Background: HIV-1 Vpu counteracts the cellular antiviral factor BST-2 via an interaction that maps to the transmembrane domains of each protein.
Results: This interaction is detectable by NMR spectroscopy and involves conserved faces of each helix.
Conclusion: HIV-1 avoids an innate host defense via a lipid-embedded helix-helix interface.
Significance: Intermolecular interactions within the lipid bilayer can be highly specific and shape the host-pathogen relationship.
Keywords: HIV-1, Innate Immunity, Membrane Proteins, NMR, Protein-Protein Interactions, BST-2, Vpu, Transmembrane Helical Proteins
Abstract
The Vpu protein of HIV-1 antagonizes BST-2 (tetherin), a broad spectrum effector of the innate immune response to viral infection, by an intermolecular interaction that maps genetically to the α-helical transmembrane domains (TMDs) of each protein. Here we utilize NMR spectroscopy to describe key features of the helix-helix pairing that underlies this interaction. The antagonism of BST-2 involves a sequence of three alanines and a tryptophan spaced at four residue intervals within the Vpu TMD helix. Responsiveness to Vpu involves bulky hydrophobic residues in the C-terminal region of the BST-2 TMD helix that likely fit between the alanines on the interactive face of Vpu. These aspects of Vpu and BST-2 form an anti-parallel, lipid-embedded helix-helix interface. Changes in human BST-2 that mimic sequences found in nonhuman primate orthologs unresponsive to Vpu change the tilt angle of the TMD in the lipid bilayer without abrogating its intrinsic ability to interact with Vpu. These data explain the mechanism by which HIV-1 evades a key aspect of innate immunity and the species specificity of Vpu using an anti-parallel helix-helix packing model.
Introduction
The host-pathogen relationship is replete with examples of host defenses that are specifically antagonized by viral proteins. The HIV-1 protein Vpu antagonizes the interferon-induced restriction factor BST-2 (tetherin/CD317) by a protein-protein interaction that maps genetically to the helical transmembrane domains (TMDs)4 of each protein (1–3). This interaction serves to remove BST-2 from the plasma membrane, where it would otherwise directly prevent the release of newly formed, enveloped virus particles from the surface of the infected cell (2, 4, 5).
Helix-helix packing arrangements have been well studied in membrane proteins that contain two or more membrane-spanning TMDs. These arrangements fall into a relatively small number of structurally similar groups (6). Vpu is a type I transmembrane protein with a single TMD (7). BST-2 is a type II transmembrane protein also with a single TMD (8). Consequently, we hypothesized that the TMD-based interaction between Vpu and BST-2 would be anti-parallel in orientation and suspected that it would fall into an established structurally defined group.
We used a combination of NMR spectroscopy and mutational analyses to develop plausible models of the interaction between the TMDs of Vpu and BST-2. Solution NMR experiments provided evidence of a direct interaction between the two TMD helices in a lipid environment, whereas solid state NMR allowed determination of the tilt angles of the helices in lipid bilayers. Together, these data document an anti-parallel interaction in which an alanine repeat on the Vpu TMD faces bulky hydrophic residues on the BST-2 TMD. Moreover, the data suggest that the specificity of HIV-1 Vpu for human BST-2 resides in the tilt angle of the BST-2 TMD within the lipid bilayer rather than in an intrinsic inability of nonhuman primate orthologs to bind the Vpu TMD.
EXPERIMENTAL PROCEDURES
Plasmids, Cells, and Reagents
Most plasmids, cells, and reagents have been previously described (2, 9–11). Mutations were introduced in pVphu, pcDNA3.1-BST2, and pET31b(+) protein expression vectors using the QuikChange kit (Stratagene) and verified by nucleotide sequencing. The BST-2 TMD sequence, encoding KRSKLLLGIGILVLLIIVILGVPLIIFTIKKKKKKM, was synthesized by EZBiolab and inserted into pET31b(+) (Novagen) for expression as a ketosteroid isomerase fusion. The membrane proximal cysteine of the BST-2 cytoplasmic domain was substituted with serine to avoid aggregation, and C-terminal lysines were added to improve the solubility of the peptide. The sequence of the TMD of human BST-2 is underlined and was unmodified.
Virion Release Assays
HeLa cell-based assays were performed as described (2) using an ELISA (PerkinElmer Life Sciences) to measure the concentration of viral p24 capsid antigen in both the supernatants and cellular fractions of the cultures. The percentage of release is the fraction of the total p24 antigen produced by the culture that was secreted into the supernatant. HEK293 cell-based assays were performed as described (9), using an ELISA to measure the concentration of p24 in the culture supernatants and immunoblot to confirm equal expression of Gag protein in the cells. Virion-associated p24 was separated from soluble p24 by centrifugation through a 20% sucrose cushion before measurement by ELISA. The virion release data are the averages of duplicate transfections; error bars are the standard deviation.
Immunoprecipitations
Immunoprecipitations were performed as described using HEK293T cells transfected transiently to express Vpu and BST-2 (9). The immunoprecipitation used murine monoclonal antibody to the BST-2 ectodomain; immunoprecipitates were analyzed by immunoblot for BST-2 and Vpu.
Immunofluorescence
Immunofluorescence to co-localize BST-2 and Vpu in transfected HeLa cells was performed as previously described (2, 10).
Flow Cytometry
Flow cytometry to detect the down-regulation of surface BST-2 by Vpu was performed as previously described, using co-transfection of a GFP expression plasmid to gate on the transfected cells (2, 9, 10). HeLa cells, which express BST-2 constitutively, were used to study the Vpu mutants, and HEK293T cells, which do not express BST-2, were used to study the BST-2 mutants.
Immunoblots
Immunoblots were performed as previously described (9) and developed using enhanced chemiluminescence.
Protein Expression
The expression and purification of BST2 TMD were performed as described previously (11). The pET31b(+) vector containing the BST-2 TMD sequence was expressed in OverExpress C41(DE3) cells (Lucigen) in LB or M9 medium for unlabeled or isotopically labeled samples, respectively. Purification was performed by nickel column chromatography followed by cyanogen bromide cleavage. Lyophilized protein powder was then dissolved in 1:4 (v/v) hexafluoro-isopropanol:dichloromethane, and final pure protein was obtained by reverse phase HPLC.
Sample Preparation for NMR
Lipids were purchased from Avanti Polar Lipids, Inc. Samples in micelles were prepared by dissolving lyophilized protein powder of either construct or both constructs together in an aqueous solution of 150 mm 1,2-dihexanoyl-sn-glycero-3-phosphocholine (DHPC) with 10% (v/v) deuterium oxide (Sigma) with pH adjusted to 7.4. The molar ratio of labeled to unlabeled protein was ∼1:4. Weakly aligned protein samples were prepared in 6.5% polyacrylamide gels and stressed as previously described (12). For the paramagnetic relaxation enhancement (PRE) experiments, the spin label S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-l)methyl methanesulfonothioate (MTSL) was purchased from Toronto Research Chemicals. MTSL was added to proteins dissolved in SDS and reduced by DTT (Sigma), and the solution was incubated at room temperature overnight. Excess MTSL was removed using Sephadex G-25 quick spin protein columns (Roche Applied Science). SDS and residual-free MTSL were removed by extensive dialysis. Bicelle samples were prepared by first dissolving protein in an aqueous solution of DHPC and then adding this to 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) powder. The final DMPC/DHPC molar ratio was 3:2, and the lipid concentration was 15% (w/v). The mixture was repeatedly heated to 42 °C and cooled on ice with vigorous vortexing until it became clear.
Solution NMR Spectroscopy
The solution NMR experiments were performed on a Bruker 600-MHz spectrometer equipped with a 5-mm triple resonance cryogenic probe and z axis gradient. 1H-15N HSQC spectra were acquired at 37 °C with 2048 and 256 points in the direct and indirect dimensions, respectively (13). Triple resonance HNCA (14) experiments were performed on 13C and 15N uniformly labeled protein for the backbone amide resonance assignment in-phare/antiphase-HSQC (15) spectra were obtained on isotropic and weakly aligned samples to measure the 1H-15N dipolar couplings. For the PRE experiments, after acquiring the initial HSQC spectra using MTSL-labeled protein, the MTSL label was reduced by the addition of 10 mm l-ascorbic acid (Sigma) and incubation at room temperature for 1 h to ensure complete reaction before acquiring the reference spectra. For the extraction of resonance intensities, Lorenzian line shape fitting was performed using Sparky (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco).
Solid State NMR Spectroscopy
Solid state NMR experiments were performed on a Bruker Avance 700 MHz spectrometer equipped with a home-built 1H/15N double-resonance probe with a 5-mm solenoid coil and a strip shield (16). Two-dimensional 15N-H-N dipolar coupling spectra were acquired at 37 °C using SAMMY pulse sequence for the separated local field experiment (17) with 512 and 64 points in the direct and indirect dimensions, respectively.
Data Processing and Structure Calculation and Docking
All of the NMR data were processed using NMRPipe (18) and displayed using NMRView (One Moon Scientific). Chemical shift change was calculated using the following equation.
 |
Fitting of polarity index slant angle (PISA) wheels was performed using MATLAB program as previously described (17). The calculation of the structure of the BST2-TMD complex was performed using a simulated annealing protocol in XPLOR-NIH (19) with the temperature cooled from 3000 to 25 K in steps of 12.5 K with experimentally determined RDC values as the structural constraints. The structure is the average of 10-structure ensemble and displayed by MolMol (20). The docking calculation was performed using Rosetta 3.1 (21) with the NMR structures of Vpu-TMD and BST2-TMD. Two structures were allowed to assemble from an initial distance of 8 Å apart, keeping the backbone rigid while varying the rotation of the helices and side chains. A total of 5000 docking poses were searched during the calculation. Structures of the Vpu-BST2 complex were displayed by Chimera (22).
RESULTS
The AXXXAXXXAXXXW Sequence of Vpu Directs the Interaction with BST-2
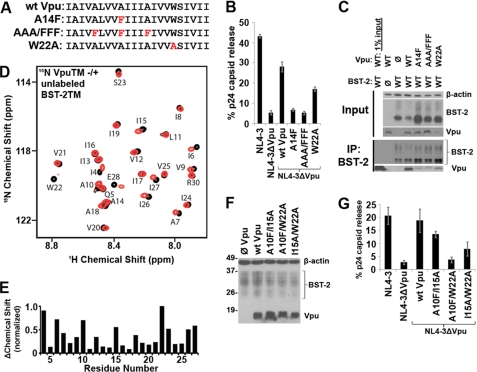
The TMD of HIV-1 group M Vpu contains a well conserved AXXXAXXXA motif, reminiscent of the GXXXG motif first identified in the TMD of glycophorin (Fig. 1A) (23). This motif, in which small residues such as glycines, alanines, or serines (G/A/S) are spaced four residues apart, allows the close approach of transmembrane helices (24). We replaced these alanines in Vpu (Ala10, Ala14, and Ala18) with phenylalanines, whose bulky side groups might disrupt the putative interaction with BST-2. Vpu-AAA/FFF was well expressed and co-localized normally with BST-2 within endosomes (supplemental Fig. S1), but it was almost completely unable to either remove BST-2 from the cell surface (supplemental Fig. S2) or enhance the release of HIV-1 virions from cells that express BST-2 (Fig. 1B). The substitution of the single residue Ala14 by phenylalanine was nearly sufficient for these effects (Fig. 1B and supplemental Fig. S2). When the invariant tryptophan (Trp22) near the cytoplasmic end of the Vpu TMD was substituted with alanine (Fig. 1A), the activity of the protein was moderately impaired (Fig. 1B and supplemental Fig. S2). Moreover, Vpu-AAA/FFF, Vpu-A14F, and to a lesser extent Vpu-W22A interacted poorly with BST-2 in human cells as measured by co-immunoprecipitation (Fig. 1C). These data corroborated a recent mutational analysis of the VpuTMD (25). The data also supported the notion that the AXXXAXXXA repeat of pandemic HIV-1 Vpu is biologically important.
FIGURE 1.
The AXXXAXXXAXXXW face of the Vpu TMD directs the interaction with BST-2. A, sequence of the wild type Vpu TMD. Red letters indicate substituted residues in the initial set of Vpu mutant proteins. B, HIV-1 virion release measured as the fractional secretion of viral capsid (p24) from HeLa cells transfected to express the complete HIV-1 genome (group M, clade B clone NL4-3). Vpu expression was provided in trans to the genome lacking Vpu (ΔVpu) to analyze the indicated mutants. C, immunoprecipitation (IP) of BST-2 from cells (HEK293T) transfected to express BST-2 and Vpu. D, assigned 1H-15N HSQC NMR spectrum of uniformly labeled 15N-Vpu TMD peptide in DHPC lipid micelles either without (black) or with (red) added unlabeled BST-2 TMD peptide. E, chemical shift change induced in Vpu residues by BST-2. F, immunoblot confirming expression of Vpu proteins containing A10F, I15A, and W22A substitutions following transfection of BST-2 positive cells (HeLa) with Vpu expression plasmids. G, virion release measured as the fractional secretion of viral capsid (p24) from HeLa cells transfected to express the complete HIV-1 genome. Vpu expression was provided in trans to the genome lacking Vpu (ΔVpu) to analyze the indicated mutants.
The Transmembrane Domains of Vpu and BST-2 Interact Directly in a Lipid Environment
We tested the hypothesis that the interaction between Vpu and BST-2 protein constructs was direct and could be mapped using NMR spectroscopy, which enables the structural and dynamic features of membrane proteins to be assessed within a lipid environment. We expressed uniformly 15N-labeled Vpu TMD and obtained a well resolved, assigned 1H-15N HSQC NMR spectrum when the peptide was solubilized in DHPC lipid micelles (Fig. 1D and Ref. 11). When unlabeled BST-2 TMD peptide was added to 15N-labeled Vpu TMD in micelles, perturbations of chemical shift indicating a change in the local environment of backbone amides were induced in several Vpu residues including Ile6, Ala10, Ile15, Trp22, and Ile26, suggesting a direct interaction between the TMDs of the two proteins (Fig. 1, D and E). The structurally well defined helical region of the Vpu-TMD spans residues 8–25 (11); therefore the changes in residues Ala10, Ile15, and Trp22 are likely to serve as monitors of the helix-helix interaction. The signals in the NMR spectrum corresponding to residues Ala14 and Ala18 in Vpu were less affected. As noted above, alanine substitution of Trp22, the residue in which the greatest change in chemical shift was induced by BST-2, moderately impaired the activity of Vpu (Fig. 1B and supplemental S2). The individual substitutions V6A, A10F, I15A, A18F, or I26A did not impair activity (data not shown), but the combination of A10F and W22A, and to a lesser extent I15A and W22A, rendered Vpu almost completely inactive (Fig. 1, F and G, and supplemental S2). Overall, these functional results supported the NMR data and suggested that the key residues directing the interaction with BST-2 extend along one face of the Vpu TMD, including Ala10 of the conserved AXXXAXXXA repeat and Trp22 at the cytoplasmic end of the helix.
Large Hydrophobic Residues in the C-terminal Region of the BST-2 TMD Direct the Interaction with Vpu
The TMD of BST-2 does not contain a (G/A/S)XXX(G/A/S) motif but is instead formed predominantly by residues with large hydrophobic side chains. We looked for chemical shifts in the NMR spectra to identify directly the face of the BST-2 TMD helix that interacts with Vpu. Like Vpu, the TMD of BST-2 yielded a well resolved 1H-15N HSQC spectrum when solubilized in DHPC lipid micelles (Fig. 2A). The resonances were assigned using standard methods. When unlabeled Vpu TMD polypeptide was added to the uniformly 15N-labeled BST-2 TMD, chemical shift changes were induced in several BST-2 residues including Val30, Ile34, Leu37, and Leu41 (Fig. 2, A and B), and the signal intensities of Leu37 and Leu41 were markedly reduced, suggesting that these residues are affected by the interaction with Vpu. To confirm the interactive face of the BST-2 helix, we substituted the residues in BST-2 identified by the NMR data with alanines and measured the responsiveness of these proteins to Vpu. Substitution of Ile34, Leu37, or Leu41 rendered human BST-2 relatively resistant to removal from the cell surface and to virologic antagonism by Vpu, whereas the V30A mutant was too poorly expressed to assess (Fig. 2, C–E, and data not shown). In contrast, substitution of Ile26 did not confer decreased responsiveness to Vpu (Fig. 2, C–E). Consistent with these results, substitution of Ile34, Leu37, or Leu41 in BST-2 disrupted the interaction with Vpu detected by co-immunoprecipitation (Fig. 2F). These functional data corroborated a recent mutational analysis of the BST-2 TMD (26). The data also suggested that Vpu indeed targets the hydrophobic face of the BST-2 TMD helix identified by the NMR experiments. Moreover, unlabeled Vpu TMD containing the A10F, A14F, and A18F substitutions induced only minimal chemical shift changes in the spectrum of the BST-2 TMD (Fig. 3). This lack of interaction between the TMD of Vpu-AAA/FFF with the TMD of BST-2 is consistent with the observation that this Vpu mutant has limited biological activity and does not interact strongly with BST-2 in human cells (Fig. 1, B and C).
FIGURE 2.
Large hydrophobic residues in the C-terminal region of the BST-2 TMD direct the interaction with Vpu. A, assigned 1H-15N HSQC NMR spectrum of uniformly labeled 15N-BST-2 TMD peptide in DHPC lipid micelles either without (black) or with (red) added unlabeled Vpu TMD peptide. B, chemical shift change induced in BST-2 residues by Vpu. C, down-regulation of wild type or mutated BST-2 proteins from the cell surface by Vpu. HEK293T cells transfected to express BST-2, Vpu, and GFP (as a transfection marker) were analyzed by flow cytometry for surface levels of BST-2; data for GFP-positive cells are shown. D, virion release measured as the amount of viral capsid (p24) secreted from HEK293T cells transfected to express the complete HIV-1 genome, either with or without an intact vpu gene, together with wild type BST-2 or the indicated BST-2 mutants. E, immunoblot of the virus producer cells from the experiment shown in D. F, immunoprecipitation (IP) of BST-2 from cells (HEK293T) transfected to express wild type BST-2 or the indicated mutants and Vpu.
FIGURE 3.
A Vpu TMD containing the A10F, A14F, and A18F substitutions induces minimal chemical shift changes in the spectrum of the BST-2 TMD, with the exception of residue Leu23. A, overlay of 1H-15N HSQC spectra of uniformly 15N-labeled BST2-TMD with (red) and without (black) unlabeled Vpu-TMD A10F/A14F/A18F. B, chemical shift change plotted against residue number of the BST2-TMD.
The Interaction Detected between TMD Helices of Vpu and BST-2 in Lipid Micelles Is Anti-parallel
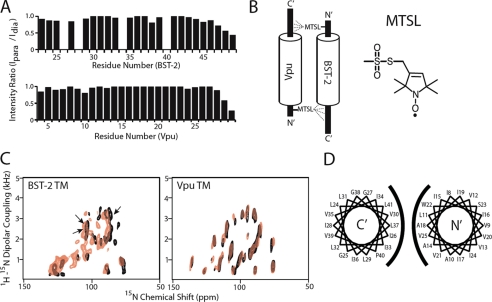
We used PRE to determine the orientation of the BST-2 and Vpu helices in the solution NMR experiments. In this approach (Fig. 4, A and B), the small molecule MTSL is covalently attached to a cysteine side chain of one of the helices by a disulfide linkage. MTSL contains an unpaired electron on its nitroxide group, which enhances the relaxation rates of the surrounding nuclei through magnetic dipolar interactions. The PRE effect can influence nuclei separated by up to 25 Å from the unpaired electron. By contrast, the effective range of the NOE is less than 6 Å. Like the NOE, the magnitude of the PRE effect is proportional to the inverse of the distance to the sixth power. PRE has been used in a variety of applications including the analysis of protein folding as well as of protein-protein and protein-nucleic acid complexes (27). The PRE effect is measured experimentally by comparing the resonance intensities of the HSQC spectrum in the presence of MTSL with those of a reference spectrum acquired after the MTSL is reduced by ascorbic acid and thus becomes diamagnetic. Here, the MTSL spin label was attached to the terminal regions of Vpu and BST-2 to minimize potential disturbance of the helix-helix interaction by cysteine substitution and/or the presence of the spin-label itself. The correct attachment of the label was verified by the HSQC spectrum of MTSL-labeled 15N cysteine-containing proteins (data not shown).
FIGURE 4.
The interaction between BST-2 and Vpu is anti-parallel, involves helices with slightly different tilt angles in the lipid bilayer, and is oriented such that residue Leu37 of BST-2 faces residue Ala18 of Vpu. A, PRE experiments indicate an anti-parallel orientation of the helices when solubilized in DHPC lipid micelles. Top panel, peak intensities of the HSQC resonances of 15N-labeled BST2 were measured with (Ipara) and without (Idia) paramagnetic contribution of a spin label placed on the N-terminal side of Vpu TMD (residue Gln5 substituted with Cys). The ratios of the two values are plotted against the residue number of BST-2. The missing bars correspond to peaks that were not sufficiently resolved for accurate measurement, as well as to proline 40, which contains no amide hydrogen. Bottom panel, intensity ratios (with and without paramagnetic contribution) of 15N-labeled Vpu TMD in the presence of a BST2 TMD in which the N-terminal residue Cys20 was spin-labeled. B, diagram summarizing the findings of the PRE experiments. The spin label (MTSL) at the N terminus of Vpu selectively decreases the intensity of C-terminal residues of BST-2, whereas the spin label at the N terminus of BST-2 selectively decreases the intensity of C-terminal residues of Vpu. C, solid state 15N chemical shift/1H-15N dipolar coupling separated local field spectrum of 15N uniformly labeled BST-2 and Vpu TMDs embedded in magnetically aligned phospholipid bicelles containing long chain (DMPC) and short chain (DHPC) phospholipids. The overlaid spectra correspond to the indicated protein either alone (black) or with its unlabeled binding partner (red). Based on PISA wheel fitting, the tilt angle of the Vpu helix is 28° either with or without BST-2, whereas the tilt angle of the BST-2 helix is 21° without Vpu and 24° with Vpu. The arrows indicate the positions of valines in BST-2. D, helical wheel diagrams of the BST-2 (left diagram) and Vpu (right diagram) TMDs; the rotation of the BST-2 helix was determined using [15N]valine labeling to assign the solid state spectra. The TMDs of BST-2 and Vpu are depicted in their anti-parallel orientation.
The signal intensity changes of 15N-labeled BST-2 TMD resulting from binding with an unlabeled Vpu-Q5C mutant to which MTSL was attached are shown in Fig. 4A (top panel). The residues on the C-terminal side of Thr45–Lys49 experienced a stronger effect from the spin label. Signals from amide sites from residues beyond 49 were not observable because of deuterium/hydrogen exchange under the sample conditions. Distance calculations placed the region affected by the MTSL within a 20 Å radius of the paramagnetic center (28). Because the paramagnetic label on the N-terminal side of Vpu affects the C-terminal region of BST-2, the helix-helix interaction detected in these solution NMR experiments is in an anti-parallel orientation. To confirm this, MTSL was attached to the unlabeled BST2 protein on the native Cys20 residue of the cytoplasmic, N-terminal region, and the effect on the spectrum of 15N-labeled Vpu was measured (Fig. 4A, bottom panel). The residues affected by the N-terminal MTSL on BST-2 are within the C-terminal region of Vpu (residues 28–30), confirming the anti-parallel orientation.
Solid State NMR Confirms Interaction Faces and Reveals Tilt Angles of Vpu and BST2 TMDs within Bilayer
We used oriented sample solid state NMR to determine the spatial relationship of the Vpu and BST-2 TMDs in aligned DMPC/DHPC phospholipid bicelles, which contain dimyristoyl phosphatidylcholine as the long chain lipid. Two-dimensional 15N chemical shift/1H-15N dipolar coupling spectra revealed that the BST-2 TMD, like that of Vpu, displayed a well defined wheel-like pattern of resonances characteristic of a tilted transmembrane helix (Fig. 4C). The tilt angle of the BST-2 TMD was determined from the fitting of the PISA wheel and was similar to that of the Vpu TMD (BST-2, 21°; Vpu, 28°), consistent with the notion that the two helices interact along their length but with a slightly crossed orientation. We used selective (by residue type) [15N]valine labeling to partially assign the solid state spectrum of the BST-2 TMD and to determine its rotation angle within the phospholipid bilayer (supplemental Fig. S3). When the helical wheel for BST-2 is shown side-by-side with that previously determined for Vpu (11), the data indicate a helix-helix interface between BST-2 and Vpu consistent with the HSQC and functional data, including residues Val30, Ile34, Leu37, and Leu41 of BST-2 and Ala14, Ala18, and Trp22 of Vpu (Fig. 4D and supplemental Fig. S3).
To investigate the protein-protein interaction in lipid bilayers, two-dimensional spectra were recorded for each 15N-labeled protein with its unlabeled partner (red in Fig. 4C), and the results were superimposed on the spectrum of the labeled protein alone (black in Fig. 4C). The Vpu TMD did not experience major structural or topological changes in the presence of the BST2 TMD, based on the almost perfect overlap of the spectra obtained with and without unlabeled BST-2. Moreover, the two spectra displayed similar line widths in both dimensions. In contrast, in the case of BST-2, a small change in the overall wheel pattern of the spectrum was detected, characterized by a decrease in the dipolar coupling value and a downfield shift in the 15N chemical shift. These spectral changes indicated a change in the PISA wheel fitting to 24° for the tilt angle of the BST-2 helix in the membrane. In addition, the overall quality of the BST-2 spectrum was significantly affected, as indicated by the broadening and disappearance of the peaks in the presence of unlabeled Vpu. This suggests that the stability of BST-2 within the bilayer was perturbed by Vpu. Overall, these data support the notion that the TMDs of Vpu and BST-2 interact directly within the lipid bilayer. The data also suggest that the interaction with Vpu induces a slight increase in the tilt angle of the BST-2 TMD within the membrane.
Lack of Responsiveness to Vpu Is Associated with Altered Tilt Angle of the BST-2 Transmembrane Helix
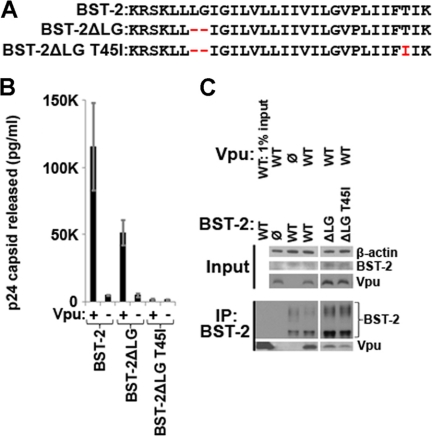
Certain BST-2 proteins from nonhuman primates, such as pig-tailed macaques, are relatively unresponsive to HIV-1 Vpu (3, 29). This has been attributed to the absence of two large hydrophobic residues near the cytoplasmic end of the BST-2 TMD and to the substitution of a threonine (Thr45) with an isoleucine near the lumenal end of the TMD (Fig. 5A) (3, 30). We confirmed that mutation at or near these sites can impair Vpu responsiveness; the mutated human BST-2 protein ΔLG,T45I was not removed from the cell surface by Vpu (data not shown), nor could Vpu antagonize its ability to restrict virion release (Fig. 5B). We also confirmed that the biological effects correlated with protein-protein interactions; the co-immunoprecipitation of Vpu with BST-2 was much less efficient when the human BST-2 protein ΔLG,T45I was expressed (Fig. 5C).
FIGURE 5.
An N-terminal deletion and T45I substitution in the TMD of human BST-2 render the protein unresponsive to Vpu. A, sequence of the human BST-2 TMD. Red, substituted or deleted residues that conform to protein sequences from nonhuman primates such as pig-tailed macaques. B, virion release measured as the amount of viral capsid (p24) secreted from HEK293T cells transfected to express the complete HIV-1 genome, either with or without an intact vpu gene, together with wild type BST-2 or the indicated BST-2 mutants. C, immunoprecipitation (IP) of BST-2 from cells (HEK293T) transfected to express BST-2 and Vpu. (Controls are identical to those shown in Fig. 1C.)
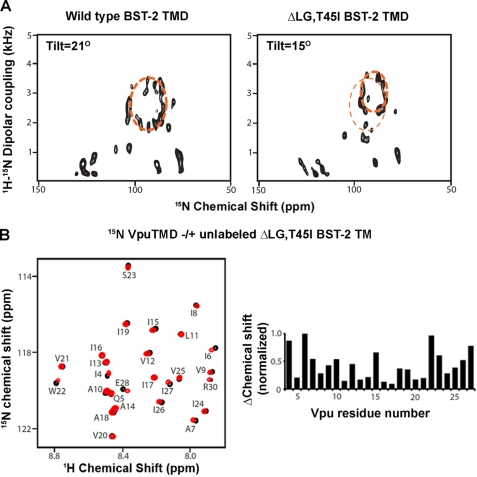
We used oriented sample, solid state NMR to determine whether the ΔLG,T45I BST-2 TMD might be tilted abnormally in the lipid bilayer when compared with wild type BST-2 (Fig. 6A). The tilt angle of the ΔLG,T45I BST-2 TMD determined from the fitting of the PISA wheel was 15°, substantially less than that of the wild type protein. Notably, the Vpu TMD minimally affected the signals in the HSQC spectrum corresponding to residues Leu24, Gly25, and Thr45 in wild type BST-2 (Fig. 2, A and B), suggesting that these residues might not be critical to the interaction in solution. Moreover, the ΔLG,T45I BST-2 TMD induced chemical shift changes in the spectrum of the Vpu TMD that were similar to those induced by the wild type BST-2 TMD (Fig. 6B; compare with Fig. 1D), further supporting the notion that Leu24, Gly25, and Thr45 do not directly mediate the interaction with Vpu. Together, these data suggest that the ΔLG,T45I BST-2 TMD is intrinsically able to interact with the TMD of Vpu, but when constrained in a lipid bilayer, its altered tilt angle is nonpermissive for binding.
FIGURE 6.
NMR characterization of the ΔLG,T45I BST-2 mutant indicates an altered tilt angle in the lipid bilayer but a preserved intrinsic ability to interact with Vpu. A, solid state NMR indicating a difference in tilt angle between the TMD of BST2 ΔLG,T45I and that of wild type BST-2. Left panel, two-dimensional 15N chemical shift/1H-15N dipolar coupling spectrum of the wild type BST-2 TMD in a DMPC/DHPC lipid bicelle. A PISA wheel fitting is shown as the dashed ellipse; the tilt angle of the helix determined from the fitting is 21° with respect to the normal of the bilayer. Right panel, two-dimensional 15N chemical shift/1H-15N dipolar spectrum of BST-2 ΔLG,T45I. PISA wheel fitting indicates that the tilt angle of the mutant is 15°. The fit for the wild type BST2-TMD is superimposed on the spectrum as the thinner dashed ellipse for comparison. B, the ΔLG,T45I BST-2 TMD induced chemical shift changes in the spectrum of the Vpu TMD similar to those induced by the wild type BST-2 TMD. Left panel, overlay of 1H-15N HSQC spectra of 15N-labeled Vpu-TMD with (red) and without (black) unlabeled BST2-TMD ΔLG,T45I in DHPC micelles. Right panel, chemical shift change plotted against residue number of the Vpu-TMD.
NMR-determined Structure of the BST-2 TMD Backbone and Minimal Energy Docking Simulations of Helix-Helix Interaction with Vpu
We next created a three-dimensional model of the BST-2 TMD in a lipid environment. We measured the residual dipolar coupling (RDC) values of the backbone amides of uniformly 15N-labeled BST-2 TMD in DHPC micelles when weakly aligned within stretched polyacrylamide gels (12). The RDC value is dependent on the angle between the amide bond and the axis of the magnetic field and displays characteristic periodicity in the case of a well defined helix (31). For the BST-2 TMD, the plot of RDC versus residue number revealed a dipolar wave that fitted well with residues 24–45 (Fig. 7A). The change in amplitude but not the phase of the wave at Pro40 indicated a kink in the helix introduced by the proline residue. The three-dimensional structure produced by an annealing protocol using the RDC values as constraints revealed that the BST-2 TMD is α-helical, with an 18° kink introduced by the proline at position 40 (Fig. 7A).
FIGURE 7.
Structure of the BST-2 TMD helix and simulated interactions with the TMD of Vpu. A, left panel, RDC values for uniformly labeled 15N BST-2 TMD in DHPC micelles and weakly aligned in stretched polyacrylamide gels. Right panel, three-dimensional backbone structure of the BST-2 TMD averaged from a 10-structure ensemble derived using RDC and dihedral angle restraints. The structures were produced using the annealing protocol of Xplor-NIH (19). B, docking simulation of the BST-2/Vpu interaction. Left panel, backbone helix ensemble of 20 lowest energy Vpu (red) and BST-2 (blue) helix-helix interactions constrained only by the NMR-determined structure of each helix. Right panel, surface structure of a typical low energy simulation highlighting residues Val30, Ile34, Leu37, and Leu41 of BST-2 and residues Ala-10, Ala-14, Ala-18, and Trp-22 of Vpu; the side chains of BST-2 residues Ile-34 and Leu-37 are juxtaposed with crevices formed by the AXXXAXXXA motif of Vpu. C, models summarizing the Vpu-BST-2 interaction and perturbations that affect it. Left panel, model of the BST-2 TMD binding with the Vpu TMD. The interaction face of BST-2 is lined by residues 30, 34, and 37 (colored green with the side chains shown) as well as by residue 41 (not shown). The interaction face of Vpu is lined by residues 10, 14, 18, and 22 (colored red with the side chains shown). The helices are oriented anti-parallel, and they are tilted slightly differently within the lipid bilayer. Middle panel, when two residues are removed (represented as dashed lines) from the N-terminal side of the BST-2 TMD and when residue threonine 45 is substituted with isoleucine (not shown), the tilt angle of the BST-2 helix in the bilayer changes, and BST-2 can no longer interact with Vpu. Right panel, when the small residues Ala-10, Ala-14, and Ala-18 of Vpu are replaced with bulky phenylalanines, the interaction with BST-2 is lost because of steric hindrance.
Finally, we used a computational approach to examine the interaction by performing docking simulations on the experimentally determined structures of the BST-2 and Vpu TMD backbone helices. The 20 lowest energy models of the complex (of 5000) depicted an anti-parallel helix-helix interaction in which the bulky hydrophobic residues Ile34, Leu37, and Leu41 of the BST-2 TMD are juxtaposed between residues of the AXXXAXXXA repeat of Vpu (Fig. 7B). The region centered around residue Leu37 showed less deviation than either end of the BST-2 TMD, suggesting a key role for this residue in the interaction. When the same docking simulation was performed using the Vpu-AAA/FFF and BST-2 TMDs, the calculation did not converge well, indicated by a backbone root mean square deviation of 12 Å for the 20 lowest energy models; in comparison, the wild type complexes yielded a backbone root mean square deviation of 2 Å. Overall, the features of these simulations are consistent with results from the solution and solid state NMR and the biological data obtained by mutational analysis.
DISCUSSION
We have identified key features of an intermolecular, direct helix-helix pairing that occurs between the TMDs of the HIV-1 Vpu protein and the host defense factor BST-2. This pairing occurs within a lipid environment in vitro, consistent with its formation within lipid bilayers in vivo. As expected, the helix-helix interaction is anti-parallel, and it involves single faces of each helix: the 10AXXXAXXXAXXXW22 sequence of Vpu and the 30VXXXIXXLXXXL41 sequence of BST-2.
The AXXXAXXXA motif in Vpu is well conserved among pandemic, group M HIV-1 isolates. The sequence itself suggests that the interaction between the TMDs of Vpu and BST-2 is of the right-handed anti-parallel variety, in which small residues in one helix (Vpu) are spaced at four-residue intervals (6). In comparison to previously characterized helical packing structures of membrane proteins, these alanine residues in the Vpu TMD are predicted to fit into ridges formed by large hydrophobic residues in the BST-2 TMD (6). These residues in the BST-2 TMD should also be spaced at four-residue intervals, but the residues forming each ridge should be shifted by one residue relative to each other (6). These criteria are fulfilled based on the data herein: one ridge in the BST-2 TMD would be formed by Val30 and Ile34, whereas the other ridge would be formed by Leu37 and Leu41. BST-2 residue Leu37 appears to play an especially key role in binding the alanine motif of Vpu, potentially by inserting into the crevice between alanine residues 14 and 18. This model explains how substitution of the key alanines in Vpu with bulky residues such as phenylalanines inhibits binding to BST-2 and impairs Vpu activity (Fig. 7C, right panel).
Trp22 is invariant among Vpu proteins, and the experimental data indicate that it plays an important role in the interaction with BST-2. In the solution NMR experiments, the presence of BST-2 significantly reduced the intensity of the Trp22 backbone amide signal caused by line broadening. This suggests a decrease in the internal motion of the residue, likely resulting from its participation in stabilizing interactions. Tryptophan residues at the ends of hydrophobic, membrane-spanning sequences can act as anchors because of the propensity of their side chains to align along the polar-apolar interface near lipid carbonyl groups (32). In this manner, Trp22 could ensure the correct positioning of the Vpu helix within the bilayer for interaction with BST-2. The membrane anchoring effect of Trp-22 might also account for the lack of effect of BST-2 on the solid state NMR spectrum of Vpu in the lipid bilayer; in contrast, the spectrum of the BST-2 TMD, which lacks a membrane anchoring tryptophan, was affected by Vpu in the bilayer experiments.
Although the W22A substitution impaired Vpu activity, the combination of A10F and W22A substitutions nearly abolished it. These data, along with the effect of replacing all three residues in the alanine repeat with phenylalanines, support the notion that correct membrane anchoring and multiple contacts along the alanine face of Vpu are required for activity.
The VXXXIXXLXXXL sequence of BST-2 is relatively well conserved among primates, with the exception of residue Val30, which is a glycine in rhesus macaques (3). This suggests that the helical faces of most nonhuman primate BST-2 orthologs are intrinsically able to bind Vpu. Why, then, are these orthologs biologically unresponsive to Vpu and unable to bind Vpu when expressed in human cells? We found that the key differences between the TMDs of human BST-2 and the nonresponsive BST-2 proteins of nonhuman primates (deletion of the Leu24-Gly25 sequence and a T45I substitution) induce a change in the tilt angle of the BST-2 TMD within lipid bilayers. This altered tilt angle might be sufficient to impair the interaction between the BST-2 and Vpu helices (Fig. 7C, middle panel), despite their preserved intrinsic ability to interact with each other as documented by the solution NMR experiments. This notion is also supported by the increase in the tilt angle of human BST-2 within the bilayer induced by Vpu; the shorter TMD of nonhuman primate BST-2 might not be able to accommodate this increased tilt without forcing hydrophilic residues flanking the TMD region into the lipid bilayer, an energetically unfavorable event.
Although the PRE experiments indicate an anti-parallel orientation of the Vpu and BST-2 TMD helices, it is not yet possible to generate distance restraints from these data with sufficient accuracy to refine a structural model of the complex. The interaction observed in the artificial lipid environment is weak, as suggested by the relatively small chemical shift changes, and this may contribute to the inability to generate converged structures. This finding is not entirely unexpected because one construct must overcome the energetic barrier of leaving its own stabilizing lipid environment to enter that of the other construct. The difficulty of determining intermolecular distances between two proteins with low binding affinity is compounded by the slow tumbling of protein-micelle complexes, which hinders magnetization transfer. Nonetheless, the biological, spectroscopic, and computational approaches used herein are consistent in their support of a direct intermolecular interaction between specific, conserved faces of the Vpu and BST-2 transmembrane helices. In summary, the interaction between Vpu and BST-2 and the consequent antagonism of an innate host defense by HIV-1 can be explained by a helix-helix interaction in which the TMDs of these proteins bind directly to each other within the lipid bilayer.
Supplementary Material
Acknowledgments
Monoclonal antibody to BST-2 (HM1.24) was a gift from Chugai Pharmaceutical Co. (Kanagawa, Japan). Polyclonal antibodies to BST-2 and Vpu were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program and contributed by Klaus Strebel. We thank Sang Ho Park for helpful discussions on protein purification and NMR spectroscopy and Marissa Suarez for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants AI081668 and related American Reinvestment and Recovery Act supplements (to J. G.), P41EB002031 and GM066978 (to S. J. O.), and AI051981 (to E. B. S.). This work was also supported by the University of California, San Diego Center for AIDS Research.

This article contains supplemental Figs. S1–S3.
- TMD
- transmembrane domain
- DHPC
- 1,2-dihexanoyl-sn-glycero-3-phosphocholine
- PRE
- paramagnetic relaxation enhancement
- MTSL
- S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-l)methyl methanesulfonothioate
- DMPC
- 1,2-dimyristoyl-sn-glycero-3-phosphocholine
- HSQC
- heteronuclear single quantum coherence
- PISA
- polarity index slant angle
- RDC
- residual dipolar coupling.
REFERENCES
- 1. Neil S. J., Zang T., Bieniasz P. D. (2008) Nature 451, 425–430 [DOI] [PubMed] [Google Scholar]
- 2. Van Damme N., Goff D., Katsura C., Jorgenson R. L., Mitchell R., Johnson M. C., Stephens E. B., Guatelli J. (2008) Cell Host Microbe. 3, 245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McNatt M. W., Zang T., Hatziioannou T., Bartlett M., Fofana I. B., Johnson W. E., Neil S. J., Bieniasz P. D. (2009) PLoS Pathog. 5, e1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Perez-Caballero D., Zang T., Ebrahimi A., McNatt M. W., Gregory D. A., Johnson M. C., Bieniasz P. D. (2009) Cell 139, 499–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fitzpatrick K., Skasko M., Deerinck T. J., Crum J., Ellisman M. H., Guatelli J. (2010) PLoS Pathog. 6, e1000701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Walters R. F., DeGrado W. F. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13658–13663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Park S. H., De Angelis A. A., Nevzorov A. A., Wu C. H., Opella S. J. (2006) Biophys. J. 91, 3032–3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kupzig S., Korolchuk V., Rollason R., Sugden A., Wilde A., Banting G. (2003) Traffic 4, 694–709 [DOI] [PubMed] [Google Scholar]
- 9. Tokarev A. A., Munguia J., Guatelli J. C. (2011) J. Virol. 85, 51–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Skasko M., Tokarev A., Chen C. C., Fischer W. B., Pillai S. K., Guatelli J. (2011) Virology 411, 65–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Park S. H., Mrse A. A., Nevzorov A. A., Mesleh M. F., Oblatt-Montal M., Montal M., Opella S. J. (2003) J. Mol. Biol. 333, 409–424 [DOI] [PubMed] [Google Scholar]
- 12. Chou J. J., Gaemers S., Howder B., Louis J. M., Bax A. (2001) J. Biomol. NMR 21, 377–382 [DOI] [PubMed] [Google Scholar]
- 13. Mori S., Abeygunawardana C., Johnson M. O., van Zijl P. C. (1995) J. Magn Reson. B 108, 94–98 [DOI] [PubMed] [Google Scholar]
- 14. Grzesiek S., Döbeli H., Gentz R., Garotta G., Labhardt A. M., Bax A. (1992) Biochemistry 31, 8180–8190 [DOI] [PubMed] [Google Scholar]
- 15. Ding K., Gronenborn A. M. (2003) J. Magn Reson. 163, 208–214 [DOI] [PubMed] [Google Scholar]
- 16. Wu C. H., Grant C. V., Cook G. A., Park S. H., Opella S. J. (2009) J. Magn Reson. 200, 74–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nevzorov A. A., Opella S. J. (2003) J. Magn Reson. 160, 33–39 [DOI] [PubMed] [Google Scholar]
- 18. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 19. Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. (2003) J. Magn Reson. 160, 65–73 [DOI] [PubMed] [Google Scholar]
- 20. Koradi R., Billeter M., Wüthrich K. (1996) J. Mol. Graph. 14, 51–55, 29–32 [DOI] [PubMed] [Google Scholar]
- 21. Gray J. J., Moughon S., Wang C., Schueler-Furman O., Kuhlman B., Rohl C. A., Baker D. (2003) J. Mol. Biol. 331, 281–299 [DOI] [PubMed] [Google Scholar]
- 22. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 23. Lemmon M. A., Flanagan J. M., Treutlein H. R., Zhang J., Engelman D. M. (1992) Biochemistry 31, 12719–12725 [DOI] [PubMed] [Google Scholar]
- 24. Senes A., Engel D. E., DeGrado W. F. (2004) Curr. Opin. Struct. Biol. 14, 465–479 [DOI] [PubMed] [Google Scholar]
- 25. Vigan R., Neil S. J. (2010) J. Virol. 84, 12958–12970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kobayashi T., Ode H., Yoshida T., Sato K., Gee P., Yamamoto S. P., Ebina H., Strebel K., Sato H., Koyanagi Y. (2011) J. Virol. 85, 932–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Clore G. M., Tang C., Iwahara J. (2007) Curr. Opin. Struct. Biol. 17, 603–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Battiste J. L., Wagner G. (2000) Biochemistry 39, 5355–5365 [DOI] [PubMed] [Google Scholar]
- 29. Ruiz A., Lau D., Mitchell R. S., Hill M. S., Schmitt K., Guatelli J. C., Stephens E. B. (2010) Virology 406, 312–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gupta R. K., Hué S., Schaller T., Verschoor E., Pillay D., Towers G. J. (2009) PLoS Pathog. 5, e1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mesleh M. F., Opella S. J. (2003) J. Magn Reson. 163, 288–299 [DOI] [PubMed] [Google Scholar]
- 32. de Planque M. R., Bonev B. B., Demmers J. A., Greathouse D. V., Koeppe R. E., 2nd, Separovic F., Watts A., Killian J. A. (2003) Biochemistry 42, 5341–5348 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.