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Freire-Picos M. A., Krishnamurthy S., Sun Z. W., Hampsey M. (2005) Nucleic Acids Res. 33, 5045–5052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sun Z. W., Hampsey M. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 3127–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ghazy M. A., Brodie S. A., Ammerman M. L., Ziegler L. M., Ponticelli A. S. (2004) Mol. Cell Biol. 24, 10975–10985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sun Z. W., Tessmer A., Hampsey M. (1996) Nucleic Acids Res. 24, 2560–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen B. S., Hampsey M. (2004) Mol. Cell Biol. 24, 3983–3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Egly J. M., Coin F. (2011) DNA Repair 10, 714–721 [DOI] [PubMed] [Google Scholar]
- 33. Tirode F., Busso D., Coin F., Egly J. M. (1999) Mol. Cell 3, 87–95 [DOI] [PubMed] [Google Scholar]
- 34. Singh B. N., Hampsey M. (2007) Mol. Cell 27, 806–816 [DOI] [PubMed] [Google Scholar]
- 35. Lainé J. P., Singh B. N., Krishnamurthy S., Hampsey M. (2009) Genes Dev. 23, 2604–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tan-Wong S. M., Wijayatilake H. D., Proudfoot N. J. (2009) Genes Dev. 23, 2610–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 38. Sherman F. (1991) Methods Enzymol. 194, 3–21 [DOI] [PubMed] [Google Scholar]
- 39. Rose M. D., Broach J. R. (1991) Methods Enzymol. 194, 195–230 [DOI] [PubMed] [Google Scholar]
- 40. Toyn J. H., Gunyuzlu P. L., White W. H., Thompson L. A., Hollis G. F. (2000) Yeast 16, 553–560 [DOI] [PubMed] [Google Scholar]
- 41. Guarente L. (1983) Methods Enzymol. 101, 181–191 [DOI] [PubMed] [Google Scholar]
- 42. Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. (1987) Gene 60, 237–243 [DOI] [PubMed] [Google Scholar]
- 43. Guzder S. N., Sung P., Bailly V., Prakash L., Prakash S. (1994) Nature 369, 578–581 [DOI] [PubMed] [Google Scholar]
- 44. Rothstein R. (1991) Methods Enzymol. 194, 281–301 [DOI] [PubMed] [Google Scholar]
- 45. Pinto I., Na J. G., Sherman F., Hampsey M. (1992) Genetics 132, 97–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Komarnitsky P., Cho E. J., Buratowski S. (2000) Genes Dev. 14, 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ahn S. H., Kim M., Buratowski S. (2004) Mol. Cell 13, 67–76 [DOI] [PubMed] [Google Scholar]
- 48. Chang C. R., Wu C. S., Hom Y., Gartenberg M. R. (2005) Genes Dev. 19, 3031–3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dekker J. (2006) Nat. Methods 3, 17–21 [DOI] [PubMed] [Google Scholar]
- 50. Dekker J., Rippe K., Dekker M., Kleckner N. (2002) Science 295, 1306–1311 [DOI] [PubMed] [Google Scholar]
- 51. Singh B. N., Ansari A., Hampsey M. (2009) Methods 48, 361–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fan L., Arvai A. S., Cooper P. K., Iwai S., Hanaoka F., Tainer J. A. (2006) Mol. Cell 22, 27–37 [DOI] [PubMed] [Google Scholar]
- 53. Greger I. H., Proudfoot N. J. (1998) EMBO J. 17, 4771–4779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Exinger F., Lacroute F. (1992) Curr. Genet. 22, 9–11 [DOI] [PubMed] [Google Scholar]
- 55. Powell W., Reines D. (1996) J. Biol. Chem. 271, 6866–6873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee B. S., Lichtenstein C. P., Faiola B., Rinckel L. A., Wysock W., Curcio M. J., Garfinkel D. J. (1998) Genetics 148, 1743–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gulyas K. D., Donahue T. F. (1992) Cell 69, 1031–1042 [DOI] [PubMed] [Google Scholar]
- 58. Sweder K. S., Hanawalt P. C. (1994) J. Biol. Chem. 269, 1852–1857 [PubMed] [Google Scholar]
- 59. Mavrich T. N., Ioshikhes I. P., Venters B. J., Jiang C., Tomsho L. P., Qi J., Schuster S. C., Albert I., Pugh B. F. (2008) Genome Res. 18, 1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mapendano C. K., Lykke-Andersen S., Kjems J., Bertrand E., Jensen T. H. (2010) Mol. Cell 40, 410–422 [DOI] [PubMed] [Google Scholar]
- 61. O'Sullivan J. M., Tan-Wong S. M., Morillon A., Lee B., Coles J., Mellor J., Proudfoot N. J. (2004) Nat. Genet. 36, 1014–1018 [DOI] [PubMed] [Google Scholar]
- 62. Ansari A., Hampsey M. (2005) Genes Dev. 19, 2969–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Singleton M. R., Dillingham M. S., Wigley D. B. (2007) Annu. Rev. Biochem. 76, 23–50 [DOI] [PubMed] [Google Scholar]
- 64. Fairman-Williams M. E., Guenther U. P., Jankowsky E. (2010) Curr. Opin. Struct. Biol. 20, 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bushnell D. A., Kornberg R. D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 6969–6973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Armache K. J., Kettenberger H., Cramer P. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 6964–6968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kapanidis A. N., Margeat E., Ho S. O., Kortkhonjia E., Weiss S., Ebright R. H. (2006) Science 314, 1144–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kuehner J. N., Brow D. A. (2006) J. Biol. Chem. 281, 14119–14128 [DOI] [PubMed] [Google Scholar]
- 69. Lykke-Andersen S., Mapendano C. K., Jensen T. H. (2011) Cell Cycle 10, 863–865 [DOI] [PubMed] [Google Scholar]
- 70. Kawauchi J., Mischo H., Braglia P., Rondon A., Proudfoot N. J. (2008) Genes Dev. 22, 1082–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Steinmetz E. J., Brow D. A. (1996) Mol. Cell Biol. 16, 6993–7003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Steinmetz E. J., Warren C. L., Kuehner J. N., Panbehi B., Ansari A. Z., Brow D. A. (2006) Mol. Cell 24, 735–746 [DOI] [PubMed] [Google Scholar]
- 73. Vasiljeva L., Kim M., Mutschler H., Buratowski S., Meinhart A. (2008) Nat. Struct. Mol. Biol. 15, 795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mischo H. E., Gómez-González B., Grzechnik P., Rondón A. G., Wei W., Steinmetz L., Aguilera A., Proudfoot N. J. (2011) Mol. Cell 41, 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Skourti-Stathaki K., Proudfoot N. J., Gromak N. (2011) Mol. Cell 42, 794–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.