Background: Hepatitis C virus replication requires that non-structural (NS) proteins be derived from a cleavable polyprotein precursor.
Results: Increasing the rate of processing at an NS cleavage boundary inhibits replication.
Conclusion: Some transient NS precursors have a time-dependent function.
Significance: The findings are a key reference point for future investigations establishing NS precursor function.
Keywords: Hepatitis C Virus, Viral Genetics, Viral Protease, Viral Protein, Viral Replication
Abstract
In hepatitis C virus, non-structural proteins are cleaved from the viral polyprotein by viral encoded proteases. Although proteolytic processing goes to completion, the rate of cleavage differs between different boundaries, primarily due to the sequence at these positions. However, it is not known whether slow cleavage is important for viral replication or a consequence of restrictions on sequences that can be tolerated at the cleaved ends of non-structural proteins. To address this question, mutations were introduced into the NS4B side of the NS4B5A boundary, and their effect on replication and polyprotein processing was examined in the context of a subgenomic replicon. Single mutations that modestly increased the rate of boundary processing were phenotypically silent, but a double mutation, which further increased the rate of boundary cleavage, was lethal. Rescue experiments relying on viral RNA polymerase-induced error failed to identify second site compensatory mutations. Use of a replicon library with codon degeneracy did allow identification of second site compensatory mutations, some of which fell exclusively within the NS5A side of the boundary. These mutations slowed boundary cleavage and only enhanced replication in the context of the original lethal NS4B double mutation. Overall, the data indicate that slow cleavage of the NS4B5A boundary is important and identify a previously unrecognized role for NS4B5A-containing precursors requiring them to exist for a minimum finite period of time.
Introduction
Hepatitis C virus (HCV),3 a member of the flaviviridae family, is a positive sense, single-stranded RNA virus with a genome of 9.6 kb containing a single ORF that is flanked by 5′- and 3′-UTRs. The first third of the viral ORF encodes for both structural proteins (core E1 and E2) and a viroporin-like ion channel (p7), whereas the latter two-thirds encodes for non-structural proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) (1–3). Release and processing of the structural proteins from the viral polyprotein is brought about by the action of host cell signal peptidases and signal peptide peptidase (3–6), but all NS boundaries are cleaved by virus-encoded proteases. Cleavage of the first NS protein boundary at the NS23 junction is an autocatalytic event directed by the NS2/3 protease (3, 4, 7, 8). Cleavage of all remaining downstream NS protein boundaries is mediated by the viral protease, NS3, in conjunction with its co-factor NS4A (8–11). This latter set of cleavage events is responsible for release of the NS protein components that go on to form replication complexes.
Although NS3/4A-mediated cleavage of the HCV polyprotein goes to completion and does not result in the formation of stable precursor intermediates, boundaries are cleaved at different rates. Processing at the NS34A junction is extremely rapid, occurs in cis, precedes all other NS3/4A-dependent cleavage events, and is required for correct association of the NS3 protease with its co-factor sequence in NS4A (12, 13). Cleavage at the NS5A5B boundary is also very rapid. However, the remaining two boundaries flanking NS4B are cleaved less rapidly, leading to formation of detectable transient precursors NS4A4B, NS4A4B5A, and NS4B5A (9, 11, 14). The rate of cleavage at each boundary is thought to be dictated by the primary amino acid sequence at these same locations (12, 15–19), but whether there is a need for slowly processed boundaries flanking NS4B is unclear. One possibility is that slow cleavage reflects the functional importance of the N- and C-terminal sequences at these boundaries to the mature proteins generated from the longer precursor proteins. Alternatively, NS4B-containing precursors may possess an activity or activities necessary for replication that are not supplied by their mature, cleaved products, and the delay in boundary cleavage provides sufficient time for these functions to operate. Evidence to support such a hypothesis is currently limited. Certainly, an inability to trans-complement most but not all genetic defects in HCV NS proteins indicates the importance of expressing NS proteins from a functional precursor and a replication-competent RNA transcript (20–23). Because some functions of NS5A can only be complemented in trans by expression of an NS3–5A precursor (20), it is conceivable that NS4B5A-containing precursors might have a functional role. Alternatively, HCV NS proteins may need to be expressed as a polyprotein precursor to ensure a high localized concentration of viral protein, akin to models proposed for enterovirus replication (24). There is at least a precedent for a role for transient polyprotein precursors in the replication of other positive strand RNA viruses, given that the substrate for poliovirus 3B (VpG) uridylation is a transient 3BC-containing precursor (25).
Here, we wished to test whether delayed cleavage between NS4B and NS5A contributes to replication. To achieve this, specific mutations on the NS4B side of the NS4B5A boundary were introduced to examine the relationship between the rate of boundary cleavage and replication. Should changes be identified that were detrimental to replication but enhanced boundary cleavage, it was anticipated that the location of compensatory mutations might provide insight into the nature of the selective pressures imposed by the original lethal mutations. Results suggest that transient NS4B5A precursors do have a distinct role in replication, which extends beyond the minimum requirements that can be provided to the virus by expression of NS proteins in cis.
EXPERIMENTAL PROCEDURES
Cell Culture
Huh7 and Huh7.5 cells were kindly provided by Ralf Bartenschlager (University of Heidelberg) and Charles Rice (Rockefeller University, New York). HepG2 cells and Spodoptera frugiperda (Sf9) insect cells were obtained from Mark Harris (University of Leeds). All cells were maintained as described previously (26, 27).
Baculovirus Production
Baculovirus was produced using the Bac-to-Bac system (Invitrogen) in combination with pFB-based transfer vectors as described previously (27).
Plasmid Constructs
Details relating to the generation of constructs and the sequence of primers are provided as supplemental material.
Western Blot Analysis
This was carried out as described previously (27, 28). Primary antibodies used were sheep anti-NS3, sheep anti-NS5A, sheep anti-NS5B (all gifts from Mark Harris, University of Leeds), and rabbit anti-NS4B (22). Donkey anti-sheep and goat anti-rabbit HRP conjugates (Sigma) were employed as secondary reagents.
Pulse-Chase Analysis
HepG2 cells were seeded in 6-well dishes precoated with rat tail collagen at a density of 4 × 104 cells/cm2 and allowed to recover for 24 h. Transduction was performed at a concentration of 1.25 × 107 pfu/ml of both BACtTA (26) and the relevant replicon-containing baculovirus for 4 h. Cells were left a further 15 h before incubating at 37 °C in Met−/Cys− serum-free DMEM (Invitrogen) supplemented with 25 mm HEPES. After 1 h, the medium was replaced with Met−/Cys− serum-free DMEM supplemented with 25 mm HEPES and 200 μCi/ml Trans-label (MP Biomedical), and the cells were incubated at 37 °C for 15 min. They were then left in complete growth medium at 37 °C for defined periods of between 0 to 60 min before lysis in RIPA buffer (0.5 ml/well) supplemented with 2× complete protease inhibitor, 1 mm Na3VO4, and 1 mm NaF. Radiolabeled supernatants were recovered after centrifugation at 14,000 × g for 10 min at 4 °C and stored at −80 °C. To immunoprecipitate FLAG-tagged NS5A, 0.4 ml of lysate was precleared for 1 h using 10 μl of protein A/G-agarose beads (Thermo Scientific). 4 μl of M2 anti-FLAG mAb (Sigma) was added the precleared supernatant, which was left for 30 min on ice before the addition of a further 20 μl of protein A/G-agarose beads. After incubation for 2 h at 4 °C, the beads were pelleted, washed four times using RIPA buffer, and boiled in 20 μl of 2× Laemmli buffer to release bound proteins.
In Vitro Cleavage Assay
The protocol used is comparable with that described in other reports (29, 30). Briefly, DNA encoding for a truncated NS4B5A substrate, 291 amino acids in length and representing HCV Con1 residues 1758–2048, was generated from the relevant replicon-containing vector by PCR using primers 4B/5A(prec_fwd) and 4B/5A(prec_rev). This served as a template for production of a 5′ capped RNA transcript using the Maxiscript kit (Promega) according to the manufacturer's recommendations. To generate radiolabeled NS4B5A substrate, 1 μg of RNA was introduced into a 20-μl reaction containing 12.5 μl of rabbit reticulolysate (Promega), 0.5 μl of 1 mm amino acids (Met−), 0.5 μl of [35S]methionine (>1000 Ci/mmol at 10 mCi/ml), and 0.5 μl RiboLock RNase inhibitor (Fermentas). The reaction was left for 2 h at 30 °C before being terminated by the addition of 0.5 μl of 20 mg/ml RNase and 0.5 μl of 0.5 mg/ml cycloheximide. Cleavage of the NS4B5A substrate was then performed over 2 h at 30 °C using 1 μl of the translation reaction in a 10-μl reaction supplemented with 1× cleavage buffer (25 mm Tris/HCl, pH 7.5, 300 mm NaCl, 0.05% (w/v) dodecyl maltoside, 0.05 mm tris(2-carboxyethyl)phosphine, 10% (v/v) glycerol) and 10 μg/ml single chain genotype 1b NS3/4A (AnaSpec). The reaction was stopped by the addition of 10 μl of 2× Laemmli loading buffer.
Colony Forming and Luciferase Replicon Assays
Replicon transcripts were prepared as described previously, as was their introduction into cells via electroporation (27). For colony formation assays, viable electroporated cells were seeded at a range of concentrations between 103 and 105 cells/well of a 6-well dish and supplemented with non-electroporated cells, so the total cell density was 2 × 104 cells/cm2. After a 48-h recovery, G418 was added to a final concentration of 750 μg/ml G418, and selection continued for a further 2–3 weeks with twice weekly changes of medium. For luciferase assays, ∼1.5 × 105 viable cells were seeded per well of a 6-well plate, and enzyme activity was measured using a luciferase assay system (Promega) with the Lumat LB 9507 luminometer (Berthold).
Amplification and Sequencing of HCV NS Coding Region
Total RNA was extracted from replicon cell lines using TRIzol (Invitrogen). cDNA was generated from 1 μg of this RNA using SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's guidelines. For all clonal cell lines, a DNA fragment representing nucleotides 6029–6688 of the HCV Con1 genome was amplified by PCR using primers 4B5Ajct(fwd) + 4B5Ajct(rvs) and sequenced across the NS4B5A boundary using primer NS5A(Mlu)rep. For amplification of NS3/4A, primers FwdseqNS3rep + Revseq4Arep were used, and the resultant product was sequenced using primers Int_4A_seq, Int_prot_seq, Con1(5082–5063), and 5.1(3940–3959).
PCR amplification of a product encompassing the NS4B5A boundary (nucleotides 6096–6582) from the polyclonal cell line was performed using Phusion polymerase (New England Biolabs) and primers NS4B(Bst)fwd and NS5A(Mlu)rev. The DNA was ligated into pCR-Blunt (Invitrogen), and individual clones were sequenced using the primer M13(Rev).
Fluorescent Microscopy
Huh7.5 cells seeded onto glass coverslips were transduced with baculovirus constructs encoding NS4B-green fluorescent protein (GFP), NS4B5A-GFP, or NS5A-GFP at 2 × 107 pfu/ml for 4 h. Cells were left a further 16 h and then fixed with 4% paraformaldehyde and counterstained with 0.5 nm TO-PRO-3 (Invitrogen). Coverslips were mounted onto slides with ProLongGold (Invitrogen). Images were captured using a Leica TCP SP5 confocal microscope.
RESULTS
Amino Acid Sequence at NS4B5A Boundary Represents Suboptimal Recognition Site for HCV NS3/4A Protease
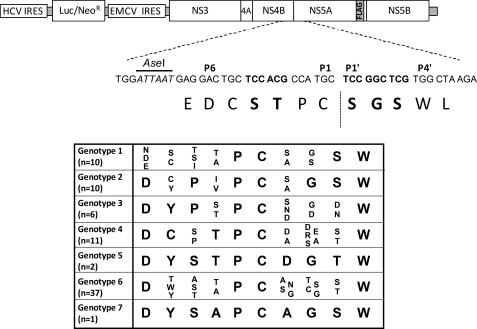
Previous studies using peptides derived from the HCV NS boundaries of genotype 1 isolates have demonstrated that the NS4B5A boundary is less efficiently cleaved by NS3/4A compared with other boundaries (12, 15–19). Changes to specific amino acid residues within the boundary sequence that may lead to more efficient proteolysis include those at the P3 and P4 positions because conversion to hydrophobic residues, such as Val, at these positions enhances cleavage of NS4B5A-based peptides (16). Sequence analysis both between and within genotypes suggests that HCV can tolerate considerable amino acid variation at the P3 and P4 positions within the NS4B5A boundary, yet a double Val motif has never been observed, although Ile and Val do sometimes occur in one or the other of these two positions (see table in Fig. 1). Assuming that in vitro data with peptides accurately reflects NS3/4A cleavage efficiency at polyprotein boundary sequences, one possible reason for these observations is that there is selective pressure on the virus to avoid rapid cleavage of the NS4B5A boundary. Alternatively, certain combinations of amino acids at the P3 and P4 positions may result in loss of NS4B function.
FIGURE 1.
Schematic representation of the bicistronic HCV replicon constructs used in study. Shown is the genotype 1 [FA] subgenomic replicon, which contains two internal ribosome entry sites (IRES) and possesses a FLAG tag sequence at the C-terminal end of NS5A. The nucleotide and amino acid sequence found at the NS4B5A boundary is displayed, with those residues targeted for mutation marked in boldface type. The inserted table depicts amino acid variability at the NS4B5A boundary based across a reference set of HCV genome sequences found at the Viral Bioinformatics Resource Center Web site.
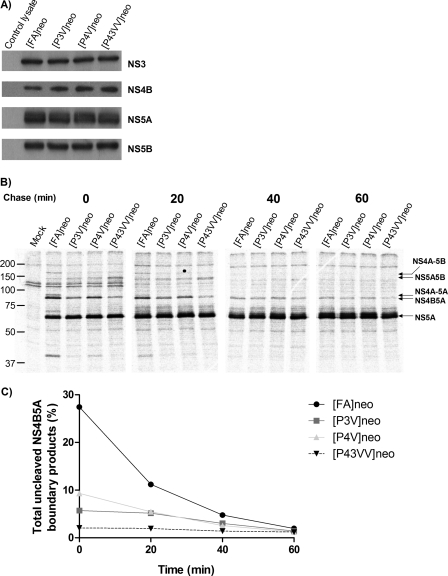
To examine these two possibilities, it was first necessary to establish the consequences of introducing Val residues at the P3 and/or P4 positions of the NS4B5A boundary to polyprotein processing. Hence, we modified a culture-adapted bicistronic genotype 1b replicon (FK5.1neo), which expressed neomycin phosphotransferase, by introducing a FLAG tag sequence into a region of NS5A known to tolerate insertions; this strategy generated a construct from which NS5A could be readily immunoprecipitated. In addition, a unique AseI site that did not alter the encoded amino acid sequence was introduced to facilitate site-directed mutagenesis of the NS4B5A boundary (Fig. 1). The resultant replicon construct [FA]neo was further altered by introduction of Val codons at the P3 position [P3V]neo, the P4 position [P4V]neo or both the P3 and P4 positions [P43VV]neo at the NS4B5A boundary. To express detectable levels of NS protein in the absence of replication, all four replicons were introduced into HepG2 cells using baculovirus transduction. After transduction for 16 h to allow expression of the NS proteins, Western blot analysis revealed that cell lysates contained comparable levels of NS3, NS4B, NS5A, and NS5B for all constructs (Fig. 2A). Levels of NS5A hyperphosphorylation were also similar between different experimental groups. From these data, we concluded that polyprotein processing reached completion, irrespective of modifications to the NS4B5A boundary. Moreover, the modifications did not appear to reduce stability of either NS4B or the other NS proteins or lead to any noticeable change in NS5A hyperphosphorylation. Using the baculovirus delivery system, pulse-chase analysis was utilized to investigate the effect of mutations on the rate of NS4B5A boundary processing. In these experiments, HepG2 cells were metabolically labeled for 15 min and then incubated for a further 60 min following removal of radioactivity. NS5A and NS5A-containing precursors were immunoprecipitated from cell lysates with an anti-FLAG mAb, separated by SDS-PAGE, and quantified by PhosphorImager analysis (Fig. 2, B and C). In [FA]neo-transduced cells, there were considerable amounts of polyprotein products with an uncleaved NS4B5A boundary at the start of the chase period (28% of total NS5A-containing proteins), which declined to a base-line level of 3% over the course of the following 60 min. By comparison, the proportion of NS4B5A-containing products in cells transduced with the [P3V]neo and [P4V]neo baculovirus constructs was lower compared with [FA]neo at all time points except for the final time point at the end of the chase period, demonstrating that the single Val substitutions resulted in similar and detectable increases in processing rates at the NS4B5A boundary. In [P43VV]neo-transduced cells, the abundance of NS4B5A-containing products was even lower than that seen in [P3V]neo or [P4V]neo-transduced cells and was effectively at base-line levels for the entire chase period. Interestingly, there appeared to be a slight transient increase in the amount of NS5A5B precursor present during the earlier time points in [P43VV]neo-transduced cells. However, the key finding was that single valine substitutions at the P3 and P4 positions enhanced the rate of cleavage at the NS4B5A boundary, but the presence of Val residues at both positions further increased the rapidity of cleavage.
FIGURE 2.
Analysis of polyprotein processing in [FA]neo, [P3V]neo, [P4V]neo, and [P43VV]neo constructs. A, Western blot of lysates from HepG2 cells transduced with baculovirus vectors engineered to deliver transcripts of [FA]neo, [P3V]neo, [P4V]neo, and [P43VV]neo, under the control of a tetracycline-responsive promoter. Promoter activity required co-transduction with BACtTA. A control lysate from cells transduced with BACtTA only was included. Lysates were probed for expression of HCV proteins (NS5B, NS5A, NS4B, and NS3) using appropriate antisera. B, HepG2 cells were transduced with the same baculovirus constructs as in A and used for a pulse-chase experiment where [35S]Met/Cys was used to label cells for 15 min, and cell lysates were then harvested 0, 20, 40, and 60 min later. NS5A-FLAG-containing precursors were immunoprecipitated by anti-FLAG mAb, separated on 10% SDS-PAGE, and visualized by a PhosphorImager. The positions of different HCV polyprotein products are indicated by arrows. Mock lysate comes from BACtTA-only-transduced cells. C, graphical representation of the data from B showing the proportion of uncleaved NS4B5A precursor products in cells over time and expressed as a percentage of total NS5A-containing products.
Mutations that Promote Rapid Cleavage of NS4B5A Boundary Suppress Replication
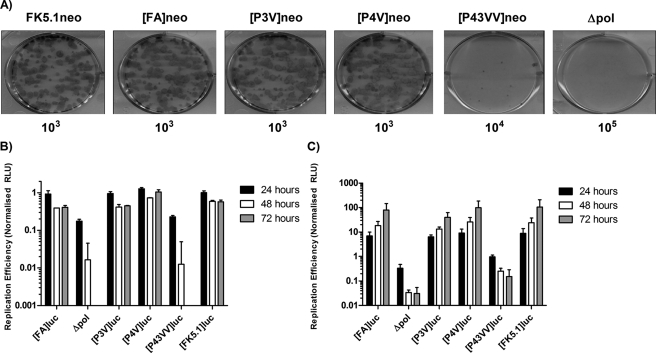
Having confirmed that Val residues at the P3 and P4 positions increased the rate of NS4B5A cleavage, we examined the effect of the mutations on HCV RNA replication. All four neomycin phosphotransferase-expressing replicon constructs were electroporated into Huh7 cells, and the cells were maintained under G418 selection for a period of 3 weeks to allow formation of replicon-containing colonies. Additional experimental groups included Huh7 cells transfected with FK5.1neo, the original construct from which [FA] had been derived, and a polymerase-defective replicon construct as a negative control (Fig. 3A). The efficiency of colony formation was similar for both FK5.1neo and [FA]neo, each resulting in ∼100 G418-resistant colonies in dishes seeded with 103 transfected cells. Therefore, the modifications that had been introduced into [FA]neo, including the introduction of a FLAG tag sequence in NS5A, did not appear to affect replication. There was also no difference in colony formation between [P3V]neo and [P4V]neo compared with [FA]neo-transfected cells, suggesting that both the P3→V and P4→V mutations had no impact on replication, despite both mutations enhancing the rate of cleavage at the NS4B5A boundary. In contrast, transfection of [P43VV]neo, which had the most rapidly cleaved NS4B5A boundary, yielded ∼1000-fold fewer colonies compared with [FA]neo, indicating that the presence of Val at both the P3 and P4 positions severely impaired replication. However, there were more colonies with the [P43VV]neo construct than observed for cells transfected with the polymerase-defective replicon, suggesting that limited replication did occur.
FIGURE 3.
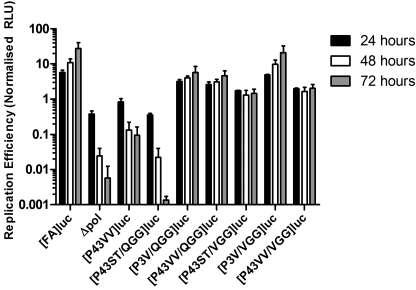
The effect of mutations that increase the rate of NS4B5A boundary cleavage on HCV RNA replication. A, Huh7 cells were electroporated with neomycin-expressing replicons [FA]neo, [P3V]neo, [P4V]neo, and [P43VV]neo. Additional experimental groups included a replication-defective replicon transcript containing a GDD→GND mutation in the polymerase active site (Δpol) and the parental FK5.1neo replicon lacking a FLAG tag sequence in NS5A. Cells were plated at 10-fold serial dilutions into the wells of a 6-well plate and selected for 2 weeks with 750 μg/ml G418 before colony formation was assessed. Images show representative wells after seeding 103 ([FK5.1]neo, [FA]neo, [P3V]nep, and [P4V]neo), 104 ([P43VV]luc), or 105 (Δpol) electroporated cells/well, as indicated below the image. B, Huh7 cells were electroporated with replicon transcripts FK5.1luc, [FA]luc, [P3V]luc, [P4V]luc, and [P43VV]luc. A luciferase-expressing polymerase knock-out control transcript (Δpol) was also included. Cell lysates were prepared at 4, 24, 48, and 72 h postelectroporation, and luciferase activity was determined. Data shown represent mean luciferase activity (n = 2, ± S.D.) at 24, 48, and 72 h normalized to the 4 h time point. C, the same experiment in B repeated using Huh7.5 cells (n = 2); error bars, S.D.
To confirm these observations in transient replication assays, the neomycin phosphotransferase gene in the replicon constructs was exchanged for firefly luciferase, and the resultant constructs were transfected into Huh7 cells (Fig. 3B). For all constructs, levels of luciferase activity were comparable at 4 h post-transfection, a time point that represents translation resulting from input RNA (data not shown). Luciferase activity produced by the polymerase-defective construct rapidly dropped after this point, becoming undetectable by 48 h. By contrast, luciferase activity declined less rapidly and reached a plateau between 24 and 72 h in FK5.1luc-, [FA]luc-, [P3V]luc-, and [P4V]luc-transfected cells, indicating that all of these constructs replicated to a similar extent. For [P43VV]luc, the pattern of luciferase activity closely matched that for the polymerase-defective control, thereby confirming that the double Val substitution blocked replication. Replication competence was also assessed in Huh7.5 cells, derivatives of Huh7 cells that support greater levels of HCV RNA synthesis. All of the constructs gave a similar pattern in these cells compared with Huh7 cells, although some residual replication was detected for [P43VV]luc (Fig. 3C). That the P43VV mutation was detrimental for replication was also established using the JFH-1 genotype 2a replicon (see supplemental Fig. 1). Furthermore, an in cellulo NS4B5A cleavage assay indicated that the JFH-1 protease exhibited a preference for hydrophobic residues (e.g. Val and Iso) at the P3 and P4 positions (supplemental Fig. 2), consistent with what has also been observed at the NS5A5B boundary (31). From both the luciferase and colony-forming assays, we concluded that the P43→VV mutation, which gives rapid cleavage of the NS4B5A boundary, severely impairs replication. By contrast, individual Val mutations at P3 and P4 result in a modest increase in boundary cleavage and have negligible impact on replication.
Presence of P43→VV Mutation in NS4B5A Boundary Results in Strong Selective Pressures for Further Modification at These Sites
Although [P43VV]neo gave barely detected replication, it was possible to isolate neomycin-resistant colonies, which could be expanded as stable replicon cell lines. Expansion of these colonies enabled analysis of [P43VV]neo replicons for additional mutations that might overcome their defect in replication. Ten G418-resistant colonies were recovered from [P43VV]neo-transfected cells, and for each clone, the segment encompassing the NS4B5A boundary coding region was amplified by RT-PCR. The sequences of these PCR products were determined across a region that represents nucleotides 6111–6492 of the HCV Con1 genome, which includes the NS4B5A boundary region. All of the clones contained a single nucleotide substitution, which resulted in loss of the Val residue at either the P3 or P4 position, but no other mutations were found within the sequenced region (Table 1). Interestingly, we found considerable flexibility in the amino acids that could be tolerated at the P3 and P4 positions. At the P4 position, two (Ala and Asp) of a possible six (Ala, Asp, Phe, Leu, Ile, and Gly) single nucleotide coding changes were observed, whereas at the P3 position, four (Leu, Met, Ala, and Glu) of a possible five (Leu, Met, Ala, Glu, and Gly) were encountered. Although sequences at other segments of the NS coding region were not determined, our findings strongly indicated that either one of the Val residues at P3 and P4 at the NS4B5A boundary had to mutate for replication to be restored. Indeed, introduction of one of these clonal sequences (P43→AV) into the [FA]luc replicon did not impair replication (see Fig. 4), leading us to conclude that further adaptive changes were not necessary to restore replication competence to the [P43VV] replicon.
TABLE 1.
Amino acid sequence of NS4B5A boundaries from replicon clones derived from transfection of Huh7 cells with [P43VV]neo
| Clone | Sequence |
|---|---|
| 1 | DCVAPCSGSWLR |
| 2 | DCVAPCSGSWLR |
| 3 | DCAVPCSGSWLR |
| 4 | DCAVPCSGSWLR |
| 5 | DCVLPCSGSWLR |
| 6 | DCVLPCSGSWLR |
| 7 | DCVEPCSGSWLR |
| 8 | DCVEPCSGSWLR |
| 9 | DCDVPCSGSWLR |
| 10 | DCVMPCSGSWLR |
FIGURE 4.
Replication of constructs with potential P43VV second site compensatory mutations. Huh7.5 cells were electroporated with replicons [FA]luc and [P43VV]luc as well as luciferase-expressing replicons containing the P43VV mutation in addition to further alterations at the NS4B5A boundary. Both a polymerase-defective construct (Δpol) and a construct containing a P4V→A alteration ([P43AV]luc) that had been identified in initial P43VV rescue experiments (see Table 1) were included. Cell extracts were harvested at 4, 24, 48, and 72 h postelectroporation, and luciferase activity was determined. Data shown represent mean luciferase activity (n = 2; error bars, S.D.) at 24, 48, and 72 h, normalized to luciferase activity at the 4 h time point.
Changes to N Terminus of NS5A Preserve P43→VV Mutation at NS4B5A Boundary in Stable Replicon Cell Lines
The above data showed that reliance on the viral RNA polymerase was not an effective approach to identify mutations capable of generating compensatory mutations outside of the P3 and P4 positions at the NS4B5A boundary. Because we wished to discriminate between the effect of the P43→VV mutations on NS4B function and whether delayed cleavage at the NS4B5A boundary was important for efficient replication, amino acid changes were introduced on the P′ side of the boundary (i.e. within NS5A), and their ability to act as second site compensatory mutations was tested. A random PCR mutagenesis approach was adopted to construct a replicon library with Val residues at both the P3 and P4 positions but nucleotide degeneracy at the P1′, P2′, and P3′ coding positions of the NS4B5A boundary. This strategy was employed because there is considerable diversity at these three positions across different genotypes, and the P1′ position is thought to influence cleavage kinetics (17, 18). RNA transcripts from this library were electroporated into Huh7 cells, along with appropriate controls, and selected with G418. Following selection, the number of colonies recovered from cells transfected with the library transcripts was ∼10,000-fold less than that of cells transfected with the control [FA]neo transcript. Nonetheless, colony formation was 5-fold greater in cells transfected with the degenerate library transcripts than in those transfected with [P43VV]neo, suggesting selection of compensatory mutations in the P1′ to P3′ coding region. Nine colonies were selected to create individual clonal cell lines. RNA from each clonal line was amplified by RT-PCR to generate DNA fragments possessing an NS4B5A boundary consensus sequence, and the fragments were directly sequenced. The remaining G418-resistant colonies were pooled and maintained as a polyclonal cell line. DNA fragments encoding the NS4B5A boundary sequences in this pool were also amplified, but in this instance, PCR fragments were cloned into a plasmid vector, and sequences from individual clones were determined.
Analysis of the clonal (Table 2) and polyclonal (Table 3) DNA fragments indicated that the NS4B5A boundary sequences could be broadly categorized into three groups. The first group contained those that had lost one of the two Val residues at either the P3 or P4 position, irrespective of other changes to the boundary sequence. This group accounted for one (clone 5) of nine sequences obtained from the clonal lines and 6 of 25 plasmids from the polyclonal cell line. The second group consisted of sequences that retained Val residues at both the P3 and P4 positions (8 of 9 sequences from the clonal lines; 17 of 25 sequences from the polyclonal line). This second group had an altered coding region at the P1′–P3′ positions but possessed no additional changes within the boundary sequence. Although no strict consensus sequence was found among these clones, there was a preference for amino acids with larger side chains at the P1′ position and for a Gly residue at the P3′ position. The final group also maintained the two Val residues at the P3 and P4 positions, had an alternative coding region at the P1′–3′ positions, and included further coding changes in the boundary sequence, specifically P6 Asp → Gly and P2 Pro → Ser mutations (Table 3); this group was exclusively obtained from the polyclonal cell line. We consider it unlikely that these sequences arise from PCR-induced errors because only two additional silent mutations and one additional coding mutation were found in all 25 sequences across a stretch of 486 nucleotides (positions 6096–6582 of the Con1 genome). The one additional coding mutation, representing a Q1946R coding change within the C-terminal amphipathic helix of NS4B, was found on investigation not to compensate for the P43→VV mutation (data not shown). Overall, this strategy indicated that maintenance of Val residues at both the P3 and P4 positions of the NS4B5A boundary was possible but relied on coding changes to the NS5A side of the boundary.
TABLE 2.
Amino acid sequence of NS4B5A boundaries from replicon clones derived from transfection of Huh7 cells with a [P43VV]neo-based replicon library with nucleotide degeneracy at the P1′–P3′ positions
| Clone | Sequence |
|---|---|
| 1 | DCVVPCKVDWLR |
| 2 | DCVVPCVGGWLR |
| 3 | DCVVPCVCTWLR |
| 4 | DCVVPCDGIWLR |
| 5 | DCVMPCKVDWLR |
| 6 | DCVVPCREGWLR |
| 7 | DCVVPCRNLWLR |
| 8 | DCVVPCVGGWLR |
| 9 | DCVVPCAGLWLR |
TABLE 3.
Results from analysis of the NS4B5A boundary sequences obtained by sequencing the replicon polyclonal library derived from transfection of Huh7 cells with a [P43VV]neo-based replicon library with nucleotide degeneracy at the P1′–P3′ positions
| P1′–P3′ coding sequence | Number of clones | Number of alternative nucleotide sequences encoding P1′–P3′ positions | Different boundary sequences observed across the P6–P4′ region |
|---|---|---|---|
| QGG | 1 | 1 | DCVVPC QGGW |
| VGG | 2 | 2 | DCVVPC VGGW |
| HLG | 3 | 1 | DCVVPC HLGW |
| RVG | 6 | 1 | DCVVPC RVGW |
| RNL | 8 | 1 | DCVVPC RNLW |
| DCDVPC RNLW | |||
| DCAVPC RNLW | |||
| DCVEPC RNLW | |||
| QAE | 2 | 1 | DCVVSC QAEW |
| DCAVPC QAEW | |||
| GQN | 2 | 1 | DCAVPC GQNW |
| TNI | 1 | 1 | GCVVPC TNIW |
Alterations to NS5A Side of NS4B5A Boundary Compensate for Presence of Val at both P3 and P4 Positions in Replication Assays
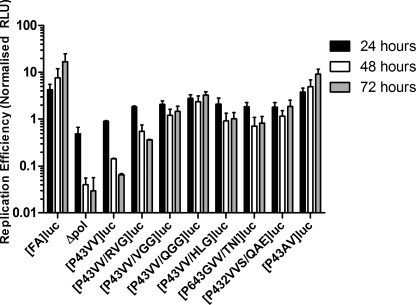
The above data suggested that coding alterations outside the P3 and P4 positions of the NS4B5A boundary compensated for the lethal P43→VV mutation. To confirm that these mutations could rescue the P43→VV replicon, boundary sequences from six plasmid clones, obtained from the polyclonal cell lines ([P43VV/QGG]luc, [P43VV/VGG]luc, [P43VV/HLG]luc, [P43VV/RVG]luc, [P432VVS/QAE]luc and [P643GVV/TNI]luc), were introduced into the [FA]luc replicon. RNA transcripts generated from these plasmids were electroporated into Huh7.5 cells along with [FA]luc, [P43VV]luc, a polymerase knock-out control replicon transcript, and [P43AV]luc (a coding change identified in the initial rescue experiment using [P43VV]neo-transfected cells; see Table 1), and luciferase activities were monitored over 72 h (Fig. 4).
Consistent with previous experiments, replication of [P43VV]luc was severely impaired compared with [FA]luc, indicated by a 200-fold difference in luciferase activity between the two constructs after 72 h. The luciferase activities from the constructs containing NS4B5A boundary sequences obtained from the selection experiment were much higher than those observed with [P43VV]luc at all time points after 4 h, although the enzyme levels varied between constructs (Fig. 4). From these results, we concluded that additional changes in the NS4B5A boundary sequence could compensate for the deleterious P43→VV mutation. Among the compensatory sequences, [P43VV/QGG]luc replicated with the highest efficiency, luciferase activity being only 5-fold lower than [FA]luc at 72 h post-transfection. Replication of [P43VV/VGG]luc, [P43VV/HLG]luc, [P432VVS/QAE]luc, and [P643GVV/TNI]luc was only slightly less, at ∼10-fold lower than [FA]luc. By comparison, replication of [P43VV/RVG]luc was the least robust, at 28-fold lower compared with [FA]luc-transfected cells. Thus, sequences on the NS5A side of the NS4B5A boundary could rescue replication of the P43→VV mutant, suggesting that the presence of Val residues at positions P3 and P4 in NS4B are unlikely to disrupt the function of NS4B.
Second Site Compensatory Mutations Reduce Rate of Cleavage at NS4B5A Boundary by Viral Protease
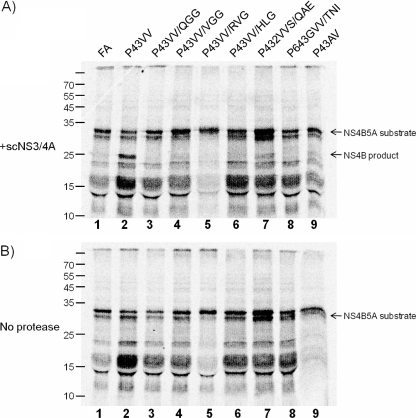
To examine whether the alterations to the P1′–P3′ sequences in the P43→VV mutant affected boundary cleavage, we used an in vitro-based cleavage assay because it allowed cleavage efficiency to be determined for relatively large numbers of boundary sequences. DNA templates encoding a truncated form of the NS4B5A precursor for all constructs tested in Fig. 4 were amplified, including [FA]luc and [P43VV]luc as controls. Radiolabeled protein was produced by in vitro transcription and translation of each DNA template and incubated with a single chain NS3/4A protease. Approximately 60% of the substrate containing the P43→VV mutation gave a readily observed NS5A cleavage product (Fig. 5, lane 2). In contrast, little or no cleaved product was detected from the wild-type NS4B5A boundary substrate (lane 1), even when the concentration of the protease or incubation times were extended (data not shown). For each substrate that contained the P43→VV mutation alongside the second site compensatory mutations at P1′–P3′ positions, as well as that derived from the [P43AV]luc replicon, little or no cleavage was observed (lanes 3–9).
FIGURE 5.
Single chain NS3/4A processing of in vitro translated NS4B5A substrates. A 35S-labeled truncated NS4B5A substrate (residues 1758–2048) derived from each of the rescue mutations analyzed for their capacity to replicate (see Fig. 4) was generated by in vitro translation. This was incubated in the presence (A) or absence (B) of 10 μg/ml scNS3/4A protease for 2 h. Reaction products were separated by SDS-PAGE, and the results were visualized by a PhosphorImager. A failure to observe both cleavage products relates to the small size and the limited number of Cys and Met residues in the NS5A-derived product.
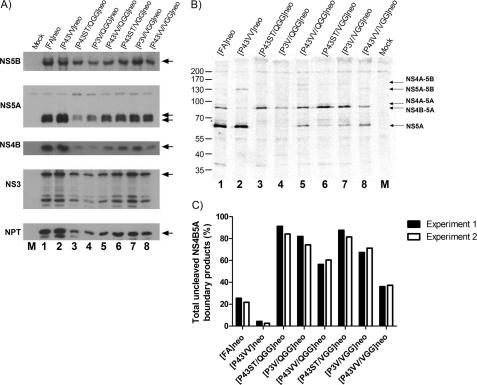
To validate these data in cellulo, neomycin phosphotransferase-expressing versions of [P43VV/QGG]luc and [P43VV/VGG]luc replicon constructs, which were most effective at promoting replication in the context of a P43→VV mutation, were introduced into HepG2 cells by baculovirus transduction. Expressed protein was radiolabeled for 15 min, and the polyprotein products were immunoprecipitated and processed for PhosphorImager analysis as in Fig. 2B. For comparison purposes, baculoviruses expressing [FA]neo and [P43VV]neo were also included (Fig. 6, B and C). As in the earlier pulse-chase experiments, introduction of the P43→VV mutation dramatically reduced the abundance of NS4B5A precursor compared with [FA]neo, further confirming that these mutations enhanced cleavage (Fig. 6B, compare lane 2 with lane 1). In contrast, the ratio of precursor NS4B5A to mature NS5A was greater for [P43VV/QGG]neo and [P43VV/VGG]neo (lanes 5 and 8) compared with both [FA]neo and [P43VV]neo, with the proportion of precursor being slightly higher in the former of the two constructs (Fig. 6, B and C). As observed earlier, constructs with the P43→VV mutation had slightly delayed cleavage of the NS5A5B precursor, irrespective of the sequence on the NS5A side of the boundary (see Fig. 6B, lanes 2, 5, and 8). In summary, there is a correlation between the ability of second site compensatory mutations at P1′–P3′ positions to restore replication and reduced cleavage efficiency at the NS4B5A boundary.
FIGURE 6.
Analysis of polyprotein processing in constructs containing NS5A second site compensatory mutations. A, Western blot analysis of lysates from HepG2 cells transduced with baculovirus vectors engineered to deliver replicon transcripts into cells using a tetracycline-responsive promoter. Transcripts introduced include [FA]neo and [P43VV]neo (lanes 1 and 2) as well as those that possess the P1′–P3′ QGG and VGG second site compensatory mutations, either alone ([P43ST/QGG]neo and [P43ST/VGG]neo) (lanes 3 and 6), in combination with the P43VV mutation ([P43VV/QGG]neo and [P43VV/VGG]neo) (lanes 5 and 8), or in combination with the P3V mutation ([P43SV/QGG]neo and [P43SV/VGG]neo) (lanes 4 and 7). A control lysate was derived from cells transduced with BACtTA only (lane M). Cell extracts were probed by Western blot for expression of HCV proteins (NS5B, NS5A, NS4B, and NS3) or neomycin phosphotransferase (NPT). Where present, arrows indicate the position of the full-length NS3 and NS5A, with other bands present representing products from internal cleavage or cross reactive cell antigens. B, HepG2 cells were transduced with the same baculovirus constructs as in A and metabolically labeled for 15 min with [35S]Met/Cys prior to cell lysis. NS5A-FLAG- and NS5A-FLAG-containing precursors were immunoprecipitated by anti-FLAG mAb, separated on 10% SDS-PAGE, and visualized by a PhosphorImager. The positions of different HCV polyprotein products are indicated by arrows. Mock lysate comes from BACtTA-only-transduced cells. C, graphical representation of the data from B showing the proportion of uncleaved NS4B5A precursor products in cells from two separate experiments and expressed as a percentage of total NS5A-containing products.
Mutations at N Terminus of NS5A That Compensate for P43→VV Mutation Are Not Culture-adaptive
Second site mutations that compensate for mutations deleterious to replication can do so either directly or indirectly by nonspecific enhancement of replication (32). If secondary site mutations operated by slowing boundary cleavage, then their effect should be direct, and their ability to enhance replication would be restricted to replicons possessing the P43→VV mutation. To examine this, both the P1′–P3′ QGG and VGG sequences were engineered into luciferase-expressing replicons either containing parental Ser and Thr residues at the P4 and P3 positions to create [P43ST/QGG]luc and [P43ST/VGG]luc or containing the P3→V mutation to create [P3V/QGG]luc and [P3V/VGG]luc, respectively. These four new transcripts were transfected into Huh7.5 cells along with relevant controls (Fig. 7).
FIGURE 7.
The effect of NS5A second site compensatory mutations on replicon replication in the presence and absence of NS4B mutations that promote boundary cleavage. Huh7.5 cells were electroporated with RNA replicons expressing either a P1′–P3′ QGG or VGG mutation, either alone ([P43ST/QGG]neo and [P43ST/VGG]neo), in combination with the P43VV mutation ([P43VV/QGG]neo and [P43VV/VGG]neo), or in combination with the P3V mutation ([P43SV/QGG]neo and [P43SV/VGG]neo). Also included were [FA]luc, [P43VV]luc, and a luciferase-expressing polymerase-defective control replicon (Δpol). Extracts were prepared at 4, 24, 48, and 72 h postelectroporation, and luciferase activity was determined. Data shown represent mean luciferase activity (n = 2; error bars, S.D.) at 24, 48, and 72 h normalized to luciferase activity at the 4 h time point. RLU, relative luciferase units.
As previously observed, both [P43VV/VGG]luc and [P43VV/QGG]luc gave a marked increase in replication compared with [P43VV]luc, although replication of these two constructs was lower than for the parental construct [FA]luc. In replicons with the P3→V mutation and either P1′–P3′ QGG or VGG, replication was similar ([P3V/VGG]luc) or slightly reduced ([P3V/QGG]luc) compared with [FA]luc (which consistently replicates to the same level as [P3V]luc). Thus, in the P3→V background, the second site compensatory mutations were either neutral (VGG) or detrimental (QGG). For the [P43ST/VGG]luc replicon, there was a modest reduction in replication compared with [FA]luc. In contrast, no replication could be detected in [P43ST/QGG]luc-transfected cells. Although the lack of any replication of the [P43ST/QGG]luc construct was not predicted, the above data indicate that P1′–P3′ QGG and VGG only enhance replication in the context of P43→VV mutations.
Given that second site compensatory sequences were potentially detrimental to replication, depending on the sequence at the end of NS4B, we examined how the various NS5A and NS4B sequences at the NS4B5A boundary affected cleavage. Neomycin phosphotransferase versions of the luciferase-based replicons were created and delivered into HepG2 cells by baculovirus transduction. From Western blot analysis, polyprotein processing went to completion, and the levels of NS5A hyperphosphorylation were similar irrespective of the NS4B5A boundary sequence (Fig. 6A). Interestingly, there were reduced levels of NS proteins as well as neomycin phosphotransferase in all P1′–P3′ QGG experimental groups. To examine the rate of NS4B5A boundary processing, proteins were radiolabeled for 15 min before NS5A products were immunoprecipitated and quantified (Fig. 6, B and C). Similar to previous observations with constructs containing the P43→VV mutations, introduction of QGG and VGG at the P1′–P3′ positions yielded a greater proportion of uncleaved NS4B5A-containing precursors. This occurred both in the context of the parental NS4B P43ST sequence (Fig. 6B, lanes 3 and 6) and the P3→V mutation (lanes 4 and 7). Therefore, the P1′–P3′ QGG and VGG second site mutations slow processing at the NS4B5A boundary not only when P43→VV mutations are present but also in other sequence contexts.
Existence of Possible Compensatory Mutations in NS3/4A
Although all P1′–P3′ mutations tested were able to rescue replication of HCV replicons, none were able to entirely compensate for the presence of the P43→VV mutation. Although our data indicated that changes to the P1′–P3′ positions within the NS4B5A boundary were required to maintain the P43→VV mutation, it was possible for additional minor compensatory mutations to occur elsewhere in the genome. Because the rate of cleavage of the modified NS4B5A boundary sequences could be affected by alterations to the HCV protease, we examined whether this region might also be under selective pressure. Therefore, the NS3/4A coding region was sequenced from three clonal replicon cell lines that had maintained the P43→VV mutation but also possessed second site compensatory mutations in NS5A (VGG, RNL, or KVD at the P1′–P3′ positions; see Table 2). Only the clone with a P1′–P3′ VGG boundary sequence contained a mutation, resulting in a Ser to Cys substitution at residue 22 within NS4A (S22C). This is a naturally occurring variation found in different genotype 1 isolates, which is located in the NS4A cofactor sequence; based on the structure of the NS3/4A complex, this amino acid alteration is unlikely to alter protease substrate recognition. Nonetheless, the NS4A S22C mutation was introduced into [FA]luc and [P43VV/VGG]luc (the latter has the same NS4B5A boundary sequence as that of the cell line harboring the S22C mutant replicon) to generate [S22C/FA]luc and [S22C/P43VV/VGG]luc, respectively. RNA from these constructs and their parent plasmids was then used in a transient luciferase-based replication assay. The NS4A S22C mutation gave a slight enhancement in replication of about 2-fold, but this increase was observed for both [S22C/FA]luc and [S22C/P43VV/VGG]luc constructs (see supplemental Fig. 3). Hence, the S22C mutation appears to have no specific compensatory effect on replication of transcripts with modified NS4B5A boundaries.
Possible Roles for NS4B5A Precursor
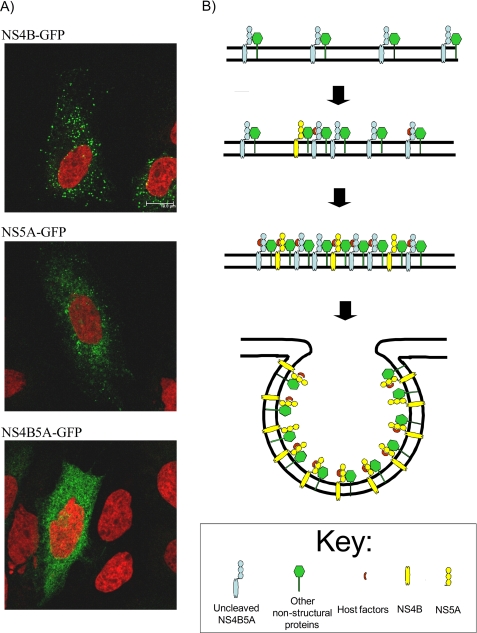
Numerous mechanisms can be proposed to explain why a delay in NS4B5A boundary cleavage might benefit viral replication. For example, delayed cleavage of the NS5A5B boundary has been implicated in facilitating peptidylprolyl isomerase modification of this precursor (31). Because mature NS5A function requires peptidylprolyl isomerase activity (33–35), it is conceivable that the NS4B5A precursor might play a similar role. However, replicon constructs with differing rates of NS4B5A boundary cleavage show sensitivities comparable with those of the peptidylprolyl isomerase inhibitor, cyclosporin A, suggesting that this is unlikely (supplemental Fig. 4). In a further attempt to determine the role of the precursor, NS4B, NS5A, and NS4B5A were tagged with GFP and expressed in Huh7.5 cells using the baculovirus system. In agreement with previous studies (36–38), NS4B formed small cytoplasmic foci, whereas NS5A gave a more complex localization, consistent with targeting to the ER membrane and lipid droplets (Fig. 8A). The NS4B5A precursor had a reticular staining pattern that is typical for ER-bound proteins. Hence, coupling NS5A to NS4B blocked the ability of NS4B to form foci. These data are consistent with a mechanism in which an NS4B5A precursor might regulate the ability of NS4B to induce formation of sites for HCV RNA replication.
FIGURE 8.
NS4B is capable of focus formation, but the NS4B5A precursor is not. A, GFP-tagged NS4B, NS5A, and NS4B5A were expressed in Huh7.5 cells using recombinant baculovirus transduction and visualized by confocal microscopy. For NS4B-GFP, the GFP tag was inserted at the COOH terminus of the protein; for the other constructs, it was inserted at a position within domain 3 of NS5A capable of tolerating insertion without loss of NS5A replicative function. Cell nuclei are shown counterstained red. B, a proposed model for the role of the NS4B5A precursor in regulating NS4B-induced curvature of the membrane to form the replication complex. In this model, cleavage of the NS4B5A boundary is required to go to completion for maturation of the replication complex. Slow cleavage allows for efficient recruitment of host cell proteins.
DISCUSSION
The HCV NS3/4A protease facilitates release of all HCV NS proteins from the immature HCV polyprotein except for NS2. Although cleavage of the polyprotein by NS3/4A goes to completion, more rapid processing occurs at certain NS boundaries, resulting in the appearance of transient precursors, such as NS4A4B, NS4B5A, and NS4A4B5A (9, 11, 14). Although stable virus precursors play roles in virus replication, only recently has it been suggested that transient polyprotein precursors might contribute to positive strand RNA virus replication (25). The situation differs for retroviruses, where it is well established that replication exhibits a dependence on ordered GAG processing and, by implication, the transient precursors generated during this process (39, 40). In this study, we have shown that a P43→VV mutation (at the C-terminal end of NS4B), which enhances the rate of NS4B5A boundary cleavage, also attenuates replication. Furthermore, we have identified mutations within the P1′–P3′ positions of the boundary (at the 5′ terminus of NS5A) that can compensate for the otherwise nearly lethal P43→VV mutation. Importantly, these second site mutations do not nonspecifically enhance replication but instead reduce the rate of NS4B5A cleavage and, in doing so, alleviate the effects of the P43→VV mutation. Based on analysis of boundary changes, which concomitantly alter the rate of proteolysis and viral replication, studies have suggested that speed of processing at different boundaries within the NS polyprotein region is important for some positive strand RNA viruses (31, 41–43). For HCV, this includes the NS34A and NS5A5B boundaries (31, 43). However, to our knowledge, this is the first study to demonstrate that altered rates of cleavage affecting replication can be compensated by further mutations on the other side of the boundary.
Our findings support the notion that the detrimental effect of the P43→VV mutation on replication arises from the increased rate of cleavage at the NS4B5A boundary. First, all second site compensatory mutations at the P1′–P3′ sites, in the context of the P43→VV substitutions, restored levels of replication close to those achieved with an unmodified NS4B5A boundary. These P1′–P3′ mutations also slowed cleavage at the NS4B5A boundary. Furthermore, they did not nonspecifically increase replication, but their ability to enhance replication was restricted to constructs carrying the P43→VV mutations. One exception was the [P43ST/QGG] construct, which unexpectedly failed to replicate, yet the QGG motif did delay cleavage. Two possible explanations might account for this. First, failure of the [P43ST/QGG] construct to replicate may reflect some further aspect of the NS4B5A precursor that extends beyond simply maintaining physical covalent linkage between NS4B and NS5A. Alternatively, whereas a delay in cleavage at the NS4B5A boundary is necessary, it may also be the case that boundary cleavage needs to go to completion within a certain time frame, and this does not happen with the [P43ST/QGG] construct. Irrespective of this issue, our overall data are consistent with a link between the rate of cleavage at the NS4B5A boundary and replication competence.
It is clear from the pulse-chase results that rates of polyprotein processing at the NS4B5A boundary can be substantially altered by changes to the boundary residues other than those at the consensus positions (i.e. P6, P1, P1′ and P4′; see Fig. 1). Although this observation fits with data from peptide- and fusion protein-based cleavage studies, there is limited information regarding how applicable these findings are to replication in an intracellular environment, where long range interactions and precursor tertiary structure might be predominating factors. An early study in cellulo indicated that only single codon changes to the P1 and P1′ positions had significant impact on polyprotein processing (12). However, in a more recent report, changes to the P3 and P4 positions at the NS5A5B boundary dramatically influenced rate of cleavage (31). Our observations extend this finding to the NS4B5A boundary. Nevertheless, it remains plausible that the context of interactions between NS3/4A and precursor substrates and the topology of the uncleaved boundaries might still be influential. Structural data suggest that the NS4B5A boundary is flanked by two membrane-binding amphipathic helices (44–46). It is also thought that the NS3/4A active site lies face down in close apposition to the ER membrane (47). Therefore, the NS4B5A boundary could be topologically well positioned for association with NS3/4A, both before and after cleavage, but the primary amino acid sequence at the boundary is suboptimal for docking with the protease active site. With this in mind, it is interesting that constructs containing the P43→VV mutation had a slight reduction in the rate of processing of the NS5A5B boundary, perhaps indicating low level inhibition of NS3/4A protease activity due to end product inhibition (43).
Although second site compensatory mutations affected the rate of NS4B5A boundary cleavage, it was also noticeable that many of the alterations included amino acid residues at the P1′ position, which, based on the N-end rule, would be expected to be destabilizing (48, 49). Indeed, of the clones where compensatory changes were restricted to the NS5A side of the NS4B5A boundary, seven different amino acids were seen at the P1′ position, of which three (Arg, Lys, and His) were primary destabilizing residues, one was a secondary destabilizing residues (Asp), and one was a tertiary destabilizing residue (Gln). The most likely explanation is that bulky residues at this position suppress cleavage by the NS3/4A protease without blocking it completely. This is consistent with the accepted consensus sequence for optimal NS3/4A recognition, which ideally includes an amino acid with a small side chain at the P1′ position, and would also fit with data from peptide-based cleavage studies (12, 15–18). Any effects on stability of NS5A were not investigated, although we did observe reduced abundance of NS5A, along with the other HCV NS proteins, and neomycin phosphotransferase in cells transduced with baculoviruses carrying the P1′–P3′ QGG constructs. Because NS proteins are produced in excess of the amounts thought to be needed to sustain viral RNA replication (50), decreases in steady state levels of NS5A may be tolerated in virus-infected cells. Interestingly, a small number of HCV isolates in the GenBankTM database also have a destabilizing residue (Asp and Gln) at the P1′ position of the NS4B5A boundary.
From our results, we propose that NS4B5A-containing precursors promote replication. The role of NS4B5A may be analogous to that described for precursors in the picornavirus field where they can influence subcellular targeting of viral proteins (51) or act as a preferred substrate for post-translational modification (25). However, our data suggest that the rate of cleavage of the precursor is unimportant for hyperphosphorylation and peptidylprolyl isomerase modification of NS5A. Instead, our studies suggest that covalent linkage of NS5A to NS4B compromises the ability of NS4B to form foci. Based on this evidence, our preferred model for a function for NS4B5A is to regulate the ability of NS4B to induce deformation of the ER membrane to create sites for viral RNA replication (Fig. 8B) (52). Such a mechanism is analogous to that proposed for regulation of NS4A-induced membrane curvature in flavivirus infection (53, 54). Further studies are necessary to explore the interactions in the NS4B5A precursor that prevent focus formation by NS4B and any other activities associated with the NS4B5A precursor.
In conclusion, this study highlights the need for the NS4B5A precursor to exist for a finite period of time prior to cleavage to promote HCV RNA replication, indicating a functional role in this process. We tentatively suggest that this role involves regulating the ability of NS4B to induce structural changes within the ER membrane, which is needed for HCV RNA replication. It is anticipated that identification of second site compensatory mutations in this study will prove particularly useful in future work aimed at further investigating the mechanistic role of polyprotein precursors in HCV RNA replication.
Supplementary Material
Acknowledgments
We thank Mark Harris, Ralf Bartenschlager, and Charles Rice for providing reagents and Antonello Pessi for provision of information regarding NS3-mediated cleavage of peptides. We are grateful to David Johnson for technical assistance with the confocal microscope.
This work was supported by Medical Research Council Grant G0701215 and a Royal Society research grant.

This article contains supplemental Table I and Figs. 1–4.
- HCV
- hepatitis C virus
- NS
- non-structural.
REFERENCES
- 1. Grakoui A., Wychowski C., Lin C., Feinstone S. M., Rice C. M. (1993) J. Virol. 67, 1385–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Grakoui A., McCourt D. W., Wychowski C., Feinstone S. M., Rice C. M. (1993) J. Virol. 67, 2832–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lin C., Lindenbach B. D., Prágai B. M., McCourt D. W., Rice C. M. (1994) J. Virol. 68, 5063–5073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mizushima H., Hijikata M., Asabe S., Hirota M., Kimura K., Shimotohno K. (1994) J. Virol. 68, 6215–6222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hüssy P., Langen H., Mous J., Jacobsen H. (1996) Virology 224, 93–104 [DOI] [PubMed] [Google Scholar]
- 6. Hijikata M., Kato N., Ootsuyama Y., Nakagawa M., Shimotohno K. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 5547–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grakoui A., McCourt D. W., Wychowski C., Feinstone S. M., Rice C. M. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 10583–10587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hijikata M., Mizushima H., Akagi T., Mori S., Kakiuchi N., Kato N., Tanaka T., Kimura K., Shimotohno K. (1993) J. Virol. 67, 4665–4675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bartenschlager R., Ahlborn-Laake L., Mous J., Jacobsen H. (1994) J. Virol. 68, 5045–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Failla C., Tomei L., De Francesco R. (1994) J. Virol. 68, 3753–3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lin C., Prágai B. M., Grakoui A., Xu J., Rice C. M. (1994) J. Virol. 68, 8147–8157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kolykhalov A. A., Agapov E. V., Rice C. M. (1994) J. Virol. 68, 7525–7533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tomei L., Failla C., Santolini E., De Francesco R., La Monica N. (1993) J. Virol. 67, 4017–4026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tanji Y., Hijikata M., Hirowatari Y., Shimotohno K. (1994) J. Virol. 68, 8418–8422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ingallinella P., Altamura S., Bianchi E., Taliani M., Ingenito R., Cortese R., De Francesco R., Steinkühler C., Pessi A. (1998) Biochemistry 37, 8906–8914 [DOI] [PubMed] [Google Scholar]
- 16. Kim S. Y., Park K. W., Lee Y. J., Back S. H., Goo J. H., Park O. K., Jang S. K., Park W. J. (2000) Anal. Biochem. 284, 42–48 [DOI] [PubMed] [Google Scholar]
- 17. Landro J. A., Raybuck S. A., Luong Y. P., O'Malley E. T., Harbeson S. L., Morgenstern K. A., Rao G., Livingston D. J. (1997) Biochemistry 36, 9340–9348 [DOI] [PubMed] [Google Scholar]
- 18. Zhang R., Durkin J., Windsor W. T., McNemar C., Ramanathan L., Le H. V. (1997) J. Virol. 71, 6208–6213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Steinkühler C., Urbani A., Tomei L., Biasiol G., Sardana M., Bianchi E., Pessi A., De Francesco R. (1996) J. Virol. 70, 6694–6700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Appel N., Herian U., Bartenschlager R. (2005) J. Virol. 79, 896–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Evans M. J., Rice C. M., Goff S. P. (2004) J. Virol. 78, 12085–12089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jones D. M., Patel A. H., Targett-Adams P., McLauchlan J. (2009) J. Virol. 83, 2163–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tong X., Malcolm B. A. (2006) Virus Res. 115, 122–130 [DOI] [PubMed] [Google Scholar]
- 24. Towner J. S., Mazanet M. M., Semler B. L. (1998) J. Virol. 72, 7191–7200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pathak H. B., Oh H. S., Goodfellow I. G., Arnold J. J., Cameron C. E. (2008) J. Biol. Chem. 283, 30677–30688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McCormick C. J., Rowlands D. J., Harris M. (2002) J. Gen. Virol. 83, 383–394 [DOI] [PubMed] [Google Scholar]
- 27. McCormick C. J., Brown D., Griffin S., Challinor L., Rowlands D. J., Harris M. (2006) J. Gen. Virol. 87, 93–102 [DOI] [PubMed] [Google Scholar]
- 28. McCormick C. J., Maucourant S., Griffin S., Rowlands D. J., Harris M. (2006) J. Gen. Virol. 87, 635–640 [DOI] [PubMed] [Google Scholar]
- 29. Beyer B. M., Zhang R., Hong Z., Madison V., Malcolm B. A. (2001) Proteins 43, 82–88 [PubMed] [Google Scholar]
- 30. Butkiewicz N. J., Wendel M., Zhang R., Jubin R., Pichardo J., Smith E. B., Hart A. M., Ingram R., Durkin J., Mui P. W., Murray M. G., Ramanathan L., Dasmahapatra B. (1996) Virology 225, 328–338 [DOI] [PubMed] [Google Scholar]
- 31. Kaul A., Stauffer S., Berger C., Pertel T., Schmitt J., Kallis S., Zayas M., Lopez M. Z., Lohmann V., Luban J., Bartenschlager R. (2009) PLoS. Pathog. 5, e1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paredes A. M., Blight K. J. (2008) J. Virol. 82, 10671–10683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang F., Robotham J. M., Grise H., Frausto S., Madan V., Zayas M., Bartenschlager R., Robinson M., Greenstein A. E., Nag A., Logan T. M., Bienkiewicz E., Tang H. (2010) PLoS Pathog. 6, e1001118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fernandes F., Poole D. S., Hoover S., Middleton R., Andrei A. C., Gerstner J., Striker R. (2007) Hepatology 46, 1026–1033 [DOI] [PubMed] [Google Scholar]
- 35. Hanoulle X., Badillo A., Wieruszeski J. M., Verdegem D., Landrieu I., Bartenschlager R., Penin F., Lippens G. (2009) J. Biol. Chem. 284, 13589–13601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gretton S. N., Taylor A. I., McLauchlan J. (2005) J. Gen. Virol. 86, 1415–1421 [DOI] [PubMed] [Google Scholar]
- 37. Kim J. E., Song W. K., Chung K. M., Back S. H., Jang S. K. (1999) Arch. Virol. 144, 329–343 [DOI] [PubMed] [Google Scholar]
- 38. Lundin M., Monné M., Widell A., Von Heijne G., Persson M. A. (2003) J. Virol. 77, 5428–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pettit S. C., Moody M. D., Wehbie R. S., Kaplan A. H., Nantermet P. V., Klein C. A., Swanstrom R. (1994) J. Virol. 68, 8017–8027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wiegers K., Rutter G., Kottler H., Tessmer U., Hohenberg H., Kräusslich H. G. (1998) J. Virol. 72, 2846–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kusov Y., Gauss-Müller V. (1999) J. Virol. 73, 9867–9878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. van Kuppeveld F. J., van den Hurk P. J., Zoll J., Galama J. M., Melchers W. J. (1996) J. Virol. 70, 7632–7640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang W., Lahser F. C., Yi M., Wright-Minogue J., Xia E., Weber P. C., Lemon S. M., Malcolm B. A. (2004) J. Virol. 78, 700–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brass V., Bieck E., Montserret R., Wölk B., Hellings J. A., Blum H. E., Penin F., Moradpour D. (2002) J. Biol. Chem. 277, 8130–8139 [DOI] [PubMed] [Google Scholar]
- 45. Elazar M., Cheong K. H., Liu P., Greenberg H. B., Rice C. M., Glenn J. S. (2003) J. Virol. 77, 6055–6061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gouttenoire J., Montserret R., Kennel A., Penin F., Moradpour D. (2009) J. Virol. 83, 11378–11384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brass V., Berke J. M., Montserret R., Blum H. E., Penin F., Moradpour D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 14545–14550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gonda D. K., Bachmair A., Wünning I., Tobias J. W., Lane W. S., Varshavsky A. (1989) J. Biol. Chem. 264, 16700–16712 [PubMed] [Google Scholar]
- 49. Bachmair A., Finley D., Varshavsky A. (1986) Science 234, 179–186 [DOI] [PubMed] [Google Scholar]
- 50. Quinkert D., Bartenschlager R., Lohmann V. (2005) J. Virol. 79, 13594–13605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yang Y., Liang Y., Qu L., Chen Z., Yi M., Li K., Lemon S. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7253–7258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Egger D., Wölk B., Gosert R., Bianchi L., Blum H. E., Moradpour D., Bienz K. (2002) J. Virol. 76, 5974–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ambrose R. L., Mackenzie J. M. (2011) J. Virol. 85, 11274–11282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Miller S., Kastner S., Krijnse-Locker J., Bühler S., Bartenschlager R. (2007) J. Biol. Chem. 282, 8873–8882 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.