Background: Rictor is an essential component of the mammalian target of rapamycin complex 2 (mTORC2).
Results: Rictor contains two central regions that (i) bind mSin1 and LST8 and (ii) function in multisite acetylation.
Conclusion: Rictor acetylation is a post-translational modification that potentiates mTORC2 activity.
Significance: Understanding the molecular mechanisms by which acetylation potentiates mTORC2 activity links nutrient signaling with critical metabolic kinases.
Keywords: Metabolism, mTOR Complex (mTORC), Protein Acylation, Signal Transduction, Akt, Mammalian Target of Rapamycin Complex 2, Protein Acetylation, Rictor, Sirtuin
Abstract
The serine/threonine protein kinase Akt is a critical regulator of cell growth and survival in response to growth factors. A key step in Akt activation is phosphorylation at Ser-473 by the mammalian target of rapamycin (mTOR) complex 2 (mTORC2). Although Rictor is required for the stability and activity of mTORC2, little is known about functional regions or post-translational modifications within Rictor that are responsible for regulating mTORC2. Here, we demonstrate that Rictor contains two distinct central regions critical for mTORC2 function. One we refer to as the stability region because it is critical for interaction with Sin1.1 and LST8, and a second adjacent region is required for multisite acetylation. p300-mediated acetylation of Rictor increases mTORC2 activity toward Akt, whereas site-directed mutants within the acetylation region of Rictor exhibit reduced insulin-like growth factor 1 (IGF-1)-stimulated mTORC2 kinase activity. Inhibition of deacetylases, including the NAD+-dependent sirtuins, promotes Rictor acetylation and IGF-1-mediated Akt phosphorylation. These results suggest that multiple-site acetylation of Rictor signals for increased activation of mTORC2, providing a critical link between nutrient-sensitive deacetylases and mTORC2 signaling to Akt.
Introduction
Insulin-like growth factor 1 (IGF-1)2 is an essential hormone that regulates cell growth and survival. Upon binding to the IGF receptor, IGF-1 stimulates key signaling molecules, including the protein kinase mammalian target of rapamycin (mTOR), the lipid kinase phosphatidylinositol 3-kinase (PI3K), and Akt (1, 2). Many of the cellular responses that occur in response to IGF-1 are mediated by the activation of Akt (3, 4). Akt is a family of serine/threonine kinases (Akt1–3) that phosphorylate >60 proteins impacting signaling, survival, metabolism, and proliferation (5). Akt1, the best characterized isoform, requires phosphorylation by two distinct IGF-1-responsive signaling pathways. PI3K-generated phosphatidylinositol 3,4,5-triphosphate recruits Akt1 and 3′-phosphoinositide-dependent kinase-1 (PDK1) to the membrane, where PDK1 phosphorylates Akt1 at Thr-308 (6, 7). In a parallel signaling cascade, activated mTOR complex 2 (mTORC2) phosphorylates Akt1 at Ser-473 (8–10). PDK1- and mTORC2-mediated phosphorylation of Akt1 at Thr-308 and Ser-473 greatly potentiates kinase activity (9, 11).
Although mTOR functions in two multiprotein complexes, mTOR complex 1 (mTORC1) and mTORC2 (12), only mTORC2 is able to phosphorylate Akt. mTORC2 consists of the catalytic subunit, mTOR, the mammalian lethal with Sec13 protein 8 (LST8), the rapamycin-insensitive companion of mTOR (Rictor), and mammalian stress-activated map kinase-interacting protein 1 (mSin1). Rictor is required for mSin1 stability and mTORC2 activity (9, 13–15). Genetic disruption of either Rictor or mSin1 results in the loss of Akt phosphorylation following growth factor addition (16).
Acetylation is a reversible post-translational modification that regulates signaling molecules in response to growth factors (17, 18). Acetyltransferases utilize the metabolite acetyl-coenzyme A (acetyl-CoA) to form N-ϵ-acetyllysine on polypeptides. The generation of acetyl-CoA is directly linked to intracellular glucose concentrations, fatty acid catabolism, and to the level of histone acetylation (19). Acetyl moieties are removed from lysine residues by histone deacetylases (HDACs). In mammals HDACs are divided into Class I, II, IV, and the nutrient-responsive Class III, called sirtuins. Sirtuins utilize NAD+ as a co-substrate to deacetylate protein targets and therefore are activated in response to nutrient depletion (20, 21).
Despite the importance of mTORC2 for Akt phosphorylation, little is known about post-translational modifications that regulate mTORC2 activity. Here, we provide evidence that acetylation of Rictor up-regulates the ability of mTORC2 to phosphorylate Akt Ser-473 in response to IGF-1 stimulation. Our findings demonstrate that Rictor mutants that can no longer be acetylated are deficient in mTORC2 activity, providing a link between nutrient-sensitive signal pathways and metabolism.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
HeLa and HEK 293T cells were obtained from ATCC and maintained in DMEM (CellGro), 10% FBS (Invitrogen), and penicillin/streptomycin (Invitrogen). The antibodies used were: α-M2 FLAG-agarose (Sigma), α-HA.11 and α-HA.11-Sepharose (Covance), and α-GST (Santa Cruz Biotechnology). All other antibodies were from Cell Signaling. Glutathione-Sepharose 4B was from GE Healthcare. Recombinant proteins included SIRT1 (Enzo), inactive recombinant human Akt1 (Millipore), and IGF-1 (Invitrogen). Nicotinamide (NAM), splitomicin, trichostatin A (TSA), and wortmannin were from Calbiochem. ATP was from Roche Applied Science. Baculogold protease inhibitors were from BD Biosciences. All other chemicals were from Sigma.
Plasmid Constructs
The plasmid encoding HA-Akt was described previously (30). Plasmids encoding myc-mSin1.1 (Addgene plasmid 12576), myc-mSin1.2 (Addgene plasmid 12577), myc-mSin1.4 (Addgene plasmid 12578), myc-mSin1.5 (Addgene plasmid 12579), HA-mSin1.1 (Addgene plasmid 12582), and HA-LST8 (Addgene plasmid 1865) were from D. M. Sabatini through Addgene. pcDNA3-HA/FLAG-Rictor, pcDNA3-HA-mTOR, and pcDNA3-HA-Raptor were a generous gift from J. C. Lawrence. Plasmids encoding V5-SIRT1 and FLAG-tagged SIRT2–7 were generous gifts from E. Verdin.
Subcloning and Site-directed Mutagenesis
The plasmid encoding p300 HAT was generated by subcloning the region encoding residues 1066–1707 of human p300 into pcDNA3. The HA-tagged pcDNA3-Rictor(1–1188) construct was generated by restriction digest of pcDNA3-HA/FLAG-Rictor with KpnI and ApaI, creation of blunt ends with Klenow, and re-ligation of the large fragment containing vector backbone and HA tag and encoding Rictor(1–1188). Rictor(Δ975–1188), Rictor(Δ1041–1188), and Rictor(Δ1141–1188) were generated by PCR of a fragment of Rictor with an engineered KpnI site immediately downstream of the nucleotides encoding amino acids 974, 1040, and 1140, respectively. These PCR fragments were subcloned into pcDNA3-HA/FLAG-Rictor using endogenous SacII and KpnI sites. All other Rictor constructs were generated by subcloning a PCR fragment into pCMV-FLAG2, or for GST-constructs, pEBG, using pcDNA3-HA/FLAG-Rictor as the template. HA/FLAG-Rictor Lys→Arg mutants were generated by site-directed mutagenesis at the following sites: M1 (Lys-1080, Lys-1082), M2 (Lys-1092, Lys-1095), M3 (Lys-1107, Lys-1108), M4 (Lys-1116, Lys-1119, Lys-1125), and M1–M4 (Lys-1080, Lys-1082, Lys-1092, Lys-1095, Lys-1107, Lys-1108, Lys-1116, Lys-1119, and Lys-1125).
Deacetylase Inhibitor and IGF-1 Treatments
Subconfluent HeLa cells were starved of serum overnight in DMEM containing 5 mm glucose. For deacetylase inhibition, cells were treated with 500 nm TSA, 500 μm NAM, 10 μm splitomicin or dimethyl sulfoxide (vehicle control). Cells were treated with deacetylase inhibitors for 4 h prior to stimulation with 10 ng/ml IGF-1 for 15 min. Deacetylase inhibitor and IGF-1 treatments were the same for HEK 293T cells, except that cells were starved of serum overnight in DMEM containing 25 mm glucose prior to treatment.
Transfections
Plasmids were transiently transfected into HEK 293T and HeLa using Polyfect reagent (Qiagen) according to the manufacturer's instructions. For HeLa cells transfected with HA/FLAG-Rictor constructs, cells were washed in PBS and fresh DMEM with 10% FBS was added 4 h post-transfection. The medium was replaced again after 24 h. Cells were incubated an additional 72 h prior to lysis and analysis of lysates by immunoblotting.
Cell Lysis, Immunoprecipitation, and Immunoblotting
For immunoprecipitation, cells were lysed in 0.3% CHAPS, 50 mm Hepes, and 200 mm NaCl, pH 7.5, with 5 mm NAM, 500 nm TSA, 1 mm DTT, 1× phosphatase and 1× Baculogold protease inhibitors. Cell lysates were precleared with mouse IgG and protein G-agarose for 30 min (Santa Cruz Biotechnology) at 4 °C with tumbling. Precleared lysates were mixed with primary antibodies and protein G-agarose or resin-conjugated primary antibodies and rotated end-over-end at 4 °C for 2 h. Immune complexes were washed three times in lysis buffer and either analyzed by immunoblotting or subjected to in vitro enzymatic assays as described below. Immunoblotting was performed using the NuPAGE system (Invitrogen). Briefly, proteins were separated on 4–12% BisTris polyacrylamide gels and transferred onto nitrocellulose membranes (Whatman). Primary antibodies were used at 1:1,000 dilution, except for α-HA, which was used at 1:5,000, in blocking solution. HRP-conjugated secondary antibodies (Promega) were used at 1:5,000 in blocking solution. Signals were detected by chemiluminescence using ECL plus reagent (Amersham Biosciences) exposed to film (Kodak).
In Cell Acetylation Assays, in Vitro Deacetylation Assay and Immunokinase Assays
In cell acetylation assays were performed as described (29). HEK 293T cells were transfected with expression plasmids encoding HA-tagged mTOR, Rictor, Raptor, mSin1.1, LST8, or Akt, with or without p300 HAT. Cells were lysed after 24 h and target proteins were immunoprecipitated with α-HA antibodies (mTOR, Rictor, Raptor, mSin1.1) or α-HA-Sepharose (Akt, LST8) and analyzed by immunoblotting with α-pan-acetyl-Lys antibody. In cell acetylation assays with GST construction were performed as described, except that glutathione-Sepharose was used for GST protein precipitation. For in cell acetylation assays with the mTORC2 complex, HEK 293T cells were co-transfected with expression plasmids encoding HA/FLAG-Rictor, HA-mTOR, HA-LST8, and HA-mSin1.1 with or without p300 HAT. 24 h later, cell lysates were subject to immunoprecipitation with α-FLAG-agarose. In vitro deacetylation assays were performed by incubating immunoprecipitated HA/FLAG-Rictor protein in deacetylase buffer (Biomol) containing 500 μm NAD+ and 5 units of recombinant SIRT1 enzyme, for 2 h at 30 °C. For kinase assays, HEK 293T cells were transfected with mTORC2 as described, and the medium was replaced after 4 h. 24 h later, cells were washed twice and incubated in serum-free DMEM overnight. Cells were treated with IGF-1 prior to lysis and immunoprecipitation. Following standard washes, immunoprecipitants were washed once in kinase assay buffer containing 25 mm Hepes, pH 7.5, 100 mm potassium acetate, 0.01% BSA, 1 mm DTT, and 1× phosphatase inhibitor mixture 1 (Sigma). Immunoprecipitants were mixed with 0.5 μg of recombinant Akt1 and 1 mm ATP in a total volume of 15 μl of kinase assay buffer and incubated at 37 °C for 30 min. For mTOR inhibition, 2 μm wortmannin was added to the kinase assay buffer. The reactions were stopped by addition of 2× SDS-PAGE sample buffer (Invitrogen) containing 100 mm DTT and boiling for 10 min. Samples containing 10 ng of Akt1 were analyzed by immunoblotting as described above.
Statistics
In the case of detecting differences in Akt phosphorylation, data were used from three independent experiments. Due to differences in transfection efficiencies, the level of Akt phosphorylation varied largely between independent experiments. Therefore, performing statistical analysis on independent data from different experiments was difficult. To overcome this, densitometry data were first normalized to pan-Akt loading control, and the -fold Akt activation was calculated for each independent experiment. All -fold changes were log2 transformed, and a one-tailed Student's t test was performed using the R statistical software. All cases in which p < 0.05, data were considered statistically significant. To graphically display data of three independent experiments in one combined graph, the mean of all replicates was calculated, and both the S.D. and p value are shown.
RESULTS
mTORC2 Adaptor Protein Rictor Is Acetylated
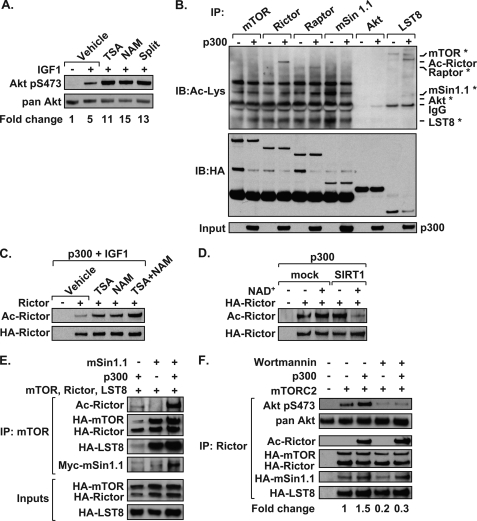
Because the mTOR pathway is intimately tied to cellular metabolism, we examined whether inhibition of HDAC activity results in increased Akt phosphorylation. Treatment of HeLa cells with the Class I/II/IV HDAC inhibitor TSA, or Class III HDAC inhibitors NAM and splitomicin, increased IGF-1-mediated Akt phosphorylation, compared with vehicle control (Fig. 1A). Expression of p300 in HEK 293T cells effectively acetylated Rictor as detected by immunoprecipitation and immunoblotting with an antibody that recognizes acetylated lysines (29) (Fig. 1B). In contrast, we found that mTOR, mSin 1.1, LST8, and Akt1 were not acetylated. p300 reproducibly acetylates Raptor; however, this effect was significantly less robust than p300-mediated acetylation of Rictor. Cells stimulated with IGF-1 and treated with TSA or NAM alone showed increased p300-mediated acetylation of Rictor (Fig. 1C). Cells treated with both TSA and NAM further increased Rictor acetylation, suggesting that Rictor is regulated by multiple classes of HDACs. SIRT1 deacetylated Rictor in vitro in an NAD+-dependent manner, indicating that Rictor was susceptible to direct regulation by the nutrient sensing HDAC (Fig. 1D). Experiments shown in Fig. 1 identify Rictor as an acetylated protein susceptible to regulation by the Class I/II/IV and Class III HDACs.
FIGURE 1.
Acetylation of Rictor promotes mTORC2 kinase activity. A, HeLa cells treated with HDAC inhibitors TSA, NAM, or splitomicin (Split) display increased Akt phosphorylation following IGF-1 treatment as detected by immunoblotting. Representative data from one of three independent experiments are shown. Densitometry results determine the -fold change of Akt Ser(P)-473 phosphorylation where values were normalized to pan-Akt. B, Rictor is an acetylated protein. HEK 293T cells were transfected with mTOR, Rictor, Raptor, mSin1.1, LST8, or Akt and with (+) or without (−) the p300 HAT domain. Proteins were immunoprecipitated (IP) and analyzed by immunoblotting (IB) using α-pan-acetyl-Lys (Ac-Lys) or α-HA antibodies. Asterisks mark the expected positions of proteins on the blot undetected by the Ac-Lys antibody. C, Rictor acetylation is regulated by HDACs. Acetylation assays were performed as in B where cells were transfected with either HA-tagged Rictor or vector control (−). Cells were treated with HDAC inhibitors for 18 h prior to stimulation with IGF-1. D, SIRT1 deacetylates Rictor in vitro. Acetylation assays were performed and immunoprecipitated Rictor proteins were mixed with recombinant SIRT1, in the presence or absence of NAD+ and analyzed as in B. E, mSin1.1 promotes Rictor acetylation. mTOR complexes were immunoprecipitated from HEK 293T cell lysates and analyzed for acetylated and total protein levels by immunoblotting. F, Rictor acetylation increases mTORC2 kinase activity. HEK 293T cells were transfected with plasmids encoding mTORC2 (mTOR, LST8, mSin1.1, and Rictor), with or without the p300 HAT domain. Rictor-containing mTORC2s were immunoprecipitated and mixed with recombinant Akt and ATP, with or without wortmannin. Reactions were analyzed by immunoblotting. The -fold change in Akt Ser(P)-473 phosphorylation was determined following normalization to the level of mTOR present in the immune complex. Data shown in Fig. 1 are representative examples of at least three independent experiments.
Rictor Acetylation Potentiates mTORC2 Activity
As reported previously (13–15), mSin1.1 expression stabilized mTORC2, resulting in a more efficient pulldown of mTOR, Rictor, and LST8 (Fig. 1E). Expression of mSin1.1 along with other members of mTORC2 greatly increased the level of acetylated Rictor. Expression of p300 significantly increased mTORC2 activity, as detected by the ability of mTORC2 to phosphorylate recombinant Akt in vitro (Fig. 1F). Akt was specifically phosphorylated by mTORC2 because the in vitro addition of the PI3K-like kinase inhibitor, wortmannin, blocked the kinase activity. Our data suggest that elevated Rictor acetylation is observed upon its assembly into mTORC2, and Rictor acetylation stimulates mTORC2-mediated phosphorylation of Akt at Ser-473. In addition to mSin1.1, mSin1.2 and mSin1.5, but not mSin1.4, forms mTORC2s that promote Rictor acetylation (supplemental Fig. 1). Thus, functional mTORC2 is associated with elevated Rictor acetylation.
Residues 957–1188 of Rictor Are Required for Acetylation
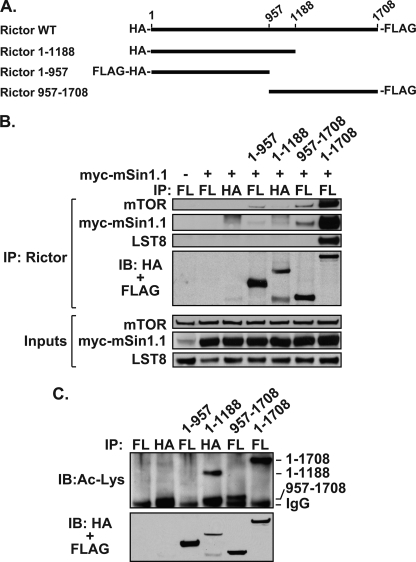
Before initiating experiments to map the acetylated regions within Rictor, we tested a series of N- and C-terminal Rictor deletions for interaction with the mTORC2 components (Fig. 2A). Although full-length Rictor and truncated Rictor proteins were effectively immunoprecipitated using either anti-HA or anti-FLAG antibodies, only wild-type Rictor interacted with mTOR, mSin1.1, and LST8 (Fig. 2B). Constructs lacking either the N or C terminus were unable to co-immunoprecipitate mTORC2 components effectively. Because it was not feasible to examine Rictor deletion constructs in the context of the mTORC2, we tested whether N- and C-terminal Rictor truncations could be acetylated by p300. Expression of p300 effectively acetylated wild-type (1–1708), as well as constructs encoding amino acid residues 957–1708 and 1–1188 (Fig. 2C). However, the Rictor C-terminal truncation consisting of amino acids 1–957 was not acetylated by p300. Because Rictor(1–1188) and Rictor(957–1708) proteins were both acetylated by p300, our results highlight an overlapping central region (residues 957–1188) as a possible acetylated region within Rictor. This region is distinct from the previously identified Rictor acetylation site, Lys-582, of unknown biological function (22).
FIGURE 2.
N terminus of Rictor is dispensable for acetylation. A, Rictor truncations are diagrammed. B, Rictor co-immunoprecipitation of mTOR, mSin1.1, and LST8 requires both termini. HEK 293T cells were transfected with or without mSin1.1 and various Rictor proteins. Rictor was immunoprecipitated (IP) using α-FLAG (FL) antibodies or α-HA antibodies as indicated. Co-immunoprecipitants were analyzed by immunoblotting (IB). C, acetylation of overlapping Rictor truncations highlights central region containing residues 957–1188. Acetylation assays were performed with various Rictor proteins. Proteins were immunoprecipitated as described in B and analyzed by immunoblotting. Data shown are representative examples of two independent experiments performed per panel.
Rictor Contains Two Regions Required for mTORC2 Stabilization and Acetylation
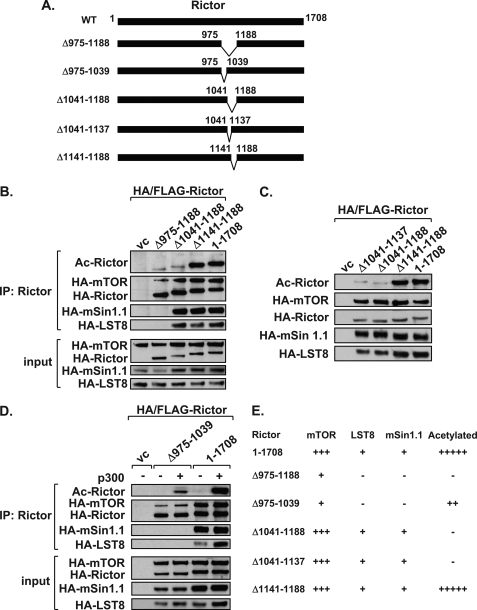
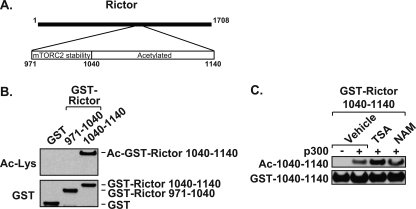
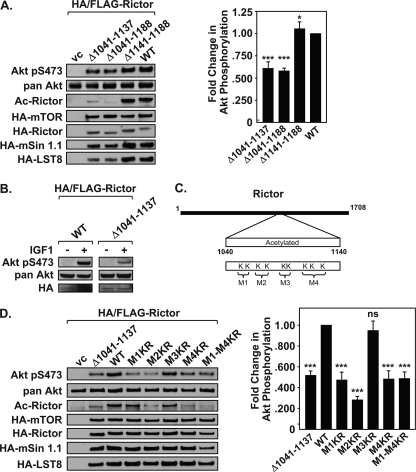
To elucidate the importance of Rictor residues 957–1188 for acetylation, a series of Rictor deletion constructs was generated (Fig. 3A). The largest deletion, Rictor Δ975–1188, showed a loss of acetylation (Fig. 3B). However, this same construct was unable to interact with mSin1.1 and LST8 and showed reduced binding to mTOR. Because mSin1.1 is required for stabilization of mTORC2, we could not discern whether the loss of mSin1.1 and/or LST8 binding was contributing to our inability to detect Rictor acetylation. To overcome this issue, we examined the primary sequence of Rictor across this region and identified six distinct lysine-rich regions located between 1041 and 1188. The deletion construct, Rictor(Δ1041–1188), retained its ability to interact with mSin1.1 and LST8, but was no longer effectively acetylated by p300 (Fig. 3B). Rictor(Δ1141–1188) was acetylated to the same extent as full-length Rictor, and as expected, retained its ability to interact with mSin1.1 and LST8 (Fig. 3B). Collectively, results from deletion analysis indicated that Rictor required residues 1041–1140 for acetylation. To address this, acetylation assays were performed using the Rictor(Δ1041–1137) construct. Similar to Rictor(Δ1041–1188), Rictor(Δ1041–1137) interacted with each of the mTORC2 members, but was no longer acetylated by p300 (Fig. 3C). These results suggest that this lysine-rich region contains the major sites of acetylation. This hypothesis is supported by the fact that GST-Rictor containing residues 1040–1140 was robustly acetylated by p300 and responsive to HDAC inhibitors (Fig. 4). However, GST-Rictor(971–1040) could not be acetylated by p300 (Fig. 4B).
FIGURE 3.
Identification of adjacent mTORC2 stabilization and acetylated regions within Rictor. A, Rictor internal deletion constructions are diagrammed. B, Rictor residues 1041–1188 are required for acetylation. Acetylation assays were performed following co-expression of mTORC2 components, p300, and with either vector control (vc) or various Rictor deletions constructs. Rictor-containing mTORC2s were immunoprecipitated (IP) and analyzed by immunoblotting (IB). C, Rictor(Δ1041–1137) maintains interaction with mTORC2 components but is no longer acetylated. mTORC2 acetylation assays were performed as described in B. D, Rictor(Δ975–1039) does not co-immunoprecipitate mSin1.1 or LST8 and shows reduced acetylation. mTORC2 acetylation assays were performed as described in B with indicated Rictor proteins. E, table summarizes results of the Rictor deletion constructs. Data shown are representative examples of three independent experiments performed per panel.
FIGURE 4.
Rictor residues 1040–1140 contain acetylated lysines that are regulated by HDACs. A, illustration depicts regions used to create GST-Rictor fusion 971–1040 and 1040–1140 proteins. B, Rictor residues 1040–1140 contain acetylated lysines. GST, GST-(971–1040) or GST-(1040–1140) were expressed in HEK 293T cells, and acetylation assays were performed. GST proteins were precipitated and analyzed by immunoblotting as indicated. C, Rictor(1040–1140) acetylation is regulated by HDACs. Acetylation and HDAC inhibitor treatment were performed as in Fig. 1C with GST-Rictor(1040–1140). Data shown in Fig. 4 are representative examples of two independent experiments performed per panel.
Because regions within Rictor responsible for mSin1.1-mediated stabilization have not been previously identified, we examined whether Rictor(Δ975–1039) contained an mSin1.1 stabilization region. Unlike wild-type Rictor, the Rictor(Δ975–1039) protein was unable to interact with either mSin1.1 or LST8 and interacted weakly with mTOR (Fig. 3D). Rictor(Δ975–1039) also showed reduced acetylation, compared with wild-type Rictor. Because p300-directed acetylation of Rictor is greatly increased in the context of the mSin1.1 containing mTORC2 (Fig. 1E), the loss of Rictor acetylation observed in Fig. 3C is most likely due to the inability of the Rictor(Δ975–1039) to interact with mSin1.1. From this point forth we will refer to this domain as the mTORC2 stabilization region because it is required for interaction with mSin1.1 and LST8.
Rictor Acetylation Region Enhances mTORC2 Activity
To evaluate whether the Rictor acetylated region was required for stimulating mTORC2 kinase activity, immunokinase assays were performed. Deletion constructs encoding Rictor(Δ1041–1137) and Rictor(Δ1041–1188) retained the ability to interact with mSin1.1 and LST8, but showed reduced mTORC2 kinase activity relative to wild-type Rictor (Fig. 5A). Rictor(Δ1141–1188), which contains a deletion outside the acetylated region, retained mTORC2 kinase activity similar to that observed for wild-type Rictor. Collectively, these data demonstrate that the acetylated region within Rictor(1041–1137) is required to promote mTORC2 kinase activity without compromising the assembly of mTORC2 components. Importantly, endogenous Akt Ser-473 phosphorylation was reduced in the HeLa cells expressing Rictor(Δ1041–1137) relative to wild-type Rictor (Fig. 5B).
FIGURE 5.
Rictor M2 and M4 contain acetylated lysines that promote mTORC2 kinase activity. A, Rictor internal deletion (Δ1041–1137) immunoprecipitates mTORC2 components but displays reduced kinase activity. Immunokinase assays and the quantification of -fold change in Akt Ser(P)-473 phosphorylation were carried out as described in Fig. 1F. Right, graphs show densitometry results of phosphorylated Akt (normalized to pan-Akt within each experiment), mean ± S.D., n = 3. Statistical analysis was performed on normalized data using standard Student's t test. *, p < 0.02; ***, p < 0.009, n = 3. B, expression of Rictor(Δ1041–1137) results in reduced Akt Ser-473 phosphorylation. HeLa cells were transfected with HA-Rictor proteins, treated with or without IGF-1, and extracts were analyzed by immunoblotting. C, lysines within Rictor(1040–1140) were arbitrarily divided into modules M1 (Lys-1080, Lys-1082), M2 (Lys-1092, Lys-1095), M3 (Lys-1107, Lys-1108), and M4 (Lys-1116, Lys-1119, Lys-1125). D, acetylation of Rictor M2 and M4 is required for enhanced mTORC2 kinase activity. Immunokinase assays were performed using Rictor mutant proteins Lys→Arg (KR) of M1, M2, M3, M4, M1–4 (inclusive), Δ1041–1137, WT, or vector control (vc). The -fold change in Akt Ser(P)-473 phosphorylation was calculated as described in Fig. 1F. Data shown are representative examples of three independent experiments performed per panel. Right, densitometry data were plotted as described in A with mean ± S.D., Student's t test, ns, not significant; ***, p < 0.005, n = 3.
Because acetyltransferases often modify Lys-rich sequences in a multisite fashion (23), we subdivided the 9 Lys into 4 Lys-rich modules (M1–M4). These included M1 (Lys-1080 and Lys-1082), M2 (Lys-1092 and Lys-1095), M3 (Lys-1107 and Lys-1108), and M4 (Lys-1116, Lys-1119, and Lys-1125; Fig. 5C). Site-directed mutagenesis was performed on the full-length Rictor, where lysine residues within each module (M1, M2, M3, or M4) were mutated to arginine or where all 9 lysine residues were converted to arginine (M1–M4; Fig. 5C). Of the single modules, Rictor Lys→Arg M2 and M4 displayed a reduction in mTORC2 kinase activity and Rictor acetylation (Fig. 5D). Consistent with these two modules controlling Rictor function, the Rictor Lys→Arg M1–M4 also showed a reduction in mTORC2 activity and acetylated Rictor. The Rictor Lys→Arg M1 showed a loss of mTORC2 activity; however, this did not correlate with a loss of Rictor acetylation. In contrast, Rictor Lys→Arg M3 failed to show changes in either mTORC2 activity or Rictor acetylation, suggesting that this region is not important for Rictor function. The Rictor(Δ1041–1137) deletion construct served as a positive control and as expected, displayed reduced mTORC2 activity and a loss of Rictor acetylation. All site-directed Rictor mutants were equally expressed, indicating that alteration of Lys→Arg does not affect the stability of Rictor or alter interaction with other members of the mTORC2 (Fig. 5D).
DISCUSSION
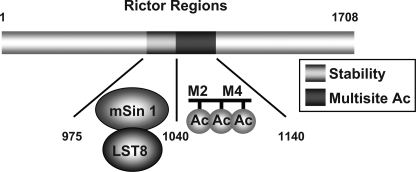
Collectively, we have identified two novel regions within Rictor; an mTORC2 stabilization region located between 975 and 1039, which is required for both mSin1.1 and LST8 interaction, and a 1041–1140 region required for multisite acetylation (Fig. 6). Our work supports a model by which the Rictor stabilization region promotes protein-protein interactions with mSin1 and LST8, stabilizing association with mTOR. However, because Rictor(971–1040) alone does not immunoprecipitate mSin1.1 or LST8 (data not shown), we conclude that Rictor(975–1039) is necessary but not sufficient for mSin1.1/LST8 binding. In contrast, the Rictor acetylation region is not required for mSin1.1/LST8 binding, but is required for mTORC2 activity, suggesting that we have identified two distinct functional regions within Rictor.
FIGURE 6.
Illustration depicting stabilization and acetylation domains within Rictor. Rictor contains: 1, a stabilization region located between 975 and 1039, which is required for both mSin1.1 and LST8 interaction, and 2, 1041–1140 region required for multisite acetylation and increased mTORC2 activity.
These observations highlight the contribution of multisite acetylation of M2 (Lys-1092 and Lys-1095) and M4 (Lys-1116, Lys-1119, and Lys-1125) for enhancing mTORC2 kinase activity. Although the mechanism by which Rictor acetylation stimulates mTORC2 activity is not known, Rictor is a unique case in which acetylation of the adaptor alters the activity of the catalytic subunit. Similar mechanisms have been shown for phosphorylation (24), but it has not been previously reported for acetylation. This is important because large scale proteomic analysis has identified a preponderance of acetylated proteins in enzymatic protein complexes, including adaptor proteins (22). Therefore, acetylation of adaptor proteins may be a common theme by which acetyltransferases impact catalytic activity of multiprotein complexes.
Our data suggest that Rictor acetylation is promoted when assembled into the mTORC2 complex. In addition to mSin1.1, mSin1.2 and mSin1.5, but not mSin1.4, form mTORC2 complexes that promote Rictor acetylation (supplemental Fig. 1). Because all mSin1 variants, except mSin1.4, have previously been shown to assemble into mTORC2 (13), this supports our finding that mTORC2 formation promotes Rictor acetylation. Importantly, the activity of mSin1.5-containing mTORC2 is growth factor-independent, suggesting that acetylated Rictor may regulate both growth factor-dependent and -independent mTORC2 pathways.
The mTORC2 pathway is activated in response to growth factors and amino acids (25). Many of the metabolic and signaling pathways that impinge on mTORC2 signaling are known to be regulated by nutrient-sensitive sirtuin HDACs (26–28). Based on this concept, we show that Rictor can be deacetylated in vitro and in vivo by SIRT1 (Fig. 1D and supplemental Fig. 2). In addition, we provide evidence that Rictor is also deacetylated by SIRT2, SIRT3, or SIRT4 (supplemental Fig. 2). Thus, it is intriguing to speculate that two nutrient-responsive enzymes, namely sirtuins and acetyltransferases, modulate the acetylation/deacetylation status of Rictor to alter mTORC2 activity dynamically based on the availability of NAD+ and acetyl-CoA.
Supplementary Material
Acknowledgments
We thank A. Sherman for editorial assistance; Drs. S. Bekiranov and X. Xu for help with the statistical analysis; and Drs. D. M. Sabatini (Whitehead Institute, MIT, via Addgene), J. C. Lawrence, Jr. (University of Virginia), R. A. Frye (Pittsburgh, PA), and E. M. Verdin (Gladstone Institute) for providing plasmids.
This work was supported, in whole or in part, by National Institutes of Health Grants CA132580 and CA104397 through the NCI (to M. W. M.) and Grant DK052753 through the NIDDK (to T. E. H.).

This article contains supplemental Figs. 1 and 2.
- IGF-1
- insulin-like growth factor 1
- Bis-Tris
- bis(2-hydroxyethyl)amino tris(hydroxymethyl)methane
- HDAC
- histone deacetylase
- LST8
- lethal with Sec13 protein 8
- mSin1
- mammalian stress-activated map kinase-interacting protein 1
- mTOR
- mammalian target of rapamycin
- mTORC2
- mTOR complex 2
- NAM
- nicotinamide
- TSA
- trichostatin A
- HAT
- histone acetyltransferase.
REFERENCES
- 1. Clemmons D. R. (2009) Role of IGF-I in skeletal muscle mass maintenance. Trends Endocrinol. Metab. 20, 349–356 [DOI] [PubMed] [Google Scholar]
- 2. Rosenzweig S. A., Atreya H. S. (2010) Defining the pathway to insulin-like growth factor system targeting in cancer. Biochem. Pharmacol. 80, 1115–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bondy C. A., Cheng C. M. (2004) Signaling by insulin-like growth factor 1 in brain. Eur. J. Pharmacol. 490, 25–31 [DOI] [PubMed] [Google Scholar]
- 4. Zhang D., Bar-Eli M., Meloche S., Brodt P. (2004) Dual regulation of MMP-2 expression by the type 1 insulin-like growth factor receptor: the phosphatidylinositol 3-kinase/Akt and Raf/ERK pathways transmit opposing signals. J. Biol. Chem. 279, 19683–19690 [DOI] [PubMed] [Google Scholar]
- 5. Manning B. D., Cantley L. C. (2007) AKT/PKB signaling: navigating downstream. Cell 129, 1261–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alessi D. R., James S. R., Downes C. P., Holmes A. B., Gaffney P. R., Reese C. B., Cohen P. (1997) Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Bα. Curr. Biol. 7, 261–269 [DOI] [PubMed] [Google Scholar]
- 7. Bellacosa A., Chan T. O., Ahmed N. N., Datta K., Malstrom S., Stokoe D., McCormick F., Feng J., Tsichlis P. (1998) Akt activation by growth factors is a multiple-step process: the role of the PH domain. Oncogene 17, 313–325 [DOI] [PubMed] [Google Scholar]
- 8. Hresko R. C., Mueckler M. (2005) mTOR.RICTOR is the Ser-473 kinase for Akt/protein kinase B in 3T3-L1 adipocytes. J. Biol. Chem. 280, 40406–40416 [DOI] [PubMed] [Google Scholar]
- 9. Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 10. Shiota C., Woo J. T., Lindner J., Shelton K. D., Magnuson M. A. (2006) Multiallelic disruption of the rictor gene in mice reveals that mTOR complex 2 is essential for fetal growth and viability. Dev. Cell 11, 583–589 [DOI] [PubMed] [Google Scholar]
- 11. Alessi D. R., Andjelkovic M., Caudwell B., Cron P., Morrice N., Cohen P., Hemmings B. A. (1996) Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 15, 6541–6551 [PMC free article] [PubMed] [Google Scholar]
- 12. Corradetti M. N., Guan K. L. (2006) Upstream of the mammalian target of rapamycin: do all roads pass through mTOR? Oncogene 25, 6347–6360 [DOI] [PubMed] [Google Scholar]
- 13. Frias M. A., Thoreen C. C., Jaffe J. D., Schroder W., Sculley T., Carr S. A., Sabatini D. M. (2006) mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr. Biol. 16, 1865–1870 [DOI] [PubMed] [Google Scholar]
- 14. Jacinto E., Facchinetti V., Liu D., Soto N., Wei S., Jung S. Y., Huang Q., Qin J., Su B. (2006) SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 127, 125–137 [DOI] [PubMed] [Google Scholar]
- 15. Yang Q., Inoki K., Ikenoue T., Guan K. L. (2006) Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 20, 2820–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zoncu R., Efeyan A., Sabatini D. M. (2011) mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12, 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang J. (2007) The direct involvement of SirT1 in insulin-induced insulin receptor substrate-2 tyrosine phosphorylation. J. Biol. Chem. 282, 34356–34364 [DOI] [PubMed] [Google Scholar]
- 18. Song H., Li C. W., Labaff A. M., Lim S. O., Li L. Y., Kan S. F., Chen Y., Zhang K., Lang J., Xie X., Wang Y., Huo L. F., Hsu S. C., Chen X., Zhao Y., Hung M. C. (2011) Acetylation of EGF receptor contributes to tumor cell resistance to histone deacetylase inhibitors. Biochem. Biophys. Res. Commun. 404, 68–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wellen K. E., Hatzivassiliou G., Sachdeva U. M., Bui T. V., Cross J. R., Thompson C. B. (2009) ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. North B. J., Verdin E. (2004) Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 5, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bao J., Sack M. N. (2010) Protein deacetylation by sirtuins: delineating a post-translational regulatory program responsive to nutrient and redox stressors. Cell Mol. Life Sci. 67, 3073–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 23. Thompson P. R., Wang D., Wang L., Fulco M., Pediconi N., Zhang D., An W., Ge Q., Roeder R. G., Wong J., Levrero M., Sartorelli V., Cotter R. J., Cole P. A. (2004) Regulation of the p300 HAT domain via a novel activation loop. Nat. Struct. Mol. Biol. 11, 308–315 [DOI] [PubMed] [Google Scholar]
- 24. Foster K. G., Acosta-Jaquez H. A., Romeo Y., Ekim B., Soliman G. A., Carriere A., Roux P. P., Ballif B. A., Fingar D. C. (2010) Regulation of mTOR complex 1 (mTORC1) by raptor Ser863 and multisite phosphorylation. J. Biol. Chem. 285, 80–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tato I., Bartrons R., Ventura F., Rosa J. L. (2011) Amino acids activate mammalian target of rapamycin complex 2 (mTORC2) via PI3K/Akt signaling. J. Biol. Chem. 286, 6128–6142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blander G., Guarente L. (2004) The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 73, 417–435 [DOI] [PubMed] [Google Scholar]
- 27. Haigis M. C., Sinclair D. A. (2010) Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 5, 253–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Verdin E., Hirschey M. D., Finley L. W., Haigis M. C. (2010) Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem. Sci. 35, 669–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yeung F., Hoberg J. E., Ramsey C. S., Keller M. D., Jones D. R., Frye R. A., Mayo M. W. (2004) Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 23, 2369–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Madrid L. V., Wang C.-Y., Guttridge D. C., Schotelius A. J. G., Baldwin A. S., Mayo M. W. (2000) Akt suppresses apoptosis by stimulating the transactivation potential of the RelA/p65 subunit of NF-κB. Mol. Cell Biol. 20, 1626–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.