Background: Staphylococcus aureus has evolved a web of mechanisms to disrupt the human complement system.
Results: We report the structures of two staphylococcal complement inhibitor proteins, SCIN-B and SCIN-D.
Conclusion: We have identified differences in C3b recognition within active SCIN proteins and suggest a physical basis for lack of C3b binding by SCIN-D.
Significance: This analysis may inform future design of complement-targeted therapeutics.
Keywords: Complement System, Innate Immunity, Protein-Protein Interactions, Staphylococcus aureus, Structural Biology
Abstract
To survive in immune-competent hosts, the pathogen Staphylococcus aureus expresses and secretes a sophisticated array of proteins that inhibit the complement system. Among these are the staphylococcal complement inhibitors (SCIN), which are composed of three active proteins (SCIN-A, -B, and -C) and one purportedly inactive member (SCIN-D or ORF-D). Because previous work has focused almost exclusively on SCIN-A, we sought to provide initial structure/function information on additional SCIN proteins. To this end we determined crystal structures of an active, N-terminal truncation mutant of SCIN-B (denoted SCIN-B18–85) both free and bound to the C3c fragment of complement component C3 at 1.5 and 3.4 Å resolution, respectively. Comparison of the C3c/SCIN-B18–85 structure with that of C3c/SCIN-A revealed that both proteins target the same functional hotspot on the C3b/C3c surface yet harbor diversity in both the type of residues and interactions formed at their C3b/C3c interfaces. Most importantly, these structures allowed identification of Arg44 and Tyr51 as residues key for SCIN-B binding to C3b and subsequent inhibition of the AP C3 convertase. In addition, we also solved several crystal structures of SCIN-D to 1.3 Å limiting resolution. This revealed an unexpected structural deviation in the N-terminal α helix relative to SCIN-A and SCIN-B. Comparative analysis of both electrostatic potentials and surface complementarity suggest a physical explanation for the inability of SCIN-D to bind C3b/C3c. Together, these studies provide a more thorough understanding of immune evasion by S. aureus and enhance potential use of SCIN proteins as templates for design of complement targeted therapeutics.
Introduction
Complement is a primary arm of innate immunity that plays essential roles in clearance of microbial invaders and immune complexes, initiation of inflammation, development, and differentiation and serves as a bridge to the adaptive immune response (1). Activation of complement may be achieved spontaneously and continuously (tick-over) or via binding of pattern recognition proteins to pathogen-specific surface structures to trigger either the classical, lectin, or alternative pathways (AP).2 Despite the mode of initiation, all three pathways of complement converge at the level of C3 convertases, which lie at the heart of the complement cascade by converting the native complement component C3 into its bioactive proteolytic fragments C3a and C3b. Whereas C3a is an anaphylatoxin that promotes several effector functions of complement, C3b covalently opsonizes nearby surfaces via its reactive thioester moiety that becomes exposed upon activation of native C3. A powerful self-amplification loop is established by surface-bound C3b and the proteolytic Bb fragment of factor B (fB), which together form the AP C3 convertase (C3bBb) that enzymatically converts native C3 into C3b. It is this self-amplification of C3b that drives the downstream formation of C5 convertases, triggers inflammation and chemotaxis of phagocytes via C5a generation, and initiates C5b-dependent assembly of the terminal complement complex. After complement activation, targeted cells may be destroyed either through opsonin-mediated phagocytic uptake or by terminal complement complex-mediated membrane disruption (1).
Because the AP exhibits a low but continuous state of activation even in the absence of infection and because the majority of complement activity is in many cases attributed to the AP C3 convertase self-amplification loop (2), it is paramount that this positive feedback system be exquisitely regulated to prevent indiscriminate complement attack and damage to healthy host cells and tissues. Whereas a number of so-called regulators of complement activation (for review, see Ref. 3) have been described, factor H (fH) is of particular importance as a negative regulator of C3 convertase activity. fH is present as a freely circulating protein within plasma; however, it is also effectively adsorbed to host cells through association with negatively charged carbohydrate markers exposed on their surfaces (4, 5). Although the inherently labile nature of AP C3 convertases clearly contributes to their regulation (6, 7), fH greatly enhances the irreversible dissociation of their Bb fragment in a process known as decay acceleration. In addition to this, fH also functions as a cofactor for the factor I-mediated degradation of C3b into iC3b. Although iC3b serves as a ligand for complement receptors exposed on the surface of lymphoid and phagocytic cells, it is no longer a substrate for further convertase assembly. In this manner fH inhibits downstream complement amplification by disrupting the multiprotein convertase enzyme and by promoting degradation of the C3b scaffold required for convertase formation (1, 3).
Interestingly, the opportunistic bacterial pathogen Staphylococcus aureus has also evolved a potent group of small secreted proteins that effectively target and disrupt the human complement response (8, 9). These proteins are both structurally divergent and mechanistically distinct from fH, and their expression and secretion (along with other immune modulators) is thought to contribute to S. aureus survival in the presence of the robust inflammatory and phagocytic response mounted by an immunocompetent host (10, 11). Although their potential antigenicity and existing antibody titers against these proteins has been suggested to prevent their direct use in treating complement-related diseases in human populations (12–14), they nevertheless present an evolutionarily optimized template for the design of therapeutic complement inhibitors (9, 15). For such long term applications to be effectively approached, however, a detailed molecular understanding of the interactions between human complement components and these bacterial inhibitors is required.
One sophisticated inhibitory mode has been recently reported for the so-called staphylococcal complement inhibitor protein (herein denoted SCIN-A) (16, 17). SCIN-A acts at the level of AP C3 convertases and blocks amplification of C3b deposition on the microbial surface. Structure/function studies revealed that SCIN-A binds a functional hotspot on C3b and that SCIN-A bound convertases (C3bBb/SCIN-A) become trapped in a catalytically inactive state (17–19). In addition to this, SCIN-A also blocks host fH binding to C3b and, in doing so, stabilizes this inactive form of the convertase against decay acceleration (17). More recent work has also shown that a second C3b binding site on SCIN-A (17–19) promotes formation of (C3bBb/SCIN-A)2 pseudo-dimers that mask the C3b recognition site of complement receptors CR1 and CRIg, thereby blocking phagocytic uptake of C3b-opsonized bacterial cells (20). In this manner, SCIN-A not only interferes with complement amplification and convertase dynamics; it also disrupts downstream immune processes initiated via complement activation.
Aside from SCIN-A, there exist two additional related proteins denoted SCIN-B and SCIN-C, with demonstrated complement inhibitor activity (12, 14). A fourth protein known as SCIN-D (also referred to as ORF-D (14)) has also been grouped with the SCIN family on the basis of sequence homology; however, it displays none of the complement inhibition or anti-phagocytic properties exhibited by the active members (12, 14). Overall, these additional proteins share 43, 47, and 32% sequence identity to SCIN-A, respectively (supplemental Fig. S1A). Still, there is little known about the structures of these remaining SCIN family proteins, and there is almost no detailed information on the molecular nature of their interactions with complement proteins. To begin addressing such questions, we report here the crystal structures of an N-terminal truncation mutant of SCIN-B, both free and in complex with the C3 fragment, C3c. Analysis of these structures has identified specific determinants in SCIN-B that are important for C3b binding and required for complement inhibition by this protein. In addition, we have also determined the crystal structure of the purportedly inactive SCIN-D protein. Interestingly, SCIN-D differs in secondary and tertiary structure in a region centered on its N-terminal α helix. Although SCIN-D maintains many of the key residues that contribute to C3b binding by its counterparts, both its electrostatic potential and surface complementarity values for C3c differ from those of SCIN-A and SCIN-B. Together, these studies expand upon our understanding of the structure and molecular basis for complement recognition and inhibition by SCIN proteins.
EXPERIMENTAL PROCEDURES
Proteins
Purified human C3c, C3b, fB, factor D (fD), fH, and cobra venom factor (CVF) were obtained from Complement Technology (Tyler, TX). A mouse monoclonal antibody (clone 030-08; catalogue number sc-58929) that recognizes the TED domain of human C3/C3b/iC3b/C3d was purchased from Santa Cruz Biotechnology, Inc.
DNA fragments encoding SCIN-A, SCIN-B, SCIN-D, and directed fragments thereof were amplified from S. aureus (strain Mu50) genomic DNA and subcloned into the prokaryotic overexpression vector pT7HMT as previously described (21, 22). Site-directed mutagenesis of SCIN-B and SCIN-D was carried out by a two-step megaprimer PCR method using their corresponding pT7HMT-based overexpression plasmids as a template (23). Individual clones were confirmed by DNA sequencing. After expression and purification, mutant proteins were analyzed for structural integrity by comparative circular dichroism spectropolarimetry with respective wild-type samples. Recombinant proteins harboring the c-myc epitope tag at their N terminus were prepared similarly to their untagged counterparts, with the exception that the tobacco etch virus protease digestion step was omitted (21, 22). Seleno-l-methionine-labeled samples were obtained using a method previously described (24).
SCIN-B was also overexpressed from a modified version of the pT7HMT vector that encodes a cysteine residue as the first amino acid of the polypeptide of interest. Site-specifically biotinylated cys-SCIN-B was produced using the EZ-Link Maleimide-PEG2-Biotin reagent (Thermo Scientific) according to the manufacturer's instructions, and the excess biotinylation reagent was removed by ultrafiltration into PBS (pH 7.4). Successful biotinylation was confirmed by Western blotting using a streptavidin-conjugated anti-HRP antibody and SuperSignal Pico Chemiluminescent substrate (Thermo Scientific).
C3b site-specifically biotinylated at Cys1010 was prepared by activating native C3 via a stable CVF-based C3 convertase at pH 7.0 (PBS with 5 mm nickel sulfate) in the presence of the biotinylation reagent described above. Briefly, 60 μl of a solution containing CVF and fB (1 μm each) were activated to CVFBb by adding factor D (0.1 μm final concentration) and incubating for 5 min at room temperature. At this time the CVFBb was added to 330 μl of a solution containing native C3 (4 μm) and EZ-Link Maleimide-PEG2-Biotin (190 μm) and incubated for 2 h further at room temperature. The reaction was then incubated overnight at 4 °C to allow free-thiol derivatization to approach completion. The next day the sample was buffer-exchanged into 20 mm Tris (pH 7.5) and applied to a 1-ml Resource Q anion-exchange column (GE Healthcare) to isolate the C3b-biotin from contaminating reactants/proteins. Successful biotinylation was confirmed by Western blotting as described above.
For preparation of samples used in crystallization trials, C3c was first treated with a 1:200 (w/w) ratio of a recombinant Escherichia coli maltose-binding protein peptide N-glycosidase F fusion protein for 2 h at 37 °C to remove N-glycans. After this, the deglycosylated C3c protein was further isolated by anion exchange chromatography on a Resource Q column (GE Healthcare). For complex formation, a solution of purified SCIN-B18–85 was mixed with deglycosylated C3c to yield an equimolar complex of C3c/SCIN18–85. The complex sample was exchanged by ultrafiltration into buffer of 10 mm Tris (pH 7.4), 50 mm NaCl, and concentrated to 5 mg/ml total protein.
Crystallization, X-ray Diffraction Data Collection, Structure Solution, and Refinement
All crystallization experiments were carried out by vapor diffusion of hanging drops. Before diffraction analysis, crystal samples were preserved by flash-cooling in a Dewar of liquid nitrogen. X-ray diffraction data were collected using either beamline 22-ID or 22-BM of the Advanced Photon Source (Argonne National Laboratory), and the individual reflections were indexed, integrated, and scaled using either HKL2000 or XDS/XSCALE (25). All aspects of structure solution and refinement were carried out using individual programs as implemented within the PHENIX suite (26). In particular, all molecular replacement solutions were obtained using the program PHASER, whereas de novo phase calculation by single wavelength anomalous dispersion was performed using PHENIX.AUTOSOL. For all structures except that of C3c/SCIN-B18–85, an initial model was built using PHENIX.AUTOBUILD. Final models were obtained after manual building in COOT (27) and subsequent rounds of refinement using PHENIX.REFINE. Models were analyzed and validated using MOLPROBITY (28) before PDB deposition. All structural representations were prepared using PyMOL, whereas comparisons of superimpositions were determined using STAMP (29). The following sections describe specific information pertaining to individual crystals and their structures.
SCIN-B18–85 crystals grew in 1 day at 4 °C using a solution of 0.1 m sodium citrate (pH 3.5), 1 m ammonium sulfate as a precipitant. Drops consisted of 1 μl of protein sample (12 mg/ml) and 1 μl of precipitant solution that had been previously diluted with three volumes of double-distilled H2O. Crystals were cryoprotected using precipitant solution containing 20% (v/v) glycerol. A polyalanine model of the previously published SCIN-A structure (PDB code 2QFF (14)) was created and used to solve this structure by molecular replacement.
C3c/SCIN-B18–85 co-crystals appeared in 2–5 days at 20 °C using 0.1 m bis-tris (pH 6.5) and 15% (w/v) polyethylene glycol 3350 as a precipitant. Drops consisted of 1 μl of complex sample (5 mg/ml) and 1 μl of precipitant solution that had been previously diluted with 1 volume of double-distilled H2O. Crystals were cryoprotected by a brief incubation in precipitant containing 20% (v/v) glycerol. The refined coordinates of SCIN-B18–85 and C3c (PDB code 2A74 (30)) were used to solve this structure by molecular replacement. Non-crystallographic symmetry restraints were employed where applicable during all steps of solution and refinement of this structure.
Both native and selenium-labeled SCIN-D8–83 crystals were obtained after 2–3 days at 20 °C using a precipitant solution of 0.1 m bis-tris (pH 5.0), 30% (w/v) polyethylene glycol 8000, and 20 mm sodium thiocyanate. Individual drops consisted of 1 μl of protein sample (10 mg/ml) and 1 μl of precipitant solution that had been previously diluted with an equal volume of double-distilled H2O. Cryoprotection was achieved by adding 10% (v/v) glycerol to the precipitant solution. The initial structure was solved by single wavelength anomalous dispersion, and this refined model was used to solve the native crystal.
Full-length SCIN-D crystals appeared after 1 month at 20 °C using 0.1 m bis-tris (pH 4.8) and 15% (w/v) polyethylene glycol 3350 as a precipitant. Drops consisted of 1 μl of protein (10 mg/ml) and 1 μl of precipitant solution that had been previously diluted with an equal volume of double-distilled H2O. Crystals were cryoprotected using precipitant solution containing 20% (v/v) glycerol. The refined coordinates of native SCIN-D8–83 were used to solve this structure by molecular replacement.
AlphaScreen Binding Assay
Assays for binding between SCIN-A/SCIN-B and various proteins were conducted by employing a luminescent microbead AlphaScreen technology (Ref. 31; PerkinElmer Life Sciences). The principle of this assay relies upon a streptavidin donor bead, which recognizes a biotinylated ligand that binds a second target protein, which itself can be adsorbed to acceptor bead via a c-Myc epitope or a protein-specific mouse monoclonal IgG. Using this approach, an equilibrium competition binding assay was established in 96-well format ½ area opaque plates using the AlphaScreen c-Myc detection kit or anti-mouse IgG kit purchased from PerkinElmer Life Sciences and carried out according to either of the following general procedures.
SCIN-A/C3b competition assays were carried out in a final reaction volume of 25 μl by adding each assay component to the following final concentrations: 50 nm myc-SCIN-A, 5 nm C3b-biotin, 20 μg/ml c-Myc AlphaScreen acceptor beads, and 20 μg/ml AlphaScreen Streptavidin Donor beads. A dilution series for each unlabeled competitor protein was prepared. The reaction was performed over the course of 2.5 h and began by mixing myc-SCIN-A, C3b-biotin, and varying concentrations of unlabeled competitor. After 1 h of incubation time at room temperature, this solution was mixed with the acceptor beads and allowed to incubate for an additional 1 h. Finally, donor beads were added and after a 30-min incubation time the evolving AlphaScreen signal (photon counts at 630 nm/s) was read on an EnSpire multimode plate reader (PerkinElmer Life Sciences).
SCIN-B/C3b competition assays were carried out using the same general protocol as described above, with the following changes in final concentrations of each component: 25 nm biotinylated-SCIN-B, 2 nm C3b, and 1 nm anti-C3d IgG. The reaction was begun by mixing, C3b, and varying concentrations of unlabeled competitor while separately mixing acceptor beads with anti-C3d IgG. After a 1-h incubation time at room temperature, these two solutions were mixed and allowed to incubate for an additional 1 h. Finally, donor beads were added, and after 30 min of incubation time the AlphaScreen signal was measured.
The AlphaScreen signal was normalized to wells containing no inhibitor, and a dose-response curve was generated by plotting normalized signal versus log[competitor (m)]. Where applicable, apparent equilibrium dissociation constants (KD) were calculated by nonlinear curve-fitting to a one-site fit Ki model using GraphPad Prism5 software (GraphPad; La Jolla, CA). IC50 values were defined by,
where [L] = [myc-SCIN-A] = 50 nm, and KD,LT = KD,SCIN-A/C3b =177 nm (as judged independently by surface plasmon resonance (SPR) (17).
SPR Binding Assay
The interaction between C3b and various SCIN proteins and their mutants was measured by SPR using a Biacore 3000 instrument (GE Healthcare) at 25 °C. HBS-ET (10 mm HEPES (pH 7.4), 150 mm NaCl, 3 mm EDTA, 0.005% (v/v) Tween 20) was used as the mobile phase throughout the experiment. Site-specifically biotinylated C3b was captured on a streptavidin sensor chip (GE Healthcare) at a density of 4400 resonance units (17). SCIN proteins were diluted to a concentration of 10 μm in HBS-ET and injected for 1 min at a 20 μl/min flow rate with a dissociation phase of 2 min. Signals from an empty streptavidin flow cell and from an ensemble of buffer blank injections were used to perform double referencing of sample responses. Data processing and evaluation was performed using Scrubber (v2.0c; BioLogic Software Pty Ltd, Campbell, Australia). Because SPR signal generation is directly dependent on the molecular weight of injected analytes, each response signal was divided by the molecular weight of the corresponding protein to allow for a ranking of SCIN mutants (the y axis was adjusted to 100% for the signal of wild-type SCIN-B).
Alternative Pathway Convertase Assay
Activation of C3 by the alternative pathway convertase was measured by monitoring proteolysis of the C3 α chain by SDS-PAGE. This reflects physiological cleavage of C3 into C3b (composed of α′ and β chains) and C3a. To begin, the initial, fluid-phase AP convertase was formed in vitro by incubating 1 volume of an equimolar mixture of C3b and fB (1 μm each) with 0.1 volumes of 1 μm factor D in a buffer of 20 mm HEPES (pH 7.4), 150 mm NaCl, 5 mm nickel sulfate. This reaction was incubated at 20 °C for 5 min, at which time 3 μl were withdrawn and added directly to 7 μl of native C3 (2 μm final concentration in the same buffer). Proteolysis of C3 was then allowed to proceed for 1 h further before quenching the reaction by adding 5 μl of reducing SDS-PAGE sample buffer and heating to 95 °C for 5 min. To assess inhibition of convertase activity by various SCIN molecules, each protein was added to the uncleaved native C3 component in a 2-fold dilution series (9 different dilutions) near their experimentally determined KD values for C3b; in the case of the SCIN-B18–85-R44A/Y51A mutant, identical concentrations to wild-type SCIN-B18–85 were used. Upon reaction completion, 50% of the denatured sample was separated by 10% tris-tricine SDS-PAGE and stained with Coomassie Brilliant Blue. The amount of both α and α′ chains present was quantitated by measuring band intensities using ImageJ (32) and normalized to the invariant β-chain (which served as an internal loading control). Data were fitted to a four-parameter (variable slope) dose-response curve.
RESULTS
Crystal Structures of a Protease-resistant Fragment of SCIN-B (SCIN-B18–85), Both Free and Bound to Human C3c
Two SCIN-A binding sites are found in all crystal structures containing SCIN-A bound to either C3b or C3c, and the residues involved in these sites were mapped onto a sequence alignment of SCIN-A and SCIN-B (supplemental Fig. S1A). Although the second binding site appears to promote formation of inhibited AP convertase (C3bBb/SCIN-A)2 pseudo-dimers and subsequent disruption of phagocytosis via certain classes of cell surface-exposed complement receptors (12), it has been shown that SCIN-A residues that comprise the first binding site are required for complement inhibition by SCIN-A (12, 19). The first binding site consists of 13 residues almost exclusively derived from the second α helix in SCIN-A; however, when compared with the homologous positions of SCIN-B, 6 of these 13 residues are non-conservative substitutions (e.g. SCIN-A-Tyr51→SCIN-B-Ala53 and SCIN-A-Gln49 → SCIN-B-Tyr51; supplemental Fig. S1A). This is intriguing because many of the amino acids in these positions make extensive side chain-mediated contacts with C3b/C3c (18, 19), and SCIN-B has been reported to have complement inhibition properties similar to those of SCIN-A (12, 14).
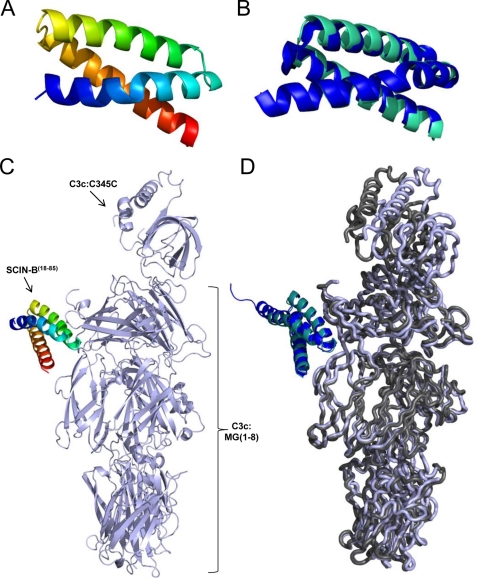
We initiated crystallographic studies on a protease-resistant fragment of SCIN-B (supplemental Fig. S1B) that lacked the first 17 residues of the predicted mature SCIN-B polypeptide (SCIN-B18–85), both free and bound to a terminal fragment of C3 activation, C3c. This approach afforded several technical advantages, including the following. (i) This truncated form of SCIN-B removed an apparently flexible N-terminal region of the molecule while leaving the putative first binding site intact. ii) Crystal structures containing C3c have historically provided higher resolution data when compared with their C3b counterparts (18, 30, 33–35), presumably due to the removal of the conformationally flexible CUB and TED domains. (iii) Previous crystallographic studies of SCIN-A bound to both C3c or C3b revealed that the C3c complex is structurally equivalent to that of C3b (18). In adopting this strategy, we succeeded in crystallizing and solving the structures of the apo form of SCIN-B18–85 and its C3c-bound complex to 1.5 and 3.4 Å limiting resolution, respectively (Fig. 1 and Table 1).
FIGURE 1.
Crystal structures of SCIN-B18–85 and C3c/SCIN-B18–85 at 1.5 and 3.4 Å resolution. A, shown is the 1.5 Å crystal structure of SCIN-B18–85 (PDB code 3T49; N terminus in blue, C terminus in red). B, shown is Cα superposition of the SCIN-B18–85 structure in panel A (cyan) with that of unbound SCIN-A (PDB 2QFF; blue). C, shown is the 3.4 Å crystal structure of human complement fragment C3c bound to SCIN-B18–85 (PDB code 3T4A; C3c in light blue, SCIN-B18–85 in rainbow). D, shown are the Cα superposition of C3c/SCIN-B18–85 (C3c in light blue ribbon, SCIN-B18–85 cyan schematic) and the first binding site of C3c/SCIN-A (PDB code 3OHX; C3c in dark gray ribbon, SCIN-A in blue schematic).
TABLE 1.
Diffraction data collection and structure refinement statistics
| SCIN-B18–85 | C3c/SCIN-B18–85 | SCIN-D8–83 (MSE) | SCIN-D8–83 | SCIN-D | |
|---|---|---|---|---|---|
| Data collectiona | |||||
| Beamline | 22-BM | 22-BM | 22-ID | 22-ID | 22-ID |
| Wavelength (Å) | 1.00 | 1.00 | 0.972 | 0.972 | 0.972 |
| Space group | P21 | P212121 | P21 | P21 | P6522 |
| Cell dimensions | |||||
| a (Å) | 44.67 | 103.30 | 26.10 | 26.15 | 28.86 |
| b (Å) | 66.94 | 165.44 | 94.28 | 94.36 | 28.86 |
| c (Å) | 60.41 | 203.13 | 29.10 | 29.20 | 262.60 |
| β (°) | 95.22 | 97.14 | 97.19 | ||
| Resolution (Å) | 50-1.5 | 50-3.4 | 50-1.5 | 50-1.3 | 50-1.5 |
| Unique reflections | 62,067 | 46,906 | 21,840 | 31,662 | 11,413 |
| Completeness (%) | 98.6 (90.3) | 97.7 (93.7) | 99.7 (91.8) | 92.2 (89.3) | 98.8 (95.4) |
| Rmerge (%)b | 5.8 (38.0) | 16.1 (57.0) | 6.7 (30.8) | 7.8 (46.0) | 6.9 (28.2) |
| Redundancy | 7.0 (4.8) | 12.6 (11.4) | 5.9 (4.2) | 18.7 (10.9) | 5.7 (4.7) |
| I/σI | 30.3 (3.2) | 14.3 (3.9) | 18.5 (5.6) | 35.6 (4.3) | 14.6 (4.5) |
| Refinement | |||||
| PDB accession code | 3T49 | 3T4A | 3T48 | 3T47 | 3T46 |
| Biological units/absorbance unit | 4 | 2 | 2 | 2 | 1 |
| Rwork/Rfree (%)c | 14.1/16.6 | 22.9/27.0 | 15.3/19.3 | 15.5/18.7 | 22.2/24.5 |
| Mean B-factors (Å2) | 19.55 | 67.19 | 25.28 | 19.62 | 16.31 |
| Root mean square deviation | |||||
| Bond lengths (Å) | 0.014 | 0.011 | 0.011 | 0.011 | 0.007 |
| Bond angles (°) | 1.53 | 1.40 | 1.14 | 1.08 | 0.99 |
| Ramachandran plot (%) | |||||
| Favored | 100.0 | 86.4 | 100.0 | 100.0 | 100.0 |
| Allowed | 0.0 | 10.3 | 0.0 | 0.0 | 0.0 |
| Outlier | 0.0 | 3.3 | 0.0 | 0.0 | 0.0 |
| No. of water molecules | 426 | 0 | 135 | 154 | 72 |
| No. of residues | 278 | 2360 | 141 | 141 | 75 |
a Numbers in parentheses are for the highest resolution shell.
b Rmerge = ΣhΣi|Ii(h) − 〈I(h)〉|/ΣhΣiIi(h), where Ii(h) is the ith measurement of reflection h, and 〈I(h)〉 is a weighted mean of all measurements of h.
c R = Σh|Fo(h) − Fc(h)|/Σh|Fo|. Rcryst and Rfree were calculated from the working and test reflection sets, respectively. The test set constituted 5% of the total reflections not used in refinement.
Nearly the entire SCIN-B18–85 protein adopts an anti-parallel, trihelical bundle fold (Fig. 1A). This fold appears to be characteristic of the SCIN family, as it has also been observed for the core portion of SCIN-A (PDB code 2QFF; Fig. 1B). In the co-crystal structure, SCIN-B18–85 is positioned near the macroglobulin 7 (MG7) domain of C3c, where it makes extensive intermolecular interactions (Fig. 1C). In total, SCIN-B18–85 buries 795 Å2 of surface area (as judged by the EBI PISA server interface analysis tool (36)) that is spread across both the first α′ chain fragment (598 Å2) and, to a lesser extent, the β chain of C3c (197 Å2). Further comparison of this C3c/SCIN-B18–85 structure with a representative C3c/SCIN-A structure (PDB code 3OHX (18)) reveals that SCIN-B18–85 binds C3c in the same position as the primary C3c/SCIN-A binding site (Fig. 1D (18)). Specifically, the root mean square deviation of superposition across all C3c Cα atoms is 1.19 Å (β-chain), 0.83 Å (α′ fragment 1-chain), and 2.20 Å (α′ fragment 2-chain), with the greatest differences found between the relative positioning of the individual C345C domains. This is not unexpected, however, as conformational flexibility of the C345C domain with respect to the so-called key-ring like core (i.e. domains MG1–8) has been reported previously for crystal structures containing either C3b or C3c (18, 19).
Two copies of the C3c/SCIN-B18–85 complex are present in the crystallographic asymmetric unit, and clear electron density is observed for all chains (supplemental Fig. S2). Interestingly, a second SCIN-B18–85 binding site is not present in the C3c/SCIN-B18–85 asymmetric unit, nor is one generated upon examining any of the formal crystallographic symmetry-related symmetry copies of C3c. Although this does not preclude the possibility that SCIN-B18–85 may form a second site interaction (as has been observed and analyzed for SCIN-A (17–20)), to our knowledge this is the first SCIN-containing co-crystal structure that lacks any pseudo-dimeric (C3c/SCIN)2 or (C3b/SCIN)2 units.
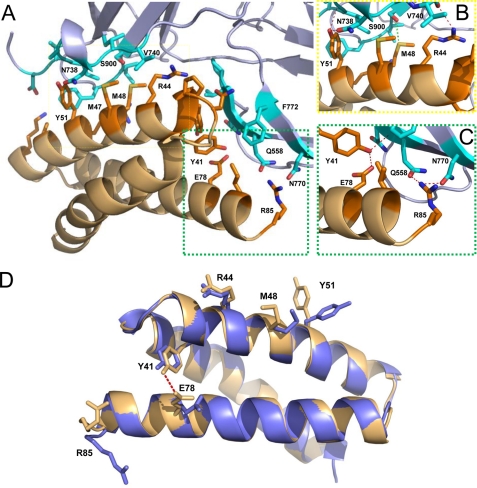
Molecular Basis for the C3c/SCIN-B18–85 Interaction
To better describe the interactions required for active SCIN proteins to bind C3b and block complement function at the level of the AP C3 convertase, we used the EBI PISA server (36) and the CCP4 program CONTACT (37) to analyze the C3c/SCIN-B18–85 interface (Fig. 2A). In total, SCIN-B18–85 donates 20-residue contacts to C3c; this value compares favorably to the 22 contacts that comprise the equivalent site in C3c/SCIN-A (18). As might be expected given their levels of sequence identity, SCIN-A and SCIN-B share some similarity in their contacts with C3c. In particular, Arg42 from SCIN-A and the analogous residue, Arg44 of SCIN-B18–85, form nearly identical interactions with Pro555, Val740, Ser741, and Asp775 of C3c (Fig. 2B). In the C3c/SCIN-A crystal structure (18), Gln49 of SCIN-A makes diverse contacts with residues Phe898, Asn738, Ile733, Ile734, and Ala735, including several side chain-mediated hydrogen bonds. Interestingly, this position in SCIN-B18–85 is non-conservatively substituted to tyrosine (Tyr51), and Phe898 of C3c must adopt an alternate rotamer to form the hydrophobic interactions between these residues in the C3c/SCIN-B18–85 complex (Fig. 2B). Yet as is the case for Gln49 of SCIN-A, this tyrosine residue buries significant surface area (>100 Å2) and appears to mediate critical interactions with C3c (similarly to Arg44 above).
FIGURE 2.
Molecular nature of the C3c/SCIN-B18–85 interface. A, shown is a depiction of the C3c/SCIN-B18–85 interface. Contact residues are drawn as stick models (PDB code 3T4A; C3c residues in cyan; SCIN-B18–85 residues in orange). B, shown is a magnified view of the area within the yellow dashed lines in panel A, which highlights key interactions between residues of C3c α′ chain fragment 1 and residues on the second α helix of SCIN-B18–85. C, shown is a magnified view of the area within the green dashed lines in panel A, which highlights key interactions between the C3c β-chain and residues on the third α helix of SCIN-B18–85. Side-chain to side-chain and side-chain to main-chain hydrogen bonds are drawn as red dashed lines, whereas sulfur-mediated hydrogen bonds of SCIN-B18–85-Met48 are drawn as green dashed lines. D, shown is a comparison of unbound SCIN-B18–85 (PDB code 3T49) with its C3c-bound form. The unbound SCIN-B18–85 structure (blue) is shown in a schematic overlaid with SCIN-B18–85 (orange) from the C3c/SCIN-B18–85 co-crystal structure. Residues that have significantly different Cβ positions are drawn in stick form. Note that the intramolecular hydrogen bond between Tyr41 and Glu78 is absent from the unbound form of SCIN-B18–85. Met47 is significantly shifted at the Cβ atom but has been removed from the figure for clarity.
One peculiar feature of the SCIN-B18–85 binding site for C3c is two sequential methionine residues that are absent from SCIN-A. One of these, Met47 of SCIN-B18–85, forms hydrophobic interactions with C3c Asn738. These contacts appear similar to those formed by Ile46 of SCIN-A, which occupies the analogous position according to sequence alignment. On the other hand, Met48 of SCIN-B18–85 forms sulfur-mediated hydrogen bonds with Ser900 of C3c (Fig. 2B). Although this sort of interaction appears relatively rare, other structural studies have also reported methionine-mediated hydrogen bonding in protein-protein interactions (38).
A final noteworthy aspect of the C3c/SCIN-B18–85 structure is found in the vicinity of the SCIN-B18–85 C terminus, where several residues, including the C-terminal residue, Arg85, form interactions with C3c (Fig. 2C). This feature is unique to SCIN-B18–85, as none of the C-terminal residues in SCIN-A appears to contribute significantly to C3c binding. Interestingly, superposition of the unbound structure with that of C3c bound SCIN-B18–85 reveals both the formation of an intramolecular hydrogen bond between Tyr41 and Glu78 and a significant movement of Arg85 in the bound protein (Fig. 2D). Aside from these and the clear rotamer differences between Tyr51 described above, relatively few changes are seen between the bound and unbound state of the SCIN-B18–85 protein (Cα root mean square deviation of 0.55 Å). This suggests that the C3b binding site in SCIN-B is preformed in much the same way as has been previously described for the S. aureus complement inhibitors Efb-C (39) and Ehp (40).
Arg44 and Tyr51 Contribute Significantly to C3b Binding and Convertase Inhibition by SCIN-B18–85
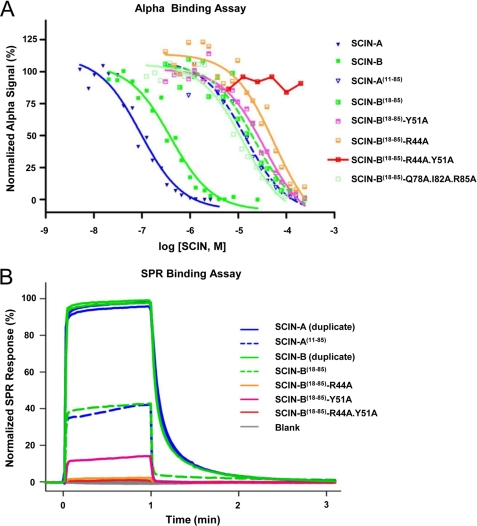
We adopted two parallel experimental approaches to quantitatively assess SCIN-B binding to its physiologically relevant C3 target, C3b (Fig. 3, supplemental Fig. S3, and Table 2). The first of these was based upon a competition AlphaScreen strategy (31), whereas the second measured binding to a site-specific-oriented C3b biosensor via SPR (17, 41). Apparent KD values of 73.5 and 308 nm were calculated from competition AlphaScreen binding curves for full-length SCIN-A and SCIN-B, respectively (Fig. 3A and Table 2). These values are comparable to those previously reported for SCIN-A (170–265 nm), as judged by both SPR and isothermal titration calorimetry (17). Furthermore, although SCIN-B18–85 has a roughly 100-fold reduced affinity for C3b in all assay formats (apparent KD of 16.9 μm; Fig. 3, supplemental Fig. S3 and Table 2), it nevertheless exhibits clearly saturable binding. This result suggests that residues 1–17 of SCIN-B must also contribute in some manner to its interaction with C3b.
FIGURE 3.
Arg44 and Tyr51 contribute significantly to SCIN-B18–85 binding to C3b. The ability of SCIN proteins to bind to C3b was assessed through two different assay formats. A, shown are results of an AlphaScreen bead-based binding assay, where competition for binding between various proteins and full-length SCIN-A was measured. SCIN-A (blue) and SCIN-B (dark green) show comparable affinity for C3b. Both SCIN-A11–85 (blue dashed lines) and SCIN-B18–85 (green dashed lines) retain binding activity but display reduced affinity for C3b when compared with their full-length counterparts. Substitution of C-terminal contact residues (Q78A/I82A/R85A; pale green) yields an affinity comparable with wild-type SCIN-B18–85. Mutants where single mutations of Arg44 and Tyr51 to alanines were made (salmon and lavender, respectively) and showed reduced affinity compared with SCIN-B18–85, whereas a double mutant (R44A/Y51A; red) fails to compete at any concentration. Data presented are representative experiments, and each experiment was repeated in duplicate or triplicate. B, shown is an SPR ranking study for various SCIN protein interactions with site-specific immobilized C3b. The SPR signals were divided by molecular weight of each protein and adjusted to 100% for wild-type SCIN-B (corresponding to 180 resonance units). These data from a well established assay format largely mirror those obtained from the high throughput AlphaScreen approach described in panel A. Legends for both panels are inset.
TABLE 2.
Solution C3b binding constants for various SCIN proteins
NA, not applicable.
| Competitor | KDa | R2 | 95% Confidence interval (KD) | Points analyzed | Outliers | S/Nb |
|---|---|---|---|---|---|---|
| SCIN-A | 73.5 nm | 0.967 | 47.6 to 113 nm | 20 | 0 | 84 |
| SCIN-B | 308 nm | 0.967 | 202 to 470 nm | 19 | 0 | 192 |
| SCIN-A11–85 | 11.0 μm | 0.970 | 6.68 to 17.9 μm | 16 | 3 | 87 |
| SCIN-B18–85 | 16.9 μm | 0.948 | 9.67 to 29.5 μm | 20 | 0 | 13 |
| SCIN-B18–85Y51A | 24.5 μm | 0.973 | 16.2 to 37.0 μm | 20 | 0 | 10 |
| SCIN-B18–85R44A | 39.5 μm | 0.942 | 20.8 to 75.1 μm | 20 | 0 | 34 |
| SCIN-B18–85 R44A/Y51A | NA | NA | NA | 8 | 0 | 50 |
| SCIN-B18–85 Q78A/I82A/R85A | 8.95 μm | 0.967 | 5.70 to 14.1 μm | 20 | 0 | 168 |
| Factor H | 20.1 nm | 0.923 | 10.9 to 37.1 nm | 20 | 0 | 169 |
a Parameters for data analysis and curve fitting are found under “Experimental Procedures.”
b S/N denotes the ratio of experimental luminescence signal to noise. Background noise levels were obtained by measuring the signal of a mock assay where SCIN-Amyc was omitted.
To test whether this feature was unique to SCIN-B, an N-terminal truncation mutant of SCIN-A (i.e. SCIN-A11–85) was also expressed and characterized. Indeed, SCIN-A11–85 also showed reduced affinity for C3b in the AlphaScreen (apparent KD of 11.0 μm) and decreased binding in SPR studies when compared with its full-length counterpart (Fig. 3 and Table 2). Thus, the N-terminal regions of active SCIN proteins contribute to high affinity C3b binding even though they are not absolutely required for binding to occur.
To directly investigate the contributions of the residues highlighted above, we generated site-directed mutants in SCIN-B18–85 and compared their abilities to bind C3b (Fig. 3 and Table 2). A SCIN-B18–85 mutant that simultaneously exchanged each of the Gln78, Ile82, and Arg85 side chains to alanine bound C3b with similar affinity to wild type. However, single mutations of Arg44 and Tyr51 to alanines resulted in reduced affinity for C3b compared with wild type, and no binding was detected for a SCIN-B18–85-R44A/Y51A double mutant. In summary, although abrogation of the C-terminal contacts does not impair SCIN-B18–85 binding to C3b, the interactions formed by Arg44 and Tyr51 are important for C3b binding by this bacterial inhibitor.
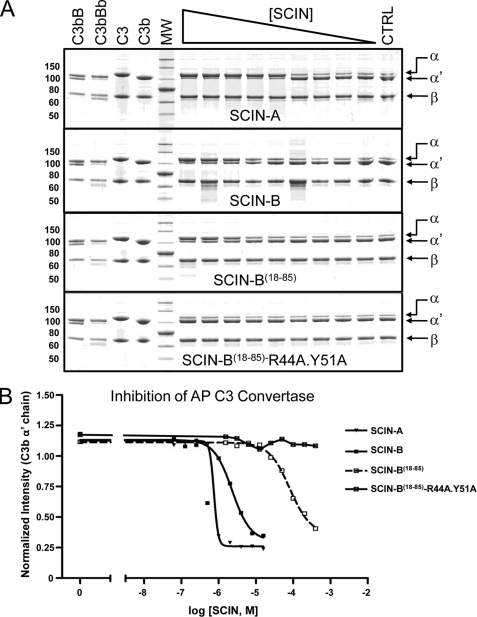
SCIN-A effectively competes with fH for binding to C3b and renders SCIN-A-bound convertases insensitive to decay acceleration (17). This behavior arises from significant overlap of the SCIN-A and fH binding sites on C3b (18) and also appears to be shared by SCIN-B as judged by the C3c/SCIN-B18–85 co-crystal (supplemental Fig. S4); not surprisingly, SCIN-B competes with fH in an AlphaScreen assay (supplemental Fig. S4). However, the principle anti-complement activity of SCIN proteins lies in their ability to inhibit C3 cleavage/activation by fully assembled convertases (17). Because convertase inhibition presumably requires direct SCIN binding to C3b, we tested whether loss of Arg44 and Tyr51 would affect the ability of SCIN-B18–85 to block C3 cleavage via the AP C3 convertase (Figs. 4 and supplemental Fig. S4). Indeed, whereas SCIN-A, SCIN-B, and SCIN-B18–85 all displayed dose-dependent inhibition of C3 conversion to C3a and C3b production, no convertase inhibitor activity was observed for the SCIN-B18–85 double mutant. In conjunction with Fig. 3, these results demonstrate that high affinity C3b binding is required for inhibition of the AP C3 convertase by SCIN-B.
FIGURE 4.
Arg44 and Tyr51 contribute significantly to SCIN-B18–85 inhibition of the AP C3 convertase. The activity of a fluid-phase AP C3 convertase was measured by monitoring proteolytic degradation of the C3 α chain by SDS-PAGE and visualized by Coomassie Brilliant Blue staining (supplemental Fig. S5). A, various SCIN proteins were examined for their respective abilities to inhibit convertase activity across a 2-fold dilution series near their apparent KD values obtained from the assay (Fig. 3A and Table 2). The dilution series employed for each protein was as follows: SCIN-A, 16 μm–62.5 nm; SCIN-B, 64 μm–250 nm; SCIN-B18–85, 400–1.56 μm; SCIN-B18–85- R44A/Y51A, 400–1.56 μm. Purified proteins (1 μg each of C3 and C3b) were included as a reference and appear at the left side of each image, as were controls demonstrating the formation of C3bBb used in each set of assays. Reactions containing no inhibitor (CTRL) served as the positive control for C3 activation to C3b and appear at the right side of each image. Data presented are representative experiments of trials repeated in triplicate. B, dose-response convertase inhibition curves for various SCIN proteins are shown in panel A. Band intensities corresponding to each α′ chain were integrated, normalized to those of the C3/C3b β chain (which served as an internal loading control), and plotted against the concentration of each inhibitor. The experimental IC50 values were 750 nm, 2.3 μm, and 80 μm for SCIN-A, SCIN-B, and SCIN-B18–85, respectively. The legend is inset.
Divergent Structure in SCIN-D
SCIN-D shares a significant amino acid sequence identity (∼30%) with SCIN-A and is predicted to have the same secondary structure as active SCIN proteins (14). However, although SCIN-D is typically considered part of the same family, it is neither an effective complement inhibitor nor does it appear to have anti-phagocytic activity; thus, SCIN-D has been used extensively as a negative control in functional studies (12, 14, 17). Because both of these functions require SCIN binding to C3b, it follows that SCIN-D is unable to interact with C3b (14, 17). Consistent with previous results, SCIN-D did not bind to C3b in any of the AlphaAssay formats described here (supplemental Fig. S3). Thus, to provide a basis for comparing the structural features of active SCIN proteins with this inactive homolog, we also undertook crystallographic studies of SCIN-D.
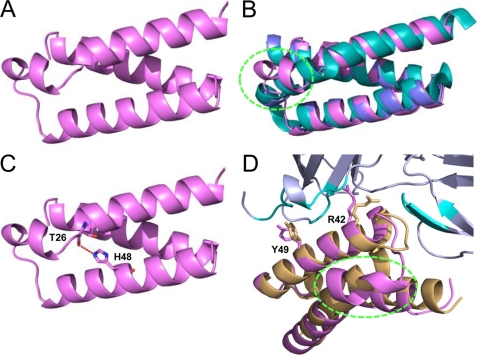
Although diffraction quality full-length SCIN-D crystals were obtained, initial attempts at structure solution via molecular replacement using SCIN-A- or SCIN-B-derived models were unsuccessful. Because these crystals took nearly a month to grow, a truncated variant (SCIN-D8–83) was designed, expressed in both native and selenium-labeled forms, and crystallized. This structure was solved by single-wavelength anomalous dispersion to 1.5 Å limiting resolution, and its refined coordinates were subsequently used to solve both native SCIN-D8–83 and full-length SCIN-D to 1.3 and 1.5 Å resolution, respectively (Figs. 5A, supplemental Fig. S6, and Table 1).
FIGURE 5.
Crystal structure of SCIN-D8–83 at 1.3 Å resolution. A, shown is a refined model of native SCIN-D8–83, drawn as violet ribbon (PDB code 3T47). B, superimposition of native SCIN-D8–83 (violet) with the crystal structures of SCIN-A (PDB code 2QFF; teal) and SCIN-B18–85 (PDB code 3T49; blue) is shown. The region of divergent secondary and tertiary structure within SCIN-D α helix 1 is highlighted by a dashed green oval. C, a hydrogen bond (dashed red line) between the backbone carbonyl of Thr26 and the side chain of His48 disrupts the canonical hydrogen bonding pattern observed in α helices. This interaction appears to be a conserved feature of the SCIN-D protein, as these residues are invariant across all SCIN-D sequences examined (Garcia, B. L., and Geisbrecht, B. V., unpublished observations). D, shown is superposition of SCIN-D8–83 (violet) onto the structure of SCIN-B18–85 (orange) as it is found in the C3c/SCIN-B18–85 co-crystal (Figs. 1 and 2). In this panel residues from C3c that contact SCIN-B18–85 are colored teal, whereas the remainder of C3c is colored gray. Residues Arg44 and Tyr51 from SCIN-B18–85 along with their corresponding positions in SCIN-D8–83 (i.e. Arg42 and Tyr49) are depicted as sticks. Note that the N terminus of SCIN-D is drawn at the right in panels A–C but appears at the left in panel D.
Whereas all SCIN-D structures are virtually identical (Cα root mean square deviation of 0.08 Å (supplemental Fig. S6)), superposition of any of these structures on to either SCIN-A or SCIN-B18–85 reveals features unique to SCIN-D (Fig. 5b). In particular, a distinct break in the first α helix of SCIN-D occurs at amino acid position Thr26, and this causes a notable deviation in SCIN-D secondary and tertiary structure. This break occurs because the backbone carbonyl of Thr26 hydrogen bonds with a side chain from the second α helix (His48) (Fig. 5C). Although this interaction appears favorable, it nevertheless disrupts the canonical hydrogen bonding pattern that is a hallmark of α helices. As a consequence, the largest region of divergence arises between SCIN-D residues 26 and 35; here, significant distances arise between corresponding residues positions in either SCIN-A or SCIN-B18–85 (e.g. ∼7.0 Å at the carbonyl position of Leu34). After this break, however, the first α helix reforms at SCIN-D-Leu28 and continues into the short loop region that connects α helix 1 to α helix 2. From the beginning of α helix 2 to the C terminus the SCIN-D structure does not differ appreciably from that of either SCIN-A and SCIN-B18–85.
Intriguingly, this peculiar structural feature of SCIN-D lies outside of either known C3c/C3b binding site of SCIN-A (18, 19) or the C3c binding site observed in its co-crystal with SCIN-B18–85 (Figs. 1 and 2). In fact, superposition of the SCIN-D8–83 structure onto that of C3c/SCIN-B18–85 suggests that this perturbation alone cannot account for the lack of C3c/C3b binding to SCIN-D (Fig. 5D). Along these lines, mutation of SCIN-D-His48 to Ala did not impart any detectable C3b binding activity to this protein even though it would be expected to eliminate the physical basis of the helical distortion described above (supplemental Fig. S3). Still, it is worth noting that two residues that contribute important interactions for forming the C3b/SCIN-B18–85 complex (i.e. Arg44 and Tyr51; Figs. 2 and 3) are found in identical positions on the second α helix of SCIN-D (i.e. Arg42 and Tyr49; Fig. 5D). This suggests that additional and perhaps more subtle physical/chemical properties of SCIN-D must be incompatible with binding to this site on C3c/C3b.
Examination of electrostatic surface potentials calculated for each SCIN monomer reveals no obvious differences other than perhaps that the second α helix of SCIN-D (i.e. first binding site of SCIN-A/SCIN-B18–85) exhibits less overall charge than its counterparts (supplemental Fig. S7). Although this may yet be significant, another potential explanation for the lack of SCIN-D binding to C3c/C3b comes from examining the surface complementarity indices of various C3c/SCIN complexes (supplemental Table S1). This suggests that both SCIN-A and SCIN-B18–85 more closely match this particular binding site on C3c than does SCIN-D8–83, as their complementarity values are far closer to those previously reported for high affinity protein-protein interactions (surface complementarity of 0.64, 0.59, and 0.32, respectively, compared with ∼0.6 (42)). This remains the case (surface complementarity of 0.35) even after the rotamers of the SCIN-D8–83 residues that lie on the solvent-exposed face of α helix 2 are changed to best approximate those found in the C3c/SCIN-B18–85 structure (Fig. 2D). Although the possibility of structural rearrangement in SCIN-D cannot be ruled out at this time, it seems very likely then that a combination of electrostatic barriers and lack of surface complementarity may prevent significant SCIN-D binding to C3c/C3b.
DISCUSSION
Until this point the structure and anti-complement properties of SCIN proteins have been investigated almost exclusively through study of the founding member of this protein family, SCIN-A (13, 14, 18–20). Because of this, our analysis of SCIN-B presented here significantly expands understanding of the molecular determinants required for SCIN-mediated complement inhibition. Although the C3c/SCIN-B18–85 interface contains many unique interactions that are lacking in any of the C3c/SCIN-A structures (18), the C3c/C3b residues targeted by both SCIN proteins themselves are largely similar. These include residues 555–558 from the C3c/C3b β chain, a large stretch of α′ chain fragment 1 residues near the Arg726-Ser727 scissile bond of C3b (732–741) and small patch of residues within the C3c/C3b MG7 domain (898–900). The presence of these interactions in separate C3c co-crystals with SCIN-A and SCIN-B18–85 underscores their importance for specific C3b binding. Furthermore, it is also consistent with the premise that these particular immune evasion proteins have evolved to target a C3b hotspot that is vital to C3 convertase structure, function, and dynamics (17–19).
Although several residues near the SCIN-B18–85 C terminus make significant contact with C3c in the co-crystal structure, exchange of these residues to alanines did not impair C3b binding of SCIN-B18–85 in solution. This observation along with a lack of equivalent interactions in any C3c/SCIN-A structures (18) suggests that these contacts are most likely an adventitious byproduct of the C3c/SCIN-B18–85 co-crystal. On the other hand, several lines of evidence presented here indicate that the AP convertase inhibition properties of SCIN-B18–85 require interactions formed by residues located within its second α helix. In particular, our structure-guided mutagenesis studies identified contributions of both Arg44 and Tyr51 as critical for high affinity SCIN-B18–85 binding to C3b but also that loss of Arg44 is more detrimental to C3b binding than is loss of Tyr51. It is noteworthy that these two residues and their corresponding positions in SCIN-A (i.e. Arg42 and Gln49) both make extensive contact with C3c/C3b residues 733–735, 738, and 898 as mentioned above. Thus, although targeting the same hotspot of C3b is critical for function of active SCIN proteins, some degree of side chain variation, even non-conservatively so, appears permissible within this SCIN active site.
The structure/function studies presented here made extensive use of a protease-stable form of SCIN-B (i.e. SCIN-B18–85) that retained both C3b binding and complement inhibitor activity. However, our solution binding studies also clearly show that such N-terminal-truncated forms of SCIN proteins display a nearly 2 order of magnitude reduction in affinity for C3b (Fig. 3 and Table 2). Whereas mutational analyses of SCIN-B18–85 Arg44 and Tyr51 can be reconciled easily by either the C3c co-crystal or sequence/structure comparisons to SCIN-A, the contribution of the flexible N-terminal region to C3b/SCIN-B binding is harder to explain. Although crystallographic analysis of full-length SCIN-B bound to C3b would be of tremendous value in this regard, our attempts to crystallize the C3b/SCIN-B complex have been largely unsuccessful so far. Furthermore, little insight can be gained by inspection of any related SCIN-A co-crystal structures as (i) SCIN proteins share little sequence similarity within their N termini (supplemental Fig. S1), and (ii) the structure of this region in SCIN-B remains unknown. Thus, it appears that a detailed, comparative investigation into the structure and function of full-length SCIN proteins will be needed to answer questions regarding the N terminus of SCIN-B specifically and all SCIN proteins in general.
We have also provided the first structural information on another SCIN family member, SCIN-D. The gene for SCIN-D is found in 98% of S. aureus strains examined (12), lies adjacent to that for staphylcoagulase (which itself results inappropriate activation of the clotting response (43)), and encodes a secreted protein related to a group of potent complement inhibitors. These observations suggest, at least circumstantially, that SCIN-D may impart an important function in the initiation or propagation of staphylococcal infection. Despite its modest but significant level of sequence identity to the remaining SCIN family members (supplemental Fig. S1A), SCIN-D does not directly bind to C3b (Refs. 12, 14, and 17 and supplemental Fig. S3). Furthermore, it seems to lack the complement inhibitor and anti-phagocytic properties of bona fide SCINs (12, 20). In light of these peculiar features, SCIN-D has often found use as a negative control in functional analysis of SCIN proteins (12, 17, 20); however, this tacitly assumes that all SCIN proteins share sufficient sequence homology as to be structurally identical. Our structure demonstrates that this is not the case, as the N-terminal α helix of SCIN-D deviates substantially in both secondary and tertiary structure when compared with either SCIN-A or SCIN-B. With these differences now documented, it may be worthwhile to reevaluate previous work on SCIN-D, especially that involving functional analysis of SCIN-A/SCIN-D protein chimeras (14, 19).
On the basis of its unique structure and inability to bind C3c/C3b, we suggest that it may now be more appropriate to consider SCIN-D a distinct protein rather than a true member of the SCIN family. Nevertheless, it is clear that SCIN-D contains several essential mediators of protein-protein interactions found in other SCIN molecules (i.e. residues corresponding to SCIN-B18–85-Arg44 and Tyr51). Although the divergent N-terminal α helix in SCIN-D alone cannot account for its lack of binding to C3b (Fig. 5D), variations in both the electrostatic potential and shape complementarity at its surface very well may (supplemental Fig. S7 and Table S1). With this in mind, it is tempting to speculate that SCIN-D might instead bind to more distantly related complement components, such as C4b or C5b, or specific protein-protein complexes thereof. Such functions may be advantageous in terms of staphylococcal immune evasion, as they could support inhibition of either the classical/lectin complement pathway (1) or downstream inflammation via sublytic levels of terminal complement complex (44), respectively. Thus, the structure of SCIN-D may serve as an important starting point for elucidating the true nature of its function through future work.
In light of the tenuous balance between complement activation and regulation, it is not surprising that uncontrolled or inappropriate complement activation either causes or directly contributes to number of severe human diseases (45). Indeed this list still continues to expand, even beyond inflammatory/immune diseases, ischemia/reperfusion injuries, and sepsis where the pathophysiological contributions of complement have been well established. The development of complement-targeted therapeutics has long held great potential for treatment of these conditions (45). Still, drug discovery efforts in this area have been complicated by the multipronged nature of the complement system, which is predicated upon an integrated network of protein-protein interactions that underlie its initiation, amplification, and regulation. With significant challenges remaining to the development of peptides or other small molecules capable of disrupting these interactions, an unexpected contribution toward complement-directed therapeutics may yet come from thoroughly understanding the naturally occurring inhibitors produced by pathogenic microorganisms such as S. aureus (9). Much of the work presented here focused on understanding the molecular basis for complement inhibition by SCIN-B, which by itself is only a single member of a multiprotein inhibitor family. Yet when considered along with previous studies of SCIN-A (18), this work on SCIN-B has provided further insight on the nature and extent of interactions that give rise to high affinity/specificity C3b binding and convertase inhibition. In addition, it has cast further light on a hotspot region of C3b that nature has already shown can be effectively targeted through relatively small stretches of polypeptide sequence. Although much more study is clearly needed, this type of information is potentially valuable as it may hold important clues to either optimization or synthetic mimicry of SCIN binding/activity toward future complement-targeted therapeutic applications.
Supplementary Material
Acknowledgments
We acknowledge the generous technical assistance of Drs. Rod Salazar and Andy Howard during x-ray diffraction data collection and Dr. Samuel Bouyain for use of the EnSpire plate reader. Use of the Advanced Photon Source was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract W-31-109-Eng-38. Data were collected at Southeast Regional Collaborative Access Team (SER-CAT) beamlines at the Advanced Photon Source, Argonne National Laboratory. A list of supporting member institutions may be found at the SER-CAT Institutional Members website.
This work was supported by National Institutes of Health Grants AI071028, AI030040, AI068730, and AI072106 (to B. V. G. and J. D. L.). This work was also supported by an American Heart Association Predoctoral Fellowship (to B. L. G.).

This article contains supplemental Table S1 and Figs. S1–S7.
- AP
- alternative pathway
- fB
- factor B
- fH
- factor H
- fD
- factor D
- CVF
- cobra venom factor
- MG
- macroglobulin
- bis-tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- SPR
- surface plasmon resonance.
REFERENCES
- 1. Ricklin D., Hajishengallis G., Yang K., Lambris J. D. (2010) Nat. Immunol. 11, 785–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Harboe M., Ulvund G., Vien L., Fung M., Mollnes T. E. (2004) Clin. Exp. Immunol. 138, 439–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zipfel P. F., Skerka C. (2009) Nat. Rev. Immunol. 9, 729–740 [DOI] [PubMed] [Google Scholar]
- 4. Meri S., Pangburn M. K. (1994) Biochem. Biophys. Res. Comm. 198, 52–59 [DOI] [PubMed] [Google Scholar]
- 5. Pangburn M. K. (2000) Immunopharmacology 49, 149–157 [DOI] [PubMed] [Google Scholar]
- 6. Pangburn M. K., Müller-Eberhard H. J. (1986) Biochem. J. 235, 723–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu J., Wu Y. Q., Ricklin D., Janssen B. J., Lambris J. D., Gros P. (2009) Nat. Immunol. 10, 728–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lambris J. D., Ricklin D., Geisbrecht B. V. (2008) Nat. Rev. Microbiol. 6, 132–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Geisbrecht B. V. (2008) Adv. Exp. Med. Biol. 632, 221–236 [PubMed] [Google Scholar]
- 10. Foster T. J. (2005) Nat. Rev. Microbiol. 3, 948–958 [DOI] [PubMed] [Google Scholar]
- 11. Chavakis T., Preissner K. T., Herrmann M. (2007) Trends Immunol. 28, 408–418 [DOI] [PubMed] [Google Scholar]
- 12. Jongerius I., Köhl J., Pandey M. K., Ruyken M., van Kessel K. P., van Strijp J. A., Rooijakkers S. H. (2007) J. Exp. Med. 204, 2461–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rooijakkers S. H., Ruyken M., Roos A., Daha M. R., Presanis J. S., Sim R. B., van Wamel W. J., van Kessel K. P., van Strijp J. A. (2005) Nat. Immunol. 6, 920–927 [DOI] [PubMed] [Google Scholar]
- 14. Rooijakkers S. H., Milder F. J., Bardoel B. W., Ruyken M., van Strijp J. A., Gros P. (2007) J. Immunol. 179, 2989–2998 [DOI] [PubMed] [Google Scholar]
- 15. Laarman A., Milder F., van Strijp J., Rooijakkers S. (2010) J. Mol. Med. 88, 115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rooijakkers S. H., van Kessel K. P., van Strijp J. A. (2005) Trends Microbiol. 13, 596–601 [DOI] [PubMed] [Google Scholar]
- 17. Ricklin D., Tzekou A., Garcia B. L., Hammel M., McWhorter W. J., Sfyroera G., Wu Y. Q., Holers V. M., Herbert A. P., Barlow P. N., Geisbrecht B. V., Lambris J. D. (2009) J. Immunol. 183, 2565–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garcia B. L., Ramyar K. X., Tzekou A., Ricklin D., McWhorter W. J., Lambris J. D., Geisbrecht B. V. (2010) J. Mol. Biol. 402, 17–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rooijakkers S. H., Wu J., Ruyken M., van Domselaar R., Planken K. L., Tzekou A., Ricklin D., Lambris J. D., Janssen B. J., van Strijp J. A., Gros P. (2009) Nat. Immunol. 10, 721–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jongerius I., Puister M., Wu J., Ruyken M., van Strijp J. A., Rooijakkers S. H. (2010) J. Immunol. 184, 420–425 [DOI] [PubMed] [Google Scholar]
- 21. Garcia B. L., Tzekou A., Ramyar K. X., McWhorter W. J., Ricklin D., Lambris J. D., Geisbrecht B. V. (2009) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65, 482–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Geisbrecht B. V., Bouyain S., Pop M. (2006) Protein Expr. Purif. 46, 23–32 [DOI] [PubMed] [Google Scholar]
- 23. Sarkar G., Sommer S. S. (1990) Biotechniques 8, 404–407 [PubMed] [Google Scholar]
- 24. Beneken J., Tu J. C., Xiao B., Nuriya M., Yuan J. P., Worley P. F., Leahy D. J. (2000) Neuron 26, 143–154 [DOI] [PubMed] [Google Scholar]
- 25. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 26. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 28. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Russell R. B., Barton G. J. (1992) Proteins 14, 309–323 [DOI] [PubMed] [Google Scholar]
- 30. Janssen B. J., Huizinga E. G., Raaijmakers H. C., Roos A., Daha M. R., Nilsson-Ekdahl K., Nilsson B., Gros P. (2005) Nature 437, 505–511 [DOI] [PubMed] [Google Scholar]
- 31. Bielefeld-Sevigny M. (2009) Assay Drug Dev. Technol. 7, 90–92 [DOI] [PubMed] [Google Scholar]
- 32. Abramoff M. D., Magalhaes P. J., Ram S. J. (2004) Biophotonics International 11, 36–42 [Google Scholar]
- 33. Janssen B. J., Halff E. F., Lambris J. D., Gros P. (2007) J. Biol. Chem. 282, 29241–29247 [DOI] [PubMed] [Google Scholar]
- 34. Janssen B. J., Christodoulidou A., McCarthy A., Lambris J. D., Gros P. (2006) Nature 444, 213–216 [DOI] [PubMed] [Google Scholar]
- 35. Wiesmann C., Katschke K. J., Yin J., Helmy K. Y., Steffek M., Fairbrother W. J., McCallum S. A., Embuscado L., DeForge L., Hass P. E., van Lookeren Campagne M. (2006) Nature 444, 217–220 [DOI] [PubMed] [Google Scholar]
- 36. Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 37. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou P., Tian F., Lv F., Shang Z. (2009) Proteins 76, 151–163 [DOI] [PubMed] [Google Scholar]
- 39. Hammel M., Sfyroera G., Ricklin D., Magotti P., Lambris J. D., Geisbrecht B. V. (2007) Nat. Immunol. 8, 430–437 [DOI] [PubMed] [Google Scholar]
- 40. Hammel M., Sfyroera G., Pyrpassopoulos S., Ricklin D., Ramyar K. X., Pop M., Jin Z., Lambris J. D., Geisbrecht B. V. (2007) J. Biol. Chem. 282, 30051–30061 [DOI] [PubMed] [Google Scholar]
- 41. Chen H., Ricklin D., Hammel M., Garcia B. L., McWhorter W. J., Sfyroera G., Wu Y. Q., Tzekou A., Li S., Geisbrecht B. V., Woods V. L., Jr., Lambris J. D. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 17621–17626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Garcia K. C., Teyton L., Wilson I. A. (1999) Annu. Rev. Immunol. 17, 369–397 [DOI] [PubMed] [Google Scholar]
- 43. Hemker H. C., Bas B. M., Muller A. D. (1975) Biochim. Biophys. Acta 379, 180–188 [DOI] [PubMed] [Google Scholar]
- 44. Tedesco F., Bulla R., Fischetti F. (2004) in The Complement System: Novel Roles in Health and Disease (Szebeni J., ed) pp. 97–127, Kluwer Academic Publishers, Boston [Google Scholar]
- 45. Ricklin D., Lambris J. D. (2007) Nat. Biotechnol. 25, 1265–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.